SUMMARY
Cancer cells hijack BCL-2 family survival proteins to suppress the death effectors and thereby enforce an immortal state. This is accomplished biochemically by an anti-apoptotic surface groove that neutralizes the pro-apoptotic BH3 α-helix of death proteins. Anti-apoptotic MCL-1 in particular has emerged as a ubiquitous resistance factor in cancer. Whereas targeting the BCL-2 anti-apoptotic subclass effectively restores the death pathway in BCL-2-dependent cancer, the development of molecules tailored to the binding specificity of MCL-1 has lagged. We previously discovered that a hydrocarbon-stapled MCL-1 BH3 helix is an exquisitely selective MCL-1 antagonist. By deploying this unique reagent in a competitive screen, we identified an MCL-1 inhibitor molecule that selectively targets the BH3-binding groove of MCL-1, neutralizes its biochemical lockhold on apoptosis, and induces caspase activation and leukemia cell death in the specific context of MCL-1 dependence.
INTRODUCTION
BCL-2 is the founding member of a family of anti- and pro-apoptotic proteins that form an interaction network to regulate the critical homeostatic balance between cellular life and death(Llambi and Green, 2011; Youle and Strasser, 2008). The original discovery of BCL-2 at the t(14;18) chromosomal breakpoint of follicular lymphoma expanded the paradigm for cancer pathogenesis to include the inability to undergo programmed cell death(Tsujimoto et al., 1985; Tsujimoto et al., 1984; Vaux et al., 1988). The pathologic overexpression of anti-apoptotic BCL-2 and its functional homologues such as BCL-XL and MCL-1 has emerged as a causative mechanism for the development, maintenance, and chemoresistance of many human cancers(Frenzel et al., 2009; Kang and Reynolds, 2009). As such, these proteins are high priority targets for therapeutic development.
The anti-apoptotic proteins contain a surface groove that can bind and sequester - with differential potency and specificity - the BH3 death domains of pro-apoptotic members(Chen et al., 2005; Sattler et al., 1997). BH3-only pro-apoptotics such as BID and BIM contain a single BH3 domain and function as afferent sensors of cellular stress, delivering their death message to the “multidomain” anti- and pro-apoptotic members, which regulate the cellular life-death decision at the level of the mitochondrion(Shamas-Din et al., 2011). When activated directly by BH3-only interaction and/or indirectly by BH3-only-mediated competitive displacement from anti-apoptotics, the multidomain pro-apoptotic proteins BAX and BAK undergo a monomer-to-oligomer transformation that results in outer mitochondrial membrane poration and release of apoptogenic factors(Walensky and Gavathiotis, 2011). Whereas cancer cells deploy the anti-apoptotic proteins to silence this pro-apoptotic pathway, pharmacologic antagonists of anti-apoptotic proteins hold promise to restore the death pathway in cancer. Thus, a series of small molecule screens and structure-based methodologies were initially applied to target BCL-2, yielding an eclectic array of small molecules and peptides with various degrees of biochemical, cellular, and in vivo activity(Degterev et al., 2001; Enyedy et al., 2001; Kitada et al., 2003; Nguyen et al., 2007; Oltersdorf et al., 2005; Petros et al., 2010; Tzung et al., 2001; Walensky et al., 2004; Wang et al., 2006; Wang et al., 2000). The breakthrough molecule ABT-263 is an orally bioavailable and selective BCL-2/BCL-XL inhibitor, which is advancing through the clinical trials process, manifesting both safety and preliminary efficacy in BCL-2-dependent cancers(Gandhi et al., 2011; Roberts et al., 2011; Tse et al., 2008; Wilson et al., 2010).
Broad experimentation with the ABT-263 molecule and its progenitor ABT-737 revealed that expression of anti-apoptotic proteins lying outside their binding spectra caused resistance(Konopleva et al., 2006; Lin et al., 2007; van Delft et al., 2006; Yecies et al., 2010), compelling the development of alternative or complementary agents that would either harbor broader anti-apoptotic targeting capacity or inherent selectivity for anti-apoptotics like MCL-1 that evade ABT-263/737 antagonism. The small molecule obatoclax(Nguyen et al., 2007) and the peptidic Stabilized Alpha-Helix of BCL-2 domains (SAHBs) modeled after the BID and BIM BH3 domains(Gavathiotis et al., 2008; Labelle et al., 2012; Walensky et al., 2004; Walensky et al., 2006) are examples of novel agents that more broadly target the BCL-2 family anti-apoptotic proteins. Given the emergence of MCL-1 as a “top ten” pathologic factor across the diversity of human cancers(Beroukhim et al., 2010), elucidating the blueprint for selective MCL-1 inhibition has also become a major focus of academic and pharmaceutical researchers.
We recently generated a library of SAHBs modeled after the diversity of natural BH3 domains and discovered that the BH3 helix of MCL-1 was itself the most potent and selective natural BH3 inhibitor of MCL-1(Stewart et al., 2010). Whereas the unmodified MCL-1 BH3 peptide was predominantly unstructured and showed little MCL-1 binding activity, we sought to determine if the structurally-fortified and MCL-1-selective stapled peptide could be deployed in a competitive binding screen to in turn identify a selective small molecule antagonist for reactivating apoptosis in MCL-1-dependent cancer. Thus, in addition to serving as a novel class of therapeutics in their own right, the development and application of high affinity/high selectivity stapled peptides for competitive screening could likewise be an effective and generalizable strategy for small molecule drug discovery.
RESULTS
From Selective Stapled Peptide to Selective Small Molecule
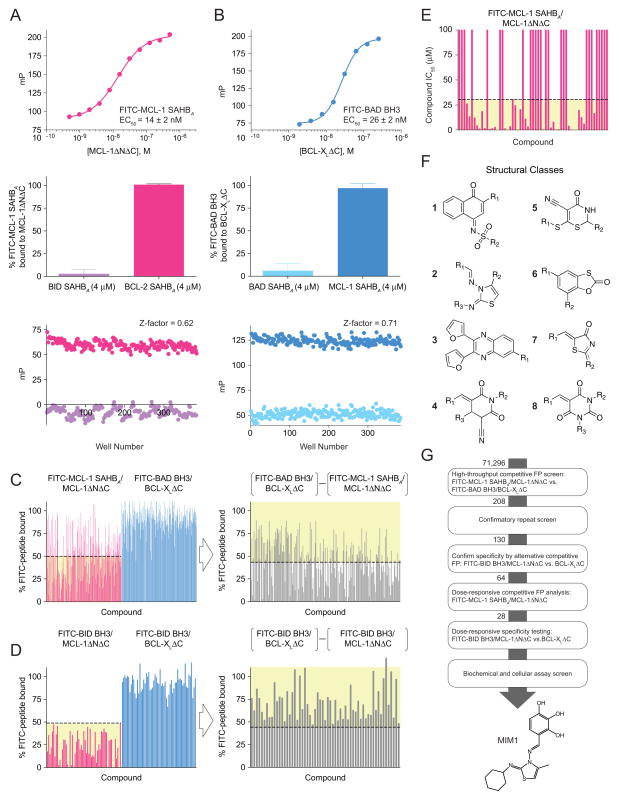
MCL-1 SAHBs are hydrocarbon-stapled MCL-1 BH3 helices that were previously shown using chemical, structural, and biological methods to selectively target MCL-1 and sensitize cancer cells to caspase-dependent apoptosis(Stewart et al., 2010). Here, we deployed MCL-1 SAHBA as a high fidelity screening tool to determine if its potency and specificity-of-action could be harnessed for small molecule discovery. A high-throughput competitive fluorescence polarization (FP) screening assay (Z-factor, 0.62) was developed based on the direct binding interaction between FITC-MCL-1 SAHBA and MCL-1ΔNΔC (EC50, 14 nM) (Figure 1A; Figure S1A). A compilation of 71,296 small molecules was screened for the capacity to displace FITC-MCL-1 SAHBA from recombinant MCL-1ΔNΔC (aa 172–327). To enrich for MCL-1-selective molecules by detecting binding activity for the BCL-XL subclass of anti-apoptotic proteins, the library was also counterscreened using a competitive FP assay (Z-factor, 0.71) developed based on the direct and selective interaction between FITC-BAD BH3 and BCL-XLΔC (EC50, 26 nM) (Figure 1B). Small molecules with an apparent preference for MCL-1ΔNΔC (208 compounds, 0.3% hit rate), as defined both by >50% displacement of the FITC-MCL-1 SAHBA/MCL-1ΔNΔC interaction and a >45% difference in peptide displacement from MCL-1ΔNΔC vs. BCL-XLΔC, were advanced to increasingly stringent confirmatory in vitro binding assays including: (1) repeat single-dose testing of 208 molecules in the differential competitive FP screen (Figure 1C); (2) alternative single-dose selectivity screen for 130 confirmed MCL-1-directed antagonists comparing relative displacement of FITC-BID BH3, a dual binder(Zhai et al., 2006), from MCL-1ΔNΔC vs. BCL-XLΔC (Figure 1D); and then (3) dose-responsive competitive binding of the 64 most selective molecules against the FITC-MCL-1 SAHBA/MCL-1ΔNΔC complex (Figure 1E). Of the 64 compounds that competed with FITC-MCL-1 SAHBA for MCL-1ΔNΔC binding at IC50 potencies of <30 μM, many of which fell into discrete structural classes (Figure 1F), 28 small molecules were subjected to dose-responsive target selectivity analysis in the comparative FITC-BID BH3/MCL-1ΔNΔC vs. FITC-BID BH3/BCL-XLΔC competition FP assay (Figure S1B), and then to screening liposomal release and Bax−/−Bak−/− mouse embryonic fibroblasts (MEFs) cytotoxicity assays. Ultimately, we selected 4-((E)-(((Z)-2-(cyclohexylimino)-4-methylthiazol-3(2H)-yl)imino)methyl)benzene-1,2,3-triol (Figure 1G; Figure S1C), termed MCL-1 Inhibitor Molecule 1 (MIM1), as our prototype compound due to a combination of favorable biophysical and biological properties that included MW>200, solubility, stability (Figure S1D), non-reactivity, MCL-1 binding potency and selectivity, compatibility with and activity in a BAX-mediated liposomal release assay, and relatively little to no toxicity in Bax−/−Bak−/− MEFs.
