Abstract
We conducted a three-stage genetic study to identify susceptibility loci for type 2 diabetes (T2D) in East Asian populations. The first stage meta-analysis of eight T2D genome-wide association studies (6,952 cases and 11,865 controls) was followed by a second stage in silico replication analysis (5,843 cases and 4,574 controls) and a stage 3 de novo replication analysis (12,284 cases and 13,172 controls). The combined analysis identified eight new T2D loci reaching genome-wide significance, which were mapped in or near GLIS3, PEPD, FITM2-R3HDML-HNF4A, KCNK16, MAEA, GCC1-PAX4, PSMD6 and ZFAND3. GLIS3, involved in pancreatic beta cell development and insulin gene expression1,2, is known for its association with fasting glucose levels3,4. The evidence of T2D association for PEPD5 and HNF4A6,7 has been detected in previous studies. KCNK16 may regulate glucose-dependent insulin secretion in the pancreas. These findings derived from East Asians provide new perspectives on the etiology of T2D.
Type 2 diabetes (T2D) is a major public health problem whose global prevalence is rapidly rising8. The development of T2D is influenced by diverse factors, and decades of epidemiological studies have linked obesity, hypertension, and dyslipidemia with the risk of T2D9. It is also known that T2D shows considerable heritability. Within only the last three years, genetic studies have yielded a rapidly lengthening list of loci harboring disease predisposing variations10. To date, genetic variants at forty-five loci have been identified for T2D10,11. Despite these advances toward a better understanding of the genetic basis of T2D, its heritability is yet to be fully explained12. In addition, most T2D loci have been detected initially in population samples of European origin, apart from KCNQ1, UBE2E2, and C2CD4A-C2CD4B, which were first identified in East Asian studies13–15. Additional efforts involving East Asian populations have identified variants at the SPRY2, PTPRD and SRR loci5,16,17. However, these have not been extensively replicated in multiple populations. A large meta-analysis in East Asians is expected to identify novel genetic associations and provide insights into T2D pathogenesis. In addition to differences in the allele frequencies between East Asians and Europeans, which may affect the power to detect associations, T2D epidemiology also differs considerably among different European populations and East Asian populations. In East Asians, the rates of diabetes are often higher at a lower average BMI’s18, suggesting that some different pathways may be involved in pathogenesis of T2D.
To discover new T2D loci, we conducted a three-stage association study in individuals of East Asian descent (Supplementary Fig. 1). The stage 1 meta-analysis combining 8 T2D GWA studies participating in the Asian Genetic Epidemiology Network (AGEN) consortium (6,952 cases and 11,865 controls) was performed using association data for 2,626,356 imputed and genotyped autosomal SNPs by the inverse-variance method for fixed effects (Supplementary Table 1). All imputed and genotyped SNPs (minor allele frequency > 0.01) passed quality control filters in each of the eight stage 1 data sets prior to conducting meta-analysis (Supplementary Table 2). The genomic control inflation factor (λ) for the meta-analysis was 1.046 (less than 1.062 for the individual studies), indicating that results in stage 1 were not likely resulting from population stratification (Supplementary Fig. 2). Individuals from each component study participated in stage 1 mostly clustered together with CHB/JPT HapMap samples in the principal component analysis plot (Supplementary Fig. 3), further demonstrating the similarity in the ethnicity between stage 1 samples. Most signals showing strong evidence for T2D associations appeared in previously known T2D genes (Fig. 1). Stage 1 P-values, OR’s, and average risk allele frequencies for 45 previously reported T2D associated SNPs are shown in Supplementary Table 3.
Figure 1. Genome-wide Manhattan plot for the EA T2D stage 1 meta-analysis.

−log10(P-values) using the trend test are shown for SNPs distributed across the entire autosomal genome. Red dots at each locus indicate the signals with P-value <10−6 detected in the genome-wide meta-analysis. A total of 1,934,619 SNPs that were present in ≥5 stage 1 studies were used to generate both plots.
