Abstract
Light-oxygen-voltage (LOV) domains bind a flavin chromophore to serve as blue light sensors in a wide range of eukaryotic and prokaryotic proteins. LOV domains are associated with a variable effector domain or a separate protein signaling partner to execute a wide variety of functions that include regulation of kinases, generation of antisigma-factor antagonists, and regulation of circadian clocks. Here we present the crystal structure, photocycle kinetics, association properties, and spectroscopic features of a full-length LOV protein from Rhodobacter sphaeroides (RsLOV). RsLOV exhibits N-terminal and C-terminal helical extensions that form an unusual helical bundle at its dimer interface with some resemblance to the helical transducer of sensory rhodopsin II. The blue light-induced conformational changes of RsLOV revealed from a comparison of light and dark state crystal structures support a shared signaling mechanism of LOV domain proteins that originates with the light-induced formation of a flavin-cysteinyl photoadduct. Adduct formation disrupts hydrogen bonding in the active site and propagates structural changes through the LOV domain core to the N- and C-terminal extensions. Single residue variants in the active site and dimer interface of RsLOV alter photoadduct lifetimes and induce structural changes that perturb oligomeric state. Size exclusion chromatography, multi-angle light scattering, small-angle X-ray scattering and cross-linking studies indicate that RsLOV dimerizes in the dark, but upon light excitation, dissociates into monomers. This light-induced switch in oligomeric state may prove useful for engineering molecular associations in controlled cellular settings.
Keywords: LOV domain, PAS domain, light-state, crystal structure, photoreceptor
INTRODUCTION
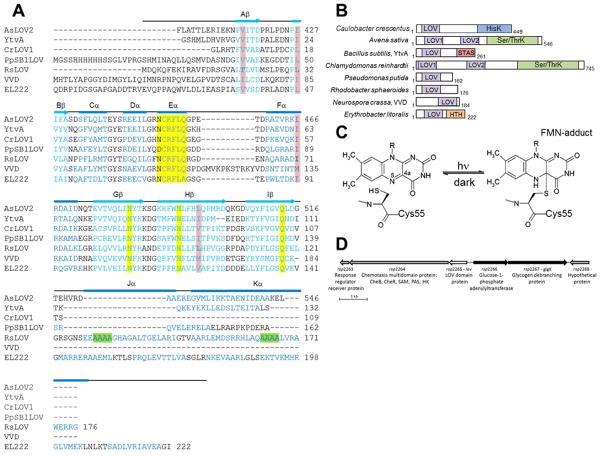
Per-ARNT-Sim (PAS) domains are key components of many photoreceptor proteins and are present in all kingdoms of life (1). PAS domains often detect signals through a cofactor that undergoes a chemical change in response to environmental stimuli (2, 3). Light, oxygen, voltage (LOV) domains are a subset of the PAS domain superfamily that exhibit a mixed α/β fold common to PAS domains but are distinguished by binding a flavin cofactor that imparts blue-light (λ = 440–480 nm) sensitivity (4, 5). LOV domains serve as sensory domains that modulate interactions in signal transduction proteins. Upon photoexcitation by blue light, the noncovalently bound flavin cofactor forms a covalent adduct between the flavin C4a carbon atom and the sulfur atom of a conserved cysteine residue in the LOV domain active center (Fig. 1)(6).
Figure 1.
LOV domain homology and chemistry (A) Structure-based sequence alignment of LOV domains, Avena sativa LOV2 (2V1A), Bacillus subtilis (2PR5), Chlamydomonas reinhardtii (1N9L), Pseudomonas putida (3SW1), Rhodobacter sphaeroides, Neurospora crassa Vivid. (2PD7), and Erythrobacter litoralis (3P7N). Secondary structure elements noted above (α-helical residues in blue, β-sheet in aqua). Residues critical to flavin coordination are highlighted in yellow, sites of RLOV single residue variants are highlighted in pink, and the four Ala repeat in Jα and Kα helices are highlighted in green. (B) Domain arrangement of sequence aligned LOV domain proteins (number of residues specified). HisK, histidine kinase; Ser/ThrK, serine/threonine kinase; STAS, sulfate transporter anti-s antagonist; HTH, helix-turn-helix. (C) Schematic of light-induced cysteinyl-FMN (C4a) covalent adduct formation. (D) Genetic context of LOV domain protein in R. sphaeroides.
LOV proteins perform many different functional roles that are determined by an associated effector domain or protein partner under control of the LOV light switch. These roles include the regulation of the phototropin serine/threonine kinases (phot1 and phot2) in Chlamydomonas reinhardtii (green algae)(7, 8) and higher plant life, Oryza sativa (rice)(9–11), regulation of histidine kinases in Caulobacter crescentus (gram-negative bacterium)(12, 13), regulation of the antisigma-factor antagonist YtvA in Bacillus subtillis (gram-positive bacterium)(14–17), and regulation of the circadian clock photosensors White collar-1 (WC-1) and Vivid (VVD) in (18–22) Neurospora crassa (filamentous fungi), (Fig. 1)(23). LOV domain proteins that do not contain an effector domain are so-called “short” LOV domains (24). Short LOV proteins include VVD and the LOV domain protein from Pseudomonas putida (PpSB1-LOV)(25) as well as the LOV protein included in this work from R. sphaeroides. VVD has been shown to signal to a protein partner in the White-Collar Complex (26–28). Thus, LOV domains propagate signals in two distinct ways: by affecting an auxiliary domain within the LOV protein, or by interactions with a protein partner.
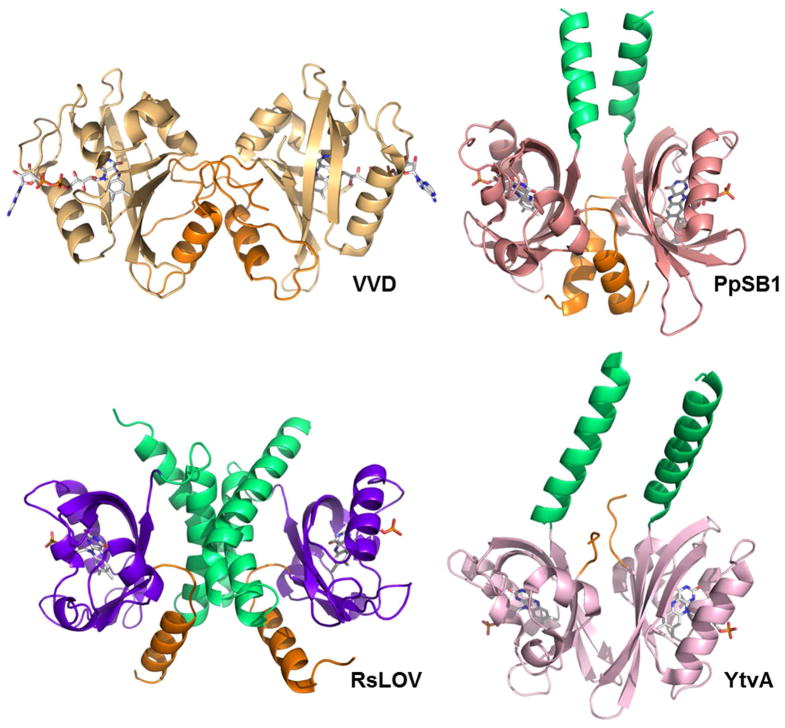
Outside of the conserved structural core (29), LOV domains are distinguished by variations of their N-terminal and C-terminal structural extensions (Fig. 2). These structural differences result in a variety of molecular mechanisms in light-dependent signaling of LOV proteins. A key attribute of LOV domains is their tendency to dimerize. There are two general association modes of PAS-PAS dimerization. In one mode, the dimer interface is the PAS domain β-scaffold, as seen in hypoxia inducible factor (30), KinA histidine kinases (31) and LOV1-type proteins (32). In the second mode, the N-terminal helices flanking the domain core (Ncap) of each subunit form intermolecular contacts that establish the dimer interface. This mode is exhibited by heme-based oxygen sensor FixL (33), nitrogen fixation regulatory protein NifL (34), and heme-regulated phosphodiesterase EcDOS (35). Interestingly, the LOV domain protein VVD exhibits the second mode of dimerization, with intermolecular contacts primarily in the Ncaps, while YtvA utilizes both modes of dimerization, using contacts in the β-scaffold and in the C-terminal extension (Jα helix) to form a dimer (Fig 3) (36). All structurally characterized tandem PAS structures have PAS-A and PAS-B associations that are head-to-tail, and form parallel or antiparallel dimers mediated by extensive contacts between exposed β-sheet and α-helical extensions (37, 38).
Figure 2.
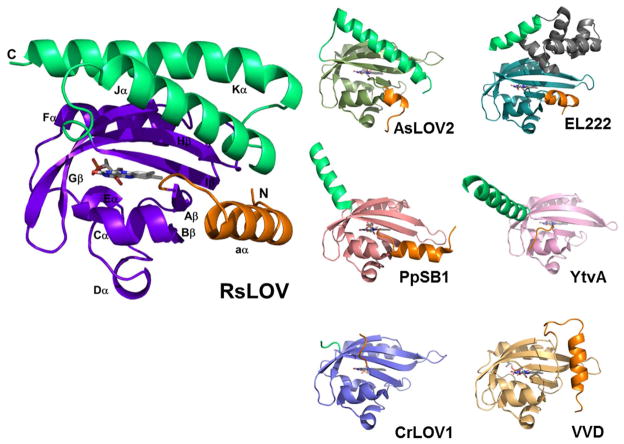
LOV subunit structures. (A) RsLOV structure in the dark state, with PAS core in violet, N-terminal extension in orange, and C-terminal extension in light green. Dark state structures of N. crassa VVD (2PD7, PAS core light orange), A.sativa LOV2 (2V1A, PAS core forest green), C. reinhardtii LOV1 (1N9L, PAS core light blue) B. subtilis LOV (2PR5, PAS core pink), EL222 (3P7N, PAS core aqua, HTH motif gray), P. putida (3SW1, PAS core salmon) with N-terminal extensions in orange, and C-terminal extensions in light green.
Figure 3.
Comparison of LOV dimers: Light state structures of N. crassa VVD (3RH8, light orange), dark state structure of B. subtilis LOV (2PR5, pink), light state structure of P. putida (3SW1, salmon), and dark state structure of RsLOV (purple), N-terminal extensions in orange, C-terminal extensions in light green.
