Summary
NPGPx is a member of the glutathione peroxidase (GPx) family; however, it lacks GPx enzymatic activity due to the absence of a critical selenocysteine residue, rendering its function an enigma. We report that NPGPx is a novel stress sensor that transmits oxidative stress signals by transferring the disulfide bond between its Cys57 and Cys86 residues to downstream effectors. Oxidized NPGPx binds and oxidizes the chaperone glucose-regulated protein (GRP)78 in the endoplasmic reticulum through covalent bonding between Cys86 of NPGPx and Cys41/Cys420 of GRP78, and facilitates the refolding of misfolded proteins by GRP78 to alleviate stress. NPGPx-deficient cells display impaired GRP78 chaperone activity, accumulate misfolded proteins, and suffer oxidative stress. Complete loss of NPGPx in animals causes systemic oxidative stress, increases carcinogenesis, and shortens lifespan. These results, for the first time, suggest that NPGPx is essential for mediating the oxidative stress response by modulating GRP78 chaperone activity to maintain physiological homeostasis.
Introduction
Reactive oxygen species (ROS) are ubiquitous in all living organisms. ROS can be produced intrinsically as by-products of mitochondrial respiration or oxidative protein folding in the endoplasmic reticulum (Malhotra and Kaufman, 2007). They can also be induced in vivo upon virus infection (Gonzalez-Dosal et al., 2011) or inflammation (Morgan and Liu, 2011). Excessive ROS production leads to oxidative stress, which has been associated with a variety of disorders including autoimmune disease (Bashir et al., 1993), cancer (Kumar et al., 2008), and aging (Rascon and Harrison, 2010). On the other hand, ROS are required for maintaining normal physiological processes including signal transduction via insulin (Goldstein et al., 2005) and proinflammatory cytokines (Ali et al., 1999). Thus, maintaining intracellular ROS homeostasis in vivo is important. Excess ROS should be released properly in vivo to alleviate stress on cells. Several enzymes are involved in the process including catalase (CAT), thioredoxin peroxidase (PRDX), and glutathione peroxidases (GPx). CAT directly decomposes H2O2 to water in peroxisomes. PRDX catalyzes the reduction of H2O2 to water coupled with thioredoxin oxidation (Rhee et al., 2005). In contrast, GPx transfers free radicals from intracellular ROS to glutathione (GSH) to reduce ROS levels in cells. So far, 8 mammalian GPxs have been identified. The first human GPx, GPx1, was identified in erythrocytes (Mills, 1957), and GPx2 to GPx8 have been subsequently identified (Chu et al., 1993; Dear et al., 1991; Ghyselinck et al., 1989; Maddipati and Marnett, 1987; Mills, 1957; Nguyen et al., 2011; Ursini et al., 1985; Utomo et al., 2004). GPxs are classified into 2 kinds based on the cysteine residues present: selenocysteine-containing GPxs (S-GPxs) such as GPx1, 2, 3, and 4, and non-selenocysteine GPxs (NS-GPx) such as GPx5, 6, 7, and 8. S-GPxs catalyze the conversion of H2O2 to water by using GSH as a substrate, while NS-GPxs do not use GSH as a major substrate. NS-GPxs, including those from plants (GPxle1 and GPxha2; (Herbette et al., 2002), P. falciparum (Sztajer et al., 2001), and B. pahanji (Tang et al., 1995) are not suitable for GSH oxidation. Instead, NS-GPxs may act as intracellular messengers that sense and transmit ROS signals to modulate redox-sensitive proteins. For example, yeast GPx3, an oxidative NS-GPx, modulates YAP1 through thiol/disulfide interactions to generate oxidized YAP1, an activated transcription factor (Delaunay et al., 2002). However, this has not been fully substantiated in mammalian cells.
Mammalian GPx7, also known as non-selenocysteine containing phospholipids hydroperoxide glutathione peroxidase (NPGPx), is an ER-resident NS-GPx (Utomo et al 2004). Knockdown of NPGPx has been shown to sensitize breast cancer cells to oxidative stress-induced cell death (Utomo et al., 2004), suggesting its importance in relieving oxidative stress. However, the detailed molecular mechanism by which NPGPx alleviates oxidative stress remains elusive.
In this study, we found that NPGPx might act as an oxidative stress sensor that transmits stress signals to activate target proteins such as GRP78. In stressed cells, NPGPx is oxidized, and then it interacts with GRP78 to facilitate GRP78 chaperone activity and it attenuates stress-induced protein misfolding. NPGPx-deficient cells accumulate ROS and become sensitive to oxidative stress. Oxidative stress-related systemic disorders were also observed in NPGPx-deficient mice, suggesting the importance of the oxidative stress sensor, NPGPx, in protecting animals from oxidative stress-induced damage.
Results
Generation of NPGPx knockout mice
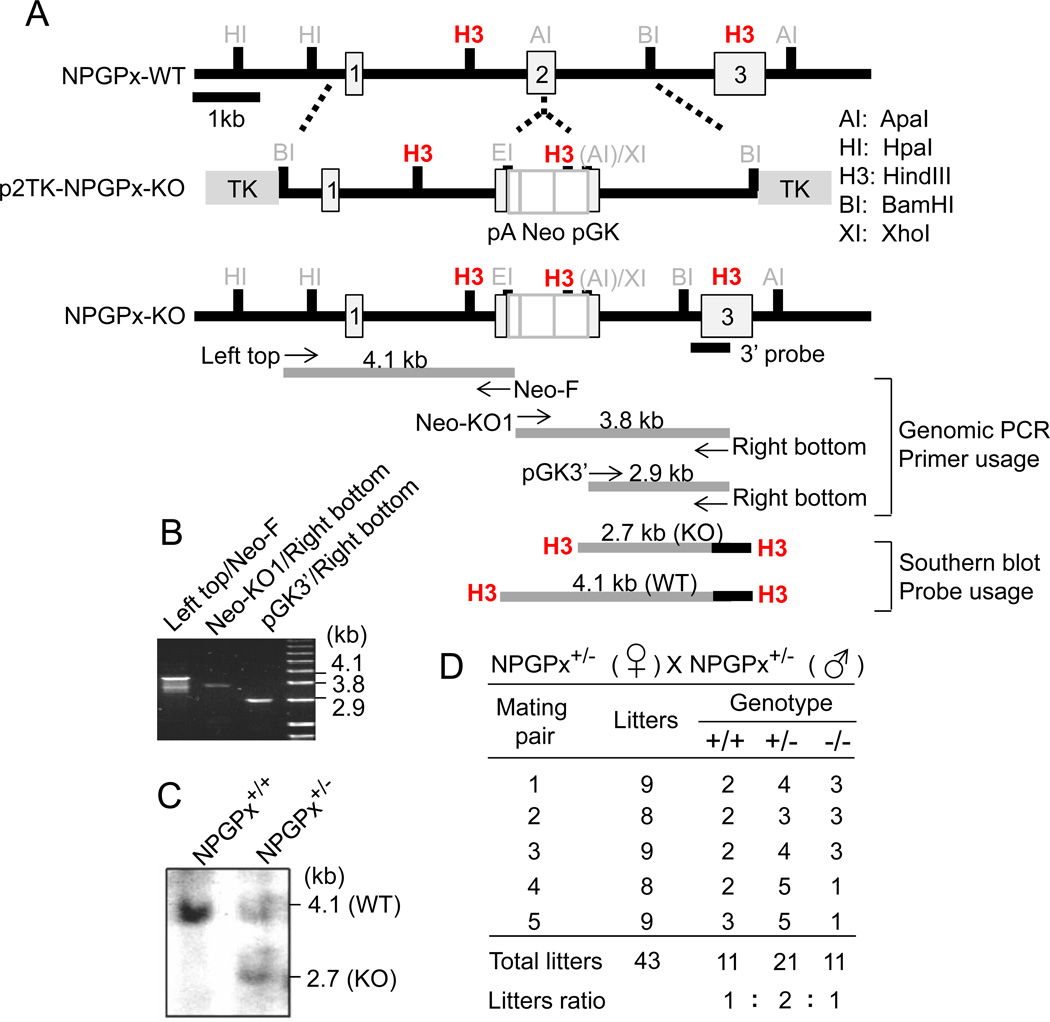
To address the potential function of NPGPx in mammalian cells, we employed a standard knockout strategy to generate NPGPx-deficient mice. As shown in Figure 1, the NPGPx gene contains 3 exons; a targeting vector containing a neomycin cassette was inserted in exon 2 by homologous recombination to inactivate the gene in 129J mouse embryonic stem (ES) cells (Figure 1A–C). NPGPx knockout mice were produced by heterozygous parents following standard procedures (see Materials and Methods). NPGPx+/+, NPGPx+/−, and NPGPx−/− pups were born with Mendelian frequencies, indicating that NPGPx deficiency did not cause embryonic lethality in mice (Figure 1D).
Figure 1. Generation of NPGPx−/− mice.
(A) Strategy for generating NPGPx targeting vector. The targeting vector (middle), WT sequences (top), and exon 2 of NPGPx (bottom) are illustrated. Primer pairs for genomic analysis and probes for genomic Southern blot are shown. (B) Genomic PCR analysis of ES cells bearing the predicted NPGPx target allele. (C) Southern blotting analysis of ES cells bearing the predicted NPGPx target allele. (D) The number and frequency of offspring with each genotype produced by crossing NPGP+/− mice. The sum of each genotype produced by 5 mates is shown.
