Abstract
Numerous candidate gene association studies of bipolar disorder (BP) have been carried out, but the results have been inconsistent. Individual studies are typically underpowered to detect associations with genes of small effect sizes. We conducted a meta-analysis of published candidate gene studies to evaluate the cumulative evidence. We systematically searched for all published candidate gene association studies of BP. We then carried out a random-effects meta-analysis on all polymorphisms that were reported on by three or more case–control studies. The results from meta-analyses of these genes were compared with the findings from a recent mega-analysis of eleven genome-wide association studies (GWAS) in BP performed by the Psychiatric GWAS Consortium (PGC). A total of 487 articles were included in our review. Among these,33 polymorphismsin 18genes were reported on by three or more case–control studies and included in the random-effects meta-analysis. Polymorphisms in BDNF, DRD4, DAOA, and TPH1, were found to be nominally significant with a P-value < 0.05. However, none of the findings were significant after correction for multiple testing. Moreover, none of these polymorphisms were nominally significant in the PGC-BP GWAS. A number of plausible candidate genes have been previously associated with BP. However, the lack of robust findings in our review of these candidate genes highlights the need for more atheoretical approaches to study the genetics of BP afforded by GWAS. The results of this meta-analysis and from other on-going genomic experiments in BP are available online at Metamoodics (http://metamoodics.igm.jhmi.edu).
Keywords: mood disorders, candidate genes, meta-analysis
INTRODUCTION
Previous research from family, twin, and adoption studies have shown that genetic factors play an important role in the etiology of bipolar disorder (BP) [Todd and Botteron, 2002; Merikangas and Low, 2004]. Since the completion of the human genome project [Venter et al., 2001] over a decade ago, there has been an explosion in genetic association studies aimed at identifying susceptibility genes for BP. Studies carried out before the era of genome-wide association studies (GWAS) typically focused on specific genes suggested by various hypotheses about the underlying biology of BP. Reports from these candidate gene studies have implicated a number of “plausible” susceptibility genes for BP, but the findings have been inconsistent, making it difficult to draw definitive conclusions. We set out to review the literature and perform a meta-analysis of existing candidate gene studies to provide a comprehensive summary of findings on the leading susceptibility genes for BP to have emerged from the field over the past decade. We compared the results from the published candidate genes studies against those from the most comprehensive GWAS of BP to date [Sklar et al., 2011] carried out by the Psychiatric GWAS Consortium (PGC) to determine if the previous findings were corroborated by more recent genome-wide effort.
MATERIALS AND METHODS
Literature Search
A literature search was conducted of the PubMed database on all articles published up through September 1, 2009 using the following keyword algorithm: (bipolar depression OR bipolar disorder OR mood disorder OR affective disorder) and (single nucleotide polymorphism OR SNP OR gene OR polymorphism). A total of 5,384 articles were returned. These were manually reviewed by looking at their titles, abstracts, keywords, and full text as needed in order to identify those that were case–control or family-based candidate gene association studies of BP meeting our inclusion/exclusion criteria described below. We further searched the references of these articles to identify any other articles that were potentially missed by the initial PubMed search. The rationale for using the September 1, 2009 cutoff for the PubMed search was so we could compare the results of our candidate gene meta-analyses to the results from the PGC, which began its efforts to conduct mega-analyses of existing GWAS in 2009 [Sklar et al., 2011].
Study Inclusion/Exclusion Criteria
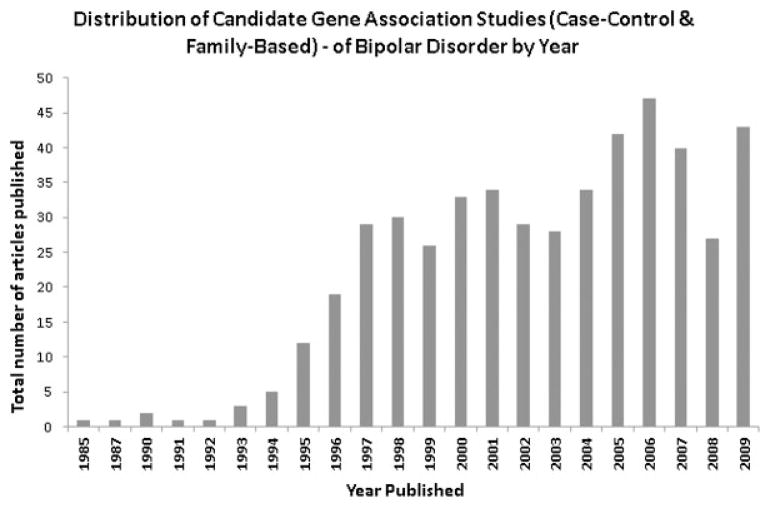
Studies were included in our review if they met the following criteria: (i) reported on candidate gene association studies (case–control or family-based) in adult humans; (ii) examined individual level genotype data (i.e., no pooled genotyping studies) with an appropriate set of controls; (iii) tested for association with a dichotomous phenotype of BP that included diagnoses of bipolar I disorder (BPI), bipolar II disorder (BPII), and/or schizoaffective disorder bipolar sub-type (SABP); (iv) no overlap with other studies (if there was overlap with another study, we included the study with the most inclusive data); and (v) published in a peer-reviewed journal in English. Studies were excluded from our review if they: (i) did not distinguish between types of mood disorders; (ii) examined drug response or other quantitative traits; (iii) examined specific clinical features and/or co-morbidities within mood disorders; (v) did not report on primary data; and (vi) did not provide any characterization of the study sample. A total of 487 articles reporting on associations with 362 unique genes met our inclusion/exclusion criteria. Figure 1 shows the distribution of publication of these 487 articles by year.
FIG. 1.
Distribution of 487 candidate gene association studies in BP (case–control and family-based) published in PubMed by year from initiation up through September 1, 2009.
Data Extraction
We extracted into evidence tables the following data from each of the published articles: (i) PubMed ID, author, journal, and year of publication; (ii) study design; (iii) sample information including country of origin, ethnicity, source of ascertainment, and subject counts; (iv) diagnoses, diagnostic instrument, and diagnostic criteria used; (v) polymorphism information including dbSNP [Sherry et al., 2001] reference SNP identification number (rsID), base pair (BP) location, major/minor allele, and genotype/allele counts for cases and controls or transmitted/untransmitted alleles for families; and (vi) sample meta-information including percentage of males for cases and controls, age at interview and age at onset. To assure the data for each polymorphism was recorded with a common orientation across all studies, we used the HapMap [International HapMap Consortium, 2003]—release 24, Phase I and II CEPH (Centre d’Etude du Polymorphisme Humain, Utah residents with ancestry from northern and western Europe) to designate the strand orientation and major/minor alleles. If a polymorphism was not genotyped in the HapMap CEPH samples, we used the SNP database of the National Center of Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/SNP/) dbSNP [Sherry et al., 2001] version 130 with human genome assembly build 36 and the University of California Santa Cruz (UCSC) Genome Browser Database [Fujita et al., 2011] (http://genome.ucsc.edu/) to determine a common orientation. Many studies did not provide rsIDs, but instead reported codes based on nucleotide or amino acid position (e.g., T102C in HTR2A or Thr25Asn also in HTR2A). To retrieve rsIDs for such polymorphisms we again used the dbSNP database. If a polymorphism still could not be mapped to an rsID we used the UCSC’s genome browser In-Silico PCR primer-BLAT [Kent, 2002] to map the polymorphisms using the provided primer sequences. When only allele counts were provided, we calculated the corresponding genotype counts assuming Hardy–Weinberg equilibrium. If the polymorphism was not in Hardy–Weinberg equilibrium in the controls, we kept the genotype counts as missing.
Meta-Analysis
We conducted a meta-analysis for all polymorphisms reported on by at least three case–control studies. We focused only on the case–control association studies in the meta-analysis in order to compare our findings with the larger case–control GWAS. Allelic odds ratios (OR), standard errors (SE), 95% confidence intervals (CI), P-values (P), and HWE for each polymorphism were calculated individually for each study included in the meta-analysis. The minor allele for the polymorphisms was used as the reference to calculate the ORs. For polymorphisms with multiple alleles (e.g. variable number of tandem repeats (VNTRs) and microsatellite markers) we used the most common allele as the reference allele and calculated separate OR’s for all other alleles relative to this. We excluded from the meta-analysis studies with genotype data that was not in HWE in controls. For each polymorphism, we calculated Woolf’s chi-squared test [WOOLF, 1955] to assess for between-study heterogeneity. Woolf’s statistic is distributed as approximately χ2 with k – 1 degrees of freedom (df), where k is the number of studies. Allelic summary OR and 95% CI were estimated under the DerSimonian–Laird random-effects model [DerSimonian and Laird, 1986] using inverse-variance weights in R statistical programming [Development Core Team, 2010]. A random-effects model allows for both between-study and within-study heterogeneity, while a fixed-effects model considers only within-study heterogeneity. When there is no evidence of heterogeneity between studies, the random-effects and fixed-effects models yield similar results. The significance of the summary OR was determined using an asymptotic Z-test. We corrected the nominal P-values by the number of candidate genes that were meta-analyzed to control for multiple testing.
Comparison With GWAS
To further explore the relevance of the BP candidate genes included in the meta-analysis, we compared our results against those from a recent mega-analysis of GWAS carried out by the PGC on BP (PGC-BP) [Sklar et al., 2011]. The PGC-BP performed a combined analysis of individual-level data (i.e., mega-analysis) from 11 case–control GWAS of BP including 16,730 subjects (7,481 BP cases and 9,249 controls). The primary analyses consisted of allelic tests of associations for ~2,427,090 directly genotyped or imputed SNPs across the genome. The untyped SNPs in the individual studies were imputed based on the HapMap—release 24, Phase I and II CEPH samples as a reference. The association tests were carried out using logistic regression with estimated allelic dosages and controlling for five principal components capturing ancestral background and indicator variables for the 11 studies.
We examined all SNPs in HWE and with imputation quality r2 > 0.30 in an area 10-kb upstream and downstream of the longest RefSeq transcript of each candidate gene We compared results for the same variant that was examined in the candidate gene meta-analysis. If the same variant was not genotyped or imputed in the PGC-BP mega-analysis, then we examined the SNP that was in highest linkage disequilibrium with it and/or nearest in physical distance.