Figure 1. Identification of a Selective Small Molecule Inhibitor of Anti-apoptotic MCL-1 by use of a Stapled Peptide-Based Screen.
(A) A high-throughput competitive fluorescence polarization (FP) binding assay was developed based on the direct binding interaction between FITC-MCL-1 SAHBA and MCL-1ΔNΔC (EC50, 14 nM), and validated for screening using a positive control for complete displacement (BID SAHBA) and a negative control for no displacement (BCL-2 SAHBA), yielding an assay Z-factor of 0.62.
(B) To enrich for MCL-1-selective small molecules, a counterscreen was developed based on the direct binding interaction between FITC-BAD BH3 and BCL-XLΔC (EC50, 26 nM), and validated for screening using a positive control for complete displacement (BAD SAHBA) and a negative control for no displacement (MCL-1 SAHBA), yielding an assay Z-factor of 0.71.
(C) Based on the results of the primary screen, 208 molecules were subjected to repeat testing in the differential competitive FP screen and 130 molecules met the selection criteria (yellow shading) of >50% displacement of the FITC-MCL-1 SAHBA/MCL-1ΔNΔC interaction (left) and a >45% difference in peptide displacement from MCL-1ΔNΔC vs. BCL-XLΔC (right).
(D) The 130 identified compounds were then tested in a second differential binding selectivity screen and 64 molecules met the selection criteria (yellow shading) of >50% displacement of the FITC-BID BH3/MCL-1ΔNΔC interaction (left) and a >45% difference in peptide displacement from MCL-1ΔNΔC vs. BCL-XLΔC (right).
(E) Dose-responsive competitive binding against the FITC-MCL-1 SAHBA/MCL-1ΔNΔC interaction identified 28 small molecules with an IC50 less than 30 μM (yellow shading).
(F) The identified MCL-1-selective molecules fell into discrete structural classes, numbered 1–8.
(G) MIM1 was chosen as the lead MCL-1-selective small molecule inhibitor based on the results of the screening algorithm and a favorable biophysical and biological properties profile. The molecular structure of MIM1 is characterized by a thiazolyl core substituted with methyl, cyclohexylimino, and benzenetriol R groups. See also Figure S1.
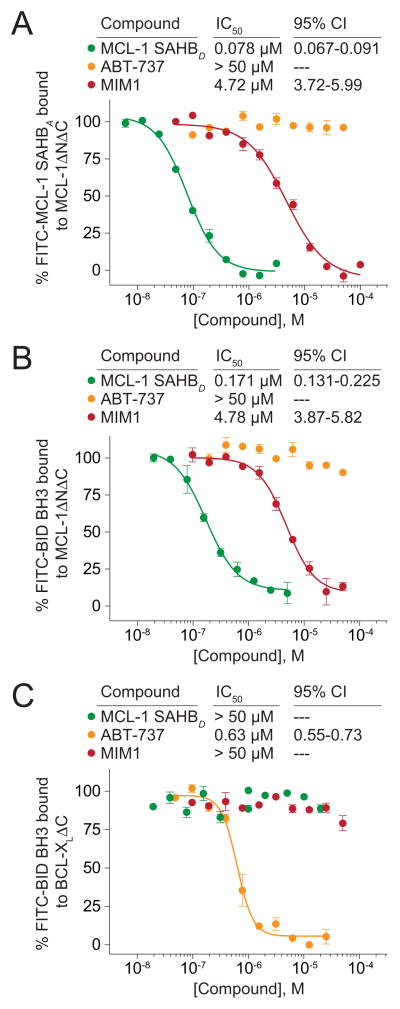
The molecular structure of MIM1 (MW 347) is characterized by a thiazolyl core substituted with methyl, cyclohexylimino, and benzenetriol R groups (Figure 1G). Preliminary SAR analysis revealed the preference for a methyl and aromatic group combination at positions R1 or R3 of the thiazolyl core and maintenance of all three hydroxyl substituents of the benzenetriol moiety, although select variations on this theme were also tolerated (Figure S1B; Figure S2). We chose to vet the anti-apoptotic binding selectivity of MIM1 in competitive FP assays by comparison with ABT-737, a selective BCL-2/BCL-XL inhibitor molecule(Oltersdorf et al., 2005). Whereas MIM1 effectively competed with FITC-MCL-1 SAHBA and FITC-BID BH3 for MCL-1ΔNΔC binding with respective IC50s of 4.7 and 4.8 μM, the compound showed no capacity to displace FITC-BID BH3 from BCL-XLΔC (IC50>50 μM), mirroring the selectivity of MCL-1 SAHBD (Figures 2A–2C). In striking contrast, ABT-737 competed with FITC-BID BH3 for BCL-XLΔC binding, but showed no activity toward MCL-1ΔNΔC (Figures 2A–2C). Although MCL-1 SAHBD was a 30 to 60-fold more potent competitor for MCL-1ΔNΔC binding than MIM1, the MCL-1-selective small molecule is one-seventh the size of the stapled peptide and exhibits an IC50 for its target (4.8 μM) that is only 7.5-fold less than that of chemically-optimized ABT-737 for BCL-XLΔC (0.63 μM) upon competition with FITC-BID BH3. Thus, MIM1 emerged from the competitive screen with a marked MCL-1ΔNΔC preference that reflects the binding specificity of the stapled peptide ligand and the opposite interaction profile of ABT-737.
Figure 2. MIM1 Displays the MCL-1 Binding Selectivity of MCL-1 SAHB and the Opposite Interaction Profile of ABT-737.
(A) MCL-1 SAHBD and MIM1 dose-responsively competed with FITC-MCL-1 SAHBA for binding to MCL-1ΔNΔC, whereas the BCL-2/BCL-XL-selective antagonist ABT-737 had no effect.
(B) Similarly, MCL-1 SAHBD and MIM1, but not ABT-737, effectively competed with FITC-BID BH3 peptide for binding to MCL-1ΔNΔC.
(C) In contrast, ABT-737 dose-responsively competed with FITC-BID BH3 for binding to BCL-XLΔC, whereas MCL-1 SAHBD and MIM1 showed no BCL-XLΔC-binding activity. Data are mean ± SEM for FP assays performed in at least duplicate and then repeated twice with independent protein preparations with similar results. CI, confidence intervals See also Figure S2.