R. sphaeroides is a purple photosynthetic gram-negative bacterium (α-proteobacterium) that contains a short LOV domain protein in addition to other light-sensing proteins that include AppA, a blue-light sensing FAD photoreceptor, and three cryptochrome-like proteins (39, 40). The LOV domain protein of R. sphaeroides (RsLOV) binds flavin mononucleotide (FMN) as its chromophore. RsLOV is 18% identical to N. crassa VVD, with a moderate pairwise score of 25 (41). Sequence alignments of RsLOV and VVD reveal structural similarities in the LOV core region, but differences in the terminal extensions (Fig. 1A). Specifically, the N-terminal latch of VVD, which participates in a “domain swapping” interaction to enable VVD light-dependent dimerization (19), is lacking in RsLOV; however, the receptor pocket for the VVD N-terminal latch is conserved. RsLOV also contains a C-terminal extension absent in VVD but present in YtvA and the LOV2 domain of Avena sativa (oat, AsLOV2)(42), which have pairwise scores from ClustalW of 28 and 25, respectively (43, 44). Other LOV domain proteins similar by sequence include the LOV proteins from Pseudomonas putida (soil bacterium, PpSB1-LOV)(45, 46) and Erythrobacter litoralis (marine bacterium, EL222). The PpSB1-LOV has a similar length N-terminal extension and a shorter C-terminal Jα helix than that of RsLOV (45), whereas the LOV domain protein from EL222 has a short N-terminal extension and a Jα helix that attaches to a helix-turn-helix (HTH) effector domain (47).
The differences in LOV domain light-dependent signaling processes are exemplified by the structural changes induced in LOV proteins VVD, YtvA, and AsLOV2 upon exposure to light. VVD forms a light-induced dimer through an N-terminal helix and flexible coiled regions (19–22). YtvA exhibits a light-induced rotation of subunits in its head-to-head dimer that involve changes in intermolecular contacts between the C-terminal Jα helices and between the individual β-scaffolds (17, 48, 49). In the isolated AsLOV2 domain of plant phototropin, light-induced rearrangements of hydrogen bonds within the structural core propagate to changes in both the N-terminal and C-terminal flanking regions that result in a partial unfolding of the Jα helix, but the protein does not homodimerize (17, 31, 50). However, the proteins display conserved aspects of signal propagation from the flavin chromophore to the variable peripheral elements despite differences in light-induced conformational changes in these peripheral regions (4).
The LOV protein of R. sphaeroides ATCC 17025 is a short-LOV (rsp2265) lacking an effector domain. Downstream on the chromosome from the RsLOV gene is a large 1165 aa protein that contains PAS, histidine kinase, S-adenosylmethionine methyltransferase, CheR methyltransferase, and CheB methylesterase domains (rsp2264, Fig. 1D). Upstream of RsLOV is a hypothetical protein of 224 residues (rsp2268), and a 409 aa glycogen-1-phosphate adenylyltransferase (rsp2266). Previous studies of RsLOV from a similar R. sphaeroides strain implicated the protein in photosynthetic gene expression in a manner similar to that of the other two photoreceptor proteins of R.sphaeroides, AppA and CryB (51). Thus, functional and gene clustering analyses potentially involve RsLOV function in carbohydrate metabolism, chemotaxis, and cellular response to photooxidative stress.
Herein, we investigate the molecular mechanism behind the light-dependent signaling of RsLOV; specifically, how the photosensor protein generates and propagates light absorption to changes in protein conformation and association. Through a combination of crystallographic and biochemical studies we define the structural features of RsLOV and describe how flavin photochemistry correlates to changes in the oligomeric state of the protein.
MATERIALS AND METHODS
Protein expression and Purification
The RsLOV (R. sphaeroides 17025) gene was cloned into the expression vector pET28a (Novagen) in frame with an N-terminal His-tag using NdeI and EndoRI restriction sites. Site-directed mutagenesis by overlap extension with the polymerase chain reaction generated point mutation constructs of RsLOV: T21V, L32V, C55A, I71L, I71V L101I, S127C, A136C, A136Y, A138Y, E142C, R145C, and A167C. All were cloned between NdeI and EndoRI restriction sites and sequenced in their entirety at the Biotechnology Resource Center of Cornell University.
RsLOV constructs were overexpressed in Escherichia coli BL21(DE3) cells. Cultures were induced with 100 μM IPTG at a cell density (OD600) of 0.6–0.8. Proteins were expressed for 20–22 h at 17 °C and shaken at 170 rpm under constant light. Cells were harvested by centrifugation (8000 rpm, 4 °C, 15 min), flash frozen in liquid N2, and the cell pellets were stored at −80 °C. Cell pellets were resuspended in buffer containing 50 mM TRIS (pH 8.5), 10 mM imidazole, and 300 mM NaCl, lysed by sonication, and cell debris was subsequently removed by centrifugation (22,000 rpm, 4 °C, 1 h). Soluble cell lysate was purified with Ni-NTA affinity chromatography. Eluted protein was concentrated and treated with thrombin overnight. Samples were then purified on a Superdex 75 Hi-load 26/60 FPLC column and concentrated to 5–30 mg/mL by centrifugation. Analytical SEC FPLC was completed using a Superdex 200 10/300 analytical column. The column was equilibrated with buffer containing 150 mM NaCl and 50 mM TRIS (pH 8.5). Samples were injected in 500 mL aliquots and run at a flow rate of 0.3 mL/min.
Crystallization, Data Collection and Structure Determination
Orthorhombic RsLOV crystals in space group P212121 and hexagonal RsLOV crystals in space group P65 were obtained at 17 °C under constant darkness by vapor diffusion. Crystals grew from an equal volume (1.5 μL) of 30 mg/mL protein in buffer containing 50 mM TRIS (pH 8.5) and 150 mM NaCl and the reservoir solution: 30% PEG400, 0.15 M MgCl26H2O, 0.1 M HEPES, pH 8.5 (orthorhombic), 0.1 M HEPES pH 7.5, 4.3 M NaCl (hexagonal). RsLOV crystallizes as single rods, which diffract to ~2.4 Å resolution. L32V RsLOV crystals in space group P212121 grew from LiSO4 in 0.1 M HEPES pH 7.9 and diffract to 1.9 Å resolution. A138Y RsLOV crystals in space group P65 grew from 0.1 M HEPES pH 7.5, 4.3 M NaCl and diffract to 2.6 Å resolution. Data were collected at 100 K on beamlines A1 and F1 at the Cornell High Energy Synchrotron Source (CHESS). Dark-state crystals were excited to the light-state by white light illumination while still in solution prior to cryo-cooling to 100 K. To minimize reduction of the covalent adduct formed in the light-adapted protein by the X-ray beam, two strategies were employed to stabilize the adduct as reported previously (19, 21). To reduce radiation exposure, each frame was collected over a 1–5° oscillation per second (X-ray flux 3x1011 photons sec−1 per 0.2 x 2.00 mm2 focus at 0.979 Å) and secondly, data were collected from several different spots on a single crystal and then merged with data collected on additional crystals in the same manner, to generate the complete dataset. Data were reduced and scaled using HKL2000 (52). Initial phases were determined by molecular replacement in AutoMR (Phenix) (53) using VVD (Protein Data Bank code 2PD7 (21)) as a search model. Models were rebuilt and fit using XFIT (54) followed by refinement using CNS (50) (Table S1) and Phenix (55).
SAXS Data Collection and Analysis
SAXS data were collected at the F2 beamline at the CHESS at 9.881 keV. RsLOV solutions at concentrations of 2.5, 5, and 10 mg/mL in a buffer of 2 mM DTT, 1% glycerol, 50 mM TRIS pH 8, 150 mM NaCl were used for the SAXS analysis. The X-ray beam was collimated to 250 × 250 mm and was centered on a 2 mm diameter vertical quartz capillary tube with 10 mm thick walls (Hampton Research, Aliso Viejo, California, USA). The protein samples were centrifuged at 14,000 rpm for 10 min prior to data collection. Samples aliquots of approximately 15–20 μl were delivered from a 96-well plate to the capillary using a Hudson SOLO single-channel pipetting robot (Hudson Robotics Inc., Springfield, New Jersey, USA). To reduce radiation damage, the sample plugs were oscillated in the X-ray beam using a computer-controlled syringe pump (Aurora Biomed, Vancouver, Canada). Images were collected on a Quantum 1 CCD detector (Area Detector Systems Corporation, Poway, California, USA) with 6 sequential 30 s exposures being used to assess possible radiation damage. Dark samples were wrapped in aluminum foil up until placement in the 96-well plate to minimize light activation. Light samples were exposed to white light prior to addition to the plate, and data collection was initiated within a short time frame (seconds to minutes) to minimize the contribution from recovered dark state sample.
Scattering data was processed according to previously described protocols (56) utilizing BioXTAS RAW (57), and programs from the ATSAS (58–60) and SITUS (61) packages. SAXS of hen egg white lysozyme (MW of 14700 Da) and glucose isomerase (MW of 173000 Da) were collected as references for determining the apparent MW of RsLOV. Additionally, the SAXS MoW tool developed by H. Fischer, et al (62), was applied to the data for MW calculations. The MW values derived from SAXS MoW utilizing the gnom output files are reported herein (Table S3). Fits to the experimental SAXS data were generated using CRYSOL (63) and the combination of RANCH13 and GAJOE13 for the EOM approximations (64). In the CRYSOL fits, the dimer model was the P65 dark state structure, and the monomer model was chain A from the P65 dark state structure. In the EOM approximation, the monomer model included residues 20 through 125 of the P65 dark structure chain B as the folded domain, whereas the rest of the chain was treated as random coil; the dimer model included residues 5 (chain A) through 172 (chain B) as the folded domain, with the remaining residues treated as random coil. Envelopes were generated using the output of DAMFILT preceded by DAMAVER (65) completed with ten DAMMIF (66) calculated reconstructions, as input for the pdb2vol utility in the SITUS package. Rigid body search to fit crystallographic models into SAXS envelopes was completed using the COLLAGE utility in the SITUS package.
Light Scattering
Multi-angle light scattering (MALS) was conducted on RsLOV with a Protein Solutions Dynapro dynamic light scattering instrument over a concentration range of 100–500 μM. Dark- and light state samples were injected into a Sephadex 75 10/300 analytical column, which was connected to a Wyatt miniDAWN Treos multiangle light scattering system. Absolute MW calculations were completed using ASTRA version 5.0 from Wyatt Technologies.
Crosslinking
Crosslinking studies of RsLOV variants with cysteine residues introduced at positions 127, 129, 136, 138, 142, and 145 were carried under native and oxidizing conditions. For oxidizing conditions, crosslinking was induced with the addition of 0.5 mM Cu(II)(1,10 phenanthroline)3 to dark and light state protein samples at concentrations of 4–8 mg/mL. Changes in oligomerization were assessed by SDS-PAGE and analytical SEC FPLC.
Time-Resolved Absorption Spectroscopy
Time-resolved electronic absorption spectroscopy measurements on RsLOV and RsLOV mutants were performed using an Agilent 8453 spectrophotometer. Samples were excited to the light state using white light from a Xe arc lamp. Measurements of dark-state recovery rates were taken every 30 s for 2–4 h. Absorbance traces at 450 and 310 nm were fit assuming first-order kinetics with a single rate constant, using a monoexponential function (Abs = B + Ce−kt). Average rate constants were determined from multiple measurements.