RESULTS
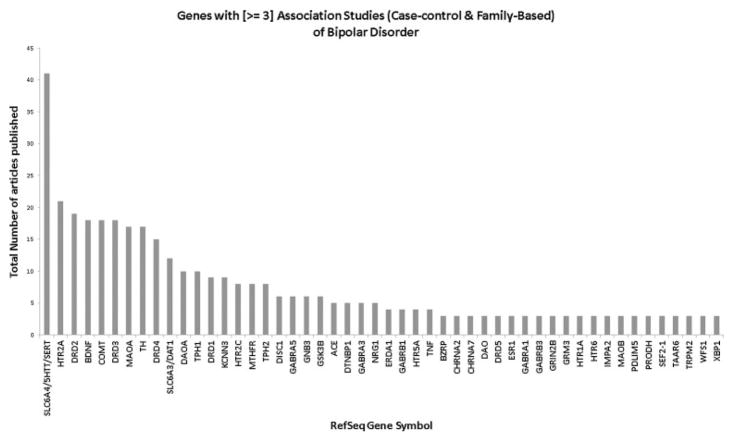
Of the 362 unique genes reported on by at least one of the 487 candidate gene association studies of BP, a total of 50 were examined by three or more such studies (case–control or family-based). These genes and the number of studies reporting on each of them are shown in Figure 2. The most widely studied gene by far was the serotonin transporter gene (SLC6A4 gene) with 41 different genetic studies of BP. The second most widely studied gene, 5-hydroxytryptamine (serotonin) receptor 2A (HTR2A), was examined in half as many studies (n = 21).
FIG. 2.
Distribution of candidate genes published with three or more association studies (case–control or family-based) in BP.
Eighteen of the 50 candidate genes identified here had 33 different polymorphisms that were examined in three or more case–control studies and therefore included in the meta-analysis. The 33 polymorphisms were investigated by a total of 140 unique studies (see Supplementary Table I for a list of references) and included 25 SNPs, two insertion/deletions (indels), and six VNTRs including four microsatellites, one minisatellite and one short tandem repeat (STR). Table I lists the top findings (i.e., lowest meta-analysis P-value) in each of the 18 candidate genes from the random-effects meta-analysis. Single polymorphisms in four different genes, BDNF, DRD4, DAOA, and TPH1, were found to be nominally significant with a P-value < 0.05. There was no evidence of heterogeneity between studies of these four polymorphisms (P > 0.05). The most significant polymorphism study-wide was rs1800532 (G/T) in tryptophan hydroxylase 1 (TPH1) (OR for the minor T allele = 1.24, 95% CI 1.09–1.41, P = 0.001). The meta-analysis of rs1800532 included data on a total of 1,848 cases and 2,075 controls from eight individual case–control studies. However, none of the findings, including with rs1800532, were significant after correcting for the 18 different genes that were meta-analyzed. We re-did the meta-analyses for these top 18 polymorphisms after removing non-Caucasian samples, but the results did not change meaningfully.
TABLE I.
Candidate Genes and Top Polymorphisms From the Random-Effects Meta-Analysis Based on the Systematic Literature Search
| RefSeq gene symbol | Chromosome position | Varianta | Variant type | bp locationb | MAFc | Total variants studiedd | No. of studiese | No. of casese | No. of controlse | Meta-analysis OR | 95% CI | Meta-analysis P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5HTT/SLC6A4 | 17q11.2 | rs57098334 (12/10) | Microsatellite | 25572722 | 0.30 | 2 | 18 | 1809 | 2693 | 0.92 | 0.80–1.06 | 0.249 |
| HTR2A | 13q14.2 | rs2070040 (G/A) | SNP | 46365627 | 0.44 | 5 | 6 | 955 | 1474 | 0.94 | 0.76–1.16 | 0.564 |
| BDNF | 11p14.1 | rs6265 (C/T) | SNP | 27636492 | 0.25 | 1 | 12 | 3897 | 6807 | 0.93 | 0.87–1.00 | 0.050 |
| DRD2 | 11q23.1-q23.2 | rs1799732 (C/-) | In-Del | 112851463 | 0.10 | 3 | 4 | 815 | 1257 | 1.12 | 0.87–1.44 | 0.377 |
| COMT | 22q11.21 | rs737865 (A/G) | SNP | 18310121 | 0.56 | 3 | 4 | 547 | 4651 | 0.93 | 0.81–1.08 | 0.341 |
| DRD3 | 3q13.31 | rs6280 (T/C) | SNP | 115373505 | 0.32 | 1 | 14 | 1456 | 1762 | 1.01 | 0.91–1.12 | 0.850 |
| TH | 11p15.5 | HUMTH01 (5/2) | STR | 2148853 | 0.40 | 1 | 9 | 480 | 525 | 1.13 | 0.94–1.37 | 0.214 |
| DRD4 | 11p15.5 | 48bp-repeat (4/2) | VNTR | ExonIII | 0.14 | 1 | 6 | 483 | 812 | 1.27 | 1.01–1.59 | 0.037 |
| TPH1 | 11p15.1 | rs1800532 (G/T) | SNP | 18004392 | 0.43 | 1 | 8 | 1848 | 2075 | 1.24 | 1.09–1.41 | 0.001 |
| MAOA | Xp11.3 | MAOA-VNTR (3/4) | Minisatellite | Intron2 | 0.53 | 3 | 4 | 194 | 229 | 0.67 | 0.35–1.29 | 0.231 |
| DAT1/SLC6A3 | 5p15.333 | rs2975226 (A/T) | SNP | 1498616 | 0.39 | 1 | 3 | 481 | 575 | 1.22 | 0.78–1.92 | 0.390 |
| DAOA | chr13q33.2 | rs3918342 (T/C) | SNP | 104983750 | 0.50 | 4 | 3 | 1051 | 1197 | 1.14 | 1.01–1.30 | 0.050 |
| HTR2C | Xq23 | rs6318 (G/C) | SNP | 13871991 | 0.14 | 1 | 4 | 716 | 753 | 1.31 | 0.84–2.03 | 0.227 |
| MTHFR | chr1p36.22 | rs1801133 (C/T) | SNP | 11778965 | 0.34 | 1 | 7 | 1260 | 1911 | 1.12 | 0.89–1.40 | 0.320 |
| GNB3 | chr12p13.31 | rs5443 (C/T) | SNP | 6825136 | 0.43 | 1 | 3 | 282 | 346 | 1.1 | 0.68–1.80 | 0.704 |
| GSK3B | chr3q13.33 | rs334558 (A/G) | SNP | 121295972 | 0.37 | 2 | 4 | 994 | 1509 | 0.96 | 0.84–1.09 | 0.529 |
| ACE | chr17q23.3 | rs4340 (ALU/—) | In-Del | 58919625 | 0.42 | 1 | 5 | 497 | 1351 | 0.96 | 0.77–1.20 | 0.720 |
| TNF-alpha | chr6p21.3 | rs1800629 (G/A) | SNP | 2790616 | 0.14 | 1 | 4 | 667 | 1205 | 1.15 | 0.64–2.09 | 0.647 |
Annotated using the dbSNP version 130 database, UCSC Genome Browser and HapMap, release 24, build 36, phase I, II (major allele/minor allele).
Starting base pair location of each polymorphism, build 36, human genome 18.
Minor allele frequency calculated based on the N cases and N controls.
Total number of polymorphisms meta-analyzed in each candidate gene.
N studies, N cases, and N controls reported for the top polymorphism only.
We compared our meta-analysis results against the recent mega-analysis of GWAS from the PGC-BP. We inspected the results from the same SNP if it was present in the PGC-BP data, or if not the one in highest linkage disequilibrium and/or nearest to the target SNP. Of the 18 top polymorphisms in 18 genes, there were 12 SNPs of which 8 of the same SNPs were present in the PGC-BP data. Of the remaining four SNPs, one of these was on the X chromosome (data not available from the PGC-BP) and for the remaining three the nearest neighbors were extracted. None of the six non-SNP polymorphisms were genotyped in the PGC-BP data, thus only the closest/nearest neighbors were extracted. Of the four polymorphisms that were nominally significant in our gene meta-analysis, none were nominally significant in PGC-BP. In fact, only one SNP of the 18 candidate genes (nearest neighbor to rs334558) included in our meta-analysis was nominally significant in the PGC-BP data (rs4688059 in GSK3β, OR = 1.15, P = 0.025).
DISCUSSION
Although family, twin, and adoption studies strongly implicate the role of genetic factors in the etiology of BP, genetic association studies of the disorder have been inconclusive. To provide a comprehensive assessment of the existing evidence, we conducted a literature review and meta-analysis of published candidate gene association studies. We identified 18 candidate genes with a total of 33 polymorphisms that were reported by three or more case–control association studies. Of these, there were four polymorphisms in four different genes that were found to be nominally significant upon meta-analysis. However, these did not remain significant after correcting for the 18 genes included in the meta-analysis. It should be noted that the Bonferroni correction method used in this context may be considered too conservative, in that each gene (and in many cases individual variants) represent specific hypotheses. Nevertheless, none of these polymorphisms were even nominally significant in the recently reported GWAS from the PGC-BP.
The lack of significant findings with previously implicated candidate genes in BP is in some ways telling. These genes were largely selected for investigation based on various hypotheses about the etiology of BP, and they emerged from the existing literature as leading candidates and attracted considerable attention in the field. However, that none appear to be associated with BP upon closer inspection suggests our understanding of the etiology of BP was inaccurate. However, these candidate genes should not be ruled out and it is still possible these genes do contribute to BP through rare variants or other types of variants not previously studied or captured by GWAS, but the overall evidence is not encouraging. These findings highlight what most have already come to accept, that is, that more atheoretical genome-wide approaches that are not constrained by our limited understanding of the disorder will be needed to shed light on its complex etiology.
This review provides a comprehensive evaluation of the existing literature on candidate genes studies of BP. Studies reported in the literature may be subject to publication biases in which positive studies are more likely to be published and this may unduly influence the inferences drawn in summarizing the findings. However, this is not a particular concern here, because all the results were essentially null. This review also provides a comparison with more recent GWAS findings, which were consistent with the conclusions from the meta-analysis. One concern with this comparison is that subjects included in the candidate gene studies may also have been included in the later GWAS, especially since investigators have attempted to GWAS every available BP sample, leading to non-independence in the comparison. Again, because the results were null, this is less of a concern.
It is clear that the etiology of BP is very complex. An increasing number of genomic experiments are being carried out to make sense of the complexity. It will be important to continue to monitor and evaluate the accumulating evidence about the role of different genetic factors that are identified by these studies using systematic reviews similar to the one carried out here. We have developed an online resource available to the research community called Metamoodics (http://metamoodics.igm.jhmi.edu) which will continue to gather and systematically review the results from these studies and provide tools for their quantitative analyses.