After removing known T2D variants, 297 SNPs from independent loci were selected from the stage 1 meta-analysis based on our arbitrary inclusion criteria for follow-up in silico replication: meta-analysis P-value < 5×10−4 (based on the divergence between the observed and expected P-values on the Q-Q plot (Supplementary Fig. 2)), heterogeneity P-value>0.01 and the number of studies included in meta-analysis ≥ 7 (Supplementary Table 4). A total of 3,756 SNPs including the 297 selected SNPs and their proxies (r2 >0.8 based on phase2 CHB/JPT HapMap data) were taken forward to stage 2, an in silico replication, in three independent GWA studies (5,843 cases and 4,574 controls). After meta-analysis combining stage 1 and stage 2 data for 3,756 SNPs, we selected the 19 SNPs showing the most compelling evidence for association (stage 1 and 2 combined P-value<10−5) (Supplementary Table 5) for stage 3 de novo genotyping in up to 12,284 cases and 13,172 controls recruited from five independent studies (Supplementary Tables 1 & 2). This resulted in 8 novel T2D loci reaching genome-wide significance from the combined meta-analysis across all three stages (Table 1 & Fig. 2).
Table 1.
Eight new T2D loci reaching genome-wide significance from combined meta-analysis of stage 1, 2 and 3.
| SNP | Chr | Position (bp) | Nearby gene | Risk allele | Other allele | Stage 1 (discovery)
|
Stage 2 (in silico replication)
|
Stage3 (de novo replication)
|
Combined (stage 1+2+3)
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (CI) | P-value | OR (CI) | P-value | OR (CI) | P-value | OR (CI) | P-value | ||||||
| up to 6,952 cases and 11,865 controls | up to 5,843 cases and 4,574 controls | up to 12,284 cases and 13,172 controls | up to 25,079 cases and 29,611 controls | ||||||||||
| Loci showing strong evidence of association with T2D | |||||||||||||
| rs6815464 | 4 | 1299901 | MAEA | c | g | 1.09 (1.04–1.14) | 8.21E-04 | 1.13 (1.07–1.20) | 3.67E-05 | 1.16 (1.11–1.20) | 4.15E-15 | 1.13 (1.10–1.16) | 1.57E-20 |
| rs7041847 | 9 | 4277466 | GLIS3 | a | g | 1.09 (1.04–1.14) | 1.29E-04 | 1.09 (1.03–1.15) | 2.20E-03 | 1.11 (1.07–1.15) | 2.89E-09 | 1.10 (1.07–1.13) | 1.99E-14 |
| rs6017317 | 20 | 42380380 | FITM2-R3HDML-HNF4A | g | t | 1.10 (1.05–1.15) | 2.43E-05 | 1.07 (0.99–1.15) | 8.42E-02 | 1.10 (1.06–1.14) | 3.96E-07 | 1.09 (1.07–1.12) | 1.12E-11 |
| rs6467136 | 7 | 126952194 | GCC1-PAX4 | g | a | 1.12 (1.06–1.18) | 6.47E-05 | 1.11 (1.04–1.18) | 2.09E-03 | 1.10 (1.05–1.15) | 2.31E-05 | 1.11 (1.07–1.14) | 4.96E-11 |
| rs831571 | 3 | 64023337 | PSMD6 | c | t | 1.11 (1.06–1.17) | 4.85E-06 | 1.06 (1.00–1.13) | 4.46E-02 | 1.08 (1.05–1.12) | 1.41E-05 | 1.09 (1.06–1.12) | 8.41E-11 |
| rs9470794 | 6 | 38214822 | ZFAND3 | c | t | 1.11 (1.05–1.17) | 1.45E-04 | 1.09 (1.02–1.17) | 1.48E-02 | 1.16 (1.09–1.23) | 3.20E-06 | 1.12 (1.08–1.16) | 2.06E-10 |
| rs3786897 | 19 | 38584848 | PEPD | a | g | 1.14 (1.08–1.20) | 3.74E-06 | 1.05 (0.99–1.12) | 1.28E-01 | 1.11 (1.04–1.17) | 5.46E-04 | 1.10 (1.07–1.14) | 1.30E-08 |
| rs1535500 | 6 | 39392028 | KCNK16 | t | g | 1.11 (1.06–1.16) | 5.34E-06 | 1.07 (1.01–1.15) | 3.33E-02 | 1.06 (1.02–1.10) | 3.50E-03 | 1.08 (1.05–1.11) | 2.30E-08 |
| Loci showing moderate evidence of association with T2D | |||||||||||||
| rs16955379* | 16 | 80046874 | CMIP | c | t | 1.13 (1.07–1.20) | 2.20E-05 | 1.10 (1.03–1.17) | 6.59E-03 | 1.05 (1.01–1.10) | 2.19E-02 | 1.08 (1.05–1.12) | 2.84E-07 |
| rs17797882 | 16 | 77964419 | WWOX | t | c | 1.12 (1.05–1.18) | 1.76E-04 | 1.09 (1.02–1.16) | 1.21E-02 | 1.06 (1.01–1.11) | 1.61E-02 | 1.08 (1.05–1.12) | 9.49E-07 |
Proxy SNP, rs9930117 (r2 =1), was genotyped in the stage3 CAGE study.