Accesion Codes
Protein Data Bank: The crystal structures of RsLOV in the dark-(P212121 and P65) and light-adapted states, L32V RsLOV, and A138Y RsLOV in the dark state were deposited under accession codes 4HJ4, 4HJ6, 4HNB, 4HIA and 4HJ3, respectively.
RESULTS
Dark State of R. sphaeroides LOV
Orthorhombic crystals of RsLOV that were grown in the dark diffracted to 2.4 Å resolution (Table S1) at the synchrotron. The crystallographic structure was determined by molecular replacement using dark state VVD (PDB 2PD7) as the probe. The two molecules of RsLOV in the asymmetric unit form a tight dimer that is related by non-crystallographic symmetry. RsLOV adopts the PAS fold (1) with the conserved core of a five-stranded antiparallel β-sheet, Aβ (Leu20-Met25), Bβ (Pro31-Ala35), Gβ (Glu80-Arg89), Hβ (Glu93-Gly106), and Iβ (Pro111-Leu121), and the helical connector elements Cα (Pro37-Thr43), Dα (Glu47-Gly52), Eα (Cys55-Gln59), and Fα (Ala65-Leu77).
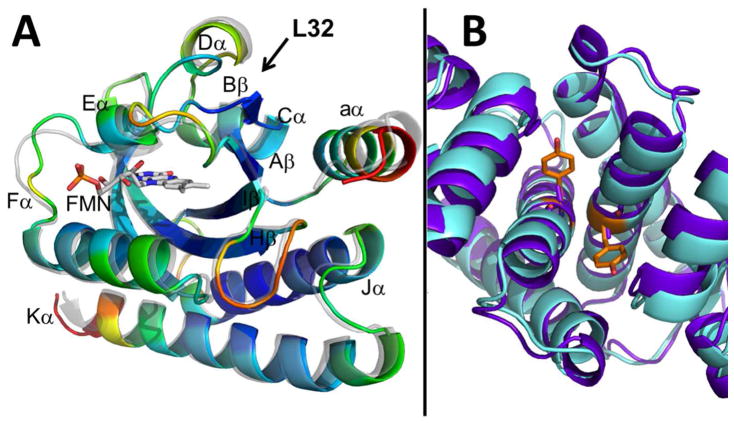
Auxiliary to the PAS core, RsLOV has an additional N-terminal aα helix (Met1-Arg15) and a C-terminal helix-turn-helix motif composed of Jα (Glu128-Ile146) and Kα (Leu153-Gly176) (Fig. 2). The 15-residue N-terminal helix (aα) stretches 21 Å in length, and attaches to Aβ through a short 5 residue loop that includes Gly17 in the hinge. The aα helix is adjacent to the end of the Jα helix, and flanks the β-scaffold for most of its length until the Hβ-Iβ loop is reached. The aα helix is amphipathic; the solvent exposed side of the helix is made up of hydrophilic and charged residues, whereas the inner side that flanks the β-sheet is generally hydrophobic. Asn36 mediates a direct interaction between aα helix and Bβ.
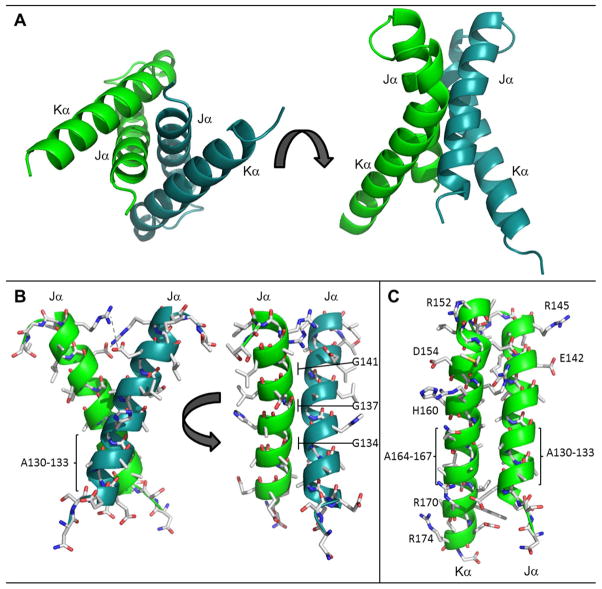
The C-terminal helix-turn-helix motif composes the dimer interface from roughly 48 residues on each subunit. The shorter Jα helix stretches 29 Å in length, whereas the longer Kα helix extends 40 Å. The helices are at a ~30° angle to one another and make a close central crossing of 3.8 Å in the region of Ala136, Ala162, Thr140 and Arg158, and Val169 and Glu129. The close proximity of the helices is largely due to four alanine repeat sequences at their centers and several glycine residues (Figs. 1 and 4). His135 anchors the Jα helix to the LOV domain via the Iβ Ser117. The Jα and β-scaffold interface is primarily hydrophobic, with Jα nonpolar residues Ala132, Leu139, L143, and Ile146, extended toward the sheet, and the nonpolar side chains of Leu115 and Phe119 of Iβ extended toward the helix. Direct interactions between Jα and aα occur via hydrogen bonding residues Arg15 and/or Ser16 and Glu142. The Kα helix has Leu161, Leu168, Trp172 directed toward the β-sheet, whereas Hβ has Leu98 and Phe100 directed toward the Kα helices.
Figure 4.
The RsLOV dimerization motif. (A) Four helix bundle at interface of the RsLOV crystallographic dimer, A and B subunits distinguished by color. (B) Side chain contacts at Jα interface of chain A and chain B (C) Side chain contacts at the Jα and Kα interface. There are salt bridge between Arg145 and Glu142 (same monomer) and between Arg15 and Glu154 (opposing monomer). Hydrogen bonding interactions range from 2.45 to 3.66 Å and also include the amide H of Arg158 to the backbone O of Gly17 (opposing monomer), and the amide H of Arg159 to the side chain O of Ser124 (opposing monomer). The residues with the most buried surface area are within the crossing point of the two Jα helices at the region of the four alanine repeat: Ala130–133, Gly134, Gly137, Ala138, and the cross-section of Kα including Glu154, Arg158, and Ala162.
RsLOV is unique among short-LOVs in that it has both N-terminal and C-terminal flanking helices that could interact with an effector protein. At the center of the crystallized dimer interface are the C-terminal helix-turn-helix motifs (Jα, Kα) of the two subunits (A and B) that pack against each other in a coiled-coil to form a distorted four-helix bundle with a highly unusual packing interface. The Jα/Kα helix dimer interface has a highly complementary solvent exclusive core formed primarily by close symmetric interactions of three Jα Gly residues (134, 137 and 141) that allow the 137–138 peptide bonds from each subunit to stack against each other (~3.5Å). Packing of the Ala4 repeats at the beginning of Jα (130–133) and end of Kα (162–167) in roughly perpendicular helices also mediates an especially tight interface (Fig. 4). Long side chains at the ends of the helices stabilize the interface periphery with salt bridges and hydrogen bonds. In total, the dimer interface involves 34 residues and buries 1100 Å2 per subunit, but due to the predominance of Ala and Gly residues does not contain the branched hydrophobic side chains typical of coiled-coils. As a result, interface analysis as performed by PISA complex (67, 68) indicates that the contact buries a small amount of hydrophobic surface area (ΔG of complex formation of −0.4 kcal/mol) and therefore is unlikely to participate in solution dimerization. However, this estimate is based almost exclusively on buried hydrophobic surface area and there are also substantial hydrophilic interactions between the subunits including salt-bridges between Arg145 and Glu142, and Arg15 and Glu154. This, plus the unusual packing interactions at the bundle core mediated by the symmetric contacts of the Gly and Ala repeats suggests that the dimer interface may not simply be a result of crystallization.
The FMN binding pocket buries the isoalloxazine ring in the β-scaffold and exposes the ribityl-phosphate chain to solvent. The pocket is polar on the pyrimidine side of the isoalloxazine ring where its closest contacts are Gln59, Asn87, Asn97; nonpolar around the dimethylbenzimidazole moiety with proximal residues Val23 and Phe114; and polar along the solvent-exposed ribityl-phosphate where the phosphate interacts with Arg56 and Arg68. The S atom of Cys55 is 4.5 Å from C4a of the FMN in the dark state. At the periphery of the LOV domain Arg56, Arg68, Gln29 and Gln59 interact with the FMN phosphate moiety and inside the pocket Asn87 and Asn97 hydrogen bond to the pyrimidine ring of the flavin.
Light State of R. sphaeroides LOV
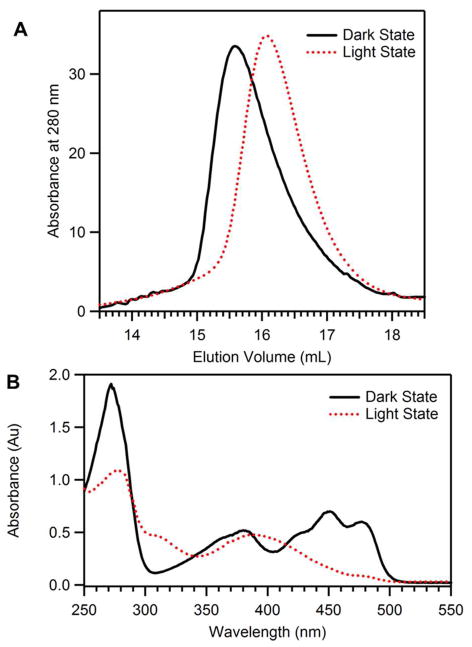
Illumination of RsLOV in solution generates a distinct photobleaching of the protein associated with formation of the cysteinyl-flavin adduct. This is seen by a change from the dark state absorption peak at 447 nm to the maxima light state absorption peak centered at 390 nm (Fig. 5B). As part of the reversible photocycle, the cysteinyl-flavin adduct (SCys55-C4a) spontaneously breaks down in a first-order reaction with a lifetime of 2357 s.
Figure 5.
Solution properties of RsLOV. (A) Elution profile of RsLOV expressed in E. coli with a Supredex 75 10/300 column. Light state (red, dashed) elutes at 16.1 mL, dark state (black, solid) elutes at 15.5 mL. (B) Electronic absorption spectra of light state RsLOV (red, dashed), and dark state RsLOV (black, solid).