NPGPx−/− mouse embryonic fibroblasts (MEFs) accumulate ROS and are sensitive to oxidative stress
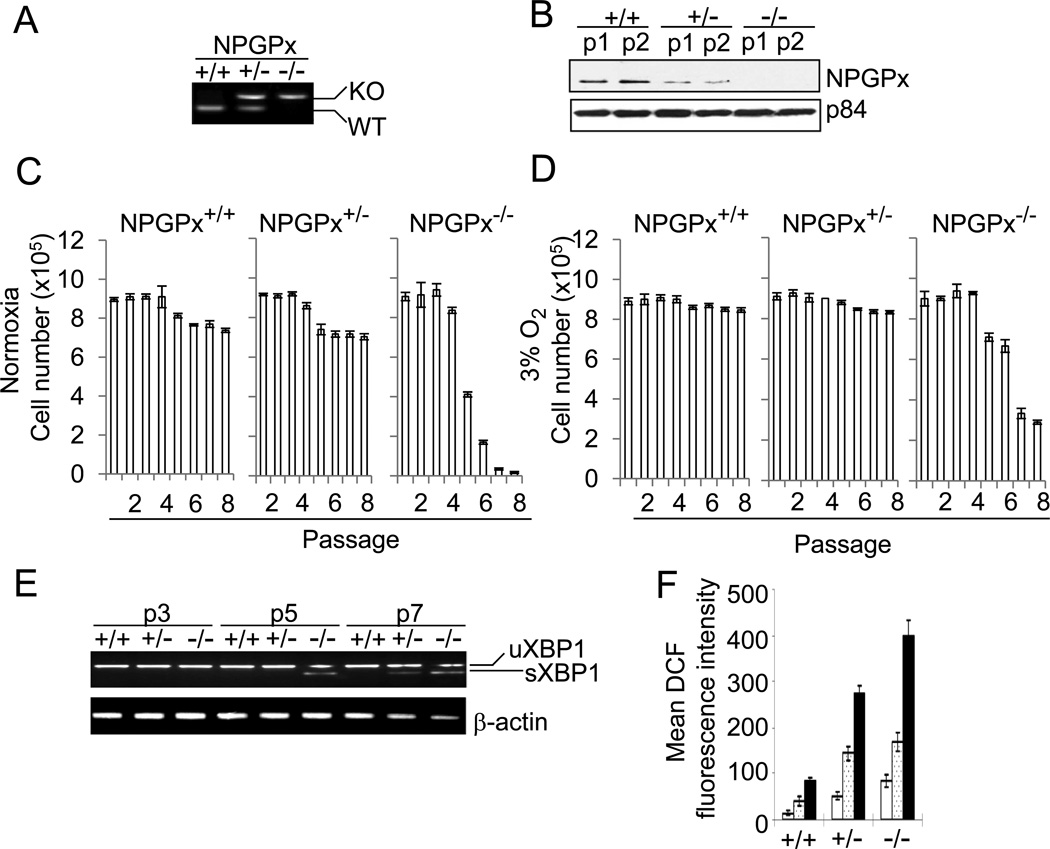
To explore NPGPx function, primary MEFs from wild-type (WT) or NPGPx-deficient embryos were prepared and genotyped (Figure 2A). Western blotting revealed that NPGPx+/− heterozygotes expressed 50% NPGPx protein compared to WT litters, and that NPGPx expression was completely abolished in NPGPx−/− MEFs (Figure 2B). Since NPGPx-depleted cells are sensitive to oxidative stress (Utomo et al., 2004), we measured the susceptibility of WT or NPGPx−/− MEFs to environmental oxygen. The number of NPGPx−/− MEFs declined drastically after the 5th passage, compared to WT MEFs, in 20% O2 (Figure 2C). In contrast, the number of NPGPx−/− MEFs after the same number of passages was much higher in 3% O2 (Figure 2D) than in 20% O2 (Figure 2C). In NPGPx−/− MEFs cultured in 20% O2, the mRNA of the oxidative stress-responsive gene X-box binding protein 1 (XBP1; (Yoshida et al., 2001), was spliced into the activated form (see sXBP1 in Figure 2E). The spliced form of XBP1 did not appear until the 5th passage (Figure 2E), and was retained in subsequent passages in NPGPx−/− MEFs (Figure 2E). In contrast, sXBP1 could not be detected in WT MEFs even after the 7th passage. Consistently, the endogenous ROS level measured by dichlorodihydrofluorescein (DCF) staining was significantly higher in NPGPx−/− and NPGPx+/− MEFs than in NPGPx+/+ MEFs cultured in normoxia (Figure 2F). Taken together, these results indicate that NPGPx is essential for cell survival in the presence of excess environmental oxygen, and that NPGPx deficiency leads to ROS accumulation and sensitizes cells to oxidative stress.
Figure 2. Compromised proliferation of NPGPx-deficient MEFs with accumulated oxidative stress.
(A) Genotyping using PCR analysis. NPGPx genotyping of WT, NPGPx+/− or NPGPx−/− MEFs is shown. (B) Western blot analysis of NPGPx protein expression in WT, NPGPx+/−, or NPGPx−/− MEFs. p1: passage 1; p2: passage 2. P84 served as an internal control. (C, D) Proliferation of MEFs in either normoxia (C) or low-oxygen (D) conditions. (E) RT-PCR analysis of XBP-1 mRNA level. Total RNA was extracted from WT, NPGPx+/−, or NPGPx−/− MEFs at different passages (passages 3, 5 and 7; labeled as p3, p5 and p7), and XBP-1 mRNA status was measured by RT-PCR. uXBP-1: unspliced XBP-1 mRNA; sXBP-1: spliced XBP-1 mRNA. β-actin was the internal control. (F) Quantification of endogenous ROS level in WT, NPGPx+/−, or NPGPx−/− MEFs. The DCF precursor is an ester that can be oxidized by ROS and that emits green florescence upon excitation. Different passages of MEFs were treated with DCF precursor and analyzed by FACS. Open bar: passage 1; dotted bar: passage 3; black bar: passage 5.
Cys57 and Cys86 of NPGPx are essential for relieving oxidative stress
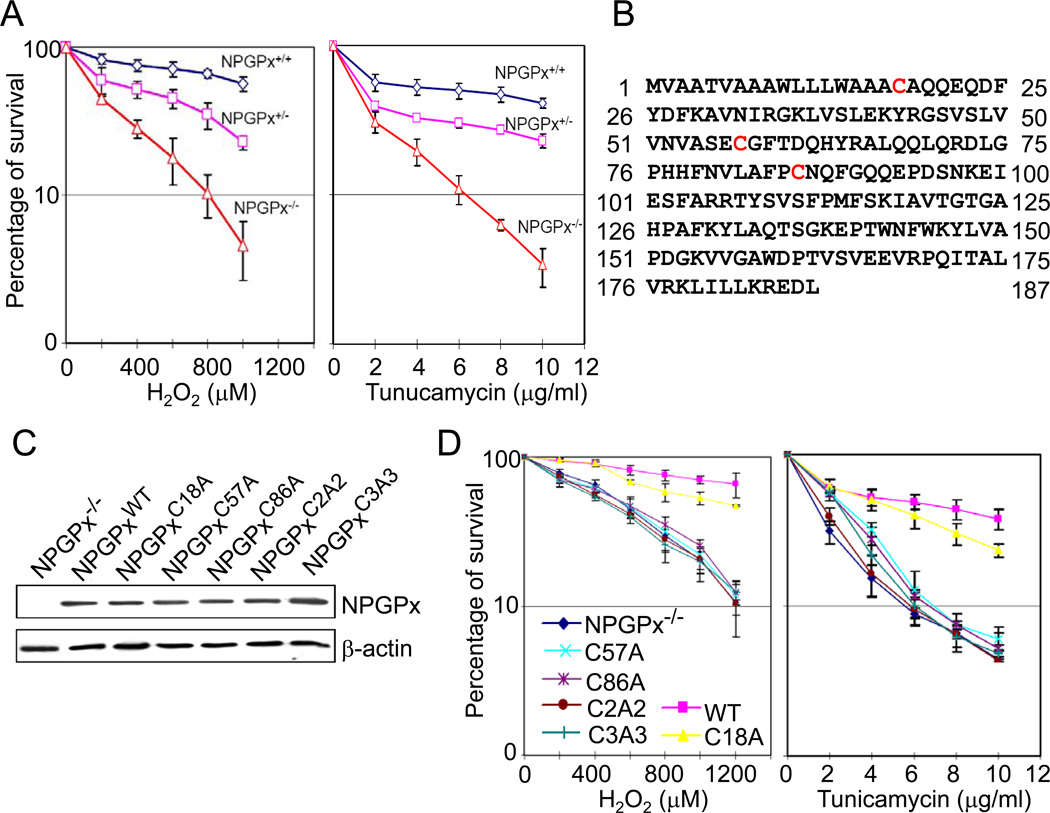
To address whether the loss of NPGPx is solely responsible for the sensitivity of cells to exogenous oxidative stress, we compared the viabilities of NPGPx+/+, NPGPx+/−, and NPGPx−/− MEFs upon treatment with H2O2 and tunicamycin, an oxidative stress inducer that inhibits protein glycosylation to cause protein misfolding (Kuo and Lampen, 1974). NPGPx−/− MEFs were significantly more sensitive to H2O2 (Figure 3A, left) and tunicamycin (Figure 3A, right) treatment than were WT MEFs, suggesting that the presence of NPGPx protects cells from excessive oxidative stress.
Figure 3. Cys 57 and 86 of NPGPx are essential for its anti-oxidative stress activity.
(A) Cell survival analysis with H2O2 or tunicamycin treatment. Untreated cells (0 point) viability was set as 100 percent. (B) NPGPx amino acid sequences. The cysteines (Cys18, Cys57, and Cys86) are highlighted in red. (C) Western blot analysis of expressed WT or mutant NPGPx protein. (D) Cell survival assay. NPGPx−/− MEFs restored with WT or mutant NPGPx were treated with H2O2 or tunicamycin for 3 hours, and their viability was measured by MTT assay 21 hours after treatment.
To further elucidate the role of NPGPx in relieving oxidative stress, we investigated the importance of 3 cysteine residues, Cys18, Cys57, and Cys86 (Figure 3B), that may be crucial for NPGPx activity. We engineered 5 NPGPx mutants with alanine substitutions at the indicated Cys sites: NPGPxC18A, NPGPxC57A, NPGPxC86A, NPGPxC2A2, and NPGPxC3A3. NPGPxC2A2 contains 2 amino acid substitutions (Cys57 and Cys86 to Ala), while NPGPxC3A3 has all cysteines replaced by alanine. The protein expression levels of these NPGPx mutants were comparable when measured by Western blot (Figure 3C). We then treated these NPGPx−/− MEFs ectopically expressing NPGPx proteins with either H2O2 (Figure 3D, left) or tunicamycin (Figure 3D, right). NPGPx−/− MEFs expressing NPGPxC57A, NPGPxC86A, NPGPxC2A2, or NPGPxC3A3 were as sensitive to H2O2 or tunicamycin treatment as were the parental MEFs, while NPGPx−/− MEFs expressing WT NPGPx or NPGPxC18A were more resistant (Figure 3D). These results confirmed that NPGPx is essential for alleviating oxidative stress and that the 2 cysteine residues, Cys57 and Cys86, but not Cys18, play important roles in this process.
