Abstract
Cell junctions are sites of intercellular adhesion that maintain the integrity of epithelial tissue and regulate signalling between cells. These adhesive junctions are comprised of protein complexes that serve to establish an intercellular cytoskeletal network for anchoring cells, in addition to regulating cell polarity, molecular transport and communication. The expression of cell adhesion molecules is tightly controlled and their downregulation is essential for epithelial-mesenchymal transition (EMT), a process that facilitates the generation of morphologically and functionally diverse cell types during embryogenesis. The characteristics of EMT are a loss of cell adhesion and increased cellular mobility. Hence, in addition to its normal role in development, dysregulated EMT has been linked to cancer progression and metastasis, the process whereby primary tumors migrate to invasive secondary sites in the body. This paper will review the current understanding of cell junctions and their role in cancer, with reference to the abnormal regulation of junction protein genes. The potential use of cell junction molecules as diagnostic and prognostic markers will also be discussed, as well as possible therapies for adhesive dysregulation.
Keywords: Cell junction, cell adhesion, epithelial-mesenchymal transition, EMT
INTRODUCTION
Dysregulation of cellular adhesion plays a critical role in the process of malignant transformation and metastasis [1]. Desmosomes, adherens junctions and tight junctions are sites of intercellular anchoring and signalling, comprised of a network of proteins and associated molecules that contribute to the maintenance and integrity of normal adhesion. The molecular events that underlie cell-cell connections are finely controlled and there is a capacity to adjust adhesion for physiological purposes. One such process is classical epithelial-mesenchymal transition (EMT), a natural biological phenomenon seen in embryogenesis (type I EMT) and wound healing (type II), which can become pathological and lead to cancer (type III). EMT is characterized by a tightly controlled system of gene regulation that permits a decrease in intercellular adhesion and enhances cell migration. Dysregulation of cell junction adhesion has been heavily implicated in the process of non-classical, pathological EMT, leading to oncogenic transformation and metastasis [2]. The deregulation of junction genes has been widely reported in breast, prostate, ovarian, endometrial, lung, liver and colorectal carcinomas [3-9] (Table 1). However, this list is certainly not exhaustive and the exploration of links between cell adhesion, gene deregulation and cancer persists.
Table 1.
Cell junction genes and cancer
| Cancer | Upregulated genes | Downregulated genes | References |
|---|---|---|---|
| Breast | SNAI1, SIP1, CLDN4 | OCLN, CDH1, CLDN1,2,7 | [6, 30, 33, 44-48] |
| Colorectal | CLDN1, CTNNB, | CDH1 | [5] |
| Endometrial | OCLN | [7] | |
| Liver | CLDN1, PKP2 | TJP1, OCLN, PKP1,3 | [5, 9, 13] |
| Lung | CLDN1-5, 7 | DSP, PKP3CLDN2 | [3, 11, 39] |
| Oral | CDH1, KRT8 | [25, 49] | |
| Ovarian | SNAI1, SIP1, SNAI2 | CDH1, CTNNB, OCLN, TJP1, DSG2 | [8, 30] |
| Pancreatic | SNAI1 | CDH1 | [29] |
| Prostate | PKP3, PKP1 | PKP3, PKP2, CTNNA, CTNNB, CTNND, CDH1 | [13, 32, 33, 36, 38] |
This table lists some of the many examples where cell junction genes are deregulated in human cancer and highlights the diversity of disease that is associated with dysregulated cell adhesion.
This paper will examine the molecular components and mechanisms of adherens junctions, desmosomes and tight junctions, and how their normal regulation can be altered, resulting in tumor transformation, invasion and ultimately, metastasis. EMT plays an integral role in the junction dysregulation associated with neoplastic progression, and thus provides a foundation for the understanding of cell adhesion and cancer spread. Conventional therapies for targeting transformation and metastasis are by no means comprehensive, and a clearer understanding of the molecular events underlying EMT and the dysregulation of cellular adhesion, will elucidate novel therapeutic approaches. In addition to this, a more holistic perception of the genetic deregulation associated with abnormal cell adhesion, should result in the development of more effective clinical diagnostic and prognostic markers for cancer treatment.