Structural Analysis of the MIM1/MCL-1ΔNΔC Interaction
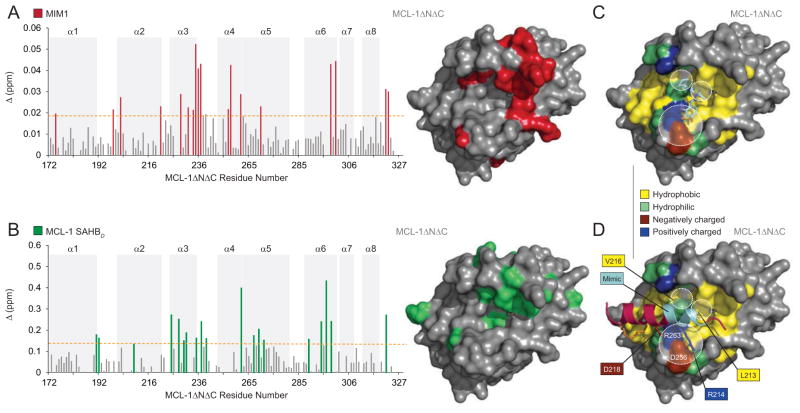
To localize the protein interaction site that accounts for competitive small molecule binding activity, we performed NMR analysis of 15N-MCL-1ΔNΔC upon MIM1 titration. The addition of MIM1 up to a 2:1 molecule:protein ratio induced significant backbone amide chemical shift changes in those MCL-1ΔNΔC residues concentrated in a subregion of the canonical BH3-binding pocket, which is comprised of residues from α2 (BH3) and portions of α3, α4, α5 (BH1) and α8 (BH2) (Figure 3A). Comparison of the 15N-MCL-1ΔNΔC chemical shift changes upon titration with MIM1 versus MCL-1 SAHBD further confirmed the colocalization of small molecule and stapled peptide binding interactions at the canonical BH3-binding site (Figure 3B), as also observed for BID BH3 by NMR analysis(Liu et al., 2010). These data are consistent with a direct interaction between MIM1 and MCL-1ΔNΔC at the very surface employed by BH3 helices to engage MCL-1.
Figure 3. MIM1 Targets the Canonical BH3-Binding Pocket of MCL-1.
(A) Measured chemical shift changes of 15N-MCL-1ΔNΔC upon MIM1 titration up to a ratio of 2:1 MIM1:MCL-1 are plotted as a function of MCL-1ΔNΔC residue. Affected residues are represented as red bars in the plot (calculated significance threshold >0.0197 p.p.m.). Residues with significant backbone amide chemical shift changes (red) are concentrated in a subregion of the canonical BH3-binding pocket. Of note, the plot does not reflect all MIM1-induced changes within the BH3-binding pocket as select residues were not previously assigned in the unliganded structure (e.g. M250, V253, F254, S255, D256, G257, G262, and R263).
(B) Measured chemical shift changes of 15N-MCL-1ΔNΔC upon MCL-1 SAHBD titration up to a ratio of 2:1 MCL-1 SAHBD:MCL-1 are plotted as a function of MCL-1ΔNΔC residue. Affected residues are represented as green bars in the plot (calculated significance threshold >0.14 p.p.m.) and predominantly localize to the canonical BH3-binding pocket, which is comprised of amino acids from α2 (BH3) and portions of α3, α4, α5 (BH1) and α8 (BH2). Of note, the plot does not reflect all MCL-1 SAHBD-induced changes within the canonical pocket as select residues could not be assigned due to large chemical shift changes or were not previously assigned in the unliganded structure.
(C, D) The docked structure of MIM1 at the canonical BH3-binding pocket of MCL-1ΔNΔC predicts that (1) the cyclohexyl group makes complementary hydrophobic contacts with the region of the protein interface flanked by MCL-1 SAHBD residues L213 and V216, (2) the thiazolyl core and its methyl substituent point directly into a deep crevice occupied by MCL-1 SAHBD L213 in the stapled peptide/protein complex, and (3) the benzene-1,2,3-triol (or pyrogallol) moiety engages in hydrophilic contacts with D256 and R263, two charged MCL-1 residues implicated in complementary electrostatic interactions with R214 and D218 of MCL-1 SAHBD.
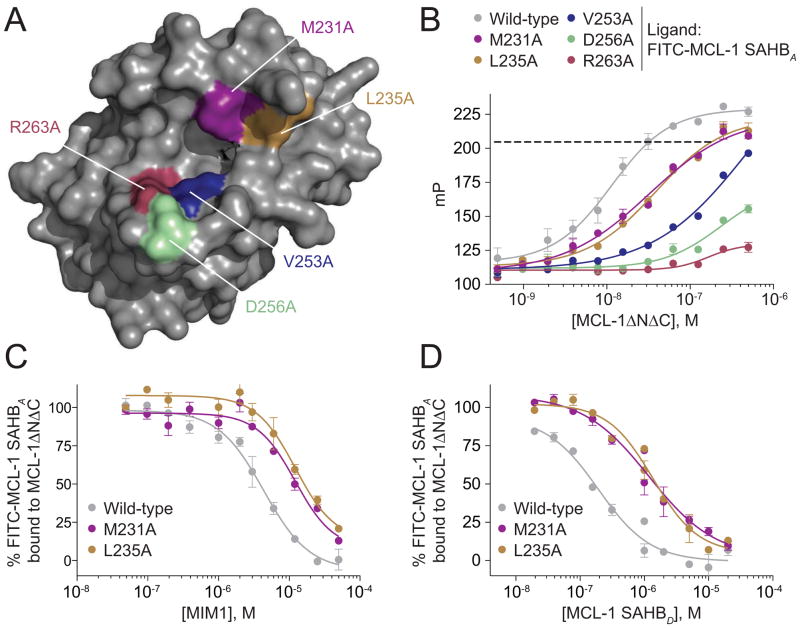
We next performed molecular docking analysis to examine the predicted interactions between MIM1 and MCL-1ΔNΔC at the BH3-binding pocket. Interestingly, MIM1 is predicted to occupy that portion of the BH3-binding site engaged by residues ETLRRV (aa 211–216) of MCL-1 SAHBD (Figures 3C and 3D) (Stewart et al., 2010). Whereas the cyclohexyl group is predicted to make complementary hydrophobic contacts with the region of the protein interface flanked by MCL-1 SAHBD residues L213 and V216, the thiazolyl core and its methyl substituent are predicted to point directly into a deep crevice occupied in the MCL-1 SAHBD/MCL-1ΔNΔC complex by the highly conserved leucine (MCL-1 SAHBD L213) of BH3 domains. Interestingly, the benzene-1,2,3-triol (or pyrogallol) moiety is predicted to engage in hydrophilic contacts with D256 and R263, two charged MCL-1 residues implicated in complementary electrostatic interactions with a variety of BH3 domain R/D pairs (e.g. aa R214, D218 of MCL-1 SAHBD). Of note, several residues predicted to interact with MIM1 based on the docking analysis are not apparent by NMR because select BH3-binding pocket residues are unassigned (e.g. M250, V253, F254, S255, D256, G257, G262, R263). Single point mutagenesis of individual residues predicted to interact with MIM1 based on the docking analysis (M231, L235, V253, D256, R263) and/or that undergo prominent chemical shift change upon MIM1 titration (M231, L235), impaired or abrogated FITC-MCL-1 SAHBA binding to MCL-1ΔNΔC (Figures 4A and 4B). For the MCL-1ΔNΔC L235A and MCL-1ΔNΔC M231A constructs that retain sufficient FITC-MCL-1 SAHBA binding to permit competitive binding analysis, both MIM1 and MCL-1 SAHBD manifested a similar degree of impairment in mutant compared to wild-type MCL-1ΔNΔC binding (Figures 4C and 4D). Taken together, these data suggest that MIM1 simulates key molecular features of approximately 1.5 turns of the MCL-1 BH3 helix at a potential selectivity hotspot on the MCL-1 binding surface.
Figure 4. Point Mutagenesis of Discrete Residues within the BH3-binding Groove of MCL-1ΔNΔC Disrupts Interaction with MCL-1 SAHB and MIM1.
(A) A series of MCL-1ΔNΔC residues predicted to interact with MIM1 based on the docking and/or NMR analyses, including M231, L235, V253, D256, R263, were mutated to alanine for comparative FP binding studies.
(B) Compared to wild-type MCL-1ΔNΔC, alanine mutagenesis of the indicated residues impaired or abrogated FITC-MCL-1 SAHBA binding. The dotted line indicates the mP and corresponding dosing levels used for MCL-1ΔNΔC, MCL-1ΔNΔC L235A, and MCL-1ΔNΔC M231A in the competitive FP assays below.
(C, D) MIM1 and MCL-1 SAHBD manifest a similar degree of impairment in binding to mutant vs. wild-type protein for the MCL-1ΔNΔC L235A and MCL-1ΔNΔC M231A constructs, which retained sufficient FITC-MCL-1 SAHBA binding (B) to enable competitive binding analyses. Data are mean ± SEM for FP assays performed in at least duplicate and then repeated twice with independent protein preparations with similar results.