NPGPx covalently binds to GRP78 through cysteine residues
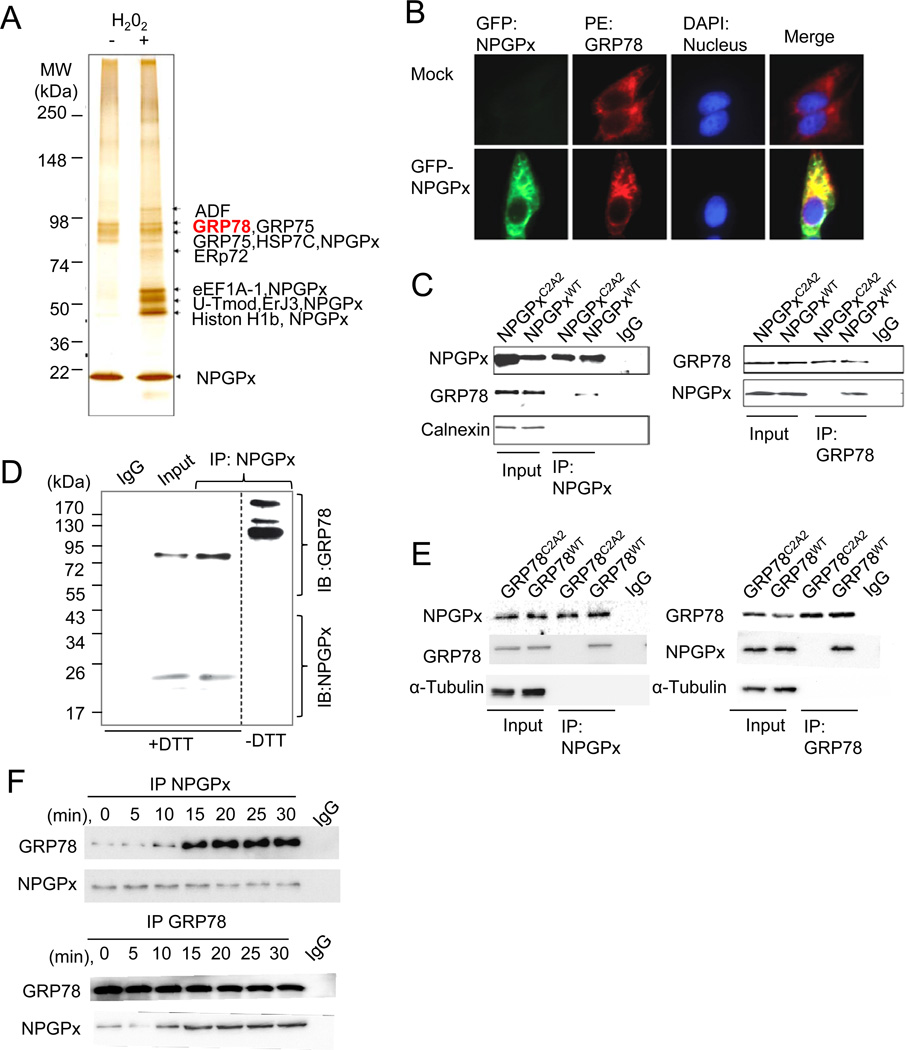
Since NPGPx does not contain the conserved GSH binding domain (Nguyen et al., 2011) and lacks enzymatic activity (Supplement Figure 1), it is likely that NPGPx may execute its anti-stress function by other means. To explore this possibility, we investigated whether NPGPx forms stress-induced protein complexes in vivo. Western blot analyses using H2O2-treated U2OS cells revealed many NPGPx-containing bands under non-reducing conditions (Supplemental Figure 2), suggesting that H2O2 treatment induced covalent interactions between NPGPx and its target proteins. To further identify H2O2-induced covalent interactions of NPGPx, we used an affinity column conjugated with an anti-NPGPx antibody to pull down NPGPx and its interacting proteins from NPGPx-expressing U2OS cells. The affinity-purified proteins were separated by non-reducing SDS-PAGE (Figure 4A) and each band was subjected to mass spectrometry (MS) analysis for identification. Protein ID results revealed that the majority of NPGPx-interacting proteins were chaperones such as GRP78 (Tokunaga et al., 1992), GRP75 (Chen et al., 1996), and endoplasmic reticulum (ER) protein (ERp)72 (Nigam et al., 1994) (Figure 4A). Among these, we focused on GRP78 because it plays a vital role in mediating oxidative stress, and loss of GRP78 is lethal (Luo et al., 2006; Riemer et al., 2009). Immunostaining of cells expressing GFP-tagged NPGPx revealed that NPGPx mainly localizes in the ER, where GRP78 is also retained (Figure 4B). Furthermore, WT NPGPx, but not NPGPxC2A2, reciprocally co-immunoprecipitated (IP) with GRP78 in lysates prepared from U2OS cells expressing either WT NPGPx or NPGPxC2A2. Protein-protein interactions were not seen between NPGPx and other ER resident proteins such as calnexin (Figure 4C). This in vivo interaction was further supported by the presence of high-molecular weight (≥90 kDa) GRP78 containing bands in the NPGPx-containing protein IP complex, upon separation by non-reducing SDS-PAGE and analysis by Western blot (Figure 4D). GRP78 contains 2 Cys residues: Cys41 and Cys420 (schematic in Supplement Figure 4), and these 2 Cys residues may be required for NPGPx interaction. To explore this possibility, we cloned human WT GRP78 or Cys-to-Ala substituted GRP78 (hGRP78C2A2) into a mammalian expression vector and transfected the constructs into endogenous GRP78-depleted MEFs (see Supplemental Materials and Methods). NPGPx and GRP78 were subsequently immunoprecipitated in lysates from MEFs expressing WT or hGRP78C2A2, and Western blot analysis revealed that hGRP78C2A2 did not interact with NPGPx (Figure 4E). Importantly, the NPGPx-GRP78 interaction was initiated within 10 minutes of H2O2 treatment and maintained for another 20 minutes (Figure 4F). The interaction declined after 30 minutes of H2O2 treatment (Supplemental Figure 5). Taken together, these data indicate that NPGPx interacts with GRP78 covalently through cysteine residues and that the interaction is enhanced by excessive oxidative stress within 10 minutes of H2O2 treatment.
Figure 4. NPGPx covalently interacts with GRP78 in cells under oxidative stress.
(A) Identification of NPGPx-interacting proteins pulled down from the NPGPx affinity column. (B) Immunofluorescent staining of MEFs transduced with or without (mock) GFP-NPGPx: MEFs were stained with GRP78 antibody (red) and DAPI (blue) (C) Reciprocal IP of NPGPx and GRP78 using NPGPx−/− MEFs ectopically expressing WT or C2A2 NPGPx. NPGPx, GRP78, and calnexin were probed. (D) IP proteins of NPGPx and GRP78 were subsequently analyzed by Western blot in reducing (+DTT) or non-reducing (−DTT) conditions. NPGPx and GRP78 were probed as indicated. Under non-reducing conditions, NPGPx covalently interacts with GRP78, resulting in high molecular weight bands containing NPGPx. (E). Reciprocal IP of NPGPx and GRP78 in MEFs reconstituted with hGRP78WT or hGRP78C2A2. Precipitated proteins were subsequently analyzed by Western blot analysis. (F) Reciprocal IP of NPGPx and GRP78 in lysates prepared using WT MEFs treated with H2O2 for the indicated duration. Immunoprecipitated proteins were subjected to Western blot analysis.