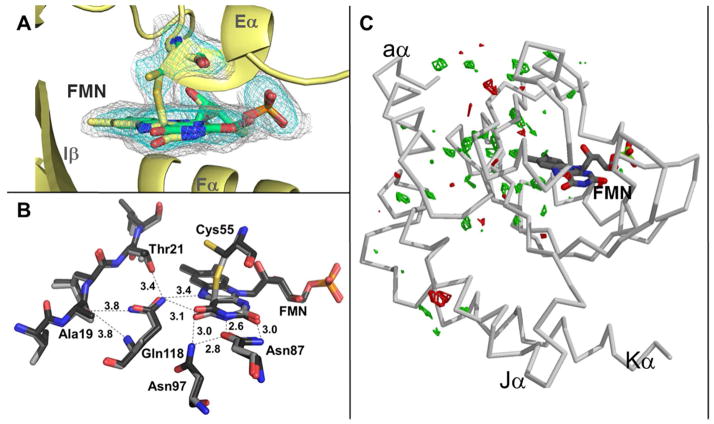
To investigate changes in structure induced by light exposure, crystals were irradiated with white light until visibly bleached and then flash frozen. It is well known that the adduct is not stable to the reducing power of the synchrotron beam (19, 21, 69) and thus the light state data set was composed from data collected on multiple crystals that were each minimally exposed. In the final 2.14 Å resolution structure, the flavin-cysteinyl adduct is present in only one subunit of the RsLOV crystallographic dimer and even in that one subunit, the light state is not fully occupied, with approximately 40% of the molecules present in an alternative (dark state) conformation. In the light-state structure, the Cys-FMN adduct forms with a SCys55–C4a bond distance of ~1.8 Å. The planarity of the reduced FMN is disrupted, with C4a pulled up above the ring plane and the N5 and O forced below the plane by the SCys55-C4a bond. This effectively breaks the conjugated π-system and induces a pucker across the isoalloxazine ring (Fig. 6A). In general, the dark and light state structures are very similar in conformation. This is not surprising because conversion of RsLOV to the light state within the confines of the dark state crystals likely eliminates the possibility of large-scale conformational changes. Nonetheless, light minus dark FL–FD Fourier difference maps identify residues that exhibit some perturbation in position or dynamics from dark to light state (Fig. 6C). Residues that show difference peaks of 3.5 σ or greater are primarily within the first sphere of the flavin, on the interior of the binding pocket; these include Thr21, Leu22, Val23, Met25, Cys55, Phe100, Leu115, and Gln118 (Fig. 6B). It appears that conformational changes propagate from the FMN N5, which will undergo protonation upon adduct formation, through the β-sheet, to proximal second sphere residues that include Leu34, Leu51, Phe13, and Arg10. Arg10 and Phe13 are residues in the N-terminal flanking helix. The furthest extent of the light-dark residue changes are observed at Ala133 and Gln49. Ala133 resides in the center of the Jα helix whereas Gln49 is in the Dα helix, a solvent exposed structural feature on the domain periphery. The potential of signal propagation through to the Jα helix has been shown before in the AsLOV2 protein (70, 71). Our data also indicates that the unique flanking terminal regions of RsLOV respond to adduct formation and that a series of conformational changes, dampened by the crystal lattice, propagate from the N5 edge of the flavin through Iβ, to the terminal helices at the dimer interface.
Figure 6.
Irridiation of RsLOV crystals. (A) Light state structure of RsLOV with electron density corresponding to the light-induced cysteinyl-C4a adduct of Cys55 and FMN (Fo-Fc simulated annealing omit map, obtained by omitting the flavin molecule and Cys55 residue, contoured at 2.2σ in cyan, 0.7σ in gray). The two conformations of Cys55B and flavin are shown in yellow (covalently bound) and green (unbound). (B) FMN binding pocket residues affected by changes in light state conversion. (C) RsLOV structure in the light state with Fo–Fo difference map of light-dark amplitudes (green is +3.5σ, red is −3.5σ).
In VVD, the conserved Gln residue in the Iβ sheet (Gln182 in VVD, Gln118 in RsLOV) flips upon blue-light excitation and thereby alters hydrogen bonding interactions with the backbone of Ala72, which precedes the Aβ strand in the hinge region of the N-terminal helical cap (19, 21). Thus, in VVD the hinge region between the Ncap and PAS β-sheet plays a major role in mediating light-activated conformational changes between the flavin center and the protein interaction regions (19). The reorientation of the Gln residue and its role in disrupting hydrogen bonding interactions that extend to the surface of the protein has also been supported in LOV2, LOV2-Jα and full-length phototropin systems (71–75). Although the Ala72 residue that couples to the N-terminal cap in VVD is conserved in RsLOV (Ala19), differences in the neighboring residues, especially the presence of Thr21 in RsLOV, suggest that the mechanism of conformational propagation may not be conserved. The side chain hydroxyl group of Thr21 is closer to the Gln118 residue (3.4 Å) than the carbonyl O of Ala19 (3.8 Å; Fig. 6B). In AsLOV2, changes in hydrogen bonding triggered by conformational change of flavin-interacting Gln513 strengthen the coupling between the Iβ and Hβ-strands. This overall tightening of the β-sheet leads to detachment of the Jα helix from its outside surface (71). In AsLOV2, Asn482 and Asn492 hydrogen bond with the flavin and Gln513. Changes in the dynamics of these residues upon adduct formation is thought to be critical for propagating a light signal to the surface of the protein (71). Both residues are conserved in RsLOV (Asn87 and Asn97, respectively). Since RsLOV has both aα and Jα/Kα extensions, it is possible that a hybrid form of these aforementioned structural changes are responsible for propagating conformational changes out from the flavin. The hinge region is likely to be affected by N5 protonation and Gln rearrangement, as is the association of Jα with the PAS β-sheet. Although the crystal lattice prevents observation of large-amplitude perturbations, the difference Fourier analysis does anticipate conformational changes in all of these regions.
Solution State Behavior of RsLOV
RsLOV elutes on SEC at a MW consistent with a dimer. To test whether the RsLOV solution dimer is the same as that observed in the crystal, we engineered Cys residues at the dimer interface and evaluated their ability to cross-link. Cys residues placed on the Jα and Kα helices cross-link the subunits effectively, even in the absence of oxidants, confirming that the crystallographic dimer is represented in solution (Figs. 7B and S3, Table 2). Moreover, mutation of E142, which participates in a salt bridge with Arg145 of the opposing subunit, completely abolished dimerization in the dark (Fig. 4B and S3). Thus, RsLOV dimerizes through the interface found in the crystals.
Figure 7.
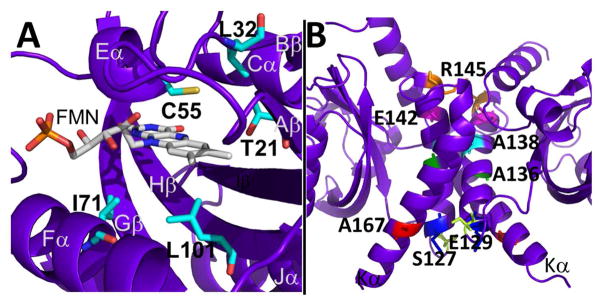
RsLOV point mutants. (A) Dark state structure of RsLOV with residues adjacent to the FMN binding pocket targeted for point mutations highlighted in cyan. (B) Residues targeted for point mutations at the RsLOV dimer interface highlighted: S127 (yellow), E129 (blue), A136 (green), A138 (aqua), E142 (pink), R145 (orange), A167 (red).
Table 2.
Results of crosslinking in RsLOV variants showing distance between Cα atoms in the relevant Cys residue, the B factor of the Cys Cα, the oligomeric state in the dark, and whether crosslinking increases with addition of Cu(phen)3 in the dark. Gel shown in Fig S3.
| Variant | Distance between Cα (Å) | B factor of Cα | Dimer or Monomer in the dark | Crosslinked by Cu(phen)3 in the dark |
|---|---|---|---|---|
| WT (C55) | 48.2 | 47.8 | Monomer | No |
| S127C | 14.3 | 66.4 | Dimer/ Monomer | Yes |
| A136C | 10.1 | 37.7 | Monomer | Yes |
| E142C | 10.1 | 46.6 | Monomer | No |
| R145C | 11.3 | 56.0 | Monomer/ Dimer | No |
| A167C | 17.9 | 55.1 | Dimer/ Monomer | Yes |
Upon conversion to the adduct state, RsLOV elutes at a much later volume on SEC compared to the dark state (Figs. 5A and S2) (39). The transparent column was exposed to white light for the duration of the SEC experiments to minimize dark-state reversion, which occurs with a half-life of 27 min. The elution profiles of the light-state are consistently shifted by ~0.6 mL, which corresponds to an 8.3 kDa reduction in MW. This is not enough of a MW shift for a full dimer to monomer conversion (which would represent a change of ~19 kDa) but the reduction in light-state elution volume was consistent across a large range of protein concentrations. The oligomeric state of RsLOV was further analyzed by multi-angle light scattering (MALS). In the case of SEC-coupled MALS, the protein could not be irradiated during chromatography which typically took ~40 min to complete. Nevertheless, the light state consistently showed a smaller MW than the dark state. In fact, the MALS determined light-state MW (20.4 kDa) was very close to that predicted for a pure monomer (19.9 kDa), whereas the dark-state MW was smaller (30.2 kDa) than that predicted for a true dimer (39.8 kDa) (Fig. S1, Table S2). The behavior of the dark state is consistent with a dimeric species that exchanges rapidly with monomer on the time scale of the size-exclusion (20) and thus the MALS data support a monomer-dimer equilibrium that is shifted toward monomer by light.