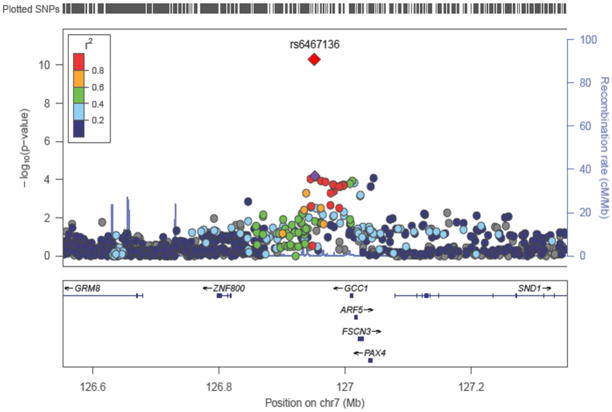
Figure 2. Regional association plots for new T2D loci.
The upper panel indicates the positions of SNPs and the middle panel, shows regional association results from the genome-wide meta-analysis. The trend test −log10(P-values) are shown for SNPs distributed in a 0.8 Mb genomic region centered on the most strongly associated signal, which is depicted as a purple diamond in the stage 1 results and a red diamond for the combined stage 1+2+3 results. The lower panel shows the locations of known genes in the region. Genetic information is taken from Human Genome build hg18 and LD structure was based on 1000 Genomes (June 2010) HapMap JPT+CHB.
Three of these eight loci have been associated previously with metabolic traits or related diseases, or suggestively with T2D itself. One such locus was detected within an intron of GLIS3, a gene that is highly expressed in islet beta cells. The coding product of this gene, a Krüppel-like zinc finger transcription factor, has been proposed as a critical player in the regulation of pancreatic beta cell development and insulin gene expression1,2. SNPs in high LD with this locus have already been implicated in association with type 1 diabetes (T1D)19 and fasting plasma glucose3. The second locus, on 19q13, is located in an intron of PEPD. Several SNPs (lead SNP: rs10425678) in this gene were identified in association with T2D previously in Japanese 5. However, the SNP in our study (rs3786897) is not in LD with those identified in Japanese (r2 = 0.008 and D′ = 0.143 between rs3786897 and rs10425678), and our GWA data do not support an association for T2D with rs10425678 (P = 0.528). The third such signal is near FITM2-R3HDML-HNF4A. FITM2 may be involved in lipid droplet accumulation20, while the function of R3HDML is not known. Mutations in HNF4A cause maturity onset diabetes of the young type 1 (MODY1)21. Common variants, rs1884613 and rs2144908, in the P2 promoter region of this gene have been associated with T2D in a population-specific manner6,22. The SNPs in our study (rs6017317) are not in strong LD with HNF4A P2 promoter SNPs (r2 = 0.23~0.25, D′ = 0.50~0.54 based on phase2 CHB/JPT HapMap data), indicating that rs6017317 is a new T2D signal in the 20q13.12 region where HNF4A resides.
The remaining five loci reaching genome-wide significance in our study have not previously been reported in the context of any metabolic traits, including the loci mapped in or near KCNK16, MAEA, GCC1-PAX4, PSMD6 and ZFAND3. KCNK16, expressed predominantly in the pancreas, encodes a potassium channel protein containing two pore-forming P domains23. In pancreatic β cells, potassium channels that are inhibited by ATP regulate glucose-dependent insulin secretion. Among the variants in strong LD with this signal (rs1535500) is rs11756091 (r2 = 0.977, D′ = 1.0 based on phase2 CHB/JPT HapMap data), which encodes a proline-to-histidine substitution in two isoforms of KCNK16. This or other variants influencing this gene may result in defective regulation of potassium channel activity contributing to the etiology of T2D24. MAEA encodes a protein that plays a role in erythroblast enucleation and in the development of mature macrophages25. The gene-set analysis of the stage 1 P-values using GSA-SNP26 indicated that MAEA belongs to a group of genes that showed significant association with T2D and includes IDE located at a known T2D susceptibility locus27 (stage 1 P = 1.41×10−7 for rs6583826 at the IDE locus in this study). The GRIP domain containing protein encoded by GCC1 may play a role in the organization of the trans-Golgi network involved in membrane transport 28. PAX4 that is only 30kb further away from GCC1 is an outstanding candidate for T2D given its involvement in pancreatic islet development. This gene was recently implicated in a Japanese case of MODY29. The expression product of PSMD6 that acts as a regulatory subunit of the 26S proteasome is likely involved in the ATP-dependent degradation of ubiquitinated proteins 30. Although the function of ZFAND3 has not been fully elucidated, it is noteworthy that family member ZFAND6 is present along with FAH at a T2D locus detected previously31. We examined whether 8 novel loci may be associated with T2D through an effect on obesity, as seen with FTO32. All T2D association signals remained after the adjustment with BMI (Supplementary Table 6), indicating that the association with T2D for 8 loci are not mediated through an effect on obesity.