NPGPx is required for GRP78 to relieve stress due to accumulated misfolded proteins
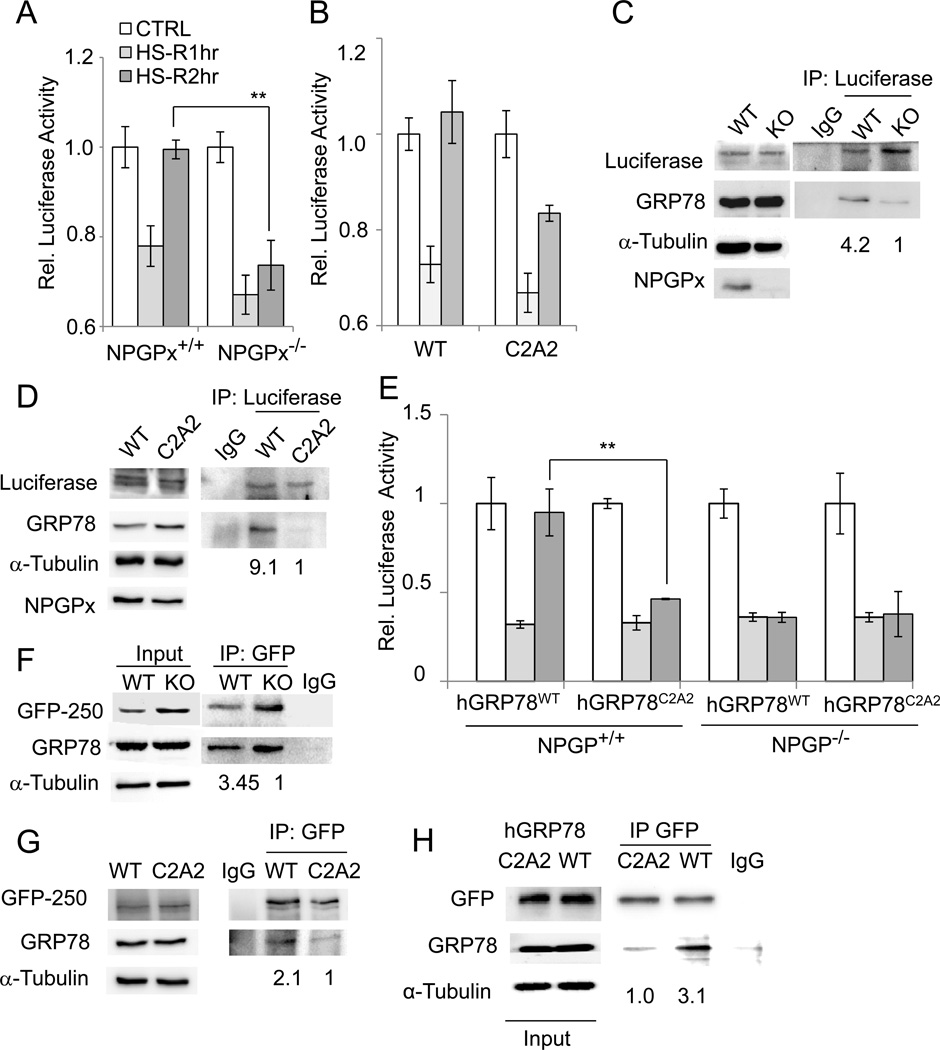
The interaction between NPGPx and GRP78 upon H2O2 treatment implies that NPGPx may be required for GRP78 activation under oxidative stress. Since one of the major functions of GRP78 in the ER is to serve as a chaperone to bind the hydrophobic regions of misfolded protein and facilitate protein refolding (Luo et al., 2006; Riemer et al., 2009), we tested whether NPGPx has an impact on such activity. Two distinctive systems were employed; first, we expressed fly luciferase in NPGPx-expressing and-deficient MEFs, incubated the cells at 42 °C for 30 minutes and returned them to 37 °C for recovery. This heat shock process causes denaturation of proteins, including the ectopically expressed luciferase (Nollen et al., 1999). One hour after heat shock, luciferase activity significantly decreased in WT and NPGPx−/− MEFs. However, luciferase activity nearly completely recovered in WT MEFs within 2 hours, while it remained repressed in NPGPx−/− MEFs (Figure 5A). NPGPx-deficient cells expressing WT NPGPx, but not NPGPxC2A2, exhibited recovery of luciferase activity (Figure 5B). Consistently, IP experiments revealed that the amount of luciferase-interacting GRP78 was about 4- to 9-fold higher in NPGPx-proficient cells than in NPGPx-deficient cells (Figure 5C) or cells with ectopic NPGPxC2A2 (Fig. 5D). On the other hand, the luciferase activity in heat-shocked MEFs expressing mutant human GRP78 (Figure 5E and Supplement Figure 6) was not restored after the 2-hour recovery, suggesting the importance of Cys bonding in GRP78. Moreover, luciferase activity did not recover in heat-shocked NPGPx−/− MEFs expressing WT hGRP78, suggesting that NPGPx serves as a GRP78 regulator prior to the heat-shock response.
Figure 5. NPGPx is required to facilitate the binding of GRP78 to misfolded proteins.
(A,B) In vivo chaperone activity assay. Luciferase-expressing WT or NPGPx−/− MEFs (A) or NPGPx−/− MEFs restored with the NPGPx WT or C2A2 mutant (B) were heat-shocked at 42°C for 30 minutes followed by recovery at 37 °C for 1 hour (HS-R1hr) or 2 hours (HS-R2hr). Cell harvested from these time points were subjected to the luciferase activity assay to detect the activity of refolded luciferase. Control: without heat-shock. (C, D) IP and Western blot analysis of GRP78 interacting with luciferase. Luciferase protein was immunoprecipitated from either HS-R1hr MEFs with WT or NPGPx−/− genotype (C) or from NPGPx−/− MEFs restored with the NPGPx WT or C2A2 mutant (D), and precipitated proteins were subjected to Western blot analysis. The quantity of co-precipitated GRP78 was normalized to luciferase, and the relative amount of GRP78 (NPGPx−/− versus WT) is shown. (E) In vivo chaperone activity assay. WT or NPGPx−/− MEFs reconstituted with WT hGRP78 or hGRP78C2A2 were used in this assay following the procedure described in (A). (F) IP using MEFs transduced with GFP-250. GFP-250 was immunoprecipitated using an antibody against GFP, and then the IP protein complexes were subjected to Western blot analysis. The quantity of co-precipitated GRP78 was normalized to GFP-250, and the relative amount of GFP-250 (NPGPx−/− versus WT) is shown. (G) IP experiment using GRP-250-expressing MEFs rescued with WT NPGPx or the C2A2 mutant following the procedure used in (F). The relative amount of co-precipitated GRP78 (C2A2 versus WT) is provided. (H) IP experiment using GRP-250 expressing MEFs reconstituted with WT hGRP78 or hGRP78C2A2. The relative amount of GFP-250/GRP78 was indicated. The IP experiments were repeated 3 times with similar results.
In the other system, we expressed a misfolded protein, GFP-250, a chimera protein obtained by fusing GFP with the cytosolic protein p115 in MEFs (Kawaguchi et al., 2003). GFP-250 is freshly synthesized in the ER (Garcia-Mata et al., 1999) and is not easily folded; therefore, the misfolded GFP-250 is shuttled into the cytoplasm, and it forms an aggresome in the microtubule-organizing center. GRP78 interacts with misfolded proteins (Marquardt and Helenius, 1992); therefore, we examined how efficiently GRP78 interacts with GFP-250 in the presence or absence of NPGPx. Using this system, we found that the amount of GFP-250 interacting with GRP78 is about 3-fold higher in WT MEFs than in NPGPx−/− MEFs (Fig. 5F). The GRP-250/GRP78 ratio is 2-fold higher in WT NPGPx proficient MEFs than in NPGPxC2A2-restored MEFs (Fig. 5G), suggesting that NPGPx facilitates the binding of GRP78 to misfolded GFP-250. On the other hand, hGRP78C2A2 showed lower interaction with GRP-250 (Fig. 5H), indicating the importance of Cys bonding of GRP78 in its protein-binding ability. The formation of GFP-250 aggresome was faster and bigger in NPGPx-deficient MEFs than in NPGPx-proficient cells (Supplemental Figure 7A, B). Additionally time-lapse microscopy revealed that many NPGPx−/−, but not WT, MEFs expressing GFP-250 underwent apoptosis at ~15 hour (Supplemental Figure 7C). Consistently, a higher percentage of NPGPx−/− MEFs positive for annexin V staining was seen by fluorescence-activated cell sorting (FACS) analysis (Supplemental Figure 7D, E), indicating that GFP-250 overexpression increases the apoptosis of NPGPx−/− MEFs. The enlarged aggresome was also found in hGRP78C2A2-proficient MEFs (Supplemental Figure 8A), and the percentage of these cells containing the aggresome was about 2-fold higher than in WT GRP78-reconstituted MEFs (Supplemental Fig. 8B). These data collectively suggest that the loss of NPGPx attenuates the efficiency of GRP78 chaperone activity for misfolded proteins and exacerbates cellular stress responses such as aggresome formation and apoptosis.
NPGPx servers as an oxidative stress sensor to activate GRP78
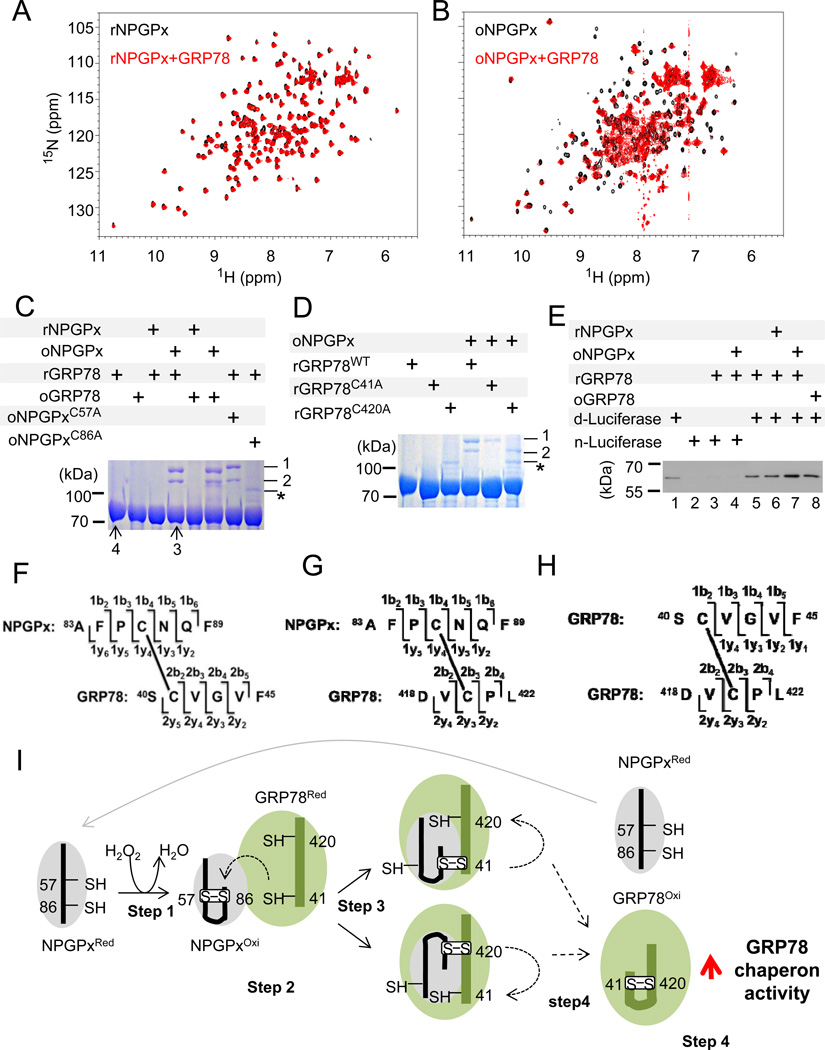