Supplementary Material
Footnotes
The authors declare that there are no conflicts of interest.
Additional supporting information may be found in the online version of this article.
References
- Arinami T, Li L, Mitsushio H, Itokawa M, Hamaguchi H, Toru M. An insertion/deletion polymorphism in the angiotensin converting enzyme gene is associated with both brain substance P contents and affective disorders. Biol Psychiatry. 1996;40:1122–1127. doi: 10.1016/s0006-3223(95)00597-8. [DOI] [PubMed] [Google Scholar]
- Arinami T, Yamada N, Yamakawa-Kobayashi K, Hamaguchi H, Toru M. Methylenetetrahydrofolate reductase variant and schizophrenia/depression. Am J Med Genet. 1997;74:526–528. doi: 10.1002/(sici)1096-8628(19970919)74:5<526::aid-ajmg14>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Arranz MJ, Erdmann J, Kirov G, Rietschel M, Sodhi M, Albus M, Ball D, Maier W, Davies N, Franzek E, Abusaad I, Weigelt B, Murray R, Shimron-Abarbanell D, Kerwin R, Propping P, Sham P, Nothen MM, Collier DA. 5-HT2A receptor and bipolar affective disorder: Association studies in affected patients. Neurosci Lett. 1997;224:95–98. doi: 10.1016/s0304-3940(97)13456-5. [DOI] [PubMed] [Google Scholar]
- Battersby S, Ogilvie AD, Smith CA, Blackwood DH, Muir WJ, Quinn JP, Fink G, Goodwin GM, Harmar AJ. Structure of a variable number tandem repeat of the serotonin transporter gene and association with affective disorder. Psychiatr Genet. 1996;6:177–181. doi: 10.1097/00041444-199624000-00001. [DOI] [PubMed] [Google Scholar]
- Bellivier F, Laplanche JL, Leboyer M, Feingold J, Bottos C, Allilaire JF, Launay JM. Serotonin transporter gene and manic depressive illness: An association study. Biol Psychiatry. 1997;41:750–752. doi: 10.1016/S0006-3223(96)00524-0. [DOI] [PubMed] [Google Scholar]
- Bellivier F, Henry C, Szoke A, Schurhoff F, Nosten-Bertrand M, Feingold J, Launay JM, Leboyer M, Laplanche JL. Serotonin transporter gene polymorphisms in patients with unipolar or bipolar depression. Neurosci Lett. 1998a;255:143–146. doi: 10.1016/s0304-3940(98)00677-6. [DOI] [PubMed] [Google Scholar]
- Bellivier F, Leboyer M, Courtet P, Buresi C, Beaufils B, Samolyk D, Allilaire JF, Feingold J, Mallet J, Malafosse A. Association between the tryptophan hydroxylase gene and manic-depressive illness. Arch Gen Psychiatry. 1998b;55:33–37. doi: 10.1001/archpsyc.55.1.33. [DOI] [PubMed] [Google Scholar]
- Benedetti F, Bernasconi A, Lorenzi C, Pontiggia A, Serretti A, Colombo C, Smeraldi E. A single nucleotide polymorphism in glycogen synthase kinase 3-beta promoter gene influences onset of illness in patients affected by bipolar disorder. Neurosci Lett. 2004;355:37–40. doi: 10.1016/j.neulet.2003.10.021. [DOI] [PubMed] [Google Scholar]
- BIOMED European Bipolar Collaborative Group. No association between bipolar disorder and alleles at a functional polymorphism in the COMT gene. Biomed European Bipolar Collaborative Group. Br J Psychiatry. 1997;170:526–528. doi: 10.1192/bjp.170.6.526. [DOI] [PubMed] [Google Scholar]
- Blairy S, Massat I, Staner L, Le Bon O, Van Gestel S, Van Broeckhoven C, Hilger C, Hentges F, Souery D, Mendlewicz J. 5-HT2a receptor polymorphism gene in bipolar disorder and harm avoidance personality trait. Am J Med Genet. 2000;96:360–364. doi: 10.1002/1096-8628(20000612)96:3<360::aid-ajmg24>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Bonnier B, Gorwood P, Hamon M, Sarfati Y, Boni C, Hardy-Bayle MC. Association of 5-HT(2A) receptor gene polymorphism with major affective disorders: The case of a subgroup of bipolar disorder with low suicide risk. Biol Psychiatry. 2002;51:762–765. doi: 10.1016/s0006-3223(01)01228-8. [DOI] [PubMed] [Google Scholar]
- Burdick KE, Funke B, Goldberg JF, Bates JA, Jaeger J, Kucherlapati R, Malhotra AK. COMT genotype increases risk for bipolar I disorder and influences neurocognitive performance. Bipolar Disord. 2007;9:370–376. doi: 10.1111/j.1399-5618.2007.00384.x. [DOI] [PubMed] [Google Scholar]
- Burgert E, Crocq MA, Bausch E, Macher JP, Morris-Rosendahl DJ. No association between the tyrosine hydroxylase microsatellite marker HUMTH01 and schizophrenia or bipolar I disorder. Psychiatr Genet. 1998;8:45–48. doi: 10.1097/00041444-199800820-00002. [DOI] [PubMed] [Google Scholar]
- Chee IS, Lee SW, Kim JL, Wang SK, Shin YO, Shin SC, Lee YH, Hwang HM, Lim MR. 5-HT2A receptor gene promoter polymorphism – 1438A/G and bipolar disorder. Psychiatr Genet. 2001;11:111–114. doi: 10.1097/00041444-200109000-00001. [DOI] [PubMed] [Google Scholar]
- Chen YS, Akula N, Detera-Wadleigh SD, Schulze TG, Thomas J, Potash JB, DePaulo JR, McInnis MG, Cox NJ, McMahon FJ. Findings in an independent sample support an association between bipolar affective disorder and the G72/G30 locus on chromosome 13q33. Mol Psychiatry. 2004;9:87–92. doi: 10.1038/sj.mp.4001453. image 5. [DOI] [PubMed] [Google Scholar]
- Chen Z, Liu Y, Zhang D, Liu Z, Wang P, Zhou D, Zhao T, Wang T, Xu H, Li S, Feng G, He L, Yu L. C677T methylenetetrahydrofolate reductase gene polymorphisms in bipolar disorder: An association study in the Chinese population and a meta-analysis of genetic association studies. Neurosci Lett. 2009;449:48–51. doi: 10.1016/j.neulet.2008.10.077. [DOI] [PubMed] [Google Scholar]
- Chiaroni P, Azorin JM, Dassa D, Henry JM, Giudicelli S, Malthiery Y, Planells R. Possible involvement of the dopamine D3 receptor locus in subtypes of bipolar affective disorder. Psychiatr Genet. 2000;10:43–49. doi: 10.1097/00041444-200010010-00008. [DOI] [PubMed] [Google Scholar]
- Cichon S, Winge I, Mattheisen M, Georgi A, Karpushova A, Freudenberg J, Freudenberg-Hua Y, Babadjanova G, Van Den Bogaert A, Abramova LI, Kapiletti S, Knappskog PM, McKinney J, Maier W, Jamra RA, Schulze TG, Schumacher J, Propping P, Rietschel M, Haavik J, Nothen MM. Brain-specific tryptophan hydroxylase 2 (TPH2): A functional Pro206Ser substitution and variation in the 5′-region are associated with bipolar affective disorder. Hum Mol Genet. 2008;17:87–97. doi: 10.1093/hmg/ddm286. [DOI] [PubMed] [Google Scholar]
- Collier DA, Arranz MJ, Sham P, Battersby S, Vallada H, Gill P, Aitchison KJ, Sodhi M, Li T, Roberts GW, Smith B, Morton J, Murray RM, Smith D, Kirov G. The serotonin transporter is a potential susceptibility factor for bipolar affective disorder. Neuroreport. 1996a;7:1675–1679. doi: 10.1097/00001756-199607080-00030. [DOI] [PubMed] [Google Scholar]
- Collier DA, Stober G, Li T, Heils A, Catalano M, Di Bella D, Arranz MJ, Murray RM, Vallada HP, Bengel D, Muller CR, Roberts GW, Smeraldi E, Kirov G, Sham P, Lesch KP. A novel functional polymorphism within the promoter of the serotonin transporter gene: Possible role in susceptibility to affective disorders. Mol Psychiatry. 1996b;1:453–460. [PubMed] [Google Scholar]
- Corson TW, Li PP, Kennedy JL, Macciardi F, Cooke RG, Parikh SV, Warsh JJ. Association analysis of G-protein beta 3 subunit gene with altered ca(2+) homeostasis in bipolar disorder. Mol Psychiatry. 2001;6:125–126. doi: 10.1038/sj.mp.4000775. [DOI] [PubMed] [Google Scholar]
- Craddock N, Daniels J, Roberts E, Rees M, McGuffin P, Owen MJ. No evidence for allelic association between bipolar disorder and mono-amine oxidase A gene polymorphisms. Am J Med Genet. 1995a;60:322–324. doi: 10.1002/ajmg.1320600412. [DOI] [PubMed] [Google Scholar]
- Craddock N, Roberts Q, Williams N, McGuffin P, Owen MJ. Association study of bipolar disorder using a functional polymorphism (Ser311 → Cys) in the dopamine D2 receptor gene. Psychiatr Genet. 1995b;5:63–65. doi: 10.1097/00041444-199522000-00003. [DOI] [PubMed] [Google Scholar]
- Czerski PM, Rybakowski F, Kapelski P, Rybakowski JK, Dmitrzak-Weglarz M, Leszczynska-Rodziewicz A, Slopien A, Skibinska M, Kaczmarkiewicz-Fass M, Hauser J. Association of tumor necrosis factor –308G/A promoter polymorphism with schizophrenia and bipolar affective disorder in a polish population. Neuropsychobiology. 2008;57:88–94. doi: 10.1159/000135642. [DOI] [PubMed] [Google Scholar]
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Development Core Team. R foundation for statistical computing. Vienna, Austria: 2010. R: A language and environment for statistical computing. URL http://www.R-project.org. [Google Scholar]