MIM1 Blocks MCL-1-mediated Suppression of Pro-Apoptotic BAX
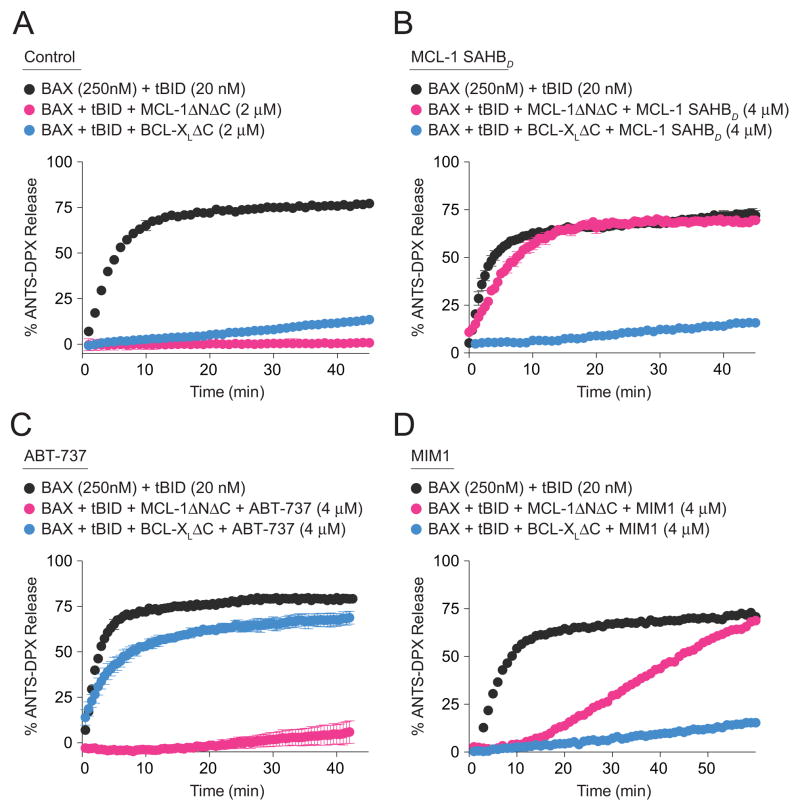
We next examined whether MIM1 could selectively block MCL-1ΔNΔC-based suppression of BAX activation, as monitored by a BAX-mediated liposomal release assay tailored to distinguish between pharmacologic regulation by MCL-1ΔNΔC vs. BCL-XLΔC. The BH3-only protein tBID directly triggers the transformation of monomeric BAX into a membrane-embedded oligomer that porates liposomal vesicles and releases encapsulated fluorophore; the addition of anti-apoptotic proteins, such as MCL-1ΔNΔC or BCL-XLΔC, blocks tBID-induced BAX activation and liposomal release (Figure 5A). Whereas the BAX-suppressive effects of MCL-1ΔNΔC were completely eliminated by pre-incubation with MCL-1 SAHBD, BCL-XLΔC-based inhibition of BAX activation was unimpeded by the MCL-1-selective stapled peptide (Figure 5B). Conversely, ABT-737, which selectively blocks BCL-XLΔC, negated BCL-XLΔC-mediated suppression of BAX activation but had no effect on MCL-1ΔNΔC activity (Figure 5C). Having documented the high fidelity of this tailored liposomal assay for distinguishing between anti-apoptotic selectivities, we next evaluated the functional activity of MIM1. Indeed, we find that MIM1 simulates the pharmacologic activity of MCL-1 SAHBD, preventing BAX suppression by MCL-1ΔNΔC but not by BCL-XLΔC (Figure 5D). Consistent with the reduced molecular weight and competitive binding activity of MIM1 compared to MCL-1 SAHBD, the kinetics of MIM1 inhibition of MCL-1ΔNΔC-mediated BAX suppression were correspondingly slower (Figures 2A and 2B; Figures 5B and 5D). Thus, these data explicitly link the selective MCL-1ΔNΔC binding activity of MIM1 with functional blockade of MCL-1ΔNΔC-mediated inhibition of BAX activation.
Figure 5. Selective Blockade of MCL-1-mediated Suppression of BAX Activation by MIM1.
(A) BH3-only protein tBID directly activates BAX-mediated liposomal release, which is effectively suppressed by treatment with anti-apoptotic MCL-1ΔNΔC and BCL-XLΔC.
(B) MCL-1 SAHBD selectively inhibited MCL-1ΔNΔC suppression of tBID-induced BAX activation.
(C) ABT-737 selectively inhibited BCL-XLΔC suppression of tBID-induced BAX activation.
(D) The activity profile of MIM1 in the liposomal release assay mirrored the MCL-1 selectivity of MCL-1 SAHBD.
Liposomal assays were conducted in triplicate for each condition and repeated with an independent preparation of recombinant BAX with similar results.
Selective Activation of MCL-1-dependent Leukemia Cell Death by MIM1
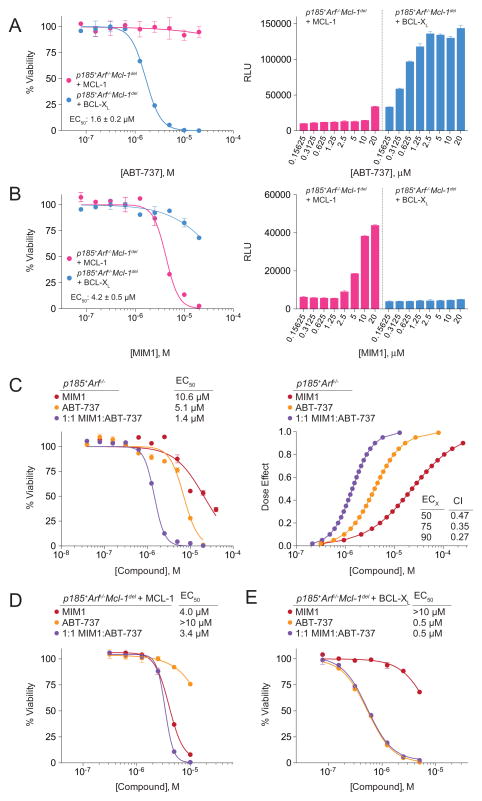
One of the key challenges in developing and applying molecular antagonists for BCL-2 family anti-apoptotic proteins is the variable expression of multiple homologues. That is, a cancer cell will only be susceptible to a selective anti-apoptotic inhibitor if the cell is especially dependent on that particular survival protein. Thus, the mere expression of MCL-1 does not predict cancer cell sensitivity to an MCL-1-selective inhibitor, as other anti-apoptotics lying outside its binding spectrum may continue to effectively suppress BAX/BAK. To test MIM1’s activity and specificity in cancer cells, we employed murine BCR-ABL(p185)-transformed, Arf-null, B-lineage acute lymphoblastic leukemia (p185+Arf−/− B-ALL) cells that are unable to survive upon Mcl-1 deletion unless reconstituted with MCL-1, reflecting a stringent system for assessing MCL-1 dependence. To validate the cellular assay, we first compared the effect of ABT-737 on p185+Arf−/−/Mcl-1-deleted B-ALL cells rescued by overexpression of MCL-1 or BCL-XL (Figure S3A) and observed dose-responsive impairment of cancer cell viability (IC50, 1.6 μM) that coincided with dose-responsive caspase 3/7 activation in the BCL-XL-dependent cells, but no effect on the MCL-1-dependent cells (Figure 6A). Strikingly, MIM1 had the exact opposite effect, negatively impacting the viability of the MCL-1-dependent cells (IC50, 4.2 μM), including dose-dependent induction of caspase 3/7 activity, but having little to no effect on the BCL-XL-dependent cells (Figure 6B). MIM1’scytotoxic effect on the MCL-1-dependent cells likewise corresponded to dose-dependent dissociation of the inhibitory MCL-1/BAK complex, as assessed by co-immunoprecipitation analysis (Figure S3B). Importantly, ABT-737 and MIM1 had no significant effect on the viability of wild-type or Bax−/−Bak−/− MEFs over the same dose range, with no observed caspase 3/7 activation (Figures S3C and S3D).
Figure 6. MCL-1-dependent Anti-leukemia Activity of MIM1 and Synergy with ABT-737.
(A) ABT-737 selectively impaired the viability of p185+Arf−/−Mcl-1-deleted B-ALL cells rescued by BCL-XL, but not MCL-1, as measured by CellTiter-Glo assay at 24 h. The selective cytotoxic effect of ABT-737 was accompanied by dose-responsive caspase 3/7 activation in the BCL-XL-rescued leukemia cell line, as measured at 8 h post-treatment.
(B) MIM1 exhibited the opposite cellular activity profile of ABT-737, selectively impairing the viability of p185+Arf−/−Mcl-1-deleted B-ALL cells rescued by MCL-1, but not BCL-XL. Correspondingly, the selective cytotoxic effect of MIM1 was accompanied by dose-responsive caspase 3/7 activation only in the MCL-1-rescued leukemia cell line.
(C) Combination treatment with MIM1 and ABT-737 induced synergistic killing of parental p185+Arf−/− B-ALL cells that express endogenous MCL-1 and BCL-XL, as reflected by a leftward shift of the viability isotherm (left) and the CalcuSyn dose effect curve (right), with calculated CI values of <1 at ED50, ED75, and ED90. CI, combination index; ED, effective dose.