Based on the above data, NPGPx appears to be important in activating GRP78 to bind to misfolded proteins through covalent interactions via its 2 cysteine residues. To further substantiate this observation, we investigated how NPGPx interacts with GRP78 in vitro. Purified recombinant NPGPx behaved as a monomer with free thiol groups under reducing conditions. Reduced NPGPx (rNPGPx) had lower mobility in non-reducing SDS-PAGE, but it was readily oxidized by H2O2 and its mobility was increased (Supplemental Figure 3). MS analysis of the oxidized form of NPGPx (oNPGPx) revealed a single disulfide bond between Cys57 and Cys86 (Supplemental Figure 9 also see Supplemental Materials and Methods). Since these 2 Cys residues are 11.58 Å apart according to the known crystal structure of rNPGPx (Supplemental Figure 10; PDB ID: 2p31), oNPGPx must undergo a significant conformational change, as supported by the mobility change in non-reducing SDS-PAGE. Next, we used nuclear magnetic resonance (NMR) of purified recombinant NPGPx and GRP78 proteins to detect the interaction between them. As shown in Figures 6A and 6B, oNPGPx, but not rNPGPx, can interact with reduced GRP78, causing line broadening of peaks in the 15N-HSQC spectra (Figure 6B). We then identified the complexes of oNPGPx and reduced GRP78 using non-reducing SDS-PAGE (Figure 6C, D). Each individual band was further analyzed by MS. The upper and lower bands (labeled as 1 and 2, respectively) contained NPGPx and GRP78 cross-linked via disulfide bonds between NPGPx Cys86 and GRP78 Cys420 or Cys41, respectively (Supp. Figure 11 and schematics in Figure 6F, G). Consistently, the C86A NPGPx mutant failed to completely form the complex (Figure 6C). On the other hand, mutations in GRP78 Cys420 or Cys41 (Cys to Ala) only disrupted the formation of one of the complex bands (Figure 6D), suggesting that the formation of covalently linked NPGPx-GRP78 is mediated by disulfide bonds between NPGPx Cys86 and GRP78 Cys41/Cys420 (Figure 6E, F). In addition, the monomer fraction of rGRP78 incubated with or without oNPGPx (bands 3 and 4 in Figure 6C) was analyzed by MS. The Cys41-Cys420 bond of GRP78 was identified in band 3 but was barely found in band 4 (Supplemental Figure 12 A and B), suggesting that oNPGPx mediates Cys41-Cys420 bond formation in GRP78 in vitro. Separately, we also showed that, in the absence of NPGPx, treating GRP78 with 9 mM H2O2 (10 minutes at room temperature) resulted in the formation of the Cys41-Cys420 bond (band 2 in Supplemental Figure 12C), along with a sulfonic Cys residue (Cys-SO3H, band 3 in Supplemental Figure 12C) that was not seen in band 3 of Figure 6C, suggesting that H2O2 treatment is not as efficient as oNPGPx in inducing Cys41-Cys420 formation in vitro. Next, we tested whether GRP78 binding to denatured proteins was enhanced after treating with oNPGPx. As shown in Figure 6E and Supplemental Figure 13, GRP78 bound a significantly higher amount of denatured luciferase after treatment with oNPGPx (lane 7) compared to either rGRP78 (lane 5) or H2O2-treated GRP78 (lane 8). As summarized in Figure 6I, NPGPx is oxidized, and it forms an intramolecular disulfide bond between Cys57 and Cys86 in the presence of H2O2 (demonstrated in Supp. Figure 3 and 9; also step 1 of Figure 6I). oNPGPx then interacts with rGRP78 (Figure 6B; also step 2 of Figure 6I) and forms NPGPx-GRP78 intermediates (step 3 of Figure 6I, and Figure 6C, D). At the end, the Cys41-Cys420 bond in GRP78 is formed (Supp. Fig. 12A, B) and facilitates its misfolded/denatured protein-binding activity (step 4 in Figure 6I; also see Figure 6E and Supplement Figure 13). Taken together, these results suggest that oNPGPx transfers its disulfide bond to GRP78 to facilitate its chaperone activity.
Figure 6. Oxidized NPGPx interacts with GRP78 to form a disulfide-linked complex and enhances its binding affinity to denatured luciferase.
(A) Superimposition of 2D 15N-HSQC spectra of DTT-reduced 15N-labeled NPGPx, free (black) and with GRP78 (red). (B) Superimposition of 2D 15N-HSQC spectra of H2O2-oxidized 15N-labeled NPGPx, free (black) and with GRP78 (red). Only oxidized NPGPx is able to interact with GRP78, as evidenced by significant line broadening. 0.2~0.4 mM of NPGPx and GRP78 was used in NMR analysis with a 1:1 molar ratio. (C) Non-reducing SDS-PAGE of GRP78 with NPGPx and its mutants in different conditions. The upper and lower arrows (bands 1 and 2) indicate NPGPx and GRP78 complexes with disulfide bonds between Cys86 of NPGPx and Cys420/Cys41 of GRP78. Bands 3 and 4 were GRP78 monomers, and they were also subjected to mass analysis (Supp. Fig. 12A, B). Cys41-Cys420 of GRP78 was detected in band 3 but was barely found in band 4 (ratio was about 40:1). *: impurity (E. coli thioredoxin) (D) Non-reducing SDS-PAGE of GRP78 and its mutants with NPGPx in different conditions. The arrows mark NPGPx and GRP78 complexes described previously. (E) Immunoblot of native (n-) or heat-denatured (d-) luciferase with or without GRP78 under different conditions. GRP78 was able to interact only with heat-denatured luciferase and this ability was enhanced after interaction with H2O2-oxidized NPGPx (lane 7). Lanes 5–8 were normalized to GRP78; the quantification is shown in Supplemental Figure 13. (F-H) Schematics of peptide sequences of NPGPx and GRP78 with Cys cross-linking. NPGPx Cys86-S-S-Cys41 GRP78 (F), NPGPx Cys86-S-S-Cys420 GRP78 (G), and GRP78 Cys41-S-SCys420 (H) are shown. (I) Model summarizing how NPGPx modulates the chaperone activity of GRP78. A detailed explanation is provided in the text.
Loss of NPGPx results in systemic oxidative stress damage, increased carcinogenesis, and shortened lifespan in mice
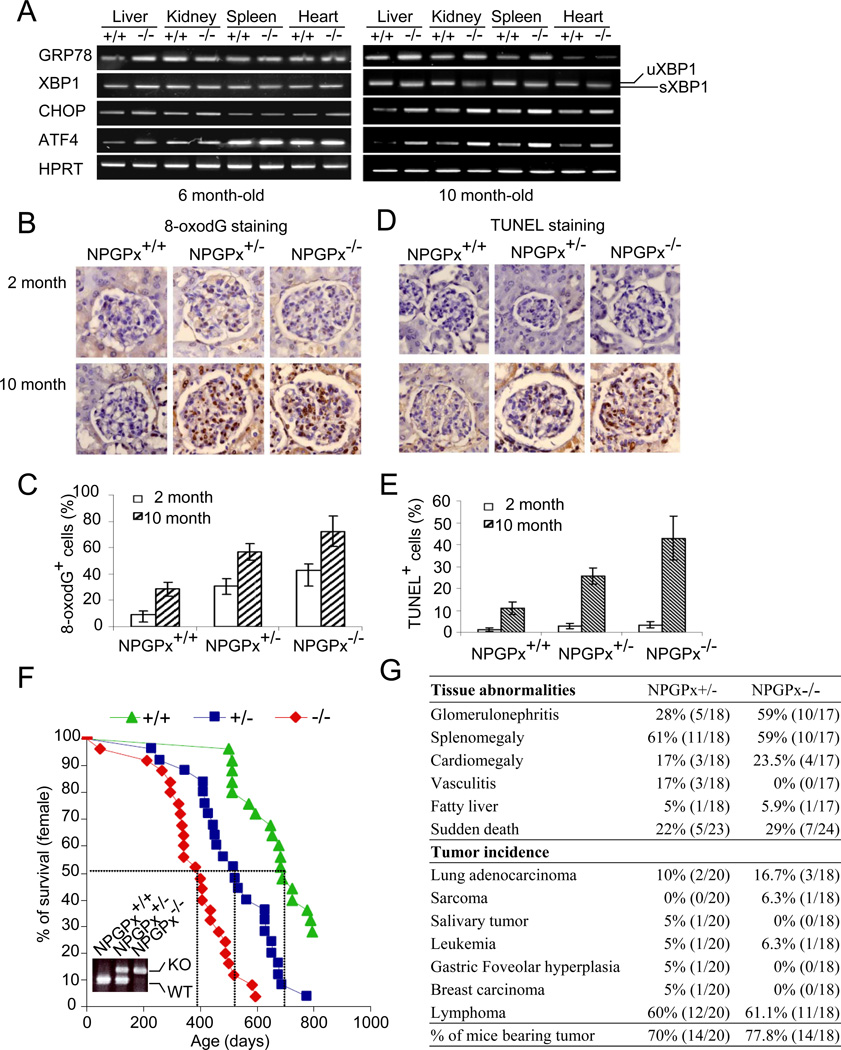
We have shown that loss of NPGPx leads to increased oxidative stress in MEFs. To explore how NPGPx affects animals, NPGPx-deficient mice were systematically characterized. We isolated and compared different organs, including the liver, kidney, spleen, and heart, from NPGPx−/− and WT mice at different ages using several ER unfolded protein response readouts including GRP78, XBP1, activating transcription factor 4 (ATF4), and C/EBP homology protein (CHOP) (Schroder and Kaufman, 2005). GRP78 and ATF4 increased in the liver of 6-month-old NPGPx−/− mice compared with WT mice, whereas the expression of GRP78, CHOP, and ATF4 and the splicing of XBP-1 (sXBP1) significantly increased in the liver, kidney, spleen, and heart of 10-month-old NPGPx−/− mice (Figure 7A). In addition, severe oxidative DNA damage was indicated by 8-oxoguanine staining accumulated in kidney sections derived from NPGPx-deficient mice at 2–10 months of age (Figure 7B, C). Importantly, a significant increase in cell apoptosis as indicated by TUNEL staining was observed in NPGPx−/− kidney sections, but few were seen in the corresponding WT mice (Figure 7D, E). These results indicate the accumulation of systemic oxidative damage in different organs with age. Consistent with the increased oxidative stress in multiple organs, the lifespan of NPGPx−/− mice was significantly shortened. The median lifespan decreased from 760 days in WT mice to 400 days in NPGPx−/− mice (Figure 7F). Autopsies of moribund mice revealed that NPGPx−/− mice carried multi-organ abnormalities, including antibody deposition in kidney (Supplemental Figure 14A), anti-nucleus antibody in serum (Supplemental Figure 14B), glomerulonephritis, splenomegaly, cardiomegaly, vasculitis, and fatty liver (Figure 7G). Further incidence of various malignant neoplasms was also observed in NPGPx−/− mice (Figure 7G) compared with WT strains with the same genetic background (Jeng et al., 2007). These observations support the notion that loss of NPGPx leads to increased systemic oxidative stress damage, high risk of carcinogenesis, and shortened lifespan.