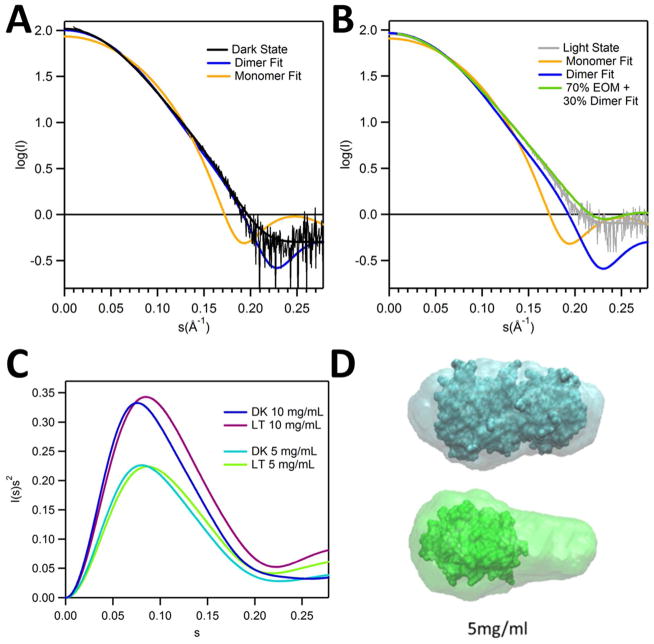
Small-angle X-ray scattering was also used to determine the oligomeric state of RsLOV in both the light and dark states. In total, from the time of light exposure the experiment takes less than 10 min for complete data collection, allowing only minimal dark state recovery. The molecular weights derived from SAXS MoW analysis of GNOM output files (76, 77) of the experimental SAXS data predict a dark state MW of 34.1 kDa and a light state MW of 27.8 kDa, which is higher than that of the smaller species observed in MALS. This is probably explained by the fact that protein concentrations used for SAXS are higher than that found on the MALS SEC column and thus favor some dimerization of the light state. It is also possible that the synchrotron beam reduces the adduct to some extent and drives the system back towards dimer. Theoretical scattering curves for the crystallographic dimer and monomer structures were generated and fit to the dark and light state experimental SAXS data using CRYSOL (63)(Fig. 9A–B). The theoretical curve generated for the dimer structure fits well to the dark state experimental data (χ2 value of 0.9). The theoretical curve generated for an RsLOV monomer does not fit either state well, χ2 value of 3.5 (dark) and 2.6 (light) indicating some contribution from dimer and possibly conformational changes in the monomer that are not modeled well using the single compact subunit of the RsLOV crystal structure. The best fit to the light state experimental data was obtained using the ensemble optimization method (EOM) via RANCH and GAJOE programs (64) with a monomeric model made from an ordered LOV domain core and disordered N-terminal aα helix and the C-terminal Jα and Kα helices (χ2 value of 0.42). However, the predicted molecular weight derived from this experimental SAXS data indicates contribution from an RsLOV dimer even in the “light-state” ensemble. Accordingly, the best fit to the experimental scattering curve came from a linear combination of calculated scattering from the EOM modeled monomer and the CRYSOL-predicted dimeric state (Fig. 9B). In further support of an altered monomer structure, Kratky plots of the SAXS data, which emphasize changes at larger q values (i.e. intramolecular), are indicative of a more unfolded light state (Fig. 9C). The ~1 Å difference in experimental Rg values between the dark and light state at three protein concentrations (Table S3) agrees well with the difference between average Rg values from the EOM models of the monomer and dimer with varying degrees of disorder (21.9 Å, and 22.6 Å, respectively). These slightly larger values than those predicted from CRYSOL with the static crystal structures, especially in the case of the monomer (17.9 Å for the monomer and 21.4 Å for the dimer) support enhanced flexibility in the light-state, and perhaps to a lesser extent in the dark state as well. The predicted molecular envelopes based on the SAXS (Fig. 9D, corresponding data, Table S3), confirm that the dark state envelope completely overlays the crystallographic dimer, whereas the light state fits a monomer with additional, unfilled volume. The unfilled volume in the light state envelope could result from a monomer with terminal helices in an extended conformation and/or contribution from the dimeric state. Although, the fits to the scattering curves themselves are the most reliable indicators of the light-state ensemble, the envelope reconstructions do reflect what we believe to be the key changes upon conversion of RsLOV to the light state: loss of dimer content and increase in subunit disorder.
Figure 9.
SAXS of RsLOV in dark and light states. (A) Fits of scaled theoretical monomer (orange) and dimer (blue) scattering curves generated with CRYSOL for the dark (black) and (B) light (gray) state RsLOV (5 mg/mL data). EOM fits generated with RANCH13/GAJOE13 of the monomer were generated from the truncated pdb file of the dimer with only the A subunit. Light state fit composed of 70% EOM monomer model and 30% dimer (lime). (C) Kratky plot of dark and light state RsLOV data of 10 and 5 mg/mL samples. (D) Scattering envelope models based on the 5 mg/mL SAXS data of dark state RsLOV (top, cyan) and light state RsLOV (bottom: green), superimposed with the crystal structure surface of RsLOV, as a complete dimer or truncated to a monomer.
Variants of RsLOV that Affect Dark State Recovery
Sequence variations in LOV proteins and related PAS domain proteins like PYP have been shown to alter thermal reversion rates and affect overall protein stability (22, 78, 79). In previous studies of LOV domain variants, substitutions that affected hydrophobicity, solvent accessibility, hydrogen bonding, and salt-bridges in the flavin pocket all altered adduct lifetime. In RsLOV we examined active site variants Thr21→Val, Leu32→Val, Ile71→Leu, Ile71→Val, Leu101→Ile, and Cys55→Ala, which were all chosen to alter the adduct state stability. Cys55 forms the adduct state with the C4a atom of FMN. Indeed UV/vis spectroscopy confirmed that the Cys55→Ala variant does not form the light-state adduct. Thr21 (5 Å to Cys55, 3.6 Å to FMN C6) and Leu32 (3.3 Å to Cys55) were expected to affect the conformational stability and solvent accessibility of Cys55. Ile71 (3.7 Å to the O3′ in FMN tail) and Leu101 (3.8 Å to FMN C7) contact the re-face of the flavin ring and influence solvent access in the FMN binding pocket (Fig. 7A).
Despite the relatively conservative residue substitutions, each variant decreased the adduct stability (Table 1). The substitution at Leu32 had the most substantial increase in thermal revision rate, which increased 14-fold in the variant. Substitutions at Thr21, Ile71 and Leu101 also increased the thermal reversion rate, though to a lesser extent, about 1.4 –3-fold. None of the FMN binding site variants decreased the reversion rate.
Table 1.
Parameters for adduct decay kinetics in RsLOV variants and comparable LOV domain proteins
| Variant | Rate constant (s−1) | Lifetime (s) |
|---|---|---|
| RLOV | 4.2 × 10−4 | 2374 ± 304 |
| T21V | 1.3 × 10−3 | 799 ± 78 |
| L32V | 6.1 × 10−3 | 164 ± 7 |
| I71L | 5.6 × 10−4 | 1801 ± 362 |
| I71V | 6.5 × 10−4 | 1544 ± 95 |
| L101I | 6.1 × 10−4 | 1647 ± 71 |
| S127C | 6.5 × 10−4 | 1536 ± 122 |
| E142C | 5.3 × 10−4 | 1904 ± 99 |
| A167C | 5.8 × 10−4 | 1720 ± 162 |
| R145C | 3.5 × 10−4 | 2849 ± 268 |
| A136C | 3.9 × 10−4 | 2587 ± 96 |
| A136Y | 4.2 × 10−4 | 2372 ± 802 |
| A138Y | 5.5 × 10−4 | 1829 ± 62 |
| WT YtvA | 2.8 × 10−4 | 3600 |
| WT AsLOV2 | 1.2 × 10−2 | 81 |
| VVD-36 | 5.6 × 10−5 | 18000 |
Leu32 resides on the bβ strand proximal to the flexible connective loop region within the FMN binding pocket and is within van der Waals contact of Cys55 (3.3 Å). Substitution of Leu to Val would increase the volume of the pocket, make additional space for solvent and disrupt the packing of Cys55. Because the most substantial changes to thermal reversion rate were due to the Leu32→Val mutation, we characterized this substitution further by determining the L32V dark state crystal structure (vida infra).
The changes in recovery rate exhibited by the remaining variants, at Thr21, Ile71 and Leu101, are modest, although their structural contexts provide possible explanations for the slightly increased recovery rates. In VVD, thermal reversion is rate limited by deprotonation of N5 (22); thus, changes that increase solvent accessibility of the pocket are likely to destabilize the adduct. Thr21 contacts FMN C6 hydrogen bonds to the conserved Gln118 and resides within the first sphere of residues adjacent to Cys55. The change from Thr21 to Val will eliminate hydrogen bonding interactions through the side chain and introduce more non-polar character to the binding pocket. Ile71 and Leu101 are situated below the re-face of FMN and changes in these residues would be expected to affect solvent access to flavin N5 and likely the flavin redox potential (19, 22). Ile71 is on the interior of Fα and within 4 Å of the FMN ribose moiety, and Leu101 is on the Hβ strand. For both positions, removal and addition of a methyl group is likely to subtly alter the accessibility, dynamics and redox properties of the adduct, thereby producing the small reductions in light state stability that were observed.
The variants that introduced Cys and Tyr residues within the C-terminal helices were also analyzed for effects on light-cycle kinetics (Fig. 7B). Like most variants proximal to the FMN binding site, the variants in the Jα and Kα helices had minimal effect on the reversion rates. The largest deviations from the WT were R145C which save a 1.2-fold longer recovery, and S127C which save a 1.6-fold shorter recovery.
Structure of L32V RsLOV Variant
To further investigate the 14-fold change in recovery rate brought-about by the Leu32Val substitution, orthorhombic crystals of the RsLOV L32V were grown. Interestingly, crystal growth required different conditions than wild-type RsLOV and the resulting cell dimensions were also different from wild-type crystals. Data were collected from a single crystal to 1.9 Å resolution, scaled in the P212121 space group and the structure was determined by molecular replacement with wt RsLOV as a probe (Table S1). Leu32 is found on Bβ, approximately 3.3 Å away from the Cys55 residue. The most substantial differences in electron density due to the Val mutation are located in three of the four consecutive helices on the si-face of the flavin: Dα, Eα, Fα, and the flexible loops that connect them (Fig. 8A). The turn that connects Aβ and Bβ sheets and the loop that connects Eα and Fα helices are all repositioned with respect to the wild-type RsLOV structure. The alterations in loop position correlate with increased B-factors in these regions (Fig. 8A). The Val32 side chain does not extend as far into the FMN pocket as Leu and thereby provides more conformational freedom for Cys55. In general, Leu32Val appears to destabilize the protein structure surrounding the active site Cys: the loop regions have opened-up, they are more mobile, and the Cys is less conformationally constrained. These factors all likely contribute to the increased reversion rates.
Figure 8.
Structures of L32V and A138Y RsLOV. (A) Structure of L32V RsLOV variant in the dark state colored by B factor (ROYGBIV spectrum coloring in which red is the maximum and violet is the minimum value) overlayed on the P65 dark state RLOV structure (gray, transparent). (B) Structure of the A138Y RsLOV variant in the dark state (aqua) overlaid on the P65 dark state RsLOV structure (violet). Dimer interface with Tyr (orange) and Ala (violet) residues shown as sticks on the Jα-helices.