(D) The addition of ABT-737 to MIM1 treatment of MCL-1-rescued p185+Arf−/−Mcl-1-deleted B-ALL cells had little to no additional cytotoxic effect, consistent with the relative inactivity of ABT-737 in the context of MCL-1-dependence.
(E) Correspondingly, the addition of MIM1 to ABT-737 treatment of BCL-XL-rescued p185+Arf−/−Mcl-1-deleted B-ALL cells provided no additional cytotoxic effect, consistent with the relative inactivity of MIM1 in the context of BCL-XL-dependence.
Data are mean ± SEM for experiments performed in at least duplicate, normalized to vehicle control, and repeated twice with independent cell cultures with similar results. See also Figure S3.
We next examined the functional impact of combining ABT-737 and MIM1 in isogenic p185+Arf−/− B-ALL cells differing only in their expression of MCL-1 and BCL-XL (Figure S3A). In parental p185+Arf−/− B-ALL cells that express both endogenous MCL-1 and BCL-XL, the combination of ABT-737 (IC50, 5.1 μM) and MIM1 (IC50, 10.6 μM) resulted in synergistic cytotoxicity, as determined by CalcuSyn analysis(Chou, 2006) (IC50, 1.4 μM; CI at ED50, 0.47) (Figure 6C). Of note, MIM1, like ABT-737, was somewhat more active in the rescued compared to the parental cell line, consistent with the presence of increased levels of displaceable BH3-only proteins, such as BIM, in complex with overexpressed anti-apoptotic protein (Figure S3E)(Certo et al., 2006; Del Gaizo Moore et al., 2007; Merino et al., 2012). Strikingly, when the MIM1/ABT-737 combination was applied to MCL-1-reconstituted p185+Arf−/−/Mcl-1-deleted B-ALL cells, the addition of ABT-737 had little effect (Figure 6D). Similarly, the cytotoxic effects of single agent ABT-737 and the combination on BCL-XL-reconstituted p185+Arf−/−/Mcl-1-deleted B-ALL cells were identical, reflecting no added benefit of MIM1 in the absence of MCL-1 (Figure 6E). These data underscore the selectivity of MIM1 and ABT-737 for their respective targets in the context of high stringency cancer cell dependence on MCL-1 or BCL-XL. The relative resistance of non-malignant, wild-type fibroblasts to MIM1 treatment (Figure S3C), as observed for ABT-737(Figure S3D)(van Delft et al., 2006), suggests that a therapeutic window may exist, with preferential toxicity to cells driven by discrete anti-apoptotic blockades.
DISCUSSION
Cancer cells are empowered by a combination of proliferative drive and apoptotic blockade. Under homeostatic conditions, BCL-2 family anti-apoptotic proteins guard against premature or unwanted cellular death, but in the context of cancer, their overexpression subverts the natural death pathway and promotes tumor development, maintenance, recurrence, and chemoresistance. To reduce the apoptotic threshold in cancer and thereby facilitate the efficacy of chemotherapy and radiation treatment, an ideal arsenal of targeted apoptotic therapies would include agents with specificities tailored to individual, subsets of, and all anti-apoptotic proteins. The BH3 helix-binding groove on the surface of anti-apoptotic proteins represents the pharmacologic bull’s-eye for such targeted therapies. The first structure of a pro-apoptotic BH3 helix in complex with anti-apoptotic BCL-XL(Sattler et al., 1997) provided the blueprint for developing such molecules and designer peptides. The bench-to-bedside learnings from ABT-737 and ABT-263 have yielded enormous insight into the remarkable potential and remaining challenges of this pharmacologic strategy. Deciphering the topographic hot spots that dictate similarities and differences among the BH3-binding sites of numerous anti-apoptotic proteins presents a considerable drug design challenge. However, the advancement of ABT-263, and a growing diversity of small molecules and peptides that address the variety of BCL-2 family targets(Arnold et al., 2008; Doi et al., 2012; Feng et al., 2010; Kazi et al., 2011; Lee et al., 2008; M.D. et al., 2012; Mohammad et al., 2007; Nguyen et al., 2007; Oh et al., 2010; Paoluzzi et al., 2008; Petros et al., 2010; Placzek et al., 2011; Stewart et al., 2010; Wang et al., 2006; Zhang et al., 2011; Zheng et al., 2012), predicts that this pharmacologic puzzle can ultimately be solved.
With BCL-2/BCL-XL-selective inhibitors having led the field, significant attention has shifted to expanding the scope of anti-apoptotic targeting, with a special emphasis on MCL-1, a ubiquitous pathogenic and resistance factor in cancer. To uncover the binding and specificity determinants for MCL-1, we generated a library of hydrocarbon-stapled BH3 helices to screen for a naturally selective MCL-1 inhibitor. Ironically, the BH3 helix of MCL-1 itself emerged as the most potent and selective antagonist. Biochemical and structural analysis of the MCL-1 SAHBD/MCL-1ΔNΔC interaction provided new insight into key distinguishing features of the binding interface and how they could potentially be harnessed for drug development(Stewart et al., 2010). Here, we applied this uniquely selective, high affinity stapled peptide to screen for small molecules that could effectively displace it from the BH3-binding groove of MCL-1ΔNΔC, yet not target BCL-XL. MIM1 emerged as a potent and selective small molecule inhibitor of MCL-1ΔNΔC, capable of targeting the canonical BH3-binding pocket of MCL-1, blocking MCL-1-mediated suppression of tBID-induced BAX activation in vitro, and inducing caspase 3/7 activation and cell death in MCL-1-dependent, but not BCL-XL-dependent, leukemia cells. In each case, ABT-737 had the opposite biochemical and cellular activity profile of MIM1, and synergized with MIM1 only in the context of dual MCL-1 and BCL-XL expression. These data indicate that MIM1 may serve as a prototype for the development of next generation small molecules that effectively reduce the apoptotic threshold in cancers specifically driven by anti-apoptotic MCL-1.
METHODS
SAHB Synthesis
Hydrocarbon-stapled peptides corresponding to BCL-2 family BH3 domains and their FITC-βAla derivatives were synthesized, purified, and characterized according to previously described methods(Bird et al., 2008; Walensky et al., 2004; Walensky et al., 2006). The sequence compositions of BH3 peptides used in this study are indicated in Figure S1A.
BCL-2 Family Protein Production
Recombinant MCL-1ΔNΔC, BCL-XLΔC, and full-length BAX were expressed and purified as previously reported(Gavathiotis et al., 2008; Pitter et al., 2008). Point mutations in MCL-1ΔNΔC were generated by PCR-based site-directed mutagenesis (QuikChange Mutagenesis Kit, Stratagene) followed by DNA sequencing to verify the constructs. Transformed Escherichia coli BL21 (DE3) were cultured in ampicillin-containing Luria Broth, and protein expression was induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). The bacterial pellets were resuspended in buffer (1% Triton X-100 in PBS, complete protease inhibitor tablet for MCL-1ΔNΔC and BCL-XLΔC, and 250 mM NaCl, 20 mM Tris, complete protease inhibitor tablet, pH 7.2 for BAX), sonicated, and after centifugation at 45,000×g for 45 min, the supernatants were applied to glutathione-sepharose columns (GE Healthcare) for GST-MCL-1ΔNΔC and BCL-XLΔC, or a chitin column (BioLabs) for Intein-BAX. On-bead digestion of GST-tagged protein was accomplished by overnight incubation at room temperature in the presence of thrombin (75 units) in PBS (3 mL), whereas the intein tag was cleaved from BAX by overnight incubation of the chitin beads at 4°C with 50 mM DTT. The tagless recombinant proteins were purified by size exclusion chromatography (SEC) using a Superdex-75 column (GE Healthcare) with 150 mM NaCl, 50 mM Tris, pH 7.4 buffer conditions for MCL-1ΔNΔC and BCL-XLΔC, and 20 mM HEPES pH 7.2, 150 mM KCl buffer conditions for full-length monomeric BAX protein.
Fluorescence Polarization Binding Assays
Fluorescence polarization assays (FPA) were performed as previously described(Bernal et al., 2010; Pitter et al., 2008). Briefly, direct binding curves were first generated by incubating FITC-MCL-1 SAHBA, FITC-BID BH3, or FITC-BAD BH3 (15 nM) with serial dilutions of anti-apoptotic protein, and fluorescence polarization measured at 5 min on a SpectraMax M5 microplate reader (Molecular Devices). For competition assays, a serial dilution of small molecule or acetylated peptide was added to recombinant protein at ~EC75 concentration, as determined by the direct binding assay (MCL-1ΔNΔC, 45 nM; BCL-XLΔC, 300 nM). Fluorescence polarization was measured at equilibrium and IC50 values calculated by nonlinear regression analysis of competitive binding curves using Prism software (Graphpad).