- Etain B, Rousseva A, Roy I, Henry C, Malafosse A, Buresi C, Preisig M, Rayah F, Leboyer M, Bellivier F. Lack of association between 5HT2A receptor gene haplotype, bipolar disorder and its clinical subtypes in a West European sample. Am J Med Genet Part B. 2004;129B:29–33. doi: 10.1002/ajmg.b.30055. [DOI] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, Diekhans M, Dreszer TR, Giardine BM, Harte RA, Hillman-Jackson J, Hsu F, Kirkup V, Kuhn RM, Learned K, Li CH, Meyer LR, Pohl A, Raney BJ, Rosenbloom KR, Smith KE, Haussler D, Kent WJ. The UCSC genome browser database: Update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke B, Malhotra AK, Finn CT, Plocik AM, Lake SL, Lencz T, DeRosse P, Kane JM, Kucherlapati R. COMT genetic variation confers risk for psychotic and affective disorders: A case control study. Behav Brain Funct. 2005;1:19. doi: 10.1186/1744-9081-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong RA, Coleman TA, Ho L, Rubinsztein JS, Walsh C, Paykel ES, Rubinsztein DC. No association of a functional polymorphism in the dopamine D2 receptor promoter region with bipolar or unipolar affective disorders. Am J Med Genet. 1998a;81:385–387. [PubMed] [Google Scholar]
- Furlong RA, Ho L, Rubinsztein JS, Walsh C, Paykel ES, Rubinsztein DC. No association of the tryptophan hydroxylase gene with bipolar affective disorder, unipolar affective disorder, or suicidal behaviour in major affective disorder. Am J Med Genet. 1998b;81:245–247. [PubMed] [Google Scholar]
- Furlong RA, Ho L, Walsh C, Rubinsztein JS, Jain S, Paykel ES, Easton DF, Rubinsztein DC. Analysis and meta-analysis of two serotonin transporter gene polymorphisms in bipolar and unipolar affective disorders. Am J Med Genet. 1998c;81:58–63. [PubMed] [Google Scholar]
- Furlong RA, Ho L, Rubinsztein JS, Walsh C, Paykel ES, Rubinsztein DC. Analysis of the monoamine oxidase A (MAOA) gene in bipolar affective disorder by association studies, meta-analyses, and sequencing of the promoter. Am J Med Genet. 1999a;88:398–406. [PubMed] [Google Scholar]
- Furlong RA, Rubinsztein JS, Ho L, Walsh C, Coleman TA, Muir WJ, Paykel ES, Blackwood DH, Rubinsztein DC. Analysis and metaanalysis of two polymorphisms within the tyrosine hydroxylase gene in bipolar and unipolar affective disorders. Am J Med Genet. 1999b;88:88–94. doi: 10.1002/(sici)1096-8628(19990205)88:1<88::aid-ajmg16>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Furlong RA, Keramatipour M, Ho LW, Rubinsztein JS, Michael A, Walsh C, Paykel ES, Rubinsztein DC. No association of an insertion/deletion polymorphism in the angiotensin I converting enzyme gene with bipolar or unipolar affective disorders. Am J Med Genet. 2000;96:733–735. doi: 10.1002/1096-8628(20001204)96:6<733::aid-ajmg7>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Gomez-Casero E, Perez de Castro I, Saiz-Ruiz J, Llinares C, Fernandez-Piqueras J. No association between particular DRD3 and DAT gene polymorphisms and manic-depressive illness in a Spanish sample. Psychiatr Genet. 1996;6:209–212. doi: 10.1097/00041444-199624000-00007. [DOI] [PubMed] [Google Scholar]
- Green EK, Raybould R, Macgregor S, Hyde S, Young AH, O’Donovan MC, Owen MJ, Kirov G, Jones L, Jones I, Craddock N. Genetic variation of brain-derived neurotrophic factor (BDNF) in bipolar disorder: Case-control study of over 3000 individuals from the UK. Br J Psychiatry. 2006;188:21–25. doi: 10.1192/bjp.bp.105.009969. [DOI] [PubMed] [Google Scholar]
- Grigoroiu-Serbanescu M, Diaconu CC, Herms S, Bleotu C, Vollmer J, Muhleisen TW, Prelipceanu D, Priebe L, Mihailescu R, Georgescu MJ, Sima D, Grimberg M, Nothen MM, Cichon S. Investigation of the tryptophan hydroxylase 2 gene in bipolar I disorder in the Romanian population. Psychiatr Genet. 2008;18:240–247. doi: 10.1097/YPG.0b013e3283053045. [DOI] [PubMed] [Google Scholar]
- Gutierrez B, Fananas L, Arranz MJ, Valles V, Guillamat R, van Os J, Collier D. Allelic association analysis of the 5-HT2C receptor gene in bipolar affective disorder. Neurosci Lett. 1996;212:65–67. doi: 10.1016/0304-3940(96)12746-4. [DOI] [PubMed] [Google Scholar]
- Gutierrez B, Bertranpetit J, Collier D, Arranz MJ, Valles V, Guillamat R, Van Os J, Fananas L. Genetic variation of the 5-HT2A receptor gene and bipolar affective disorder. Hum Genet. 1997a;100:582–584. doi: 10.1007/s004390050556. [DOI] [PubMed] [Google Scholar]
- Gutierrez B, Bertranpetit J, Guillamat R, Valles V, Arranz MJ, Kerwin R, Fananas L. Association analysis of the catechol O-methyltransferase gene and bipolar affective disorder. Am J Psychiatry. 1997b;154:113–115. doi: 10.1176/ajp.154.1.113. [DOI] [PubMed] [Google Scholar]
- Gutierrez B, Arranz MJ, Collier DA, Valles V, Guillamat R, Bertranpetit J, Murray RM, Fanas L. Serotonin transporter gene and risk for bipolar affective disorder: An association study in Spanish population. Biol Psychiatry. 1998;43:843–847. doi: 10.1016/s0006-3223(97)00540-4. [DOI] [PubMed] [Google Scholar]
- Gutierrez B, Arias B, Papiol S, Rosa A, Fananas L. Association study between novel promoter variants at the 5-HT2C receptor gene and human patients with bipolar affective disorder. Neurosci Lett. 2001;309:135–137. doi: 10.1016/s0304-3940(01)02046-8. [DOI] [PubMed] [Google Scholar]
- Gutierrez B, Arias B, Gasto C, Catalan R, Papiol S, Pintor L, Fananas L. Association analysis between a functional polymorphism in the mono-amine oxidase A gene promoter and severe mood disorders. Psychiatr Genet. 2004;14:203–208. doi: 10.1097/00041444-200412000-00007. [DOI] [PubMed] [Google Scholar]
- Hauser J, Leszczynska A, Samochowiec J, Czerski PM, Ostapowicz A, Chlopocka M, Horodnicki J, Rybakowski JK. Association analysis of the insertion/deletion polymorphism in serotonin transporter gene in patients with affective disorder. Eur Psychiatry. 2003;18:129–132. doi: 10.1016/s0924-9338(03)00026-9. [DOI] [PubMed] [Google Scholar]
- Heiden A, Schussler P, Itzlinger U, Leisch F, Scharfetter J, Gebhardt C, Fuchs K, Willeit M, Nilsson L, Miller-Reiter E, Stompe T, Meszaros K, Sieghart W, Hornik K, Kasper S, Aschauer HN. Association studies of candidate genes in bipolar disorders. Neuropsychobiology. 2000;42(Suppl1):18–21. doi: 10.1159/000054846. [DOI] [PubMed] [Google Scholar]
- Hoehe MR, Wendel B, Grunewald I, Chiaroni P, Levy N, Morris-Rosendahl D, Macher JP, Sander T, Crocq MA. Serotonin transporter (5-HTT) gene polymorphisms are not associated with susceptibility to mood disorders. Am J Med Genet. 1998;81:1–3. doi: 10.1002/(sici)1096-8628(19980207)81:1<1::aid-ajmg1>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Hong CJ, Huo SJ, Yen FC, Tung CL, Pan GM, Tsai SJ. Association study of a brain-derived neurotrophic-factor genetic polymorphism and mood disorders, age of onset and suicidal behavior. Neuropsychobiology. 2003;48:186–189. doi: 10.1159/000074636. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Iwata N, Suzuki T, Kitajima T, Yamanouchi Y, Kinoshita Y, Ozaki N. No association of serotonin transporter gene (SLC6A4) with schizophrenia and bipolar disorder in Japanese patients: Association analysis based on linkage disequilibrium. J Neural Transm. 2006;113:899–905. doi: 10.1007/s00702-005-0349-6. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium. The international HapMap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Jin UH, Lee JY, Kang SK, Kim JK, Park WH, Kim JG, Moon SK, Kim CH. A phenolic compound, 5-caffeoylquinic acid (chlorogenic acid), is a new type and strong matrix metalloproteinase-9 inhibitor: Isolation and identification from methanol extract of Euonymus alatus. Life Sci. 2005;77:2760–2769. doi: 10.1016/j.lfs.2005.02.028. [DOI] [PubMed] [Google Scholar]
- Johansson C, Smedh C, Partonen T, Pekkarinen P, Paunio T, Ekholm J, Peltonen L, Lichtermann D, Palmgren J, Adolfsson R, Schalling M. Seasonal affective disorder and serotonin-related polymorphisms. Neurobiol Dis. 2001;8:351–357. doi: 10.1006/nbdi.2000.0373. [DOI] [PubMed] [Google Scholar]
- Jonsson EG, Larsson K, Vares M, Hansen T, Wang AG, Djurovic S, Ronningen KS, Andreassen OA, Agartz I, Werge T, Terenius L, Hall H. Two methylenetetrahydrofolate reductase gene (MTHFR) polymorphisms, schizophrenia and bipolar disorder: An association study. Am J Med Genet Part B. 2008;147B:976–982. doi: 10.1002/ajmg.b.30671. [DOI] [PubMed] [Google Scholar]
- Kawada Y, Hattori M, Dai XY, Nanko S. Possible association between monoamine oxidase A gene and bipolar affective disorder. Am J Hum Genet. 1995a;56:335–336. [PMC free article] [PubMed] [Google Scholar]
- Kawada Y, Hattori M, Fukuda R, Arai H, Inoue R, Nanko S. No evidence of linkage or association between tyrosine hydroxylase gene and affective disorder. J Affect Disord. 1995b;34:89–94. doi: 10.1016/0165-0327(95)00004-7. [DOI] [PubMed] [Google Scholar]