Structure and Solution Behavior of A138Y RsLOV Variant
We also produced a variant RsLOV designed to alter the dimer interface in the dark state by mutating one of the Ala repeat residues at the center of the Jα interface to Tyr. The structure of the A138Y RsLOV variant was determined from a single crystal that formed in the same crystallization condition as the WT. Despite the same condition, the hexagonal crystal was distinctly conical in shape in contrast to the long rods that formed from the WT. However, the crystallographic data scaled to the P65 space group, with similar unit cell dimensions (a = 105.9 Å, b = 105.9 Å, c = 84.1 Å, α=90, β=90, γ=120) as the WT. The structure is the same crystallographic dimer as the WT, with the Tyr side chains forced alongside the Jα-helical interface (Fig. 8B). Despite the introduction of the bulky Tyr residues at the dimer interface, the A138Y structure produces a nearly identical subunit association as the WT RsLOV structure. There are no significant deviations of the path or direction of the Jα or Kα helices at the interface, and no conformational changes that propagate from the single residue change. Thus, crystallization strongly favors the dark-state dimer and the crystal packing forces are able to overcome a substantial perturbation to the interface. The curve generated from SEC of A138Y is included in Figure S3 and indicates that this substitution, designed to disrupt the dimer interface, in fact, predisposes the variant to form a dimer in solution. Tyrosine is well suited for mediating molecular recognition at protein–protein interfaces (80), because tyrosine uses interactions of both the side-chain hydroxyl and the aromatic ring to make specific contacts with a wide variety of residues on a protein surface. Thus, despite the large increase in side chain volume at the interface, for these other reasons Tyr138 facilitates RsLOV dimerization. Moreover, there is no significant change to the A138Y variant in solution upon exposure to light, indicating that this variant interferes with the propagation of signal from the PAS core to the flanking helices.
DISCUSSION
RsLOV is a LOV domain protein that has a moderate lifetime (2374 s) for the cysteinyl-flavin adduct state. The striking aspects of the RsLOV structure are its short N-terminal helical extension and its long C-terminal HTH dimerization motif. In the crystal structure, two RsLOV subunits pack together through the C-terminal HTH motifs to form a closely associated four-helix bundle, flanked by the N-terminal helices. A preponderance of Gly and Ala residues in the interface generates a tight knitting of the Jα helices. Evidence from chromatographic, light-scattering and chemical cross-linking indicates that light causes the RsLOV dimer to dissociate. This behavior is in marked contrast to that of VVD, where light causes dimerization. The structural changes determined from the light state X-ray crystal structure suggest that despite its unusual oligomerization, RsLOV behaves similarly to other well-characterized LOV domain proteins, such as the AsLOV2 (LOV2 domain protein of oat), with which it shares a long flanking Jα helix. Although irradiation of the dark-state crystals produces few large scale structural perturbations, difference Fourier analysis indicates that conformational changes propagate from the flavin N5 position, through the conserved Gln amide, across Iβ to the Ncap and Jα, which against the PAS β-sheet. These structural changes must ultimately disrupt contacts in the helical dimer interface and lead to subunit dissociation.
The study of several RsLOV variants revealed that minor perturbations in side chain size or polarity have minor effects on the adduct stability. Mutations localized to the FMN binding pocket, destabilized the Cys-FMN adduct and increased the rate of thermal reversion. The most impactful mutation was L32V, which increased the recovery rate 14-fold. The crystallographic structure of L32V RsLOV showed that a conservative point mutation in a small, compact domain can induce extensive structural changes. The substitution of Leu32 located in the Bβ strand, altered the positioning of the Dα and Eα helices and the flexible connecting loops on the si-face of the FMN. These changes likely cause increased domain dynamics that result in destabilization of the adduct bond and greater solvent accessibility of flavin N5 to aid in the deprotonation.
Blue-light-Induced Structural Changes
The changes exhibited in light state RsLOV crystal structure, obtained from irradiation of dark state RsLOV crystals, capture the initial structural perturbations induced by adduct formation, which was visualized in one of the two independent subunits. The majority of residues impacted by the light in the crystal structure are within the first sphere of residues of the FMN binding pocket. These residues, highlighted by the changes in the Fo-Fc omit maps between the light and dark states include Cys55 and Gln118, which have been shown in other LOV domains to be critical for altering hydrogen bonding in the flavin binding pocket and thereby causing the predicted large conformational changes. Second sphere residues in the solvent exposed regions also highlighted by the difference Fourier analysis include Ala133 in the Jα helix, Arg10 and F13 in the aα helix, and Gly48 and Leu51 in the Dα helix. Differences in the density surrounding the Jα helix of AsLOV2 indicate that movement of the Jα helix is an initial response in RsLOV, as it is in other LOV domains (72, 81). Indeed the integrity of the tightly packed helical interface of RsLOV may be especially sensitive to small changes in the position of Jα. The subunit contact relies on having virtually no side-chain contribution at the center of the interface and such an association would be incompatible with a slight shift or rotation of Jα.
Blue-light Signal Propagation in R. sphaeroides
Although large structural rearrangements in the RsLOV light-state will be prevented by crystal packing constraints and the short-lived adduct state precludes a long-lived, crystallizable intermediate, the differences observed between the light and dark state RsLOV structures in illuminated crystals reveal structural perturbations that anticipate a larger rearrangement involving the N- and C-terminal flanking regions. It is interesting that the SAXS scattering profiles of the light-excited samples are not particularly well modeled from linear combinations of the monomer and dimer scattering profiles, as calculated from the crystal structure, and are better modeled by designating regions of disorder. One explanation is for this is that the dissociated RsLOV monomer changes conformation considerably from how it appears in the dimer crystal structure.
In a recent SAXS study of light-induced movement of the LOV2 domain in Arabidopsis phot2, the linker region that is responsible for executing the light-induced conformational changes between the LOV2 and kinase domains is Jα-helix-like (82). The SAXS profiles suggest photoreversible movement of the LOV domain with Jα-linker relative to the effector domain, which produces a more extended structure in the light state (82). Molecular dynamics studies of AsLOV2 revealed a partial unfolding of the Jα helix, that precedes its detachment from the LOV2 domain interface, a rearrangement originally proposed from the initial structural characterization of AsLOV2 (71, 72, 81). The RsLOV residues attributed to the partial unfolding of the Jα helix (Ile533-Ile539, centrally located residues adjacent to the Iβ strand Gln513) are similarly affected in the observed structural changes from dark to light state in RsLOV crystals (Ala133 adjacent to Gln118). Thus, a large conformational change is also possible in the RsLOV protein that would extend the Jα and Kα helices away from the LOV domain. The SAXS data suggest that this may indeed be the case (Fig. 9C).
The RsLOV Flanking Regions
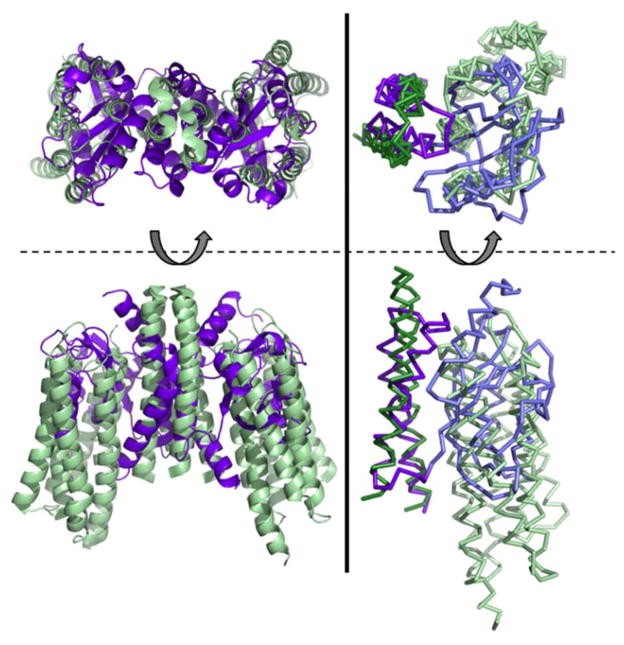
The N-terminal and C-terminal extensions of RsLOV flank the LOV domain core where they are held in place by hydrophobic interactions and side-chain mediated hydrogen bonds. The long C-terminal extension of RsLOV is a unique HTH motif (Jα and Kα) that has not been observed previously in LOV domain proteins and forms an unusual dimerization domain. A Dali search (83) of the dimerization domain finds only weak structural homology to other coiled-coils. The topology and juxtaposition of Jα and Kα are most similar to those of the membrane helices found in mechanosensitive channels, membrane transporters and the transducer of sensory rhodopsin, but the relationships are only approximate (Fig. 10). To our knowledge, the tight interaction between Jαs and helical crossing angles at the interface has not been previously observed. The overall domain architecture of RsLOV is perhaps similar to that of EL222, which has a DNA-binding HTH effector domain coordinated to the LOV domain via a Jα helix, but the HTH motif of EL222 consists of shorter helices that arrange differently, do not flank the LOV core, nor dimerize (47). However the EL222 HTH does respond to light, which alters the ability of the protein to bind DNA.
Figure 10.
Analogy between RsLOV and sensory Rhodopsin. Left: Structure of RsLOV dimer (violet) overlaid on the dimer of sensory rhodopsin II in complex with helical transducer (pale green, PDB code 1H2S). Right: Overlay of the monomers of RsLOV and sensory rhodopsin II depicted in ribbon with the C-terminal helices aligned and colored darker than the PAS core and transducer helix bundle.
Perhaps the most relevant analogs of RsLOV are the bacterial sensory rhodopsin (sR) light sensors, which are membrane proteins that convert light signals to conformational changes in a helical transducer protein. The coupling module of the transducer is a membrane embedded 4-helix bundle, composed of two antiparallel helices from each subunit (84, 85). On the periphery of each helical hairpin are two transmembrane sR proteins. Isomerization of the retinal cofactor in sR is thought to perturb the orientation of the transducer helices thereby sending conformational signals through their cytoplasmic HAMP domains (86). The sR units are similar in size to that of the RsLOV PAS domains and the transducer helices are oriented similarly as Jα and Kα within each subunit. One of the key interface Gly residues and its flanking sequence of RsLOV are also found in the transducer. Thus, RsLOV may be seen as a soluble, functional analog of bacterial sensory rhodopsin. The sR transducer remains a dimer throughout signal transduction, but the subunit association is stabilized by extensive interactions in the cytoplasmic regions. Without these additional regions, shifts in the RsLOV helices induced by the light-sensing PAS domains destabilize the dimeric interface enough to cause subunit dissociation.
The RsLOV N-terminal extension may also play an important role in the light-induced subunit association and subsequent recognition of another protein, yet to be defined. The RsLOV N-terminal extension is a short aα helix that lacks the long flexible region that enables light-induced dimerization of VVD. The aα helix also flanks the LOV core, and is held in place by hydrogen bonding with the Jα helix and hydrophobic interactions with the core β-strands. Arg10 and Phe13 of the N-terminal aα helix are perturbed when the adduct is formed in a manner similar to that of the second sphere residue in the Jα helix. VVD undergoes a large structural rearrangement of the N-terminal extension upon conversion to the light state, but the RsLOV N-terminal extension lacks the flexible hinge regions preceding the LOV domain that transduce conformational change in VVD. However, the role of the shorter aα helix in AsLOV2 has also recently been examined and was identified as a possible control element for use in optogenetic studies (87). The structural changes observed in the RsLOV aα helix may indicate that the helix is forced away from the LOV domain in a manner similar to that seen with the AsLOV2 Jα helix: first by distorting the helix at its center, which in turn disrupts the hydrophobic interactions that stabilize the aα helix against the surface of the PAS β-sheet. Should the Jα helix unwind and release from the core like that of AsLOV2, the aα helix would likely follow, and its new conformation could act as a recognition site for a signaling partner.