In addition to 8 loci reaching genome-wide significance, we identified 2 loci showing moderate evidence (combined P-value<10−6) of association with T2D including WWOX and CMIP (Table 1). We looked up the association results for these 10 loci in GWA data from up to 47,117 European samples generated by the DIAGRAM consortium (‘DIAGRAM+’ for the current version of data set)31. Results from lookup for these loci indicated that three loci including FITM2-R3HDML-HNF4A (rs6017317: P = 1.47×10−2, OR = 1.07), CMIP (rs16955379: P = 3.33×10−2, OR = 1.20 ] and MAEA (proxy SNP rs11247991 for rs6815464 (r2=0.96): P = 6.56×10−3, OR = 1.19 ) were modestly associated, while a locus in GLIS3 (rs7041847: P = 6.43×10−2, OR = 1.04 ) was nominally associated with T2D. The direction of effect was consistent in four (PSMD6, PEPD, WWOX and KCNK16) of six loci that were not replicated in DIAGRAM+ (Supplementary Table 5).
The functional connections among 10 new T2D genes and 28 known T2D genes that were replicated in this study (Supplementary Table 3) were analyzed by GRAIL33. Connection results highlighted notable biological functions of sets of genes within T2D-assocaited regions (Supplementary Fig. 4 & Supplementary Tables 7 & 8). For example, KCNK16 shows strong connections with previously known T2D genes encoding potassium channels (KCNJ11 and KCNQ1), implying its physiological role in the regulation of potassium transport in the pancreatic cells.
The association between each new T2D SNP and the expression levels of genes within 1Mb of the SNP was examined by eQTL analysis in the MuTHER consortium. One SNP (rs3786897) in an intron of PEPD was highly associated with mRNA expression levels of PEPD in the adipose tissue of 776 individuals of European ancestry (PeQTL = 2.14×10−8) (Supplementary Table 9). However, this SNP did not show an association with T2D in populations of European ancestry, and the significance of this finding is unclear.
We considered the possibility that autoimmune diabetes may be driving some of the signals observed. Firstly, cases in all studies were predominantly those with adult onset diabetes (age of disease onset ≥ 30 years), and none of the clinically diagnosed patients had T1D, defined as acute ketosis and continuous requirement of insulin within 1 year of diagnosis. Secondly, we looked up the associations for all known T1D associated variants in our study. Only a given number of loci survived from the tests applied (Supplementary Table 10). This is in distinct contrast to the situation for known T2D associated variants where many replicated in our study (Supplementary Table 3), further suggesting that our findings are most relevant to T2D. Finally, since variants close to the GLIS3 locus have been shown to be associated with T1D19, we examined the association between rs7041847 and diabetes in four studies (n=8,383) in which cases with positive GAD antibodies had been excluded (data not shown). In each study, the association between this SNP and diabetes was the same as when all samples were included (meta-analysis P-value = 3.4×10−4, OR = 1.12). This finding, along with the fact that SNPs at the GLIS3 locus also show associations with fasting plasma glucose in nondiabetic adults3 and healthy children and adolescents4, is consistent with the hypothesis that SNPs at this locus may impact fasting glucose homeostasis rather than the immune system. Taken together, it is unlikely that a significant proportion of the positive associations observed in our study were driven by autoimmune diabetes.