1. Cell adhesion junctions
1.1 Desmosomes
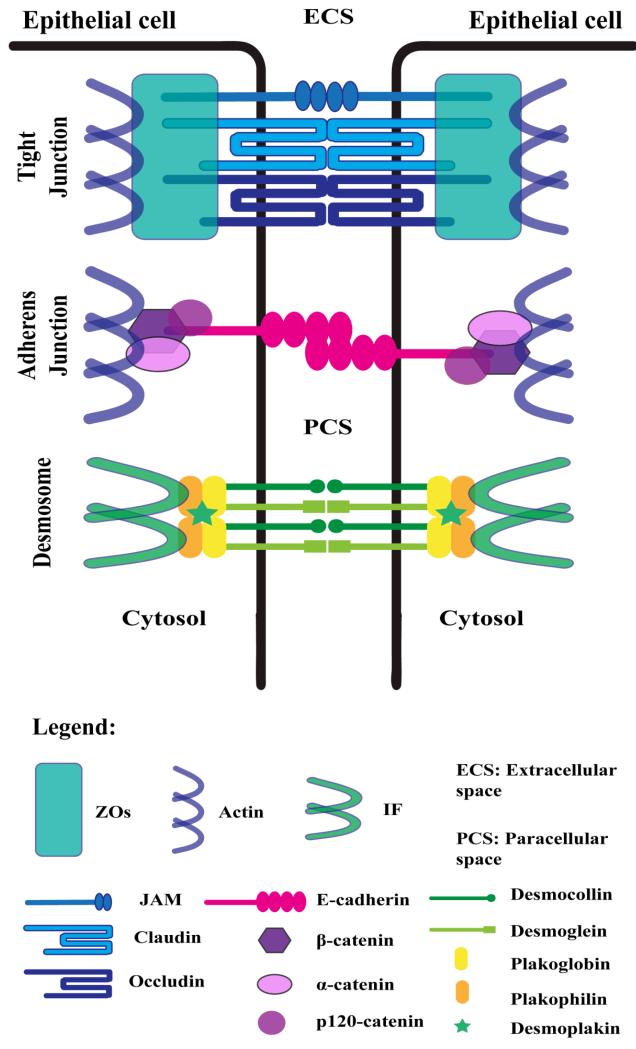
Desmosomes are a type of anchoring junction responsible for establishing an intercellular adhesive framework between the cytoskeleton and plasma membrane [10, 11] (Figure 1). A potential role of desmosomes in intercellular signalling pathways and proliferation has also been reported [12]. Desmosomes provide mechanical integrity between cells by anchoring the intermediate filaments of the cytoskeleton via a complex of proteins in the cytoplasmic and extracellular regions of the junction. Three main classes of proteins interact to bridge the intercellular space and anchor the keratin cytoskeletons of apposing cells – the cadherins, armadillo proteins and plakins. Desmocollin (DSC1-3) and desmoglein (DSG1-4) are cadherin proteins which span the transmembrane space as heterodimers, providing a hyperadhesive link between neighboring cells [10]. This provides the junction with a strong resistance to adhesive disruption and mechanical stress. In the outer dense plaque region of the cell, desmoglein and desmocollin associate with armadillo proteins. The armadillo proteins plakophilin (PKP1-4) and plakoglobin (JUP), characterized by their 41 amino acid ‘armadillo’ tandem repeats, are responsible for mediating the interaction between the transmembrane cadherins and the cytolinker plakin protein, desmoplakin (DSP) [13]. Desmoplakin facilitates the link between the junction plaque complex and the intermediate filament cytoskeleton in the inner dense plaque region of the junctional complex [14]. It has been suggested that desmosomal genes are regulated by a coordinated transcriptional program [15], directed by transcription factors prominent in the EMT process - Snail, Slug and Twist [16].
Figure 1.
Lateral Cell Junctions
Shown is a representation of the molecular components present in tight junctions, adherens junctions and desmosomes. ZOs, zonula occluden proteins; IFs, cytoskeletal intermediate filaments; JAM, junctional adhesion molecule.
1.2 Adherens Junctions
Adherens junctions are sites of lateral cell-cell adhesion, playing a similar anchoring role to desmosomes (Figure 1). As in the desmosome, three primary protein families constitute the adherens junction, cadherins, armadillo proteins, and plakins [17]. E-cadherin (CDH1) is the major transmembrane protein responsible for intercellular adhesion at the junction site, operating in a calcium-dependent, homophilic fashion [18]. Other members of the cadherin family present in cell junctions include the desmosomal proteins desmoglein and desmocollin, and N-cadherin, typically associated with epithelial cell adherens junctions in an EMT setting. Armadillo proteins α- and β-catenin (CTNNA1-3, CTNNB1) facilitate the interaction between transmembrane E-cadherin and the actin cytoskeleton, in conjunction with the cytolinker protein p120-catenin (CTTND1-2) [17]. Hyperadhesion at the junction is achieved via E-cadherin clustering, mediated by p120-catenin [18]. In addition to the anchoring role played by adherens junctions, regulation of the actin cytoskeleton, intracellular signalling and transcriptional processes also typify the biological role of these cell-cell junctions. For this reason, dysregulation of the junction system has particular implications in transformation and tumor invasion [17, 19].