Targets and interacting partners of RsLOV are currently unknown. The structural analogy between the RsLOV dimerization domain and the membrane helices of channels, photosensory transducers and transporters raise the possibility that the target for these helices, once the RsLOV dimer is dissociated by light, is a similar set of helices, perhaps in or associated with the membrane. Previous studies of RsLOV from R. sphaeroides 2.4.1 detected the protein in both the cytoplasm and the membrane fractions (39), making a signal to a transporter or a channel protein not unreasonable. RsLOV has been linked to a regulatory role in photosynthetic gene expression, carbohydrate metabolism, chemotaxis, and the response to photooxidative stress (51). RsLOV also impacts blue light-dependent gene expression and redox-dependent regulation.
LOV domain proteins are rapidly being designed and applied as tools in the emerging field of optogenetics (29, 88–90). RsLOV offers new utility as a photoswitch with a unique combination of features for controlling cellular processes. These properties include the ability to switch from dimer to monomer upon exposure to light, as well as both N-terminal and C-terminal helical extensions whose conformations change on release and may thereby serve as sites for new protein interactions. Moreover, the RsLOV change in oligomeric state can be manipulated by mutation. Applications could include oligomers of designed RsLOV domains that cage or localize interactions to be released by photoactivation, and controlled conformational switching of attached segments with either N- or C-terminal coupling.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by NIH grant GM079679 to B.R.C. and NIH grant FGM099391A to K.S.C.
We thank the Cornell High Energy Synchrotron Source (CHESS) for access to data collection facilities. We also thank J. Widom for help with mutagenesis and protein expression, and L. J. Byrnes for assistance with MALS.
Abbreviations
- LOV
light-oxygen-voltage
- PAS
Per-ARNT-Sim
- FMN
flavin mononucleotide
- FAD
flavin adenine dinucleotide
- SAXS
small-angle x-ray scattering
- Phot
phototropin
- EL
Erythrobacter litoralis
- As
Avena sativa
- Pp
Pseudomonas putida
- VVD
VIVID
- HTH
helix-turn-helix
- sR
sensory rhodopsin
- EOM
ensemble optimization method
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript
Supporting Information. Contains structural and refinement statistics for crystallographic data, data obtained by SEC-MALS and analytical SEC, SAXS data and shape reconstruction statistics, and data from crosslinking experiments. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Taylor B, Zhulin I. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayers R, Moffat K. Changes in Quaternary Structure in the Signaling Mechanisms of PAS Domains. Biochemistry. 2008;47:12078–12086. doi: 10.1021/bi801254c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Möglich A, Ayers RA, Moffat K. Structure and Signaling Mechanism of Per-ARNT-Sim Domains. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herrou J, Crosson S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat Rev Microbiol. 2011;9:713–723. doi: 10.1038/nrmicro2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci U S A. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 7.Kottke T, Dick B, Fedorov R, Schlichting I, Deutzmann R, Hegemann P. Irreversible photoreduction of flavin in a mutated Phot-LOV1 domain. Biochemistry. 2003;42:9854–9862. doi: 10.1021/bi034863r. [DOI] [PubMed] [Google Scholar]
- 8.Kottke T, Heberle J, Hehn D, Dick B, Hegemann P. Phot-LOV1: Photocycle of a Blue-Light Receptor Domain from the Green Alga Chlamydomonas reinhardtii. Biophysical Journal. 2003;84:1192–1201. doi: 10.1016/S0006-3495(03)74933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasahara M, Swartz TE, Olney MA, Onodera A, Mochizuki N, Fukuzawa H, Asamizu E, Tabata S, Kanegae H, Takano M, Christie JM, Nagatani A, Briggs WR. Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol. 2002;129:762–773. doi: 10.1104/pp.002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs WR, Huala E. Blue-light photoreceptors in higher plants. Annu Rev Cell Dev Biol. 1999;15:33–62. doi: 10.1146/annurev.cellbio.15.1.33. [DOI] [PubMed] [Google Scholar]
- 11.Jain M, Sharma P, Tyagi SB, Tyagi AK, Khurana JP. Light regulation and differential tissue-specific expression of phototropin homologues from rice (Oryza sativa ssp indica) Plant Sci. 2007;172:164–171. [Google Scholar]
- 12.Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S. A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci U S A. 2007;104:18241–18246. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purcell EB, McDonald CA, Palfey BA, Crosson S. An Analysis of the Solution Structure and Signaling Mechanism of LovK, a Sensor Histidine Kinase Integrating Light and Redox Signals. Biochemistry. 2010;49:6761–6770. doi: 10.1021/bi1006404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aravind L, Koonin EV. The STAS domain - a link between anion transporters and antisigma-factor antagonists. Curr Biol. 2000;10:R53–55. doi: 10.1016/s0960-9822(00)00335-3. [DOI] [PubMed] [Google Scholar]
- 15.Buttani V, Losi A, Eggert T, Krauss U, Jaeger K, Cao Z, Gärtner W. Conformational analysis of the blue-light sensing protein YtvA reveals a competitive interface for LOV–LOV dimerization and interdomain interactions. Photochemical & Photobiological Sciences. 2007;6:41–49. doi: 10.1039/b610375h. [DOI] [PubMed] [Google Scholar]
- 16.Losi A, Quest B, Gartner W. Listening to the blue: the time-resolved thermodynamics of the bacterial blue-light receptor YtvA and its isolated LOV domain. Photochem Photobiol Sci. 2003;2:759–766. doi: 10.1039/b301782f. [DOI] [PubMed] [Google Scholar]
- 17.Möglich A, Moffat K. Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. Journal of Molecular Biology. 2007;373:112–126. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwerdtfeger C, Linden H. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. The EMBO Journal. 2003;22:4846–4855. doi: 10.1093/emboj/cdg451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaidya AT, Chen CH, Dunlap JC, Loros JJ, Crane BR. Structure of a Light-Activated LOV Protein Dimer That Regulates Transcription. Sci Signal. 2011;4 doi: 10.1126/scisignal.2001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zoltowski BD, Crane BR. Light activation of the LOV protein Vivid generates a rapidly exchanging dimer. Biochemistry. 2008;47:7012–7019. doi: 10.1021/bi8007017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR. Conformational switching in the fungal light sensor vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zoltowski BD, Vaccaro B, Crane BR. Mechanism-based tuning of a LOV domain photoreceptor. Nature Chemical Biology. 2009;5:827–834. doi: 10.1038/nchembio.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crosson S, Rajagopal S, Moffat K. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 24.Losi A. Flavin-based blue-light photosensors: A photobiophysics update. Photochem Photobiol. 2007;83:1283–1300. doi: 10.1111/j.1751-1097.2007.00196.x. [DOI] [PubMed] [Google Scholar]
- 25.Circolone F, Granzin J, Jentzsch K, Drepper T, Jaeger KE, Willbold D, Krauss U, Batra-Safferling R. Structural Basis for the Slow Dark Recovery of a Full-Length LOV Protein from Pseudomonas putida. Journal of Molecular Biology. 2012;417:362–374. doi: 10.1016/j.jmb.2012.01.056. [DOI] [PubMed] [Google Scholar]
- 26.Chen CH, DeMay BS, Gladfelter AS, Dunlap JC, Loros JJ. Physical interaction between VIVID and white collar complex regulates photoadaptation in Neurospora. Proc Natl Acad Sci USA. 2010;107:16715–16720. doi: 10.1073/pnas.1011190107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunt SM, Thompson S, Elvin M, Heintzen C. VIVID interacts with the WHITE COLLAR complex and FREQUENCY-interacting RNA helicase to alter light and clock responses in Neurospora. P Natl Acad Sci USA. 2010;107:16709–16714. doi: 10.1073/pnas.1009474107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malzahn E, Ciprianidis S, Káldi K, Schafmeier T, Brunner M. Photoadaptation in Neurospora by Competitive Interaction of Activating and Inhibitory LOV Domains. Cell. 2010;142:762–772. doi: 10.1016/j.cell.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Strickland D, Yao XL, Gawlak G, Rosen MK, Gardner KH, Sosnick TR. Rationally improving LOV domain-based photoswitches. Nat Methods. 2010;7:623–U618. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erbel PJA, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix-loop-helix-PAS transcription factor hypoxia-inducible factor. Proc Natl Acad Sci USA. 2003;100:15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Tomchick DR, Brautigam CA, Machius M, Kort R, Hellingwerf KJ, Gardner KH. Changes at the KinA PAS-A dimerization interface influence histidine kinase function. Biochemistry. 2008;47:4051–4064. doi: 10.1021/bi7021156. [DOI] [PubMed] [Google Scholar]
- 32.Salomon M, Lempert U, Rudiger W. Dimerization of the plant photoreceptor phototropin is probably mediated by the LOV1 domain. FEBS Letters. 2004;572:8–10. doi: 10.1016/j.febslet.2004.06.081. [DOI] [PubMed] [Google Scholar]
- 33.Miyatake H, Mukai M, Park SY, Adachi S, Tamura K, Nakamura H, Nakamura K, Tsuchiya T, Iizuka T, Shiro Y. Sensory mechanism of oxygen sensor FixL from Rhizobium meliloti: Crystallographic, mutagenesis and resonance Raman spectroscopic studies. Journal of Molecular Biology. 2000;301:415–431. doi: 10.1006/jmbi.2000.3954. [DOI] [PubMed] [Google Scholar]
- 34.Key J, Hefti M, Purcell EB, Moffat K. Structure of the redox sensor domain of Azotobacter vinelandii NifL at atomic resolution: Signaling, dimerization, and mechanism. Biochemistry. 2007;46:3614–3623. doi: 10.1021/bi0620407. [DOI] [PubMed] [Google Scholar]
- 35.Kurokawa H, Lee DS, Watanabe M, Sagami I, Mikami B, Raman CS, Shimizu T. A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor. J Biol Chem. 2004;279:20186–20193. doi: 10.1074/jbc.M314199200. [DOI] [PubMed] [Google Scholar]
- 36.Jurk M, Dorn M, Schmieder P. Blue Flickers of Hope: Secondary Structure, Dynamics, and Putative Dimerization Interface of the Blue-Light Receptor YtvA from Bacillus subtilis. Biochemistry. 2011;50:8163–8171. doi: 10.1021/bi200782j. [DOI] [PubMed] [Google Scholar]
- 37.Möglich A, Ayers RA, Moffat K. Addition at the Molecular Level: Signal Integration in Designed Per–ARNT–Sim Receptor Proteins. Journal of Molecular Biology. 2010;400:477–486. doi: 10.1016/j.jmb.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 38.King HA, Hoelz A, Crane BR, Young MW. Structure of an Enclosed Dimer Formed by the Drosophila Period Protein. Journal of Molecular Biology. 2011;413:561–572. doi: 10.1016/j.jmb.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hendrischk AK, Moldt J, Fruhwirth SW, Klug G. Characterization of an Unusual LOV Domain Protein in the alpha-Proteobacterium Rhodobacter sphaeroides. Photochem Photobiol. 2009;85:1254–1259. doi: 10.1111/j.1751-1097.2009.00554.x. [DOI] [PubMed] [Google Scholar]
- 40.Hendrischk AK, Fruhwirth SW, Moldt J, Pokorny R, Metz S, Kaiser G, Jager A, Batschauer A, Klug G. A cryptochrome-like protein is involved in the regulation of photosynthesis genes in Rhodobacter sphaeroides. Molecular Microbiology. 2009;74:990–1003. doi: 10.1111/j.1365-2958.2009.06912.x. [DOI] [PubMed] [Google Scholar]
- 41.Pairwise scores are percentages which represent the number of identities between the two sequences divided by the length of the alignment.