This study is the largest GWA meta-analysis conducted for T2D in East Asians. Findings from this study highlight not only previously unknown biological pathways but also population specific loci for T2D. The association of rs9470794 in ZFAND3 with T2D looks highly specific to East Asians (Supplementary Table 5), whereas rs11634397 near ZFAND6 looks specific to Europeans (Supplementary Table 3). A significant difference in risk allele frequencies of both loci is observed between the two continental populations (rs9470794 RAF = 0.32 CHB/JPT vs 0.12 CEU; rs11634397 RAF = 0.07 CHB/JPT vs 0.64 CEU). Although these loci are related to T2D differently in the two populations, these results lead one to speculate that the broader A20 domain-containing zinc finger protein family plays a role in the etiology of T2D. Additional population specific T2D loci are further suggested, as seen in WWOX (rs17797882) (Supplementary Table 5) for East Asians and ZBED3 (rs4457053) (Supplementary Table 3) for Europeans. Despite the lack of clear physiological evidence, these findings may provide clues to understand population characteristic T2D phenotypes as exemplified by the high rates of diabetes at lower average BMI’s in East Asian and other populations.
ONLINE METHODS
Study subjects
Stage 1 subjects were drawn from 8 T2D GWA studies participating in the AGEN consortium that was organized for genetic studies on diverse complex traits in 2010. These eight studies included 6,952 T2D cases and 11,865 controls from the Korea Association Resource Study (KARE), the Singapore Diabetes Cohort Study (SDCS), the Singapore Prospective Study Program (SP2), the Singapore Malay Eye Study (SiMES), the Japan Cardiometabolic Genome Epidemiology Network(CAGE), the Shanghai Diabetes Genetic Study (SDGS), the Taiwan T2D Study (TDS), and the Cebu Longitudinal Health and Nutritional Survey (CLHNS). Stage 2 subjects included 5,843 cases and 4,574 controls from three independent GWA studies including the BioBank Japan Study (BBJ), the Health2 T2D Study (H2T2DS) and the Shanghai Jiao Tong University Diabetes Study (SJTUDS) for in silico replication analysis. Stage 3 included up to, 12,284 cases and 13,172 controls from five different studies comprised of the Japan Cardiometabolic Genome Epidemiology Network (CAGE), the Shanghai Diabetes Study I/II (SDS I/II), the Chinese University of Hong Kong Diabetes Study (CUHKDS), the National Taiwan University Hospital Diabetes Study (NTUHDS) and the Seoul National University Hospital Diabetes Study (SNUHDS) for de novo replication analysis. The study design and T2D diagnosis criteria of each study in stages 1, 2, and 3 are described in Supplementary Table 1 and Supplementary Note. Each study obtained approval from the appropriate institutional review board, and written informed consent from all participants. The three-stage design of the overall study is depicted in Supplementary Fig. 1.
Genotyping and imputation
Subjects for stage 1 and 2 analyses were genotyped with high-density SNP typing platforms covering the entire human genome. In most studies, only unrelated samples with missing genotype call-rates below 5% were included for subsequent GWA analyses. For the genome-wide association meta-analysis, each study participating in stages 1 and 2 performed SNP imputation. IMPUTE, MACH or BEAGLE were used, together with haplotype reference panels from the JPT and CHB founders (JPT+CHB+CEU and/or YRI in some studies) on the basis of HapMap build 36 (release 21, 22, 23a or 24). Only imputed SNPs with high genotype information content (proper info > 0.5 for IMPUTE and Rsq> 0.3 for MACH and BEAGLE) were used for association analysis. Genotyping for the stage 3 analysis was carried out by TaqMan, Sequenom MassARRAY, or the Beckman SNP Stream method. All SNPs included in stage 3 satisfied a genotype success rate over 98% (Supplementary Table 2).