Figure 7. NPGPx−/− mice exhibit systemic oxidative stress, increased tumor incidence, and shortened lifespan.
(A) RT-PCR analysis of ER stress pathway genes in liver, kidney, spleen, and heart. Total RNA from each tissue (from 6- or 10-month old mice) was reverse transcribed and amplified by PCR. The heart was used as an internal control. (B) 8-oxoG staining and (D) TUNEL staining of kidney from young (2-month old) and aged (10-month old) WT, NPGPx+/−, or NPGPx−/− mice. Quantitation of kidney 8-oxoguanin (8-oxoG) staining (C) and the percentage of TUNEL+ cells (E) within young or aged kidneys are shown. (F) Kaplan–Meier survival curves for WT, NPGPx+/−, and NPGPx−/− mice (G) Pathological examination of moribund mice.
Discussion
NPGPx serves as a sensor for ER oxidative stress
NPGPx is a member of the GPx family. However, it lacks GPx enzymatic activity, with either glutathione or thioredoxin as the acceptor, rendering the classification questionable. As illustrated by the proposed model in Figure 6I, we showed that NPGPx is a novel oxidative stress sensor that transmits stress signals by oxidation of its Cys57 and Cys86 to form a disulfide bond. oNPGPx covalently binds to GRP78 at Cys41/Cys420 through its Cys86 and facilitates the refolding of misfolded proteins by GRP78, thus alleviating oxidative stress. Cells without NPGPx exhibit impaired GRP78 chaperone activity, accumulate misfolded proteins in the ER, and are sensitive to oxidative stress. NPGPx deficiency leads to systemic oxidative stress, increases carcinogenesis, and shortens life span in animals. The importance of thiol/disulfide transfer between the oxidative stress sensor and chaperone activity was not previously clear; we are the first to demonstrate that disulfide transfer between NPGPx and GRP78 can increase GRP78 chaperone activity.
NPGPx acts as an oxidative sensor/transducer more than a glutathione peroxidase
GPx proteins were initially named according to their peroxidase activity in catalyzing the conversion of H2O2 coupled to the oxidation of reduced glutathione (GSH) (Awasthi et al., 1975). However, NPGPx cannot catalyze the conversion of H2O2 coupled to GSH or thioredoxin oxidation (Supp. Fugure 1). GPx1, 2, 3, 5, and 6 contain a homologous GSH-binding motif containing 1 Lys and 3 Arg residues (Nguyen et al., 2011). This motif is missing in NPGPx, rendering NPGPx unable to bind GSH. Instead, NPGPx interacts with Cys-containing redox-sensitive proteins through disulfide bond formation (Figure 4). This novel redox signaling is reminiscent of the function of GPx3 in yeast. Upon oxidative stress, GPx3 binds and oxidizes the transcription factor Yap1, which transactivates the expression of a variety of antioxidant proteins (Delaunay et al., 2002). As our results suggest, activated NPGPx via disulfide bonding is required for the NPGPx-GRP78 interaction and GRP78 function (Figure 6). H2O2-treated GRP78 contains some Cys-SO3H, which cannot bind to NPGPx, rendering it a weaker chaperone (Figure 6). These data suggest that the GRP78-NPGPx interaction is important for GRP78 chaperone function, and the oxidation of NPGPx is the critical initial step in this process.
Cys57 and Cys86 are critical for the anti-oxidative stress activity of NPGPx
NPGPx plays a critical role in relieving ER oxidative stress. Among its 3 Cys residues (Cys18, Cys57, and Cys86,), Cys57 and Cys86 are essential for its anti-oxidative stress activity (Figure 3). Based on our data, it is plausible that NPGPx senses oxidative stress first by oxidizing the thiol groups of Cys57 and Cys86 to form a reversible intra-molecular disulfide bond, which significantly changes its conformation since these 2 residues are 11.58 Å apart in its reduced state (Supplement Figure 10). oNPGPx binds to GRP78 and covalently links its Cys86 with the Cys41/Cys420 of GRP78. oNPGPx induces Cys41-Cys420 bond formation in GRP78 in vitro (Figure 6H and Suppelment Figure 12A, B), increasing the ability GRP78 to bind denatured/misfolded protein (Figure 6). Our data thus provide a mechanism for how NPGPx modulates the key stress responder GRP78 to relieve oxidative stress. Interestingly, the NPGPx coding sequence is highly homologous to yeast GPx3, which participates in hydroperoxide scavenging and sensing (Delaunay et al., 2002). Yeast GPx3 contains 3 Cys residues (Cys36, Cys64, Cys82), but only Cys36 is important for GPx3 function (Delaunay et al., 2002). In contrast, NPGPxC57A can neither protect cells from oxidative stress nor form oxidative intermediates with thiol group-containing proteins, indicating that Cys57 also has a distinctive role in NPGPx function. Thus, NPGPx is a unique mammalian peroxide sensor for regulating thiol-containing proteins.
NPGPx positively modulates GRP78 chaperone activity to reduce oxidative stress
GRP78 is the key effecter responding to ER stress (Schroder and Kaufman, 2005). However, many other chaperone proteins, especially those of the GRP family, also exert some stress-response functions (Fu and Lee, 2006). As shown in Figure 4, GRP78, GRP75, and HSP7C were identified as the main interacting proteins of NPGPx in the presence or absence excessive oxidative stress. These proteins have diverse functions ranging from chaperone activity to other stress responses in the ER, cytoplasm and mitochondria. Although NPGPx is mainly localized in the ER, it is also seen in the cytoplasm and nucleus (Figure 4B and unpublished data), perhaps due to its small size, providing an opportunity for these proteins to interact. It will be of interest to further dissect the other potential functions of those NPGPx-interacting proteins. Interestingly, the common function of these interacting proteins is to chaperone unfolded proteins. GRP78 offers the best example to illustrate the molecular basis of the interaction.
NPGPx facilitates GRP78 to obtain full function via intramolecular disulfide bond formation
The importance of thiols/disulfides in GRP78 has not been explored. As shown here, intramolecular disulfide formation appeared to be important to fully activate GRP78 (Figure 5E, H and Supplement Figure 6). Cys41 and Cys420 of GRP78 are located in distinct domains, and no full-length structure is available to allow determination of their distances. We speculate that Cys41 is distant from Cys420 in folded GRP78, and that generating an intramolecular disulfide bond GRP78 would require a major conformational change. This is supported by band shifts in non-reducing SDS-PAGE (Supplement Figure 12C). Disulfide bonds between such distant Cys residues may be difficult to form by H2O2 treatment, and oNPGPx is required to facilitate the process. The low efficiency of GRP78 oxidation by NPGPx in vitro could be due to the absence of potential cofactors for thiol/disulfide transfer. The efficiency may be much better in vivo, where these factors may be present. As expected, NPGPx deficiency compromised GRP78 activity, and feedback upregulation of GRP78 expression was seen in NPGPx−/− mice (Figure 7A). In summary, the loss of NPGPx impedes GRP78 intramolecular disulfide formation and compromises its chaperone activity in response to ER oxidative stress.
Loss of NPGPx causes systemic oxidative stress
The spectrum of oxidative damage in different organs in NPGPx−/− mice is different from that in other GPx-deficient mice. A recent report supports our finding that NPGPx expression protects cells from oxidative damage (Peng et al., 2011). Although NPGPx−/− mice develop normally, widespread oxidative damage was observed in 10 month old mice. One possible explanation is that NPGPx is expressed in a variety of tissues (Utomo et al., 2004) and may be required for general redox homeostasis. Since loss of GRP78 is lethal in mice (Luo et al., 2006); NPGPx cannot be the sole regulator for this critical effector. Recently, GPx8 was identified as a protein disulfide isomerase peroxidase that facilitates oxidative protein folding in the ER (Nguyen et al., 2011). NPGPx and GPx8 have similar structures and may partially compensate for the loss of NPGPx. This possibility is currently under vigorous investigation.
Does NPGPx participate in oxidative stress-related diseases?
In humans, several diseases including cancer, obesity, and autoimmune diseases may result from either a dysregulated unfolded-protein response or excessive oxidative stress. In NPGPx-deficient mice, oxidative stress significantly increased/accumulated with age (Figure 7 and Supplemental Figure 14). This may result from the suboptimal function of chaperones, including GRP78, consistent with the notion that disabled GRPs enhance cellular ER stress (Lee, 2001). Accumulated oxidative stress contributes to DNA damage, cell death, aging, and tumor formation (Figure 7). On the other hand, NPGPx may directly modulate other key metabolic enzymes or transcriptional regulators and contribute to disease, as many proteins associate with NPGPx under excessive stress (Figure 4A). Endogenous ROS levels were higher in NPGPx-deficient cells along with increased cancer incidence in mice, suggesting the importance of NPGPx as a ROS scavenger in inhibiting cancer development. Interestingly, excessive ROS is one of the risk factors that accelerate cancer progression by the direct inactivation of pyruvate kinase M2 (PKM2) (Anastasiou et al., 2011). In addition, we have observed a very high percentage of glomerulonephritis (Figure 7G), antibody deposition in kidneys (Supplement Figure 14A), and anti-nuclear autoantibodies in NPGPx−/− mice (Supplement Figure 14B). These findings are similar to human lupus nephritis, for which ER oxidative stress is considered to be the underlying cause (Morgan et al., 2005). Recently, several chemical chaperones such as 4-phenyl butyric acid and tauroursodeoxycholic acid have been developed to reduce ER stress (Ozcan et al., 2006). Whether these chemicals have any efficacy in preventing diseases by reducing systemic oxidative damage resulting from the loss of NPGPx is worth studying.