- Keikhaee MR, Fadai F, Sargolzaee MR, Javanbakht A, Najmabadi H, Ohadi M. Association analysis of the dopamine transporter (DAT1)-67A/T polymorphism in bipolar disorder. Am J Med Genet Part B. 2005;135B:47–49. doi: 10.1002/ajmg.b.30174. [DOI] [PubMed] [Google Scholar]
- Kempisty B, Mostowska A, Gorska I, Luczak M, Czerski P, Szczepankiewicz A, Hauser J, Jagodzinski PP. Association of 677C > T polymorphism of methylenetetrahydrofolate reductase (MTHFR) gene with bipolar disorder and schizophrenia. Neurosci Lett. 2006;400:267–271. doi: 10.1016/j.neulet.2006.02.055. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Kim CY, Hong JP, Kim SY, Lee C, Joo YH. Brain-derived neurotrophic factor Val/Met polymorphism and bipolar disorder. Association of the met allele with suicidal behavior of bipolar patients. Neuropsychobiology. 2008;58:97–103. doi: 10.1159/000162356. [DOI] [PubMed] [Google Scholar]
- Kirov G, Murphy KC, Arranz MJ, Jones I, McCandles F, Kunugi H, Murray RM, McGuffin P, Collier DA, Owen MJ, Craddock N. Low activity allele of catechol-O-methyltransferase gene associated with rapid cycling bipolar disorder. Mol Psychiatry. 1998;3:342–345. doi: 10.1038/sj.mp.4000385. [DOI] [PubMed] [Google Scholar]
- Kishi T, Kitajima T, Tsunoka T, Ikeda M, Yamanouchi Y, Kinoshita Y, Kawashima K, Okochi T, Okumura T, Inada T, Ozaki N, Iwata N. Genetic association analysis of serotonin 2A receptor gene (HTR2A) with bipolar disorder and major depressive disorder in the Japanese population. Neurosci Res. 2009;64:231–234. doi: 10.1016/j.neures.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Korner J, Rietschel M, Hunt N, Castle D, Gill M, Nothen MM, Craddock N, Daniels J, Owen M, Fimmers R. Association and haplotype analysis at the tyrosine hydroxylase locus in a combined German-British sample of manic depressive patients and controls. Psychiatr Genet. 1994;4:167–175. doi: 10.1097/00041444-199400430-00007. [DOI] [PubMed] [Google Scholar]
- Kumar HB, Purushottam M, Kubendran S, Gayathri P, Mukherjee O, Murthy AR, Ghosh S, Chandra P, Reddy YC, Benegal V, Brahmachari SK, Jain S. Serotonergic candidate genes and puerperal psychosis: An association study. Psychiatr Genet. 2007;17:253–260. doi: 10.1097/YPG.0b013e3280ae6cc3. [DOI] [PubMed] [Google Scholar]
- Kunugi H, Hattori M, Kato T, Tatsumi M, Sakai T, Sasaki T, Hirose T, Nanko S. Serotonin transporter gene polymorphisms: Ethnic difference and possible association with bipolar affective disorder. Mol Psychiatry. 1997a;2:457–462. doi: 10.1038/sj.mp.4000334. [DOI] [PubMed] [Google Scholar]
- Kunugi H, Vallada HP, Hoda F, Kirov G, Gill M, Aitchison KJ, Ball D, Arranz MJ, Murray RM, Collier DA. No evidence for an association of affective disorders with high- or low-activity allele of catechol-o-methyltransferase gene. Biol Psychiatry. 1997b;42:282–285. doi: 10.1016/S0006-3223(96)00366-6. [DOI] [PubMed] [Google Scholar]
- Kunugi H, Fukuda R, Hattori M, Kato T, Tatsumi M, Sakai T, Hirose T, Nanko S. C677T polymorphism in methylenetetrahydrofolate reductase gene and psychoses. Mol Psychiatry. 1998;3:435–437. doi: 10.1038/sj.mp.4000390. [DOI] [PubMed] [Google Scholar]
- Kunugi H, Ishida S, Kato T, Sakai T, Tatsumi M, Hirose T, Nanko S. No evidence for an association of polymorphisms of the tryptophan hydroxylase gene with affective disorders or attempted suicide among Japanese patients. Am J Psychiatry. 1999a;156:774–776. doi: 10.1176/ajp.156.5.774. [DOI] [PubMed] [Google Scholar]
- Kunugi H, Ishida S, Kato T, Tatsumi M, Sakai T, Hattori M, Hirose T, Nanko S. A functional polymorphism in the promoter region of monoamine oxidase-A gene and mood disorders. Mol Psychiatry. 1999b;4:393–395. doi: 10.1038/sj.mp.4000558. [DOI] [PubMed] [Google Scholar]
- Kunugi H, Iijima Y, Tatsumi M, Yoshida M, Hashimoto R, Kato T, Sakamoto K, Fukunaga T, Inada T, Suzuki T, Iwata N, Ozaki N, Yamada K, Yoshikawa T. No association between the Val66Met polymorphism of the brain-derived neurotrophic factor gene and bipolar disorder in a Japanese population: A multicenter study. Biol Psychiatry. 2004;56:376–378. doi: 10.1016/j.biopsych.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Lachman HM, Kelsoe J, Moreno L, Katz S, Papolos DF. Lack of association of catechol-O-methyltransferase (COMT) functional polymorphism in bipolar affective disorder. Psychiatr Genet. 1997;7:13–17. doi: 10.1097/00041444-199700710-00002. [DOI] [PubMed] [Google Scholar]
- Lai TJ, Wu CY, Tsai HW, Lin YM, Sun HS. Polymorphism screening and haplotype analysis of the tryptophan hydroxylase gene (TPH1) and association with bipolar affective disorder in Taiwan. BMC Med Genet. 2005;6:14. doi: 10.1186/1471-2350-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Ahn YM, Joo EJ, Jeong SH, Chang JS, Kim SC, Kim YS. No association of two common SNPs at position –1727 A/T, –50 C/T of GSK-3 beta polymorphisms with schizophrenia and bipolar disorder of Korean population. Neurosci Lett. 2006;395:175–178. doi: 10.1016/j.neulet.2005.10.059. [DOI] [PubMed] [Google Scholar]
- Lerer B, Macciardi F, Segman RH, Adolfsson R, Blackwood D, Blairy S, Del Favero J, Dikeos DG, Kaneva R, Lilli R, Massat I, Milanova V, Muir W, Noethen M, Oruc L, Petrova T, Papadimitriou GN, Rietschel M, Serretti A, Souery D, Van Gestel S, Van Broeckhoven C, Mendlewicz J. Variability of 5-HT2C receptor cys23ser polymorphism among European populations and vulnerability to affective disorder. Mol Psychiatry. 2001;6:579–585. doi: 10.1038/sj.mp.4000883. [DOI] [PubMed] [Google Scholar]
- Leszczynska-Rodziewicz A, Hauser J, Dmitrzak-Weglarz M, Skibinka M, Czerski P, Zakrzewska A, Kosmowska M, Rybakowski JK. Lack of association between polymorphisms of dopamine receptors, type D2, and bipolar affective illness in a polish population. Med Sci Monit. 2005;11:CR289–CR295. [PubMed] [Google Scholar]
- Li T, Vallada H, Curtis D, Arranz M, Xu K, Cai G, Deng H, Liu J, Murray R, Liu X, Collier DA. Catechol-O-methyltransferase Val158Met polymorphism: Frequency analysis in Han Chinese subjects and allelic association of the low activity allele with bipolar affective disorder. Pharmacogenetics. 1997;7:349–353. doi: 10.1097/00008571-199710000-00002. [DOI] [PubMed] [Google Scholar]
- Li T, Liu X, Sham PC, Aitchison KJ, Cai G, Arranz MJ, Deng H, Liu J, Kirov G, Murray RM, Collier DA. Association analysis between dopamine receptor genes and bipolar affective disorder. Psychiatry Res. 1999;86:193–201. doi: 10.1016/s0165-1781(99)00034-7. [DOI] [PubMed] [Google Scholar]
- Lim LC, Nothen MM, Korner J, Rietschel M, Castle D, Hunt N, Propping P, Murray R, Gill M. No evidence of association between dopamine D4 receptor variants and bipolar affective disorder. Am J Med Genet. 1994;54:259–263. doi: 10.1002/ajmg.1320540314. [DOI] [PubMed] [Google Scholar]
- Lim LC, Powell J, Sham P, Castle D, Hunt N, Murray R, Gill M. Evidence for a genetic association between alleles of monoamine oxidase A gene and bipolar affective disorder. Am J Med Genet. 1995;60:325–331. doi: 10.1002/ajmg.1320600413. [DOI] [PubMed] [Google Scholar]
- Lin S, Jiang S, Wu X, Qian Y, Wang D, Tang G, Gu N. Association analysis between mood disorder and monoamine oxidase gene. Am J Med Genet. 2000;96:12–14. doi: 10.1002/(sici)1096-8628(20000207)96:1<12::aid-ajmg4>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Liu W, Gu N, Feng G, Li S, Bai S, Zhang J, Shen T, Xue H, Breen G, St Clair D, He L. Tentative association of the serotonin transporter with schizophrenia and unipolar depression but not with bipolar disorder in Han Chinese. Pharmacogenetics. 1999;9:491–495. [PubMed] [Google Scholar]
- Lohoff FW, Sander T, Ferraro TN, Dahl JP, Gallinat J, Berrettini WH. Confirmation of association between the Val66Met polymorphism in the brain-derived neurotrophic factor (BDNF) gene and bipolar I disorder. Am J Med Genet Part B. 2005;139B:51–53. doi: 10.1002/ajmg.b.30215. [DOI] [PubMed] [Google Scholar]
- Mahieu B, Souery D, Lipp O, Mendelbaum K, Verheyen G, De Maertelaer V, Van Broeckhoven C, Mendlewicz J. No association between bipolar affective disorder and a serotonin receptor (5-HT2A) polymorphism. Psychiatry Res. 1997;70:65–69. doi: 10.1016/s0165-1781(97)03028-x. [DOI] [PubMed] [Google Scholar]
- Manki H, Kanba S, Muramatsu T, Higuchi S, Suzuki E, Matsushita S, Ono Y, Chiba H, Shintani F, Nakamura M, Yagi G, Asai M. Dopamine D2, D3 and D4 receptor and transporter gene polymorphisms and mood disorders. J Affect Disord. 1996;40:7–13. doi: 10.1016/0165-0327(96)00035-3. [DOI] [PubMed] [Google Scholar]