Statistical analyses, analysis tools, and SNP prioritization for stage 2 and 3
Associations between SNPs and T2D were tested by logistic regression with an additive model (1-d.f.) after adjustment for sex. Other adjustments were permitted according to the situation of individual studies. Meta-analysis was performed by an inverse-variance method assuming fixed effects with Cochran’s Q test to assess between-study heterogeneity. METAL software (http://www.sph.umich.edu/csg/abecasis/Metal) was used for all meta-analyses. A plot of the negative log of association results from the stage 1 meta-analysis, by chromosome, was generated by the WGAViewer software (http://people.genome.duke.edu/~dg48/WGAViewer/std.php). The quantile-quantile plot was constructed by plotting the distribution of observed P-values of given SNPs against the theoretical distribution of expected P-values for T2D34. The genomic control inflation factor, λ, was calculated by dividing median χ2 statistics by 0.45635 for individual GWA studies as well as stage 1 GWA meta-analysis. We did not correct for genomic control in the stage 1 analyses as inflation was modest, suggesting that population structure is unlikely to cause significant inflation of stage 1 results (Supplementary Table 2). Selection criteria for lead SNPs to take forward to stage 2 in silico replication analysis were (1) stage 1 meta-analysis P-value < 5 × 10−4 (based on the divergence between the observed and expected P-values on the Q-Q plot (Supplementary Fig. 2)), (2) heterogeneity P-value > 0.01 and (3) number of studies included in stage 1 meta-analysis ≥ 7 (Supplementary Table 4). Removing known variants associated with T2D proxies for each lead SNP (r2> 0.8) were selected using the SNAP software (http://www.broadinstitute.org/mpg/snap/)-HapMap. Replication genotyping for stage 3 was performed for novel SNPs with stage 2 combined P-value < 10−5. Regional association results from genome-wide meta-analysis were plotted by the LocusZoom software (http://csg.sph.umich.edu/locuszoom/) for SNPs reaching genome-wide significance from combined meta-analysis of stage 1, 2 and 3.
Principal components analysis
A list of 76,534 common SNPs across Illumina 550/610/1M and Affymetrix 5.0/6.0 were first selected. These set of SNPs on the Asian HapMap II samples CHB+JPT were then trained to generate a list of 44,524 SNPs with pairwise LD < 0.3 in a window side of 50 SNPs. Individuals from each component study and HapMap II were plotted based on the first two eigenvectors produced by PCA.
Expression quantitative trait locus (eQTL) analysis
Gene expression information in 776 adipose tissues, 667 skin tissues and 777 LCLs was obtained from the MuTHER consortium36. The eQTL data for 8 of 10 T2D loci identified in this study were filtered at MAF > 5% and INFO > 0.8 except rs16955379 that has a MAF of 1.5% in the MuTHER data set. Two loci, rs6815464 (chr. 4) and rs17797882 (chr. 16), are not included in the MuTHER data set. Association between each significant SNP for T2D and normalized mRNA expression values of genes within 1Mb of the lead SNP were performed with the GenABEL/ProbABEL package using the polygenic linear model incorporating a kinship matrix in GenABEL followed by the ProbABEL mmscore test with imputed genotypes. A multiple-testing correction was applied to the cis-association results. P-value thresholds of 5.06 × 10−5 in adipose, 3.81 × 10−5 in skin and 7.80 × 10−5 in LCL correspond to an estimated genome-wide FDR of 1%.
Gene relationships among implicated loci (GRAIL) analysis
A GRAIL analysis was performed as described previously31,33. A total of 38 genes within T2D-associated regions were selected for the analysis. Among them, 28 genes were from the previously implicated set (Supplementary Table 3), while the other 10 were from ones newly implicated in this study (Table 1). PubMed abstracts published after December, 2006 were omitted from the analysis to reduce confounding by results from T2D GWA studies.
Supplementary Material
Acknowledgments
We thank all the participants and the staff of the BioBank Japan project. The project was supported by a grant from the Leading Project of Ministry of Education, Culture, Sports, Science and Technology Japan.
CAGE: The CAGE Network Studies were supported by grants for the Program for Promotion of Fundamental Studies in Health Sciences, National Institute of Biomedical Innovation Organization (NIBIO); the Core Research for Evolutional Science and Technology (CREST) from the Japan Science Technology Agency; and the Grant of National Center for Global Health and Medicine (NCGM).
CLHNS: We thank the Office of Population Studies Foundation research and data collection teams for the Cebu Longitudinal Health and Nutrition Survey. This work was supported by National Institutes of Health grants DK078150, TW05596, HL085144, TW008288 and pilot funds from RR20649, ES10126, and DK56350.
CUHK: We acknowledge support from the Hong Kong Government Research Grants Council Central Allocation Scheme (CUHK 1/04C), Research Grants Council Earmarked Research Grant (CUHK4724/07M) and the Innovation and Technology Fund of the Government of the Hong Kong SAR (ITS/487/09FP). We acknowledge the Chinese University of Hong Kong Information Technology Services Center for support of computing resources. We would also like to thank the dedicated medical and nursing staff at the Prince of Wales Hospital Diabetes and Endocrine Centre.
KNIH: This work was supported by grants from Korea Centers for Disease Control and Prevention (4845-301, 4851-302, 4851-307) and an intramural grant from the Korea National Institute of Health (2010-N73002-00), the Republic of Korea.