Experimental Procedures
Generation of NPGPx−/− Mice
A gene targeting strategy was used to generate NPGPx+/− mice. To generate the NPGPx targeting vector, the 5.8-kb endogenous NPGPx genomic locus fragment, flanked by a neomycin cassette (Neo-cassette) at exon 2, was digested with BamHI and cloned into the targeting vector p2TK to generate p2TK-NPGPx-KO. This targeting vector was then linearized by SalI and electroporated into E14 ES cells. Colonies resistant to neomycin were selected and characterized for homologous recombination by Southern blotting and genomic PCR. For Southern blotting analysis, genomic DNA was digested by HindIII, separated by Tris-borate EDTA (TBE) PAGE, and transferred to a membrane. An isotope-labeled 3' probe (schematic in Figure 1A, short black bar beneath the NPGPx-KO genome) was used to hybridize to the NPGPx genomic locus and then the result developed on an X-ray film. In one of the correct clones, 4-H4, the target allele generated 2 bands (4.1 kb and 2,7 kb, Figure 1C), while the original allele showed 1 (4.1 kb, Figure 1C). To perform the genomic polymerase chain reaction (PCR), primers recognizing either intact NPGPx exon 2 loci (schematic in Figure 1A, top left and bottom right) or the Pgk-promoter driven Neo-cassette (Neo-F, Neo-KO1, and pGK3’ in Figure 1A) were used to amplify HindIII-digested genomic DNA. Genomic PCR of 4-H4 gave 3 products amplified by individual primer pairs: 4.1 kb with Neo-F (top left), 3.8 kb with Neo-KO1 (right bottom), and 2.9 kb from pGK3' (right bottom), respectively. Primer and probe usage are illustrated in Figure 1A. The 4-H4 ES clone was then microinjected into C57BL/6 blastocysts, which were then transferred into foster mothers. Offspring with a germline-transmitted target gene were obtained by backcrossing chimera mice with C57BL/6. Productive offspring from this backcross carrying the disrupted NPGPx allele (NPGPx+/− mice) were crossed to generate homozygous NPGPx knockout mice.
MEF preparation
MEFs were prepared using 13.5-day NPGPx+/+ and NPGPx−/− embryos as previously described (Zheng et al., 2000). In brief, embryos were washed with PBS, chopped in 1× trypsin, and digested at 37 °C for 30 minutes. Digestion was neutralized with 10% FBS-containing low-glucose DMEM. Cells were washed and plated following the 3T3 culture protocol in which 106 cells were passed into a 10 cm dish every 3 days.
Immunohistochemical staining and TUNEL assay
Mouse tissues from autopsy were collected, fixed in 4% paraformaldehyde and embedded in paraffin. Tissues were cut into 5-μm thick sections and subjected either to IHC analysis (VectaStain Elite ABC kit, Vector Laboratories, Burlingame, CA) using antibodies against 8-oxoguanin (ab26842; Abcam, Cambridge, UK), or the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay (Pierce, Rockford, IL), following manufacturers’ instructions.
Immunoflorescent staining (IF staining)
IF staining against GRP78 was performed following a general protocol (Reddy et al., 2003). For detail information, please see supplement materials and methods.
FACS analysis for ROS detection and annexin V staining
For ROS detection, primary MEFs were cultured in medium containing 5-(and-6)-carboxy-2’,7’-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA). CM-H2DCFDA is an ester probe that reacts with intracellular ROS, and the oxidative adduct of CM-H2DCFDA emits green florescence upon excitation (Eruslanov and Kusmartsev, 2010). After a 10 minutes incubation, cells were subjected to FACS analysis. For annexin V staining, the trypsinized cells were stained with annexin V (A13201; Molecular Probes, Eugene, OR) with propidium iodide (P4170; Sigma, St. Louis, MO) at 4 °C for 30 minutes. The stained cells were subjected to FACS analysis using FACS DIVA software (BD, Franklin Lakes, NJ).
Reverse transcription (RT)-PCR
Total RNA from tissues or MEFs were extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA). 1 µg total RNA was reverse-transcribed into cDNA, and 40 ng cDNA was used in each PCR reaction. The primers information was shown in the Supplemental table 1.
Western Blot
For Western blot analysis, primary MEFs were lysed with RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and 1 mM PMSF and 1 mM DTT), sonicated, and pre-clarified by centrifugation. About 20 μg of extracted protein lysate in each individual lane was separated in SDS-PAGE followed by Western blot with specific antibodies as probes. Antibodies against NPGPx (GTX70266; GeneTex, Irving, CA), GRP78 (610978; BD), Calnexin (MA3-027; Pierce), GFP (GTX113617; Genetex), luciferase (GTX27358; Genetex), p84 (GTX70220; GeneTex) and α-tubulin (GTX11302; GeneTex) were used.
Immunoprecipitation (IP)
For the IP assay, cells were washed with cold PBS and subsequently lysed with lysis-250 buffer (Chen et al., 1996) with 0.5% Triton-X 100. The protein extract was sonicated and pre-clarified by centrifugation, and the lysate was further pre-absorbed with agarose A/G beads. Antibodies against the protein of interest were added to the pre-cleaned lysate, and incubated at 4 °C for 4 hours with rotation. To pull-down the protein-antibody complex, the lysate was incubated with agarose A/G beads and incubated for 1 hour. Agarose A/G beads were pelleted by centrifugation, washed, and boiled in SDS sample buffer (1% SDS, 50 mM Tris-HCl, pH 6.8, 10% glycerol and 0.01% bromophenol blue) with 50 mM DTT. For non-reducing conditions, sample buffer without DTT was preheated and then mixed with the IP sample. Protein complexes were analyzed by 8-16% gradient SDS-PAGE followed by Western blot analysis. Antibodies against NPGPx (GTX108578, Genetex,), GRP78, luciferase, and GFP were used.
Retroviral vector construction, production, and transduction
To make the NPGPx retrovirus construct, we first amplified the NPGPx coding sequence from cDNA derived from U2OS mRNA. Primer pairs used in human NPGPx RT-PCR were hGPxL5' (CCgCggATCCggAACAAgCCATggTggC) and hGPxL3' (CCgCggATCCTTAAAgATCTTCTCgCTTCAgTAggATg). Amplified human NPGPx coding sequences were digested with BamHI and cloned into pBSK to derive pBSK-hNPGPx. To generate the NPGPx mutants pBSK-C18A, pBSK-C57A, pBSK-C86A, pBSK-C2A2, and pBSK-C3A3 hNPGPx, we used PCR-based site-directed mutagenesis to change Cys to Ala (TGC mutated into GCC). WT or mutant hNPGPx was further subcloned from pBSK into the retroviral vector pQCXIP by NotI and EcoRI restriction. The pQCXIP-hNPGPx vectors (WT and mutants) were individually co-transfected with pMD.G (Env-encoding vector) into Gp2-293 cells to generate hNPGPx-carring retrovirus. We collected Gp2-293 culture medium containing retrovirus every 12 hours after retroviral vector transfection. For retrovirus infection, we seeded passage 1 MEFs in a 10 cm culture dish to 30% confluence, and infected them with freshly harvested retrovirus 3 times, every 12 hours with 8 μg/ml polybrene. The retrovirus-infected MEFs were selected using 2 μg/ml puromycin and passaged once (passage 2) for the cell viability assay (Figure 4) and the co-immunoprecipitation assay (Figure 5).
Cell growth assay
The 3T3 protocol was used for this assay. Briefly, 3 × 105 primary MEFs from each passage were seeded in a 6-cm petri dish, and cells were cultured in a humidified CO2 incubator containing 20% or 3% O2. Cells were subcultured every 3 days and cell numbers were counted simultaneously.
MTT assay
To measure the viability of H2O2- or tunicamycin-treated MEFs, the MTT assay was used. Briefly, MEFs were seeded in 96-well plates (1 × 104 cells per well) 1 day before the experiment. Cells were then treated with H2O2 or tunicamycin for 3 h, then washed 3 times with PBS and maintained in fresh culture medium for 21 hour followed by the MTT assay according to the manufacturer’s instructions (Sigma).
Recombinant protein expression and purification
NPGPx (19–187 amino acids) and GRP78 (19–654 amino acids) were cloned into the pET48 and pET47 vectors (Novagen), respectively, with a His6-tag and tobacco etch virus (TEV) protease digestion site on the N-terminus. Cys to Ala substitutions in NPGPx were introduced by site-directed mutagenesis. Overexpression of NPGPx and GRP78 was induced by 1 mM IPTG at 25 °C for 16 hours. The purification was carried via immobilized metal affinity chromatography on a nickel affinity column (Sigma). The N-terminal tag was removed by TEV protease digestion at 4 °C for 24 hours.
Protein interaction assay
To prepare reduced or oxidized NPGPx and GRP78, the proteins were treated with 10 mM Tris (2-carboxyethyl) phosphine (TCEP; Sigma) or 20 mM H2O2 (Sigma), respectively, for 1 hour TCEP and H2O2 were removed by buffer exchange using Amicon Ultra-10 (Millipore, Billerica, CA). The protein interaction assays were carried out at 25 °C for 2 hours using the conditions shown in Figure 6C and D, and the results were analyzed on 8 % non-reducing SDS-PAGE.
NMR analysis
15N-labeled NPGPx was prepared using 15NH4Cl as the sole nitrogen source in the minimal growth media. After reduction or oxidation, the 15N-heteronuclear single-quantum correlation spectra of reduced or oxidized NPGPx with or without reduced GRP78 were acquired on a Bruker Avance 600MHz spectrometer equipped with cryogenic TCI probe. The data was processed and analyzed by Topspin 2.1.
Binding assay of GRP78 and luciferase
Luciferase was denatured at 42 °C for 20 minutes. 6× His-tagged reduced or oxidized GRP78 was mixed with reduced or oxidized NPGPx first before loading the complexes onto the nickel affinity resin. Native or denatured luciferase was added to the resin and incubated at 25 °C for 1 hour. After washing away unbound luciferase, TEV protease was added to the resin to release the complexes, which were analyzed by SDS-PAGE followed by Western blot.
Supplementary Material
Acknowledgements
We thank Chi-Fen Chen and Zhanyong Shu, for their initial contribution to this work. This work was mainly supported by funds from the Academia Sinica, Taiwan and partially from the Philips Morris Foundation (USA) to Wen-Hwa Lee. Support for the transgenic mouse facility was also received from the NIH (HD034534, CA014195, NS072031) to Kuo-Fen Lee.