1.3 Tight Junctions
Tight junctions are a type of intercellular occluding connection located within the apico-basal region of epithelial cell membranes (Figure 1) [20]. The tight junction is a multifunctional complex required as a barrier for regulating paracellular permeability and establishing cell polarity, in addition to their adhesive function [3]. The junction complex is comprised of transmembrane proteins occludin (OCLN) and claudin (CLDN1-25), in association with junctional adhesion molecules (JAM2-3) and tricellulin (MARVELD2), which interact with a nexus of accessory proteins and the actin cytoskeleton [3]. Accessory proteins, such as zonula occluden (ZO) proteins (TJP1-3) are involved in associating the transmembrane proteins with the actin cytoskeleton and in the regulation of signalling [19]. Claudins and occludins are multi-span transmembrane proteins which serve to control the apical-basolateral permeability of ions across the junction – acting as molecular barriers and gateways [21]. The multifunctional nature of tight junctions includes their role in cell communication pathways, via regulation of signalling cascades and transcription factors. Studies have reported roles for claudin-1 and claudin-11 in cell proliferation, while various accessory proteins (ZO-1,2,3, MUPP1, MAGI-1) have been related to regulatory suppression of cell proliferation and oncogenesis through oncogene inactivation [22, 23]. For instance, ZO-1 is known to reduce cell proliferation by reducing the nuclear accumulation of cell division kinase-4 (CDK4) depending on cellular density [24]. The diverse functionality of tight junctions is evident in their roles in adhesion, polarization and as a barrier; however, the exact mechanisms of their regulation of signalling pathways remain to be fully elucidated.
2. Epithelial-mesenchymal transition (EMT)
The epithelial-mesenchymal transition (EMT) is a biological process involved in embryogenesis and wound healing, characterized by a change in cell-cell adhesion gene expression and a subsequent loss of adhesion, change in cellular polarity and shape, and increased mobility [2]. EMT is regarded as a reversible process, with mesenchymal-epithelial transition (MET) being an integral complement to the EMT program in embryogenesis and wound healing [25]. A diverse profile of differential gene expression is evident in EMT, but the hallmark of transition seems to be the shift from E-cadherin to N-cadherin expression, termed ‘cadherin switching’ [26, 27]. Major factors and regulators in the EMT process include TGF-β and the Snail, Slug and Twist transcription factors [2, 26]. These initiate the shift in gene expression from an MET to an EMT-preferential state. EMT has been divided into three main types [26]. Type I accounts for EMT in an embryogenesis setting, involved in gastrulation and migration in particular of the neural crest cells, [28]. Type II EMT is observed in the wound healing and tissue regeneration process in adults [26]. Both types I and II EMT are regarded as classical EMT programs, which serve a physiological purpose in the body.
On the other hand, EMT has also been shown to be integral to the progression and metastasis of tumors – this pathological program of EMT is classified as type III. The scientific literature from the past decade has elucidated the importance of cell junctions and their components in the shift to an EMT state [2, 26, 27, 29]. Intercellular adhesion can resist the action of EMT by preventing epithelial cells from losing polarity and shape, thus maintaining the integrity of the epithelium within tissue [6]. This reduces the invasive potential of cells, with the added function of blocking tumor metastasis from underlying tissue to secondary sites in the body. Deregulation of junction genes can lead to EMT, leading to a loss of adhesion and apico-basal polarity in the epithelium [2, 29]. Ectopic expression of major transcription factors involved in EMT (Snail, Slug, Twist) results in a downregulation of desmosome, adherens and tight junctions genes, generating highly mobile and migratory cells with a high invasive potential [30].
In summary, EMT plays a vital role in the physiological processes of embryogenesis, wound healing and tissue regeneration, relying upon a loss of cell adhesion, change in cellular shape, polarity disruption and cell migration to play out such development. However, via the same processes, EMT can become pathological, underlying the process of oncogenesis and tumor metastasis.