- 42.Alexandre MT, Arents JC, van Grondelle R, Hellingwerf KJ, Kennis JT. A base-catalyzed mechanism for dark state recovery in the Avena sativa phototropin-1 LOV2 domain. Biochemistry. 2007;46:3129–3137. doi: 10.1021/bi062074e. [DOI] [PubMed] [Google Scholar]
- 43.Larkin MABG, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. ClustalW and ClustalX version 2 (2007) Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 44.Goujon MMH, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI (2010) Nucleic Acids Res. 2010;38(Suppl):W695–699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jentzsch K, Wirtz A, Circolone F, Drepper T, Losi A, Gärtner W, Jaeger K-E, Krauss U. Mutual Exchange of Kinetic Properties by Extended Mutagenesis in Two Short LOV Domain Proteins from Pseudomonas putida. Biochemistry. 2009;48:10321–10333. doi: 10.1021/bi901115z. [DOI] [PubMed] [Google Scholar]
- 46.Krauss U, Losi A, Gartner W, Jaeger KE, Eggert T. Initial characterization of a blue-light sensing, phototropin-related protein from Pseudomonas putida: a paradigm for an extended LOV construct. Phys Chem Chem Phys. 2005;7:2804–2811. doi: 10.1039/b504554a. [DOI] [PubMed] [Google Scholar]
- 47.Nash AI, McNulty R, Shillito ME, Swartz TE, Bogomolni RA, Luecke H, Gardner KH. Structural basis of photosensitivity in a bacterial light-oxygen-voltage/helix-turn-helix (LOV-HTH) DNA-binding protein. Proc Natl Acad Sci USA. 2011;108:9449–9454. doi: 10.1073/pnas.1100262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avila-Perez M, Vreede J, Tang YF, Bende O, Losi A, Gartner W, Hellingwerf K. In Vivo Mutational Analysis of YtvA from Bacillus subtilis Mechanism of Light Activation of the General Stress Rresponse. J Biol Chem. 2009;284:24958–24964. doi: 10.1074/jbc.M109.033316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Möglich A, Ayers RA, Moffat K. Design and Signaling Mechanism of Light-Regulated Histidine Kinases. Journal of Molecular Biology. 2009;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta crystallographica. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 51.Metz S, Jager A, Klug G. Role of a short light, oxygen, voltage (LOV) domain protein in blue light- and singlet oxygen-dependent gene regulation in Rhodobacter sphaeroides. Microbiology-(UK) 2012;158:368–379. doi: 10.1099/mic.0.054700-0. [DOI] [PubMed] [Google Scholar]
- 52.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter Charles W., Jr, editor. Method Enzymol. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 53.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McRee DE. XtalView: a visual protein crystallographic software system for X11/Xview. J Mol Graph. 1992;10:44–47. [Google Scholar]
- 55.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix. refine. Acta crystallographica. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ando N, Chenevier P, Novak M, Tate MW, Gruner SM. High hydrostatic pressure small-angle X-ray scattering cell for protein solution studies featuring diamond windows and disposable sample cells. J Appl Crystallogr. 2008;41:167–175. [Google Scholar]
- 57.Nielsen SS, Toft KN, Snakenborg D, Jeppesen MG, Jacobsen JK, Vestergaard B, Kutter JP, Arleth L. BioXTAS RAW, a software program for high-throughput automated small-angle X-ray scattering data reduction and preliminary analysis. J Appl Crystallogr. 2009;42:959–964. [Google Scholar]
- 58.Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. ATSAS 2.1 - towards automated and web-supported small-angle scattering data analysis. J Appl Crystallogr. 2007;40:S223–S228. [Google Scholar]
- 59.Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HDT, Konarev PV, Svergun DI. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 61.Wriggers W, Chacon P. Using Situs for the registration of protein structures with low-resolution bead models from X-ray solution scattering. J Appl Crystallogr. 2001;34:773–776. [Google Scholar]
- 62.Fischer H, Neto MD, Napolitano HB, Polikarpov I, Craievich AF. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J Appl Crystallogr. 2010;43:101–109. [Google Scholar]
- 63.Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- 64.Bernadó P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural Characterization of Flexible Proteins Using Small-Angle X-ray Scattering. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 65.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krissinel E. Crystal contacts as nature’s docking solution. J Comput Chem. 2009;31:133–143. doi: 10.1002/jcc.21303. [DOI] [PubMed] [Google Scholar]
- 68.Krissinel E, Henrick K. Inference of Macromolecular Assemblies from Crystalline State. Journal of Molecular Biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 69.Fedorov R, Schlichting I, Hartmann E, Domratcheva T, Fuhrmann M, Hegemann P. Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii. Biophys J. 2003;84:2474–2482. doi: 10.1016/S0006-3495(03)75052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Halavaty AS, Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 71.Peter E, Dick B, Baeurle SA. Mechanism of signal transduction of the LOV2-J alpha photosensor from Avena sativa. Nat Commun. 2010;1 doi: 10.1038/ncomms1121. [DOI] [PubMed] [Google Scholar]
- 72.Halavaty A, Moffat K. N-and C-Terminal Flanking Regions Modulate Light-Induced Signal Transduction in the LOV2 Domain of the Blue Light Sensor Phototropin 1 from Avena sativa. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 73.Nash AI, Ko WH, Harper SM, Gardner KH. A conserved glutamine plays a central role in LOV domain signal transmission and its duration. Biochemistry. 2008;47:13842–13849. doi: 10.1021/bi801430e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones MA, Feeney KA, Kelly SM, Christie JM. Mutational analysis of phototropin 1 provides insights into the mechanism underlying LOV2 signal transmission. J Biol Chem. 2007;282:6405–6414. doi: 10.1074/jbc.M605969200. [DOI] [PubMed] [Google Scholar]
- 75.Freddolino PL, Dittrich M, Schulten K. Dynamic switching mechanisms in LOV1 and LOV2 domains of plant phototropins. Biophysical Journal. 2006;91:3630–3639. doi: 10.1529/biophysj.106.088609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Semenyuk AV, Svergun DI. GNOM - A PROGRAM PACKAGE FOR SMALL-ANGLE SCATTERING DATA-PROCESSING. J Appl Crystallogr. 1991;24:537–540. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Svergun DI. Determination of the Regularization Parameter In Indirect-Transform Methods Using Perceptual Criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- 78.Philip AF, Kumauchi M, Hoff WD. Robustness and evolvability in the functional anatomy of a PER-ARNT-SIM (PAS) domain. Proc Natl Acad Sci USA. 2010;107:17986–17991. doi: 10.1073/pnas.1004823107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Raffelberg S, Mansurova M, Gartner W, Losi A. Modulation of the Photocycle of a LOV Domain Photoreceptor by the Hydrogen-Bonding Network. J Am Chem Soc. 2011;133:5346–5356. doi: 10.1021/ja1097379. [DOI] [PubMed] [Google Scholar]
- 80.Fellouse FA, Wiesmann C, Sidhu SS. Synthetic antibodies from a four-amino-acid code: A dominant role for tyrosine in antigen recognition. P Natl Acad Sci USA. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 82.Takayama Y, Nakasako M, Okajima K, Iwata A, Kashojiya S, Matsui Y, Tokutomi S. Light-Induced Movement of the LOV2 Domain in an Asp720Asn Mutant LOV2-Kinase Fragment of Arabidopsis Phototropin 2. Biochemistry. 2011;50:1174–1183. doi: 10.1021/bi101689b. [DOI] [PubMed] [Google Scholar]
- 83.Hasegawa H, Holm L. Advances and pitfalls of protein structural alignment. Curr Opin Struct Biol. 2009;19:341–348. doi: 10.1016/j.sbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 84.Moukhametzianov R, Klare JP, Efremov R, Baeken C, Goppner A, Labahn J, Engelhard M, Buldt G, Gordeliy VI. Development of the signal in sensory rhodopsin and its transfer to the cognate transducer. Nature. 2006;440:115–119. doi: 10.1038/nature04520. [DOI] [PubMed] [Google Scholar]
- 85.Gordeliy VI, Labahn J, Moukhametzianov R, Efremov R, Granzin J, Schlesinger R, Buldt G, Savopol T, Scheidig AJ, Klare JP, Engelhard M. Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature. 2002;419:484–487. doi: 10.1038/nature01109. [DOI] [PubMed] [Google Scholar]
- 86.Sasaki J, Tsai A-I, Spudich JL. Opposite Displacement of Helix F in Attractant and Repellent Signaling by Sensory Rhodopsin-Htr Complexes. J Biol Chem. 2011;286:18868–18877. doi: 10.1074/jbc.M110.200345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zayner JP, Antoniou C, Sosnick TR. The Amino-Terminal Helix Modulates Light-Activated Conformational Changes in AsLOV2. Journal of Molecular Biology. 2012;419:61–74. doi: 10.1016/j.jmb.2012.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lungu OI, Hallett RA, Choi EJ, Aiken MJ, Hahn KM, Kuhlman B. Designing Photoswitchable Peptides Using the AsLOV2 Domain. Chem Biol. 2012;19:507–517. doi: 10.1016/j.chembiol.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hahn KM, Kuhlman B. Hold me tightly LOV. Nat Methods. 2010;7:595–597. doi: 10.1038/nmeth0810-595. [DOI] [PubMed] [Google Scholar]
- 90.Christie JM, Hitomi K, Arvai AS, Hartfield KA, Mettlen M, Pratt AJ, Tainer JA, Getzoff ED. Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability. J Biol Chem. 2012;287:22295–22304. doi: 10.1074/jbc.M111.318881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.