References
- Ali MH, Schlidt SA, Chandel NS, Hynes KL, Schumacker PT, Gewertz BL. Endothelial permeability and IL-6 production during hypoxia: role of ROS in signal transduction. Am J Physiol. 1999;277:L1057–L1065. doi: 10.1152/ajplung.1999.277.5.L1057. [DOI] [PubMed] [Google Scholar]
- Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi YC, Beutler E, Srivastava SK. Purification and properties of human erythrocyte glutathione peroxidase. J Biol Chem. 1975;250:5144–5149. [PubMed] [Google Scholar]
- Bashir S, Harris G, Denman MA, Blake DR, Winyard PG. Oxidative DNA damage and cellular sensitivity to oxidative stress in human autoimmune diseases. Ann Rheum Dis. 1993;52:659–666. doi: 10.1136/ard.52.9.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CF, Chen Y, Dai K, Chen PL, Riley DJ, Lee WH. A new member of the hsp90 family of molecular chaperones interacts with the retinoblastoma protein during mitosis and after heat shock. Mol Cell Biol. 1996;16:4691–4699. doi: 10.1128/mcb.16.9.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu FF, Doroshow JH, Esworthy RS. Expression, characterization, and tissue distribution of a new cellular selenium-dependent glutathione peroxidase, GSHPx-GI. J Biol Chem. 1993;268:2571–2576. [PubMed] [Google Scholar]
- Dear TN, Campbell K, Rabbitts TH. Molecular cloning of putative odorant-binding and odorant-metabolizing proteins. Biochemistry. 1991;30:10376–10382. doi: 10.1021/bi00107a003. [DOI] [PubMed] [Google Scholar]
- Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- Eruslanov E, Kusmartsev S. Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol Biol. 2010;594:57–72. doi: 10.1007/978-1-60761-411-1_4. [DOI] [PubMed] [Google Scholar]
- Fu Y, Lee AS. Glucose regulated proteins in cancer progression, drug resistance and immunotherapy. Cancer Biol Ther. 2006;5:741–744. doi: 10.4161/cbt.5.7.2970. [DOI] [PubMed] [Google Scholar]
- Garcia-Mata R, Bebok Z, Sorscher EJ, Sztul ES. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J Cell Biol. 1999;146:1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghyselinck NB, Jimenez C, Courty Y, Dufaure JP. Androgen-dependent messenger RNA(s) related to secretory proteins in the mouse epididymis. J Reprod Fertil. 1989;85:631–639. doi: 10.1530/jrf.0.0850631. [DOI] [PubMed] [Google Scholar]
- Goldstein BJ, Mahadev K, Wu X, Zhu L, Motoshima H. Role of insulin-induced reactive oxygen species in the insulin signaling pathway. Antioxid Redox Signal. 2005;7:1021–1031. doi: 10.1089/ars.2005.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Dosal R, Horan KA, Rahbek SH, Ichijo H, Chen ZJ, Mieyal JJ, Hartmann R, Paludan SR. HSV infection induces production of ROS, which potentiate signaling from pattern recognition receptors: role for S-glutathionylation of TRAF3 and 6. PLoS Pathog. 2011;7:e1002250. doi: 10.1371/journal.ppat.1002250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbette S, Lenne C, Leblanc N, Julien JL, Drevet JR, Roeckel-Drevet P. Two GPX-like proteins from Lycopersicon esculentum and Helianthus annuus are antioxidant enzymes with phospholipid hydroperoxide glutathione peroxidase and thioredoxin peroxidase activities. Eur J Biochem. 2002;269:2414–2420. doi: 10.1046/j.1432-1033.2002.02905.x. [DOI] [PubMed] [Google Scholar]
- Jeng YM, Cai-Ng S, Li A, Furuta S, Chew H, Chen PL, Lee EY, Lee WH. Brca1 heterozygous mice have shortened life span and are prone to ovarian tumorigenesis with haploinsufficiency upon ionizing irradiation. Oncogene. 2007;26:6160–6166. doi: 10.1038/sj.onc.1210451. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- Kumar B, Koul S, Khandrika L, Meacham RB, Koul HK. Oxidative stress is inherent in prostate cancer cells and is required for aggressive phenotype. Cancer Res. 2008;68:1777–1785. doi: 10.1158/0008-5472.CAN-07-5259. [DOI] [PubMed] [Google Scholar]
- Kuo SC, Lampen JO. Tunicamycin--an inhibitor of yeast glycoprotein synthesis. Biochem Biophys Res Commun. 1974;58:287–295. doi: 10.1016/0006-291x(74)90925-5. [DOI] [PubMed] [Google Scholar]
- Lee AS. The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem Sci. 2001;26:504–510. doi: 10.1016/s0968-0004(01)01908-9. [DOI] [PubMed] [Google Scholar]
- Luo S, Mao C, Lee B, Lee AS. GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol Cell Biol. 2006;26:5688–5697. doi: 10.1128/MCB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddipati KR, Marnett LJ. Characterization of the major hydroperoxide-reducing activity of human plasma. Purification and properties of a selenium-dependent glutathione peroxidase. J Biol Chem. 1987;262:17398–17403. [PubMed] [Google Scholar]
- Malhotra JD, Kaufman RJ. Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid Redox Signal. 2007;9:2277–2293. doi: 10.1089/ars.2007.1782. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Helenius A. Misfolding and aggregation of newly synthesized proteins in the endoplasmic reticulum. J Cell Biol. 1992;117:505–513. doi: 10.1083/jcb.117.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills GC. Hemoglobin catabolism. I. Glutathione peroxidase, an erythrocyte enzyme which protects hemoglobin from oxidative breakdown. J Biol Chem. 1957;229:189–197. [PubMed] [Google Scholar]
- Morgan MJ, Liu ZG. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 2011;21:103–115. doi: 10.1038/cr.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan PE, Sturgess AD, Davies MJ. Increased levels of serum protein oxidation and correlation with disease activity in systemic lupus erythematosus. Arthritis Rheum. 2005;52:2069–2079. doi: 10.1002/art.21130. [DOI] [PubMed] [Google Scholar]
- Nguyen VD, Saaranen MJ, Karala AR, Lappi AK, Wang L, Raykhel IB, Alanen HI, Salo KE, Wang CC, Ruddock LW. Two endoplasmic reticulum PDI peroxidases increase the efficiency of the use of peroxide during disulfide bond formation. J Mol Biol. 2011;406:503–515. doi: 10.1016/j.jmb.2010.12.039. [DOI] [PubMed] [Google Scholar]
- Nigam SK, Goldberg AL, Ho S, Rohde MF, Bush KT, Sherman M. A set of endoplasmic reticulum proteins possessing properties of molecular chaperones includes Ca(2+)-binding proteins and members of the thioredoxin superfamily. J Biol Chem. 1994;269:1744–1749. [PubMed] [Google Scholar]
- Nollen EA, Brunsting JF, Roelofsen H, Weber LA, Kampinga HH. In vivo chaperone activity of heat shock protein 70 and thermotolerance. Mol Cell Biol. 1999;19:2069–2079. doi: 10.1128/mcb.19.3.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, Gorgun CZ, Hotamisligil GS. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D, Belkhiri A, Hu T, Chaturvedi R, Asim M, Wilson KT, Zaika A, El-Rifai W. Glutathione peroxidase 7 protects against oxidative DNA damage in oesophageal cells. Gut. 2011 doi: 10.1136/gutjnl-2011-301078. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rascon B, Harrison JF. Lifespan and oxidative stress show a non-linear response to atmospheric oxygen in Drosophila. J Exp Biol. 2010;213:3441–3448. doi: 10.1242/jeb.044867. [DOI] [PubMed] [Google Scholar]
- Reddy RK, Mao C, Baumeister P, Austin RC, Kaufman RJ, Lee AS. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J Biol Chem. 2003;278:20915–20924. doi: 10.1074/jbc.M212328200. [DOI] [PubMed] [Google Scholar]
- Rhee SG, Kang SW, Jeong W, Chang TS, Yang KS, Woo HA. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr Opin Cell Biol. 2005;17:183–189. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Riemer J, Bulleid N, Herrmann JM. Disulfide formation in the ER and mitochondria: two solutions to a common process. Science. 2009;324:1284–1287. doi: 10.1126/science.1170653. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Sztajer H, Gamain B, Aumann KD, Slomianny C, Becker K, Brigelius-Flohe R, Flohe L. The putative glutathione peroxidase gene of Plasmodium falciparum codes for a thioredoxin peroxidase. J Biol Chem. 2001;276:7397–7403. doi: 10.1074/jbc.M008631200. [DOI] [PubMed] [Google Scholar]
- Tang L, Gounaris K, Griffiths C, Selkirk ME. Heterologous expression and enzymatic properties of a selenium-independent glutathione peroxidase from the parasitic nematode Brugia pahangi. J Biol Chem. 1995;270:18313–18318. doi: 10.1074/jbc.270.31.18313. [DOI] [PubMed] [Google Scholar]
- Tokunaga M, Kawamura A, Kohno K. Purification and characterization of BiP/Kar2 protein from Saccharomyces cerevisiae. J Biol Chem. 1992;267:17553–17559. [PubMed] [Google Scholar]
- Ursini F, Maiorino M, Gregolin C. The selenoenzyme phospholipid hydroperoxide glutathione peroxidase. Biochim Biophys Acta. 1985;839:62–70. doi: 10.1016/0304-4165(85)90182-5. [DOI] [PubMed] [Google Scholar]
- Utomo A, Jiang X, Furuta S, Yun J, Levin DS, Wang YC, Desai KV, Green JE, Chen PL, Lee WH. Identification of a novel putative non-selenocysteine containing phospholipid hydroperoxide glutathione peroxidase (NPGPx) essential for alleviating oxidative stress generated from polyunsaturated fatty acids in breast cancer cells. J Biol Chem. 2004;279:43522–43529. doi: 10.1074/jbc.M407141200. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Zheng L, Pan H, Li S, Flesken-Nikitin A, Chen PL, Boyer TG, Lee WH. Sequence-specific transcriptional corepressor function for BRCA1 through a novel zinc finger protein, ZBRK1. Mol Cell. 2000;6:757–768. doi: 10.1016/s1097-2765(00)00075-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.