3. Adhesion dysregulation and cancer
3.1 Type III EMT and cadherin switching
Pathological deregulation of junction genes is integral to the dysregulation of cell junctions as adhesive and signalling structures, which can ultimately lead to cancer. Non-classical (Type III) EMT has been identified as an important mechanism underlying neoplastic progression and the process of tumor metastasis from primary to secondary sites in the body [26]. The role of intercellular integrity and adhesion in non-classical EMT is hence the subject of widespread study.
Transforming growth factor beta (TGF-β), a master regulator of EMT, drives the pathways that activate the major transcriptional repressors of junction genes, Snail, Slug, Twist and zinc finger E-box binding homeobox 1-3 (ZEB1-3) [26]. Aberrant expression of these transcription factors results in the deregulation of many genes encoding components of desmosomes, adherens and tight junctions [8, 30]. As mentioned above, the hallmark of EMT is the shift in expression from E-cadherin to N-cadherin at the lateral adherens junctions. Much research has been centered on elucidating the regulatory and signalling mechanisms associated with this shift in expression and its role in oncogenesis, transformation and ultimately, metastasis [4, 25-27, 31, 32].
The switch from expression of the invasion suppressor protein, E-cadherin, to the EMT biomarker N-cadherin, has been linked to aberrant DNA methylation of the CDH1 gene promoter in breast and prostate carcinoma cells. Indeed, hypermethylation of CpG islands in the CDH1 promoter region has been put forward as the primary epigenetic mechanism underlying the downregulation of E-cadherin, and subsequent initiation of the EMT program [33, 34]. Another explanation for a loss of E-cadherin occurs at the post-transcriptional level. A di-leucine motif at the membrane-proximal region of E-cadherin is bound and stabilized by the junction adaptor protein, p120-catenin [35]. Knocking down p120-catenin results in abnormal regulation of cadherin turnover, resulting in internal degradation of E-cadherin, thus removing its capacity as a suppressor of tumor invasion [31].
Active transcriptional repression is another major epigenetic mechanism fundamental to the down-regulation of key adhesion genes in EMT. Collective repression of adherens junction components (E-cadherin, α-catenin, β-catenin and p120-catenin) in prostatic adenocarcinomas points towards a coordinated regulation of transcription of junction genes [32]. In prostatic, ovarian, oral and renal cell cancers, reduced expression of α- and β-catenin has been directly associated with a poor clinical outcome and shortened patient survival [36]. In contrast, in small cell lung cancer, overexpression of β-catenin has been linked to unfavorable prognosis, perhaps due to nuclear accumulation and subsequent activation of the pro-metastatic target gene MMP14 via the Wnt signalling pathway [36, 37]. This suggests that deregulation of adherens junction genes may produce different outcomes depending upon the type and stage of cancer in the body. Transcriptional repression of E-cadherin by a Snail/HDAC1/HDAC2 repressor complex has been reported in pancreatic cancer samples, where Von Burstin et al. [29] demonstrated considerable up-regulation of Snail in metastatic pancreatic cancer sublines and binding of Snail to the CDH1 gene promoter during EMT. In addition to the epigenetic alterations leading to aberrant E-cadherin levels and EMT, genetic factors have been reported to predispose some individuals to E-cadherin inactivation in certain types of cancer. Genetic mutations of the CDH1 gene have been recorded with moderate frequency in diffuse gastric cancer, lobular breast cancer and synovial sarcomas, but here a role in early tumor development rather than invasion and metastasis has been purported [34].
3.2 Desmosomes and Cancer
Dysregulation of desmosomal components is fundamental to the cellular transition from an epithelial to a mesenchymal nature, and consequently, is critical to the metastatic potential of cells. As such, an inverse relationship between desmosomal integrity and tumor invasion has been proposed, where the loss of desmosomal adhesion and size due to reduced desmoplakin is associated with an invasive phenotype [15]. Several desmosomal proteins, namely the plakophilins and desmoplakin, have been implicated in cell proliferation, transformation and invasion of tumors [11, 13, 15, 37]. Desmoplakin has been identified as a tumor suppressor due to its inhibition of the Wnt signalling pathway and β-catenin expression. Similarly, decreased TCF/LEF-dependent transcription resulting from plakoglobin up-regulation has been implicated in non-small cell lung cancer [37]. Although the association of desmosomal adhesion with E-cadherin regulation is yet to be fully elucidated, it is clear that it is integral to maintaining a physiological state that is non-conducive to epithelial tumor invasion and metastasis.