High-Throughput Screening
Small molecule screening was performed at the Institute for Chemistry and Cellular Biology (ICCB) of Harvard Medical School, utilizing the commercial libraries Asinex1 (12,378 molecules), Chembridge3 (10,560 molecules), ChemDiv4 (14,677 molecules), Enamine2 (26,576), Life Chemicals1 (3,893 molecules), and Maybridge5 (3,212 molecules). High-throughput competitive FP binding assays were employed to screen for small molecules that disrupted the FITC-MCL-1 SAHBA/MCL-1ΔNΔC, but not the FITC-BAD BH3/BCL-XLΔC, interaction. SEC-purified MCL-1ΔNΔC or BCL-XLΔC (see above) was delivered by automated liquid handler (WellMate, Matrix) to 384 well plates, followed by addition of small molecule libraries (~5 mg/mL, 100 nL). After a 15 min incubation at room temperature, the corresponding FITC-SAHB (15 nM) was added to each well by liquid handler and FP read at 1 h using a PerkinElmer Envision plate reader (λex 480 nm, λem 535 nm).
MIM1 Characterization by Mass spectrometry and 1H-NMR Spectroscopy
4-((E)-(((Z)-2-(cyclohexylimino)-4-methylthiazol-3(2H)-yl)imino)methyl)benzene-1,2,3-triol. LC-MS: 348(M+1,ES+); 346(M−1, ES−). 1H NMR (300 MHz, DMSO-d6): d 11.35 (s, 1H, -OH); 9.3 (s, 1H, -OH); 8.42 (s, 1H, -OH); 8.31(s, 1H); 6.73 (d, 1H, J=8.4 Hz); 6.34(d, 1H, J=8.4 Hz); 6.01(s, 1H); 3.09–3.05 (m, 1H); 2.15 (s, 3H); 1.81–1.60 (m, 5H); 1.40–1.2 (m, 3H), 1.15 (t, 2H).
NMR Samples and Spectroscopy
Uniformly 15N-labeled MCL-1ΔNΔC was generated by modifying its expression and purification scheme in accordance with the method for producing 15N-BAX(Gavathiotis et al., 2008). Protein samples were prepared in a 20 mM HEPES, 5 mM DTT solution at pH 6.5 in 5% D2O. MIM1 (20 mM stock) and MCL-1 SAHBD (5 mM stock) in DMSO were titrated into a solution of 100 μM MCL-1ΔNΔC to achieve the indicated molar ratio concentrations. Correlation 1H-15N HSQC spectra(Grzesiek and Bax, 1993) were acquired at 25°C on a Bruker 800 MHz NMR spectrometer equipped with a cryogenic probe, processed using NMRPipe(Delaglio et al., 1995), and analyzed with NMRView(Johnson and Blevins, 1994). The weighted average chemical shift difference Δ at the indicated molar ratio was calculated as in p.p.m. The absence of a bar indicates no chemical shift difference, or the presence of a proline or residue that is overlapped or not assigned. MCL-1ΔNΔC cross-peak assignments were applied as previously reported(Suzuki et al., 2000). The significance threshold for backbone amide chemical shift changes was calculated based on the average chemical shift across all residues plus the standard deviation, in accordance with standard methods(Marintchev et al., 2007).
Structure Modeling
Docked structures of MCL-1ΔNΔC and MIM1 were generated using Glide and analyzed using PYMOL(DeLano, 2002).
Liposomal Release Assay
Liposomes were prepared and release assays performed as previously described(Lovell et al., 2008; Yethon et al., 2003). Liposomal composition reflects a mixture of the following molar percentages of lipids (Avanti Polar Lipids): phosphatidylcholine, 48%; phosphatidylethanolamine, 28%; phosphatidylinositol, 10%; dioleoyl phosphatidylserine, 10%; and tetraoleoyl cardiolipin, 4%. Aliquots of mixed lipids (1 mg total) are stored in glass at −20°C under nitrogen, and before use, resuspended in liposome assay buffer (10 mM HEPES, 200 mM KCl, 1 mM MgCl2, pH 7) containing 12.5 mM of the fluorescent dye ANTS (8-aminonaphthalene-1,3,6-trisulfonic acid, disodium salt) and 45 mM of the quencher DPX (p-xylene-bis-pyridinium bromide). The resulting slurry is vortexed for 10 min and freeze-thawed five times in liquid nitrogen and a 40°C water bath, respectively. The solution is then passed through an Avanti Mini-Extruder Set (#610000) equipped with a 100 nm filter, followed by passage through a Sepharose column (GE Healthcare) to remove residual ANTS/DPX. The liposomes are brought up to a volume of 3 mL to produce a final liposome stock. For the liposomal release assay, a total volume of 30 μL is used in 384 well black flat-bottom plates (Costar), and baseline fluorescent measurements of 8 μL liposomes are made for 10 min using a Tecan Infinite M1000 (λex 355 nm, λem 520 nm). Following the baseline read, recombinant anti-apoptotic protein, with or without pre-incubated small molecule or peptide, is added to the liposomes. Subsequently, 20 nM caspase-cleaved mouse BID (R&D systems) and 250 nM purified recombinant monomeric BAX is added, and fluorescence measurements are recorded each minute until the release measurements plateau, at which point the liposomes are quenched with 0.2% Triton X-100 (100% release). The percentage release of ANTS/DPX was calculated according to the equation ([F − F0]/[F100 − F0]) × 100, where F0 and F100 are baseline and maximal fluorescence, respectively.
Cell Viability and Caspase 3/7 Activation Assays
BCR-ABL(p185)-transformed Arf−/− B-ALL cells were generated by transducing Mcl-1+/+Arf−/− mouse bone marrow with BCR-ABL(p185)-IRES-Luciferase vector and then removing the cells from growth factor and stromal support(Williams et al., 2006). MCL-1 or BCL-XL-rescued p185+Arf−/−/Mcl-1-deleted B-ALL cells were generated by transducing Mcl-1f/fArf−/− mouse bone marrow with p185-IRES-Luciferase. Transformed p185+Mcl-1f/fArf−/− B-ALL cells were then transduced with MSCV-Puro-Mcl-1 or MSCV-Puro-Bcl-XL and selected to produce stable clones, which were subsequently transduced with MSCV-Cre-IRES-GFP to delete the endogenous Mcl-1 alleles. B-ALL cells were maintained in RPMI 1640 (ATCC) supplemented with 10% (v/v) FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 0.1 mM MEM non-essential amino acids, and 50 μM β-mercaptoethanol. Mouse embryonic fibroblasts (MEFs) cells were maintained in DMEM high glucose (Invitrogen) supplemented with 10% (v/v) FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine, 50 mM HEPES, 0.1 mM MEM non-essential amino acids, and 50 μM β-mercaptoethanol. Leukemia cells (4×104/well) were seeded in 96-well opaque plates and incubated with the indicated serial dilutions of vehicle (0.4% DMSO), MIM1, ABT-737, or the combination in DMEM at 37°C in a final volume of 100 μl. For MEF experiments, cells (5×103/well) were seeded in 96-well opaque plates for 24 h and then incubated with the indicated serial dilutions of vehicle (0.4% DMSO), MIM1, or ABT-737. Cell viability was assayed at 24 h by addition of CellTiter-Glo reagent according to the manufacturer’s protocol (Promega), and luminescence was measured using a SpectraMax M5 microplate reader (Molecular Devices). Caspase 3/7 activation was assayed at 8 h by addition of Caspase-Glo 3/7 reagent according to the manufacturer’s protocol (Promega), and luminescence measured using a SpectraMax M5 microplate reader. Synergy of the MIM1/ABT-737 combination in leukemia cells was calculated using the CalcuSyn software package(Chou, 2006).
Immunoprecipitation Assays
MCL-1-reconstituted p185+Arf−/−Mcl-1-deleted B-ALL cells (5×106) were incubated with vehicle or MIM1 in culture at the indicated concentrations for 6 h. The cells were lysed in 50 mM Tris (pH 7.4), 150 mM NaCl, 2.5 mM MgCl2, 0.5% NP40 and complete protease inhibitor tablet, followed by pelleting of cellular debris at 14,000g for 10 min at 4°C. The supernatant was exposed to pre-equilibrated protein A/G sepharose beads and the pre-cleared supernatant subsequently incubated with anti-MCL-1 antibody (Rockland) overnight at 4°C, followed by the addition of protein A/G sepharose beads for 1 h. The beads were pelleted and washed with lysis buffer 3 times for 10 min at 4°C. The washed beads were then pelleted, heated to 90°C for 10 min in SDS loading buffer, and the eluted protein subjected to SDS/PAGE and Western analysis using MCL-1 and BAK (anti-BAK NT, Millipore) antibodies.