NTUH: This work was supported in part by the grant (NSC99-3112-B-002-019) from the National Science Council of Taiwan. We would also like to acknowledge the National Genotyping Center of National Research Program for Genomic Medicine (NSC98-3112-B-001-037), Taiwan.
SDGS: This work was supported in part by US National Institutes of Health grants R01CA124558, R01CA64277, R01CA70867, R01CA90899, R01CA100374, R01CA118229, R01CA92585, UL1 RR024975, DK58845 and HG004399, DOD Idea Award BC050791, Vanderbilt Ingram professorship funds and Allen Foundation Fund. We would like to thank the dedicated investigators and staff members from research teams at Vanderbilt University, Shanghai Cancer Institute, and Shanghai Institute of Preventive Medicine, and most of all, the study participants for their contributions in the studies.
SDS: This work was supported by grants from National 973 Program (2011CB504001), Project of National Natural Science Foundation of China (30800617) and Shanghai Rising-Star Program (09QA1404400), China.
SJTUDS: This work was supported by grants from National 863 Program (2006AA02A409) and major program of Shanghai Municipality for Basic Research (08dj1400601), China
SNUH: This work was supported by grants from the Korea Health 21 R & D Project, Ministry of Health & Welfare (00-PJ3-PG6-GN07-001) and WCU project of the MEST and NRF(R31-2008-000-10103-0), Korea
Footnotes
AUTHOR CONTRIBUTIONS
The study was supervised by E.S.T., B.G.H., N.K., Y.S.C., Y.Y.T., W.Z., Q.C., X.O.S., Y.-T.C., J.-Y.W., L.S.A., K.L.M., T.K., C.H., W.J., L.-M.C., Y.M.C., K.S.P., J.-Y.L. and J.C. The experiments were conceived and designed by Y.S.C., E.S.T., N.K., D.P.-K.N., J.J.-M.L., M.S., T.Y.W., Y.Y.T., W.Z., F.H., X.O.S., C.-H.C., F.-J.T., Y.-T.C., J.-Y.W., L.S.A., K.L.M., S.M., C.H., L.-M.C., K.S.P., M.J.G. M.I.M. and R.M. The experiments were performed by J.L., M.S., J.L., J.-Y.W., S.M., R.Z., K.Y., Y.C., T.-J.C., L.-M.C. and S.H.K. Statistical analysis was performed by M.J.G., X.S., Y.J.K., R.T.H.O., W.T.T., Y.Y.T., F.T., J.L., C.-H.C., L.-C.C., Y.W., Y.L., K.H., C.H., Y.C., S.H.K. A.P.M. and R.M. The data were analyzed by M.J.G., X.S., Y.J.K., R.T.H.O., W.T.T., Y.Y.T., J.L., C.-H.C., L.-C.C., Y.W, N.R.L., Y.L., L.S.A., K.L.M., T.Y., C.H., Y.C., S.H.K., Y.S.C., S.K. A.K.H. and R.M. Reagents, materials and analysis tools were contributed by E.S.T., B.G.H., N.K., D.P.-K.N., J.J.-M.L., J.L., M.S., T.A., T.Y.W., E.N., M.Y., J.N., J.L., W.Z., Q.C., Y.G., W.L., F.B.H., X.O.S., F.-J.T., Y.-T.C., J.-Y.W., N.R.L., Y.L., K.O., H.I., R.T., C.W., Y.B., T.-J.C, L.-M.C., K.S.P., H.-L.K., N.H.C., J.-Y.L., W.Y.S. and J.C. The manuscript was written by Y.S.C., M.S. and E.S.T. All authors reviewed the manuscript.