Plakophilins, the armadillo proteins involved in desmosomal assembly and adhesion, have also been implicated in neoplastic progression and metastasis of tumor cells [11, 13, 38-40]. Downregulation of PKP3 by RNAi in epithelial cells lines yielded three notable hallmarks of oncogenic transformation and invasion – decreased desmosomal size and cell-cell adhesion, and increased cell migration [11]. In nude mice, this manifested itself through increased lung and skin tumor formation, consistent with the high rate of migration in vitro. In a follow up study [40], it was observed that increased cell migration due to a loss of PKP3 in HCT116 colorectal carcinoma cells was associated with an increase in the levels of the intermediate filament protein keratin 8 (KRT8). Desmosomal armadillo proteins also play a role in β-catenin signalling, through competition with E-cadherin. Chen et al. [41] demonstrated an association between plakophilin 2 and β-catenin, resulting in up-regulation of β-catenin signalling, associated with neoplastic progression in various tumor types [5, 36, 37].
3.3 Tight Junctions and Cancer
Dysregulation of tight junction proteins and their associated adhesion factors has also been strongly implicated in the transformation and invasion of tumors. Several studies have reported that deregulation of the transmembrane adhesion protein occludin is evident with increased progression and metastatic potential in breast, liver, endometrial and ovarian cancer [7-9]. This observation was initially thought to be more of a characteristic of tumor progression rather than an underlying cause. However, in 2004 Tobioka et al. [7] suggested that deregulated expression of occludin is directly involved in structural atypia associated with loss of tight junction adhesion, manifesting itself through increased carcinoma grade and malignancy potential in human endometrial cancers (HEC). The importance of occludin and tight junction integrity to breast cancer development was demonstrated when it was found that occludin expression was significantly decreased in metastatic breast cancer [6]. N- and C-terminal truncations of occludin were identified in these tumors, resulting in poor intercellular barrier and adhesive function, in addition to weaker interaction with accessory proteins, such as ZO-1, and the cytoskeleton [6].
The tight junction transmembrane protein claudin has also been associated with cancer. Transcription factors integral to the EMT process such as Slug, Snail, Twist and ZEB1-2 have been implicated in the deregulation of different claudins [42, 43], which show differential expression in a host of tumor types. For instance, in breast carcinomas, decreased expression of CLDN1, 2 and 7 has been related to greater tumor malignancy [44-47] whereas a link has been drawn between an increase in CLDN4 and breast cancer aggressiveness [48]. The ubiquitous differential expression of claudins in cancers regardless of tissue type reinforces the crucial role of tight junctions in tumor transformation and metastasis.
4. Diagnosis, prognosis and therapies
Dysregulation of adhesion plays an essential part in the transformation and metastasis of tumors in the body. The changes in gene expression associated with EMT and cancer can serve as therapeutic targets that may prevent or reverse neoplastic progression and transformation. Furthermore, these changes can be used as biomarkers, thus various deregulated junction genes and pathways have been identified as potential targets for therapeutic, prognostic and diagnostic use.
4.1 Desmosomal therapies
Desmosome components have more recently become the subject of therapeutic enquiry. One potential target for preventative therapy is plakophilin-3, as loss of this protein is associated with keratin stabilization and increased cell migration, and in epithelial cells, increased expression is connected to a reduction in transformation and metastasis [40]. However, in lung cancer, high expression levels of plakophilin-3 are associated with tumor progression, metastasis and poor survival, highlighting the challenge in therapeutic targeting of junction components in different tissues [39]. Desmoplakin is important in linking the cytoskeleton and desmosomal cadherins, and has recently been found to increase sensitivity to anti-cancer drugs targeting apoptosis, creating a major subject of interest for therapeutic intervention [37]. PERP (p53 apoptosis effector related to PMP-22) is involved in desmosomal assembly and has been connected to tumor suppression downstream of the p53 and p63 response, hailing it as a potential target for desmosomal adhesion therapy in cancer [14]. Diagnostics and prognostics could be improved via the use of desmosomal components such as desmoplakin, PERP and plakophilins as biomarkers in dysregulated adhesion and cancer [17]. Similarly, identifying post-translational modifications, such as phosphorylation of keratins, may have a future application in metastasis prediction [40, 49].