Supplementary Material
HIGHLIGHTS.
A stapled peptide-based screen identified a selective molecular inhibitor of MCL-1
MIM1 targets a subregion of the BH3-binding pocket of anti-apoptotic MCL-1
In contrast to ABT-737, MIM1 binds and functionally neutralizes MCL-1 but not BCL-XL
MIM1 exhibits MCL-1-dependent anti-leukemia activity and synergizes with ABT-737
SIGNIFICANCE.
Targeted inhibition of the BCL-2/BCL-XL subclass of anti-apoptotic proteins is a clinically validated approach to overcoming the survival advantage of select cancers driven by BCL-2/BCL-XL-mediated apoptotic blockade. The emerging pathogenic role of MCL-1 in a remarkably broad range of human cancers has catapulted its importance as a high priority therapeutic target. Harnessing the potency and selectivity of a natural MCL-1 antagonist, the BH3 domain of MCL-1 itself, we conducted the first competitive small molecule screen against a high affinity stapled peptide/protein complex. Applying a series of rigorous biochemical and cellular assays designed to vet MCL-1 vs. BCL-XL anti-apoptotic subclass specificity, we validated MIM1 as a potent and selective MCL-1 antagonist capable of preferentially blocking MCL-1-mediated suppression of pro-apoptotic BAX in vitro, and inducing caspase 3/7 activation and cell death of a strictly MCL-1-dependent leukemia. Thus, we find that high fidelity targeting of MCL-1 by a small molecule is achievable and can be combined with selective BCL-2/BCL-XL inhibition to exert synergistic anti-cancer activity.
Acknowledgments
We thank Eric D. Smith for editorial and graphics assistance, the Harvard Institute for Chemical and Cellular Biology for access to their chemical libraries and screening facility, Greg H. Bird for peptide synthesis, CreaGen Biosciences for MIM1 resynthesis and characterization, and the laboratory of Dr. Kalle Gehring for MCL-1 NMR assignments. This work was supported by an ARRA Supplement to NIH grant P01CA92625 and a Stand Up to Cancer Innovative Research Grant to L.D.W., and NIH grant RO1HL102175 and American Cancer Society award RSG-10-255-01-LIB to J.T.O.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnold AA, Aboukameel A, Chen J, Yang D, Wang S, Al-Katib A, Mohammad RM. Preclinical studies of Apogossypolone: a new nonpeptidic pan small-molecule inhibitor of Bcl-2, Bcl-XL and Mcl-1 proteins in follicular small cleaved cell lymphoma model. Mol Cancer. 2008 doi: 10.1186/1476-4598-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal F, Wade M, Godes M, Davis TN, Whitehead DG, Kung AL, Wahl GM, Walensky LD. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell. 2010;18:411–422. doi: 10.1016/j.ccr.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird GH, Bernal F, Pitter K, Walensky LD. Chapter 22 Synthesis and Biophysical Characterization of Stabilized alpha-Helices of BCL-2 Domains. Methods Enzymol. 2008;446:369–386. doi: 10.1016/S0076-6879(08)01622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, Letai A. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DC. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- Degterev A, Lugovskoy A, Cardone M, Mulley B, Wagner G, Mitchison T, Yuan J. Identification of small-molecule inhibitors of interaction between the BH3 domain and Bcl-xL. Nat Cell Biol. 2001;3:173–182. doi: 10.1038/35055085. [DOI] [PubMed] [Google Scholar]
- Del Gaizo Moore V, Brown JR, Certo M, Love TM, Novina CD, Letai A. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. J Clin Invest. 2007;117:112–121. doi: 10.1172/JCI28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos: DeLano Scientific; 2002. http://www.pymol.org. [Google Scholar]
- Doi K, Li R, Sung SS, Wu H, Liu Y, Manieri W, Krishnegowda G, Awwad A, Dewey A, Liu X, et al. Discovery of Marinopyrrole A (Maritoclax) as a Selective Mcl-1 Antagonist that Overcomes ABT-737 Resistance by Binding to and Targeting Mcl-1 for Proteasomal Degradation. J Biol Chem. 2012 doi: 10.1074/jbc.M111.334532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyedy IJ, Ling Y, Nacro K, Tomita Y, Wu X, Cao Y, Guo R, Li B, Zhu X, Huang Y, et al. Discovery of small-molecule inhibitors of Bcl-2 through structure-based computer screening. J Med Chem. 2001;44:4313–4324. doi: 10.1021/jm010016f. [DOI] [PubMed] [Google Scholar]
- Feng Y, Ding X, Chen T, Liu F, Jia X, Luo X, Shen X, Chen K, Jiang h, Wang G, et al. Design, synthesis and interaction study of quinazoline-2(1H)-thione derivatives as novel potential Bcl-xL inhibitors. J Med Chem. 2010;53:3465–3479. doi: 10.1021/jm901004c. [DOI] [PubMed] [Google Scholar]
- Frenzel A, Grespi F, Chmelewskij W, Villunger A. Bcl2 family proteins in carcinogenesis and the treatment of cancer. Apoptosis. 2009;14:584–596. doi: 10.1007/s10495-008-0300-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi L, Camidge DR, Ribeiro de Oliveira M, Bonomi P, Gandara D, Khaira D, Hann CL, McKeegan EM, Litvinovich E, Hemken PM, et al. Phase I study of Navitoclax (ABT-263), a novel Bcl-2 family inhibitor, in patients with small-cell lung cancer and other solid tumors. J Clin Oncol. 2011;29:909–916. doi: 10.1200/JCO.2010.31.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG, Tu HC, Kim H, Cheng EH, Tjandra N, et al. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–1081. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzesiek S, Bax A. The importance of not saturating water in protein NMR: application to sensitivity enhancement and NOE measurements. J Am Chem Soc. 1993;115:12593–12594. [Google Scholar]
- Johnson BA, Blevins RA. A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kang MH, Reynolds CP. Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clin Cancer Res. 2009;15:1126–1132. doi: 10.1158/1078-0432.CCR-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi A, Sun J, Doi K, Sung SS, Takahashi Y, Yin H, Rodriguez JM, Becerril J, Berndt N, Hamilton AD, et al. The BH3 alpha-helical mimic BH3-M6 disrupts Bcl-X(L), Bcl-2, and MCL-1 protein-protein interactions with Bax, Bak, Bad, or Bim and induced apoptosis in a Bax- and Bim- dependent manner. J Biol Chem. 2011;286:9382–9392. doi: 10.1074/jbc.M110.203638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada S, Leone M, Sareth S, Zhai D, Reed JC, Pellecchia M. Discovery, characterization, and structure-activity relationships studies of proapoptotic polyphenols targeting B-cell lymphocyte/leukemia-2 proteins. J Med Chem. 2003;46:4259–4264. doi: 10.1021/jm030190z. [DOI] [PubMed] [Google Scholar]
- Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP, Kitada S, Deng X, Zhai D, Shi YX, Sneed T, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Labelle JL, Katz SG, Bird GH, Gavathiotis E, Stewart ML, Lawrence C, Fisher JK, Godes M, Pitter K, Kung AL, et al. A stapled BIM peptide overcomes apoptotic resistance in hematologic cancers. J Clin Invest. 2012;122:2018–2031. doi: 10.1172/JCI46231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EF, Czabotar PE, van Delft MF, Michalak EM, Boyle MJ, Willis SN, Puthalakath H, Bouillet P, Colman PM, Huang DC, et al. A novel BH3 ligand the selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation. J Cell Biol. 2008;180:341–355. doi: 10.1083/jcb.200708096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Morgan-Lappe S, Huang X, Zakula DM, Vernetti SW, Fesik SW, Shen Y. “Seed” analysis of off-target siRNAs reveals an essential role of MCL-1 in resistance to the small-molecule Bcl-2/Bcl-xL inhibitor ABT-737. Oncogene. 2007;26:3972–3979. doi: 10.1038/sj.onc.1210166. [DOI] [PubMed] [Google Scholar]
- Liu Q, Moldoveanu T, Sprules T, Matta-Camacho E, Mansur-Azzam N, Gehring K. Apoptotic regulation by MCL-1 through heterodimerization. J Biol Chem. 2010;285:19615–19624. doi: 10.1074/jbc.M110.105452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llambi F, Green DR. Apoptosis and oncogenesis: give and take in the BCL-2 family. Curr Opin Genet Dev. 2011;21:12–20. doi: 10.1016/j.gde.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, Andrews DW. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- MDB, Haase HS, Peterson-Kauffman KJ, Lee EF, Clarke OB, Colman PM, Smith BJ, Horne WS, Fairlie WD, Gellman SH. Evaluation of diverse alpha/beta-backbone patterns for functional alpha helix mimicry: analogues of the Bim BH3 domain. J Am Chem Soc. 2012;134:315–323. doi: 10.1021/ja207148m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marintchev A, Frueh D, Wagner G. NMR methods for studying protein-protein interactions involved in translation initiation. Methods Enzymol. 2007;430:283–331. doi: 10.1016/S0076-6879(07)30012-8. [DOI] [PubMed] [Google Scholar]