- Massat I, Souery D, Lipp O, Blairy S, Papadimitriou G, Dikeos D, Ackenheil M, Fuchshuber S, Hilger C, Kaneva R, Milanova V, Verheyen G, Raeymaekers P, Staner L, Oruc L, Jakovljevic M, Serretti A, Macciardi F, Van Broeckhoven C, Mendlewicz J. A European multicenter association study of HTR2A receptor polymorphism in bipolar affective disorder. Am J Med Genet. 2000;96:136–140. [PubMed] [Google Scholar]
- Meira-Lima IV, Pereira AC, Mota GF, Krieger JE, Vallada H. Angiotensinogen and angiotensin converting enzyme gene polymorphisms and the risk of bipolar affective disorder in humans. Neurosci Lett. 2000;293:103–106. doi: 10.1016/s0304-3940(00)01512-3. [DOI] [PubMed] [Google Scholar]
- Meira-Lima IV, Pereira AC, Mota GF, Floriano M, Araujo F, Mansur AJ, Krieger JE, Vallada H. Analysis of a polymorphism in the promoter region of the tumor necrosis factor alpha gene in schizophrenia and bipolar disorder: Further support for an association with schizophrenia. Mol Psychiatry. 2003;8:718–720. doi: 10.1038/sj.mp.4001309. [DOI] [PubMed] [Google Scholar]
- Meira-Lima I, Michelon L, Cordeiro Q, Cho HJ, Vallada H. Allelic association analysis of the functional insertion/deletion polymorphism in the promoter region of the serotonin transporter gene in bipolar affective disorder. J Mol Neurosci. 2005;27:219–224. doi: 10.1385/JMN:27:2:219. [DOI] [PubMed] [Google Scholar]
- Mellerup E, Bennike B, Bolwig T, Dam H, Hasholt L, Jorgensen MB, Plenge P, Sorensen SA. Platelet serotonin transporters and the transporter gene in control subjects, unipolar patients and bipolar patients. Acta Psychiatr Scand. 2001;103:229–233. doi: 10.1034/j.1600-0447.2001.00173.x. [DOI] [PubMed] [Google Scholar]
- Meloni R, Leboyer M, Bellivier F, Barbe B, Samolyk D, Allilaire JF, Mallet J. Association of manic-depressive illness with tyrosine hydroxylase microsatellite marker. Lancet. 1995;345:932. doi: 10.1016/s0140-6736(95)90053-5. [DOI] [PubMed] [Google Scholar]
- Mendes de Oliveira JR, Otto PA, Vallada H, Lauriano V, Elkis H, Lafer B, Vasquez L, Gentil V, Passos-Bueno MR, Zatz M. Analysis of a novel functional polymorphism within the promoter region of the serotonin transporter gene (5-HTT) in Brazilian patients affected by bipolar disorder and schizophrenia. Am J Med Genet. 1998;81:225–227. doi: 10.1002/(sici)1096-8628(19980508)81:3<225::aid-ajmg4>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Mendlewicz J, Massat I, Souery D, Del-Favero J, Oruc L, Nothen MM, Blackwood D, Muir W, Battersby S, Lerer B, Segman RH, Kaneva R, Serretti A, Lilli R, Lorenzi C, Jakovljevic M, Ivezic S, Rietschel M, Milanova V, Van Broeckhoven C. Serotonin transporter 5HTTLPR polymorphism and affective disorders: No evidence of association in a large European multicenter study. Eur J Hum Genet. 2004;12:377–382. doi: 10.1038/sj.ejhg.5201149. [DOI] [PubMed] [Google Scholar]
- Merikangas KR, Low NC. The epidemiology of mood disorders. Curr Psychiatry Rep. 2004;6:411–421. doi: 10.1007/s11920-004-0004-1. [DOI] [PubMed] [Google Scholar]
- Middle F, Jones I, Robertson E, Lendon C, Craddock N. Tumour necrosis factor alpha and bipolar affective puerperal psychosis. Psychiatr Genet. 2000;10:195–198. doi: 10.1097/00041444-200010040-00008. [DOI] [PubMed] [Google Scholar]
- Muramatsu T, Matsushita S, Kanba S, Higuchi S, Manki H, Suzuki E, Asai M. Monoamine oxidase genes polymorphisms and mood disorder. Am J Med Genet. 1997;74:494–496. [PubMed] [Google Scholar]
- Mynett-Johnson LA, Murphy VE, Claffey E, Shields DC, McKeon P. Preliminary evidence of an association between bipolar disorder in females and the catechol-O-methyltransferase gene. Psychiatr Genet. 1998;8:221–225. doi: 10.1097/00041444-199808040-00004. [DOI] [PubMed] [Google Scholar]
- Nakata K, Ujike H, Sakai A, Uchida N, Nomura A, Imamura T, Katsu T, Tanaka Y, Hamamura T, Kuroda S. Association study of the brain-derived neurotrophic factor (BDNF) gene with bipolar disorder. Neurosci Lett. 2003;337:17–20. doi: 10.1016/s0304-3940(02)01292-2. [DOI] [PubMed] [Google Scholar]
- Nishiguchi N, Breen G, Russ C, St Clair D, Collier D. Association analysis of the glycogen synthase kinase-3beta gene in bipolar disorder. Neurosci Lett. 2006;394:243–245. doi: 10.1016/j.neulet.2005.10.042. [DOI] [PubMed] [Google Scholar]
- Nothen MM, Erdmann J, Korner J, Lanczik M, Fritze J, Fimmers R, Grandy DK, O’Dowd B, Propping P. Lack of association between dopamine D1 and D2 receptor genes and bipolar affective disorder. Am J Psychiatry. 1992;149:199–201. doi: 10.1176/ajp.149.2.199. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Ohara K, Shimizu H. Correlation between total aqueous protein concentrations and photon counts in rabbits. Nihon Ganka Gakkai Zasshi. 1990;94:1001–1006. [PubMed] [Google Scholar]
- Ogilvie AD, Battersby S, Bubb VJ, Fink G, Harmar AJ, Goodwim GM, Smith CA. Polymorphism in serotonin transporter gene associated with susceptibility to major depression. Lancet. 1996;347:731–733. doi: 10.1016/s0140-6736(96)90079-3. [DOI] [PubMed] [Google Scholar]
- Ohadi M, Keikhaee MR, Javanbakht A, Sargolzaee MR, Robabeh M, Najmabadi H. Gender dimorphism in the DAT1-67 T-allele homozygosity and predisposition to bipolar disorder. Brain Res. 2007;1144:142–145. doi: 10.1016/j.brainres.2007.01.067. [DOI] [PubMed] [Google Scholar]
- Ohara K, Nagai M, Suzuki Y, Ohara K. Low activity allele of catechol-o-methyltransferase gene and Japanese unipolar depression. Neuroreport. 1998a;9:1305–1308. doi: 10.1097/00001756-199805110-00009. [DOI] [PubMed] [Google Scholar]
- Ohara K, Nagai M, Tsukamoto T, Tani K, Suzuki Y, Ohara K. Functional polymorphism in the serotonin transporter promoter at the SLC6A4 locus and mood disorders. Biol Psychiatry. 1998b;44:550–554. doi: 10.1016/s0006-3223(98)00112-7. [DOI] [PubMed] [Google Scholar]
- Oruc L, Verheyen GR, Furac I, Jakovljevic M, Ivezic S, Raeymaekers P, Van Broeckhoven C. Analysis of the tyrosine hydroxylase and dopamine D4 receptor genes in a Croatian sample of bipolar I and unipolar patients. Am J Med Genet. 1997a;74:176–178. doi: 10.1002/(sici)1096-8628(19970418)74:2<176::aid-ajmg12>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Oruc L, Verheyen GR, Furac I, Jakovljevic M, Ivezic S, Raeymaekers P, Van Broeckhoven C. Association analysis of the 5-HT2C receptor and 5-HT transporter genes in bipolar disorder. Am J Med Genet. 1997b;74:504–506. [PubMed] [Google Scholar]
- Ospina-Duque J, Duque C, Carvajal-Carmona L, Ortiz-Barrientos D, Soto I, Pineda N, Cuartas M, Calle J, Lopez C, Ochoa L, Garcia J, Gomez J, Agudelo A, Lozano M, Montoya G, Ospina A, Lopez M, Gallo A, Miranda A, Serna L, Montoya P, Palacio C, Bedoya G, McCarthy M, Reus V, Freimer N, Ruiz-Linares A. An association study of bipolar mood disorder (type I) with the 5-HTTLPR serotonin transporter polymorphism in a human population isolate from Colombia. Neurosci Lett. 2000;292:199–202. doi: 10.1016/s0304-3940(00)01464-6. [DOI] [PubMed] [Google Scholar]
- Oswald P, Del-Favero J, Massat I, Souery D, Claes S, Van Broeckhoven C, Mendlewicz J. Non-replication of the brain-derived neurotrophic factor (BDNF) association in bipolar affective disorder: A Belgian patient-control study. Am J Med Genet Part B. 2004;129B:34–35. doi: 10.1002/ajmg.b.30056. [DOI] [PubMed] [Google Scholar]
- Ozaki N, Rosenthal NE, Pesonen U, Lappalainen J, Feldman-Naim S, Schwartz PJ, Turner EH, Goldman D. Two naturally occurring amino acid substitutions of the 5-HT2A receptor: Similar prevalence in patients with seasonal affective disorder and controls. Biol Psychiatry. 1996;40:1267–1272. doi: 10.1016/0006-3223(95)00649-4. [DOI] [PubMed] [Google Scholar]
- Pae CU, Lee KU, Han H, Serretti A, Jun TY. Tumor necrosis factor alpha gene-G308A polymorphism associated with bipolar I disorder in the Korean population. Psychiatry Res. 2004;125:65–68. doi: 10.1016/j.psychres.2003.06.002. [DOI] [PubMed] [Google Scholar]
- Parsian A, Todd RD. Genetic association between monoamine oxidase and manic-depressive illness: Comparison of relative risk and haplotype relative risk data. Am J Med Genet. 1997;74:475–479. doi: 10.1002/(sici)1096-8628(19970919)74:5<475::aid-ajmg3>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Parsian A, Chakraverty S, Todd RD. Possible association between the dopamine D3 receptor gene and bipolar affective disorder. Am J Med Genet. 1995;60:234–237. doi: 10.1002/ajmg.1320600313. [DOI] [PubMed] [Google Scholar]