4.2 Targeting EMT and the cadherin switch
The switch from E- to N-cadherin during EMT in adherens junctions holds considerable potential as a therapeutic target and diagnostic tool in different tumor types [4, 26, 27, 33]. Epigenetic regulation of the CDH1 gene by hypermethylation leads to the switch to N-cadherin, characterising EMT. Administration of the demethylating agent AzaC (5-aza-2′-deoxycytidine) has been shown to partially restore normal CDH1 promoter methylation patterns in breast cancer [33], indicating that the transcriptional machinery for CDH1 expression is still present in spite of gene silencing. Targeting the epigenetic methylation of the CDH1 promoter holds a substantial degree of potential in clinical therapy as well as in diagnostic and prognostic marking for cancer patients. In the same vein, targeting N-cadherin to reverse the phenomenon of EMT for anti-tumor therapy is another option [50]. Inhibition of Akt signalling activity by PIA (phosphatidylinositol ether lipid analogues) has been found to induce mesenchymal-epithelial reverting transition (MErT), which is characterized by a decrease in expression of EMT-associated factors such as N-cadherin, vimentin, Snail and Twist, while restoring E-cadherin expression [25]. Other adherens junction components such as β-catenin can serve as positive markers for disease progression in prostate cancer, where high expression is directly linked with advanced stage, metastatic carcinoma grades [36]. On the other hand though, low expression of β-catenin has been associated with a poor clinical outcome in oral, ovarian and renal cancer patients, complicating its predictive use as an accurate biomarker [36]. Further studies are clearly imperative and a more holistic understanding of the intricacies which underlie the dysregulation of cellular adhesion will inform more effective therapeutic, diagnostic and prognostic targeting of cancer.
4.3 Tight junction therapies
The claudins are a diverse protein family crucial to the integrity of tight junctions. Their deregulation in a cancer setting has been widely reported [5, 43-48]. The identification of claudin-1 as a mediator in early colorectal cancer transformation has highlighted it as a potential target for therapeutic intervention, and as a biomarker in colonic adenocarcinomas [5]. Targeting upstream transcription factors such as Snail and Slug (which are known to repress claudin-1 in epithelial cells) provides a therapeutic option for intervention in claudin-1 deregulated cancers [42]. A splice variant of claudin-18 has also been put forward as a target in therapeutic antibody development [51]. Further, modifying the regulation of occludin by reversing its repression could prevent or reverse the loss of tight junction integrity associated with cell migration and invasiveness [6].
CONCLUSIONS
Deregulation of the genes involved in cellular adhesion plays a pivotal role in the transformation and invasion of tumors. The molecular events involved in EMT underlie this process, facilitating the neoplastic progression associated with oncogenesis. Targeting the molecules and pathways which underpin these processes presents a plethora of opportunities for therapeutic treatment, diagnosis and prognosis of various types of cancer. A more comprehensive understanding of the mechanisms and interactions implicated in the dysregulation of cell junctions and their relevance to cancer will promote the development of more effective clinical approaches to cancer treatment.
ACKNOWLEDGEMENTS
This work was supported by funding from the Australian National Health and Medical Research Council, the Australian Research Council, and the National Institutes of Health.
REFERENCES
- 1.Morris MA, Young LS, Dawson CW. Eur. J. Cell Biol. 2008;87:677. doi: 10.1016/j.ejcb.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Talbot LJ, Bhattacharya SD, Kuo PC. Int. J. Biochem. Mol. Biol. 2012;3:117. [PMC free article] [PubMed] [Google Scholar]