- Merino D, Khaw SL, Glaser SP, Anderson DJ, Belmont LD, Wong C, Yue P, Robati M, Phipson B, Fairlie WD, et al. Bcl-2, Bcl-xL and Bcl-w are not equivalent targets of ABT-737 and Navitoclax (ABT-263) in lymphoid and leukemic cells. Blood. 2012 doi: 10.1182/blood-2011-12-400929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad RM, Goustin AS, Aboukameel A, Chen B, Banerjee S, Wang G, Nikolovksa-Coleska Z, Wang S, Al-Katib A. Preclinical studies of TW-37, a new nonpeptidic small molecule inhibitor of Bcl-2, in diffuse large cell lymphoma xenograft model reveal drug action on both Bcl-2 and Mcl-1. Clin Cancer Res. 2007;13:2226–2235. doi: 10.1158/1078-0432.CCR-06-1574. [DOI] [PubMed] [Google Scholar]
- Nguyen M, Marcellus RC, Roulston A, Watson M, Serfass L, Madiraju SRM, Goulet D, Viallet J, Belec L, Billot X, et al. Small molecule obatoclax (GX15-070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proc Natl Acad Sci USA. 2007;104:19512–19517. doi: 10.1073/pnas.0709443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Jensen PR, Murphy BT, Fiorilla C, Sullivan JF, Ramsey T, Fenical W. Cryptosphaerolide, a cytotoxic Mcl-1 inhibitor from a marine-derived ascomycete related to the genus Cryptosphaeria. J Nat Prod. 2010;73:998–1001. doi: 10.1021/np1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al. An inhbitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- Paoluzzi L, Gonen M, Gardner JR, Mastrella J, Yang D, Holmlund J, Sorensen M, Leopold L, Manova K, Marcucci G, et al. Targeting Bcl-2 family members with the BH3 mimetic AT-101 markedly enhances the therapeutic effects of chemotherapueitc agents in in vitro and in vivo models of B-cell lymphoma. Blood. 2008;111:5350–5358. doi: 10.1182/blood-2007-12-129833. [DOI] [PubMed] [Google Scholar]
- Petros AM, Huth JR, Oost T, Park CM, Ding H, Wang X, Zhang H, Nimmer P, Mendoza R, Sun C, et al. Discovery of a potent and selective Bcl-2 inhibitor using SAR by NMR. Bioorg Med Chem Lett. 2010;20:6587–6591. doi: 10.1016/j.bmcl.2010.09.033. [DOI] [PubMed] [Google Scholar]
- Pitter K, Bernal F, Labelle J, Walensky LD. Dissection of the BCL-2 family signaling network with stabilized alpha-helices of BCL-2 domains. Methods Enzymol. 2008;446:387–408. doi: 10.1016/S0076-6879(08)01623-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placzek WJ, Sturlese M, Wu B, Cellitti JF, Wei J, Pellecchia M. Identification of a novel Mcl-1 protein binding motif. J Biol Chem. 2011;286:39829–39835. doi: 10.1074/jbc.M111.305326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AW, Seymour JF, Brown JR, Wierda WG, Kipps TJ, Khaw SL, Carney DA, He SZ, Huang DCS, Xiong H, et al. Substantial susceptibility of chronic lymphocytic leukemia to BCL2 inhibitio: Results of Phase 1 study of navitoclax (ABT-263) in patients with relapsed or refractory disease. J Clin Onc. 2011 doi: 10.1200/JCO.2011.34.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, et al. Structure of Bcl-xL-Bak Peptide Complex: Recognition Between Regulators of Apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- Shamas-Din A, Brahmbhatt H, Leber B, Andrews DW. BH3-only proteins: Orchestrators of apoptosis. Biochim Biophys Acta. 2011;1813:508–520. doi: 10.1016/j.bbamcr.2010.11.024. [DOI] [PubMed] [Google Scholar]
- Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nature Chem Biol. 2010;6:595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Youle RJ, Tjandra N. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 2000;103:645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, Johnson EF, Marsh KC, Mitten MJ, Nimmer P, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y, Cossman J, Jaffe E, Croce CM. Involvement of the bcl-2 Gene in Human Follicular Lymphoma. Science. 1985;228:1440–1443. doi: 10.1126/science.3874430. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y, Finger LR, Yunis J, Nowell PC, Croce CM. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science. 1984;226:1097–1099. doi: 10.1126/science.6093263. [DOI] [PubMed] [Google Scholar]
- Tzung SP, Kim KM, Basanez G, Giedt CD, Simon J, Zimmerberg J, Zhang KY, Hockenbery DM. Antimycin A mimics a cell-death-inducing Bcl-2 homology domain 3. Nat Cell Biol. 2001;3:183–191. doi: 10.1038/35055095. [DOI] [PubMed] [Google Scholar]
- van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE, Willis SN, Scott CL, Day CL, Cory S, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux DL, Cory S, Adams JM. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1988;335:440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- Walensky LD, Gavathiotis E. BAX unleashed: the biochemical transformation of an inactive cytosolic monomer into a toxic mitochondrial pore. Trends Biochem Sci. 2011;36:642–652. doi: 10.1016/j.tibs.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Activation of apoptosis in vivo by a hydrocarbon stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J, Smith E, Verdine GL, Korsmeyer SJ. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell. 2006;24:199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Wang G, Nikolovksa-Coleska Z, Yang CY, Wang R, Tang G, Guo J, Shangary S, Qui S, Gao W, Yang D, et al. Structure-based design of potent small molecule inhibitors of anti-apoptotic Bcl-2 proteins. J Med Chem. 2006;49:6139–6142. doi: 10.1021/jm060460o. [DOI] [PubMed] [Google Scholar]
- Wang JL, Liu D, Zhang ZJ, Shan S, Han X, Srinivasula SM, Croce CM, Alnemri ES, Huang Z. Structure-based discovery of an organic compound that binds Bcl-2 protein and induces apoptosis of tumor cells. Proc Natl Acad Sci U S A. 2000;97:7124–7129. doi: 10.1073/pnas.97.13.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2006;103:6688–6693. doi: 10.1073/pnas.0602030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WH, O’Connor OA, Czuczman MS, LaCasce AS, Gerecitano JF, Leonard JP, Tulpule A, Dunleavy K, Xiong H, Chiu YL, et al. Navitoclax, a targeted high-affinity inhibitor of BCL-2, in lymphoid malignancies: a phase 1 dose-escalation study of safety, pharmacokinetics, pharmacodynamics, and antitumour activity. Lancet Oncol. 2010;11:1149–1159. doi: 10.1016/S1470-2045(10)70261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yecies D, Carlson NE, Deng J, Letai A. Acquired resistance to ABT-737 in lymphoma cells that up-regulate MCL-1 and BFL-1. Blood. 2010;115:3304–3313. doi: 10.1182/blood-2009-07-233304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yethon JA, Epand RF, Leber B, Epand RM, Andrews DW. Interaction with a membrane surface triggers a reversible conformational change in Bax normally associated with induction of apoptosis. J Biol Chem. 2003;278:48935–48941. doi: 10.1074/jbc.M306289200. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- Zhai D, Jin C, Satterthwait AC, Reed JC. Comparison of chemical inhibitors of antiapoptotic BCL-2 family proteins. Cell Death Diff. 2006;13:1419–1421. doi: 10.1038/sj.cdd.4401937. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Song T, Zhang T, Gao J, Wu G, An L, Du G. A novel BH3 mimetic S1 potently induces Bax/Bak-depended apoptosis by targeting both Bcl-2 and MCL-1. Int J Cancer. 2011;128:1724–1735. doi: 10.1002/ijc.25484. [DOI] [PubMed] [Google Scholar]
- Zheng CH, Yang H, Zhang M, Lu SH, Shi D, Wang J, Chen XH, Ren XH, Liu J, Lv JC, et al. Design, synthesis and activity evaluation of broad-spectrum small molecule inhibtors or anti-apoptotic Bcl-2 family proteins: characteristics of broad spectrum protein binding and its effects on anti-tumor activity. Bioorg Med Chem Lett. 2012;22:39–44. doi: 10.1016/j.bmcl.2011.11.101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.