- Pauls J, Bandelow B, Ruther E, Kornhuber J. Polymorphism of the gene of angiotensin converting enzyme: Lack of association with mood disorder. J Neural Transm. 2000;107:1361–1366. doi: 10.1007/s007020070023. [DOI] [PubMed] [Google Scholar]
- Perez de Castro I, Torres P, Fernandez-Piqueras J, Saiz-Ruiz J, Llinares C. No association between dopamine D4 receptor polymorphism and manic depressive illness. J Med Genet. 1994;31:897–898. doi: 10.1136/jmg.31.11.897-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez de Castro I, Santos J, Torres P, Visedo G, Saiz-Ruiz J, Llinares C, Fernandez-Piqueras J. A weak association between TH and DRD2 genes and bipolar affective disorder in a Spanish sample. J Med Genet. 1995;32:131–134. doi: 10.1136/jmg.32.2.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prata DP, Breen G, Munro J, Sinclair M, Osborne S, Li T, Kerwin R, St Clair D, Collier DA. Bipolar 1 disorder is not associated with the RGS4, PRODH, COMT and GRK3 genes. Psychiatr Genet. 2006;16:229–230. doi: 10.1097/01.ypg.0000242190.43773.ce. [DOI] [PubMed] [Google Scholar]
- Ranade SS, Mansour H, Wood J, Chowdari KV, Brar LK, Kupfer DJ, Nimgaonkar VL. Linkage and association between serotonin 2A receptor gene polymorphisms and bipolar I disorder. Am J Med Genet Part B. 2003;121B:28–34. doi: 10.1002/ajmg.b.20070. [DOI] [PubMed] [Google Scholar]
- Rees M, Norton N, Jones I, McCandless F, Scourfield J, Holmans P, Moorhead S, Feldman E, Sadler S, Cole T, Redman K, Farmer A, McGuffin P, Owen MJ, Craddock N. Association studies of bipolar disorder at the human serotonin transporter gene (hSERT; 5HTT) Mol Psychiatry. 1997;2:398–402. doi: 10.1038/sj.mp.4000256. [DOI] [PubMed] [Google Scholar]
- Reif A, Pfuhlmann B, Lesch KP. Homocysteinemia as well as methylenetetrahydrofolate reductase polymorphism are associated with affective psychoses. Prog Neuropsychopharmacol Biol Psychiatry. 2005;29:1162–1168. doi: 10.1016/j.pnpbp.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Rietschel M, Nothen MM, Lannfelt L, Sokoloff P, Schwartz JC, Lanczik M, Fritze J, Cichon S, Fimmers R, Korner J. A serine to glycine substitution at position 9 in the extracellular N-terminal part of the dopamine D3 receptor protein: No role in the genetic predisposition to bipolar affective disorder. Psychiatry Res. 1993;46:253–259. doi: 10.1016/0165-1781(93)90093-v. [DOI] [PubMed] [Google Scholar]
- Rotondo A, Mazzanti C, Dell’Osso L, Rucci P, Sullivan P, Bouanani S, Gonnelli C, Goldman D, Cassano GB. Catechol o-methyltransferase, serotonin transporter, and tryptophan hydroxylase gene polymorphisms in bipolar disorder patients with and without comorbid panic disorder. Am J Psychiatry. 2002;159:23–29. doi: 10.1176/appi.ajp.159.1.23. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Leggo J, Goodburn S, Walsh C, Jain S, Paykel ES. Genetic association between monoamine oxidase A microsatellite and RFLP alleles and bipolar affective disorder: Analysis and meta-analysis. Hum Mol Genet. 1996;5:779–782. doi: 10.1093/hmg/5.6.779. [DOI] [PubMed] [Google Scholar]
- Saleem Q, Ganesh S, Vijaykumar M, Reddy YC, Brahmachari SK, Jain S. Association analysis of 5HT transporter gene in bipolar disorder in the Indian population. Am J Med Genet. 2000;96:170–172. [PubMed] [Google Scholar]
- Sasaki T, Macciardi FM, Badri F, Verga M, Meltzer HY, Lieberman J, Howard A, Bean G, Joffe RT, Hudson CJ, Kennedy JL. No evidence for association of dopamine D2 receptor variant (Ser311/Cys311) with major psychosis. Am J Med Genet. 1996;67:415–417. doi: 10.1002/(SICI)1096-8628(19960726)67:4<415::AID-AJMG18>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Hattori M, Sakai T, Kato T, Kunugi H, Hirose T, Nanko S. The monoamine oxidase-A gene and major psychosis in Japanese subjects. Biol Psychiatry. 1998;44:922–924. doi: 10.1016/s0006-3223(97)00522-2. [DOI] [PubMed] [Google Scholar]
- Savoye C, Laurent C, Amadeo S, Gheysen F, Leboyer M, Lejeune J, Zarifian E, Mallet J. No association between dopamine D1, D2, and D3 receptor genes and manic-depressive illness. Biol Psychiatry. 1998;44:644–647. doi: 10.1016/s0006-3223(97)00441-1. [DOI] [PubMed] [Google Scholar]
- Scassellati C, Rotondo A, Bonvicini C, Rossi G, Cassano GB, Gennarelli M. Further evidence on the lack of association between glycogen synthase kinase 3beta gene polymorphisms and bipolar disorder. Psychiatr Genet. 2007;17:249–250. doi: 10.1097/YPG.0b013e328013d8d8. [DOI] [PubMed] [Google Scholar]
- Schulze TG, Ohlraun S, Czerski PM, Schumacher J, Kassem L, Deschner M, Gross M, Tullius M, Heidmann V, Kovalenko S, Jamra RA, Becker T, Leszczynska-Rodziewicz A, Hauser J, Illig T, Klopp N, Wellek S, Cichon S, Henn FA, McMahon FJ, Maier W, Propping P, Nothen MM, Rietschel M. Genotype-phenotype studies in bipolar disorder showing association between the DAOA/G30 locus and persecutory delusions: A first step toward a molecular genetic classification of psychiatric phenotypes. Am J Psychiatry. 2005;162:2101–2108. doi: 10.1176/appi.ajp.162.11.2101. [DOI] [PubMed] [Google Scholar]
- Schumacher J, Jamra RA, Freudenberg J, Becker T, Ohlraun S, Otte AC, Tullius M, Kovalenko S, Bogaert AV, Maier W, Rietschel M, Propping P, Nothen MM, Cichon S. Examination of G72 and D-amino-acid oxidase as genetic risk factors for schizophrenia and bipolar affective disorder. Mol Psychiatry. 2004;9:203–207. doi: 10.1038/sj.mp.4001421. [DOI] [PubMed] [Google Scholar]
- Segman RH, Shapira Y, Modai I, Hamdan A, Zislin J, Heresco-Levy U, Kanyas K, Hirschmann S, Karni O, Finkel B, Schlafman M, Lerner A, Shapira B, Macciardi F, Lerer B. Angiotensin converting enzyme gene insertion/deletion polymorphism: Case-control association studies in schizophrenia, major affective disorder, and tardive dyskinesia and a family-based association study in schizophrenia. Am J Med Genet. 2002;114:310–314. doi: 10.1002/ajmg.10255. [DOI] [PubMed] [Google Scholar]
- Serretti A, Macciardi F, Cusin C, Verga M, Pedrini S, Smeraldi E. Tyrosine hydroxylase gene in linkage disequilibrium with mood disorders. Mol Psychiatry. 1998;3:169–174. doi: 10.1038/sj.mp.4000373. [DOI] [PubMed] [Google Scholar]
- Serretti A, Lilli R, Di Bella D, Bertelli S, Nobile M, Novelli E, Catalano M, Smeraldi E. Dopamine receptor D4 gene is not associated with major psychoses. Am J Med Genet. 1999;88:486–491. [PubMed] [Google Scholar]
- Serretti A, Lilli R, Lorenzi C, Lattuada E, Cusin C, Smeraldi E. Tryptophan hydroxylase gene and major psychoses. Psychiatry Res. 2001;103:79–86. doi: 10.1016/s0165-1781(01)00269-4. [DOI] [PubMed] [Google Scholar]
- Serretti A, Lilli R, Lorenzi C, Lattuada E, Cusin C, Smeraldi E. Serotonin transporter gene (5-HTTLPR) and major psychoses. Mol Psychiatry. 2002;7:95–99. doi: 10.1038/sj.mp.4000936. [DOI] [PubMed] [Google Scholar]
- Serretti A, Rotondo A, Lorenzi C, Smeraldi E, Cassano GB. Catechol-O-methyltransferase gene variants in mood disorders in the Italian population. Psychiatr Genet. 2006;16:181–182. doi: 10.1097/01.ypg.0000218625.88504.f1. [DOI] [PubMed] [Google Scholar]
- Shaikh S, Ball D, Craddock N, Castle D, Hunt N, Mant R, Owen M, Collier D, Gill M. The dopamine D3 receptor gene: No association with bipolar affective disorder. J Med Genet. 1993;30:308–309. doi: 10.1136/jmg.30.4.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S, Bronstein M, Sternfeld M, Pisante A, Weizman A, Reznik I, Spivak B, Grisaru N, Karp L, Schiffer R, Kotler M, Strous RD, Swartz-Vanetik M, Knobler HY, Shinar E, Yakir B, Zak NB, Darvasi A. COMT: A common susceptibility gene in bipolar disorder and schizophrenia. Am J Med Genet Part B. 2004;128B:61–64. doi: 10.1002/ajmg.b.30032. [DOI] [PubMed] [Google Scholar]
- Skibinska M, Hauser J, Czerski PM, Leszczynska-Rodziewicz A, Kosmowska M, Kapelski P, Slopien A, Zakrzewska M, Rybakowski JK. Association analysis of brain-derived neurotrophic factor (BDNF) gene Val66Met polymorphism in schizophrenia and bipolar affective disorder. World J Biol Psychiatry. 2004;5:215–220. doi: 10.1080/15622970410029936. [DOI] [PubMed] [Google Scholar]
- Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, Edenberg HJ, Nurnberger JI, Jr, Rietschel M, Blackwood D, Corvin A, Flickinger M, Guan W, Mattingsdal M, McQuillin A, Kwan P, Wienker TF, Daly M, Dudbridge F, Holmans PA, Lin D, Burmeister M, Greenwood TA, Hamshere ML, Muglia P, Smith EN, Zandi PP, Nievergelt CM, McKinney R, Shilling PD, Schork NJ, Bloss CS, Foroud T, Koller DL, Gershon ES, Liu C, Badner JA, Scheftner WA, Lawson WB, Nwulia EA, Hipolito M, Coryell W, Rice J, Byerley W, McMahon FJ, Schulze TG, Berrettini W, Lohoff FW, Potash JB, Mahon PB, McInnis MG, Zollner S, Zhang P, Craig DW, Szelinger S, Barrett TB, Breuer R, Meier S, Strohmaier J, Witt SH, Tozzi F, Farmer A, McGuffin P, Strauss J, Xu W, Kennedy JL, Vincent JB, Matthews K, Day R, Ferreira MA, O’Dushlaine C, Perlis R, Raychaudhuri S, Ruderfer D, Hyoun PL, Smoller JW, Li J, Absher D, Thompson RC, Meng FG, Schatzberg AF, Bunney WE, Barchas JD, Jones EG, Watson SJ, Myers RM, Akil H, Boehnke M, Chambert K, Moran J, Scolnick E, Djurovic S, Melle I, Morken G, Gill M, Morris D, Quinn E, Muhleisen TW, Degenhardt FA, Mattheisen M, Schumacher J, Maier W, Steffens M, Propping P, Nothen MM, Anjorin A, Bass N, Gurling H, Kandaswamy R, Lawrence J, McGhee K, McIntosh A, McLean AW, Muir WJ, Pickard BS, Breen G, St Clair D, Caesar S, Gordon-Smith K, Jones L, Fraser C, Green EK, Grozeva D, Jones IR, Kirov G, Moskvina V, Nikolov I, O’Donovan MC, Owen MJ, Collier DA, Elkin A, Williamson R, Young AH, Ferrier IN, Stefansson K, Stefansson H, Thornorgeirsson T, Steinberg S, Gustafsson O, Bergen SE, Nimgaonkar V, Hultman C, Landen M, Lichtenstein P, Sullivan P, Schalling M, Osby U, Backlund L, Frisen L, Langstrom N, Jamain S, Leboyer M, Etain B, Bellivier F, Petursson H, Sigur Sson E, Muller-Mysok B, Lucae S, Schwarz M, Schofield PR, Martin N, Montgomery GW, Lathrop M, Oskarsson H, Bauer M, Wright A, Mitchell PB, Hautzinger M, Reif A, Kelsoe JR, Purcell SM Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souery D, Lipp O, Mahieu B, Mendelbaum K, De Martelaer V, Van Broeckhoven C, Mendlewicz J. Association study of bipolar disorder with candidate genes involved in catecholamine neurotransmission: DRD2, DRD3, DAT1, and TH genes. Am J Med Genet. 1996;67:551–555. doi: 10.1002/(SICI)1096-8628(19961122)67:6<551::AID-AJMG7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Souery D, Lipp O, Rivelli SK, Massat I, Serretti A, Cavallini C, Ackenheil M, Adolfsson R, Aschauer H, Blackwood D, Dam H, Dikeos D, Fuchshuber S, Heiden M, Jakovljevic M, Kaneva R, Kessing L, Lerer B, Lonnqvist J, Mellerup T, Milanova V, Muir W, Nylander PO, Oruc L, Mendlewicz J. Tyrosine hydroxylase polymorphism and phenotypic heterogeneity in bipolar affective disorder: A multicenter association study. Am J Med Genet. 1999;88:527–532. doi: 10.1002/(sici)1096-8628(19991015)88:5<527::aid-ajmg17>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Stober G, Heils A, Lesch KP. Serotonin transporter gene polymorphism and affective disorder. Lancet. 1996;347:1340–1341. [PubMed] [Google Scholar]
- Stober G, Jatzke S, Heils A, Jungkunz G, Knapp M, Mossner R, Riederer P, Lesch KP. Insertion/deletion variant (–141C Ins/Del) in the 5′ regulatory region of the dopamine D2 receptor gene: Lack of association with schizophrenia and bipolar affective disorder. Short communication. J Neural Transm. 1998;105:101–109. doi: 10.1007/s007020050041. [DOI] [PubMed] [Google Scholar]
- Stober G, Sprandel J, Schmidt F, Faul T, Jabs B, Knapp M. Association study of 5′-UTR polymorphisms of the human dopamine transporter gene with manic depression. Bipolar Disord. 2006;8:490–495. doi: 10.1111/j.1399-5618.2006.00352.x. [DOI] [PubMed] [Google Scholar]
- Sun HS, Wang HC, Lai TJ, Wang TJ, Li CM. Sequence variants and haplotype analysis of serotonin transporter gene and association with bipolar affective disorder in Taiwan. Pharmacogenetics. 2004;14:173–179. doi: 10.1097/00008571-200403000-00005. [DOI] [PubMed] [Google Scholar]
- Szczepankiewicz A, Skibinska M, Hauser J, Slopien A, Leszczynska-Rodziewicz A, Kapelski P, Dmitrzak-Weglarz M, Czerski PM, Rybakowski JK. Association analysis of the GSK-3beta T-50C gene polymorphism with schizophrenia and bipolar disorder. Neuropsychobiology. 2006;53:51–56. doi: 10.1159/000090704. [DOI] [PubMed] [Google Scholar]
- Tan EC, Chong SA, Lim LC, Chan AO, Teo YY, Tan CH, Mahendran R. Genetic analysis of the thermolabile methylenetetrahydrofolate reductase variant in schizophrenia and mood disorders. Psychiatr Genet. 2004;14:227–231. doi: 10.1097/00041444-200412000-00012. [DOI] [PubMed] [Google Scholar]
- Tang J, Xiao L, Shu C, Wang G, Liu Z, Wang X, Wang H, Bai X. Association of the brain-derived neurotrophic factor gene and bipolar disorder with early age of onset in Mainland China. Neurosci Lett. 2008;433:98–102. doi: 10.1016/j.neulet.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Todd RD, Botteron KN. Etiology and genetics of early-onset mood disorders. Child Adolesc Psychiatr Clin N Am. 2002;11:499–518. doi: 10.1016/s1056-4993(02)00013-5. [DOI] [PubMed] [Google Scholar]
- Tramontina J, Frey BN, Andreazza AC, Zandona M, Santin A, Kapczinski F. Val66met polymorphism and serum brain-derived neurotrophic factor levels in bipolar disorder. Mol Psychiatry. 2007;12:230–231. doi: 10.1038/sj.mp.4001941. [DOI] [PubMed] [Google Scholar]
- Tsai SJ, Hong CJ, Wang YC. Tryptophan hydroxylase gene polymorphism (A218C) and suicidal behaviors. Neuroreport. 1999;10:3773–3775. doi: 10.1097/00001756-199912160-00010. [DOI] [PubMed] [Google Scholar]
- Tut TG, Wang JL, Lim CC. Negative association between T102C polymorphism at the 5-HT2A receptor gene and bipolar affective disorders in Singaporean Chinese. J Affect Disord. 2000;58:211–214. doi: 10.1016/s0165-0327(99)00104-4. [DOI] [PubMed] [Google Scholar]
- Van Den Bogaert A, Sleegers K, De Zutter S, Heyrman L, Norrback KF, Adolfsson R, Van Broeckhoven C, Del-Favero J. No allelic association or interaction of three known functional polymorphisms with bipolar disorder in a northern Swedish isolated population. Psychiatr Genet. 2006;16:209–212. doi: 10.1097/01.ypg.0000218623.03752.e4. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vincent JB, Masellis M, Lawrence J, Choi V, Gurling HM, Parikh SV, Kennedy JL. Genetic association analysis of serotonin system genes in bipolar affective disorder. Am J Psychiatry. 1999;156:136–138. doi: 10.1176/ajp.156.1.136. [DOI] [PubMed] [Google Scholar]
- Vincze I, Perroud N, Buresi C, Baud P, Bellivier F, Etain B, Fournier C, Karege F, Matthey ML, Preisig M, Leboyer M, Malafosse A. Association between brain-derived neurotrophic factor gene and a severe form of bipolar disorder, but no interaction with the serotonin transporter gene. Bipolar Disord. 2008;10:580–587. doi: 10.1111/j.1399-5618.2008.00603.x. [DOI] [PubMed] [Google Scholar]
- Williams NM, Green EK, Macgregor S, Dwyer S, Norton N, Williams H, Raybould R, Grozeva D, Hamshere M, Zammit S, Jones L, Cardno A, Kirov G, Jones I, O’Donovan MC, Owen MJ, Craddock N. Variation at the DAOA/G30 locus influences susceptibility to major mood episodes but not psychosis in schizophrenia and bipolar disorder. Arch Gen Psychiatry. 2006;63:366–373. doi: 10.1001/archpsyc.63.4.366. [DOI] [PubMed] [Google Scholar]
- WOOLF B. On estimating the relation between blood group and disease. Ann Hum Genet. 1955;19:251–253. doi: 10.1111/j.1469-1809.1955.tb01348.x. [DOI] [PubMed] [Google Scholar]
- Yen FC, Hong CJ, Hou SJ, Wang JK, Tsai SJ. Association study of serotonin transporter gene VNTR polymorphism and mood disorders, onset age and suicide attempts in a Chinese sample. Neuropsychobiology. 2003;48:5–9. doi: 10.1159/000071821. [DOI] [PubMed] [Google Scholar]
- Zhang HY, Ishigaki T, Tani K, Chen K, Shih JC, Miyasato K, Ohara K, Ohara K. Serotonin2A receptor gene polymorphism in mood disorders. Biol Psychiatry. 1997;41:768–773. doi: 10.1016/S0006-3223(96)00160-6. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li Y, Zhao Q, Huang K, Wang P, Yang P, Li S, Feng G, Lindpaintner K, He L, Shi Y. First evidence of association between G72 and bipolar disorder in the Chinese Han population. Psychiatr Genet. 2009a;19:151–153. doi: 10.1097/YPG.0b013e32832a50f1. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lindpaintner K, Che R, He Z, Wang P, Yang P, Feng G, He L, Shi Y. The Val/Met functional polymorphism in COMT confers susceptibility to bipolar disorder: Evidence from an association study and a meta-analysis. J Neural Transm. 2009b;116:1193–1200. doi: 10.1007/s00702-009-0260-7. [DOI] [PubMed] [Google Scholar]
- Zill P, Baghai TC, Zwanzger P, Schule C, Minov C, Riedel M, Neumeier K, Rupprecht R, Bondy B. Evidence for an association between a G-protein beta3-gene variant with depression and response to antidepressant treatment. Neuroreport. 2000;11:1893–1897. doi: 10.1097/00001756-200006260-00018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.