- 3.Soini Y. Int. J. Clin. Exp. Pathol. 2012;5:126. [PMC free article] [PubMed] [Google Scholar]
- 4.Jaggi M, Johansson SL, Baker JJ, Smith LM, Galich A, Balaji KC. Urol. Oncol. 2005;23:402. doi: 10.1016/j.urolonc.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 5.Kinugasa T, Akagi Y, Ochi T, Tanaka N, Kawahara A, Ishibashi Y, Gotanda Y, Yamaguchi K, Shiratuchi I, Oka Y, Kage M, Shirouzu K. Anticancer Res. 2012;32:2309. [PubMed] [Google Scholar]
- 6.Martin TA, Mansel RE, Jiang WG. Int. J. Mol. Med. 2010;26:723. doi: 10.3892/ijmm_00000519. [DOI] [PubMed] [Google Scholar]
- 7.Tobioka H, Isomura H, Kokai Y, Tokunaga Y, Yamaguchi J, Sawada N. Hum. Pathol. 2004;35:159. doi: 10.1016/j.humpath.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Kurrey NK, K A, Bapat SA. Gynecol. Oncol. 2005;97:155. doi: 10.1016/j.ygyno.2004.12.043. [DOI] [PubMed] [Google Scholar]
- 9.Orban E, Szabo E, Lotz G, Kupcsulik P, Paska C, Schaff Z, Kiss A. Pathol. Oncol. Res. 2008;14:299. doi: 10.1007/s12253-008-9031-2. [DOI] [PubMed] [Google Scholar]
- 10.Garrod D, Chidgey M. Biochim. Biophys. Acta. 2008;1778:572. doi: 10.1016/j.bbamem.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Kundu ST, Gosavi P, Khapare N, Patel R, Hosing AS, Maru GB, Ingle A, Decaprio JA, Dalal SN. Int. J. Cancer. 2008;123:2303. doi: 10.1002/ijc.23797. [DOI] [PubMed] [Google Scholar]
- 12.Yin T, Green KJ. Semin. Cell Dev. Biol. 2004;15:665. doi: 10.1016/j.semcdb.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Schwarz J, Ayim A, Schmidt A, Jager S, Koch S, Baumann R, Dunne AA, Moll R. Hum. Pathol. 2006;37:613. doi: 10.1016/j.humpath.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Brooke MA, Nitoiu D, Kelsell DP. J. Pathol. 2012;226:158. doi: 10.1002/path.3027. [DOI] [PubMed] [Google Scholar]
- 15.Chun MG, Hanahan D. PLoS Genet. 2010;6:e1001120. doi: 10.1371/journal.pgen.1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peinado H, Olmeda D, Cano A. Nat. Rev. Cancer. 2007;7:415. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 17.Dusek RL, Attardi LD. Nat. Rev. Cancer. 2011;11:317. doi: 10.1038/nrc3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menke A, Giehl K. Arch. Biochem. Biophys. 2012;524:48. doi: 10.1016/j.abb.2012.04.019. [DOI] [PubMed] [Google Scholar]
- 19.Hartsock A, Nelson WJ. Biochim. Biophys. Acta. 2008;1778:660. doi: 10.1016/j.bbamem.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steed E, Balda MS, Matter K. Trends Cell Biol. 2010;20:142. doi: 10.1016/j.tcb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Chiba H, Osanai M, Murata M, Kojima T, Sawada N. Biochim. Biophys. Acta. 2008;1778:588. doi: 10.1016/j.bbamem.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 22.Aijaz S, Balda MS, Matter K. Int. Rev. Cytol. 2006;248:261. doi: 10.1016/S0074-7696(06)48005-0. [DOI] [PubMed] [Google Scholar]
- 23.Matter K, Aijaz S, Tsapara A, Balda MS. Curr. Opin. Cell Biol. 2005;17:453. doi: 10.1016/j.ceb.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Paris L, Tonutti L, Vannini C, Bazzoni G. Biochim. Biophys. Acta. 2008;1778:646. doi: 10.1016/j.bbamem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Hong KO, Kim JH, Hong JS, Yoon HJ, Lee JI, Hong SP, Hong SD. J. Exp. Clin. Cancer Res. 2009;28:28. doi: 10.1186/1756-9966-28-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krisanaprakornkit S, Iamaroon A. ISRN Oncol. 2012;2012:681469. doi: 10.5402/2012/681469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gravdal K, Halvorsen OJ, Haukaas SA, Akslen LA. Clin. Cancer Res. 2007;13:7003. doi: 10.1158/1078-0432.CCR-07-1263. [DOI] [PubMed] [Google Scholar]