References
- 1.Kang HS, et al. Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta-cell development and insulin gene expression. Mol Cell Biol. 2009;29:6366–79. doi: 10.1128/MCB.01259-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y, Chang BH, Samson SL, Li MV, Chan L. The Kruppel-like zinc finger protein Glis3 directly and indirectly activates insulin gene transcription. Nucleic Acids Res. 2009;37:2529–38. doi: 10.1093/nar/gkp122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker A, et al. Association of genetic Loci with glucose levels in childhood and adolescence: a meta-analysis of over 6,000 children. Diabetes. 2011;60:1805–12. doi: 10.2337/db10-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeuchi F, et al. Confirmation of multiple risk Loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes. 2009;58:1690–9. doi: 10.2337/db08-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barroso I, et al. Population-specific risk of type 2 diabetes conferred by HNF4A P2 promoter variants: a lesson for replication studies. Diabetes. 2008;57:3161–5. doi: 10.2337/db08-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silander K, et al. Genetic variation near the hepatocyte nuclear factor-4 alpha gene predicts susceptibility to type 2 diabetes. Diabetes. 2004;53:1141–9. doi: 10.2337/diabetes.53.4.1141. [DOI] [PubMed] [Google Scholar]
- 8.Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–7. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- 9.Tkac I. Metabolic syndrome in relationship to type 2 diabetes and atherosclerosis. Diabetes Res Clin Pract. 2005;68 (Suppl1):S2–9. doi: 10.1016/j.diabres.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Prokopenko I, McCarthy MI, Lindgren CM. Type 2 diabetes: new genes, new understanding. Trends Genet. 2008;24:613–21. doi: 10.1016/j.tig.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rung J, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet. 2009;41:1110–5. doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]
- 12.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasuda K, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40:1092–7. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 14.Unoki H, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40:1098–102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 15.Yamauchi T, et al. A genome-wide association study in the Japanese population identifies susceptibility loci for type 2 diabetes at UBE2E2 and C2CD4A-C2CD4B. Nat Genet. 42:864–8. doi: 10.1038/ng.660. [DOI] [PubMed] [Google Scholar]
- 16.Tsai FJ, et al. A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet. 2010;6:e1000847. doi: 10.1371/journal.pgen.1000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shu XO, et al. Identification of new genetic risk variants for type 2 diabetes. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stommel M, Schoenborn CA. Variations in BMI and prevalence of health risks in diverse racial and ethnic populations. Obesity (Silver Spring) 2010;18:1821–6. doi: 10.1038/oby.2009.472. [DOI] [PubMed] [Google Scholar]
- 19.Barrett JC, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–7. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadereit B, et al. Evolutionarily conserved gene family important for fat storage. Proc Natl Acad Sci U S A. 2008;105:94–9. doi: 10.1073/pnas.0708579105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakajima H, et al. Hepatocyte nuclear factor-4 alpha gene mutations in Japanese non-insulin dependent diabetes mellitus (NIDDM) patients. Res Commun Mol Pathol Pharmacol. 1996;94:327–30. [PubMed] [Google Scholar]
- 22.Johansson S, et al. Studies in 3,523 Norwegians and meta-analysis in 11,571 subjects indicate that variants in the hepatocyte nuclear factor 4 alpha (HNF4A) P2 region are associated with type 2 diabetes in Scandinavians. Diabetes. 2007;56:3112–7. doi: 10.2337/db07-0513. [DOI] [PubMed] [Google Scholar]
- 23.Girard C, et al. Genomic and functional characteristics of novel human pancreatic 2P domain K(+) channels. Biochem Biophys Res Commun. 2001;282:249–56. doi: 10.1006/bbrc.2001.4562. [DOI] [PubMed] [Google Scholar]
- 24.Ashcroft FM. ATP-sensitive potassium channelopathies: focus on insulin secretion. J Clin Invest. 2005;115:2047–58. doi: 10.1172/JCI25495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soni S, et al. Absence of erythroblast macrophage protein (Emp) leads to failure of erythroblast nuclear extrusion. J Biol Chem. 2006;281:20181–9. doi: 10.1074/jbc.M603226200. [DOI] [PubMed] [Google Scholar]
- 26.Nam D, Kim J, Kim SY, Kim S. GSA-SNP: a general approach for gene set analysis of polymorphisms. Nucleic Acids Res. 2010;38:W749–54. doi: 10.1093/nar/gkq428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–5. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luke MR, Houghton F, Perugini MA, Gleeson PA. The trans-Golgi network GRIP-domain proteins form alpha-helical homodimers. Biochem J. 2005;388:835–41. doi: 10.1042/BJ20041810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jo W, Endo M, Ishizu K, Nakamura A, Tajima T. A novel PAX4 mutation in a Japanese patient with maturity-onset diabetes of the young. Tohoku J Exp Med. 2011;223:113–8. doi: 10.1620/tjem.223.113. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, et al. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46:3553–65. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- 31.Voight BF, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–89. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raychaudhuri S, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hyndman RJ, Fan Y. Sample quantiles in statistical packages. Amer Stat. 1996;50:361–65. [Google Scholar]
- 35.Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theor Popul Biol. 2001;60:155–66. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- 36.Nica AC, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.