- 28.Lee JM, Dedhar S, Kalluri R, Thompson EW. J. Cell Biol. 2006;172:973. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Burstin J, Eser S, Paul MC, Seidler B, Brandl M, Messer M, von Werder A, Schmidt A, Mages J, Pagel P, Schnieke A, Schmid RM, Schneider G, Saur D. Gastroenterology. 2009;137:361. doi: 10.1053/j.gastro.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Elloul S, Elstrand MB, Nesland JM, Trope CG, Kvalheim G, Goldberg I, Reich R, Davidson B. Cancer. 2005;103:1631. doi: 10.1002/cncr.20946. [DOI] [PubMed] [Google Scholar]
- 31.Davis MA, Ireton RC, Reynolds AB. J. Cell Biol. 2003;163:525. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kallakury BV, Sheehan CE, Ross JS. Hum. Pathol. 2001;32:849. doi: 10.1053/hupa.2001.26463. [DOI] [PubMed] [Google Scholar]
- 33.Graff JR, Herman JG, Lapidus RG, Chopra H, Xu R, Jarrard DF, Isaacs WB, Pitha PM, Davidson NE, Baylin SB. Cancer Res. 1995;55:5195. [PubMed] [Google Scholar]
- 34.Strathdee G. Semin. Cancer Biol. 2002;12:373. doi: 10.1016/s1044-579x(02)00057-3. [DOI] [PubMed] [Google Scholar]
- 35.Kawauchi T. Int. J. Mol. Sci. 2012;13:4564. doi: 10.3390/ijms13044564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aaltomaa S, Karja V, Lipponen P, Isotalo T, Kankkunen JP, Talja M, Mokka R. Anticancer Res. 2005;25:4707. [PubMed] [Google Scholar]
- 37.Yang L, Chen Y, Cui T, Knosel T, Zhang Q, Albring K, Huber O, Petersen I. Carcinogenesis. 2012 doi: 10.1093/carcin/bgs226. Epub 2012/07/14. [DOI] [PubMed] [Google Scholar]
- 38.Breuninger S, Reidenbach S, Sauer CG, Strobel P, Pfitzenmaier J, Trojan L, Hofmann I. Am. J. Pathol. 2010;176:2509. doi: 10.2353/ajpath.2010.090737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furukawa C, Daigo Y, Ishikawa N, Kato T, Ito T, Tsuchiya E, Sone S, Nakamura Y. Cancer Res. 2005;65:7102. doi: 10.1158/0008-5472.CAN-04-1877. [DOI] [PubMed] [Google Scholar]
- 40.Khapare N, Kundu ST, Sehgal L, Sawant M, Priya R, Gosavi P, Gupta N, Alam H, Karkhanis M, Naik N, Vaidya MM, Dalal SN. PLoS One. 2012;7:e38561. doi: 10.1371/journal.pone.0038561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X, Bonne S, Hatzfeld M, van Roy F, Green KJ. J. Biol. Chem. 2002;277:10512. doi: 10.1074/jbc.M108765200. [DOI] [PubMed] [Google Scholar]
- 42.Martinez-Estrada OM, Culleres A, Soriano FX, Peinado H, Bolos V, Martinez FO, Reina M, Cano A, Fabre M, Vilaro S. Biochem. J. 2006;394:449. doi: 10.1042/BJ20050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ikenouchi J, Matsuda M, Furuse M, Tsukita S. J. Cell Sci. 2003;116:1959. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- 44.Kim TH, Huh JH, Lee S, Kang H, Kim GI, An HJ. Histopathology. 2008;53:48. doi: 10.1111/j.1365-2559.2008.03052.x. [DOI] [PubMed] [Google Scholar]
- 45.Morohashi S, Kusumi T, Sato F, Odagiri H, Chiba H, Yoshihara S, Hakamada K, Sasaki M, Kijima H. Int. J. Mol. Med. 2007;20:139. [PubMed] [Google Scholar]
- 46.Tokes AM, Kulka J, Paku S, Szik A, Paska C, Novak PK, Szilak L, Kiss A, Bogi K, Schaff Z. Breast Cancer Res. 2005;7:R296. doi: 10.1186/bcr983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kominsky SL, Argani P, Korz D, Evron E, Raman V, Garrett E, Rein A, Sauter G, Kallioniemi OP, Sukumar S. Oncogene. 2003;22:2021. doi: 10.1038/sj.onc.1206199. [DOI] [PubMed] [Google Scholar]
- 48.Lanigan F, McKiernan E, Brennan DJ, Hegarty S, Millikan RC, McBryan J, Jirstrom K, Landberg G, Martin F, Duffy MJ, Gallagher WM. Int. J. Cancer. 2009;124:2088. doi: 10.1002/ijc.24159. [DOI] [PubMed] [Google Scholar]
- 49.Alam H, Gangadaran P, Bhate AV, Chaukar DA, Sawant SS, Tiwari R, Bobade J, Kannan S, D’Cruz A,K, Kane S, Vaidya MM. PLoS One. 2011;6:e27767. doi: 10.1371/journal.pone.0027767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mariotti A, Perotti A, Sessa C, Ruegg C. Expert Opin. Investig. Drugs. 2007;16:451. doi: 10.1517/13543784.16.4.451. [DOI] [PubMed] [Google Scholar]
- 51.Sahin U, Koslowski M, Dhaene K, Usener D, Brandenburg G, Seitz G, Huber C, Tureci O. Clin. Cancer Res. 2008;14:7624. doi: 10.1158/1078-0432.CCR-08-1547. [DOI] [PubMed] [Google Scholar]