Abstract
Cationic shell crosslinked knedel-like nanoparticles (cSCKs) have emerged as a highly efficient transfection agent for nucleic acids delivery. In this study, a new class of cSCKs with tunable buffering capacities has been developed by altering the amounts of histamines and primary amines incorporated into their crosslinked shell regions. The effect of histamine content of these nanoparticles with a hydrodynamic diameter of ca. 20 nm, on the siRNA-binding affinity, cytotoxicity, immunogenicity, and transfection efficiency was investigated. The modification of cSCKs with histamine was found to reduce the siRNA-binding affinity and cellular binding. On the other hand, it significantly reduced the toxicity and immunogenicity of the nanoparticles with subsequent increase in the transfection efficiency. In addition, escape from endosomes was facilitated by having two species of low and high pKas (i.e. histamine and primary amine groups, respectively), as demonstrated by the potentiometric titration experiments and the effect of bafilomycin A1, an inhibitor of the endosomal acidification, on the transfection efficiency of cSCKs. Histamine modification of 15 mol% was a threshold, above which cSCKs with higher histamine content completely lost the ability to bind siRNA and to transfect cells. This study highlights the potential of histamine incorporation to augment the gene silencing activity of cationic nanoparticles, reduce their toxicity, and increase their biocompatibility, which is of particular importance in the design of nucleic acids delivery vectors.
Keywords: Cationic nanoparticles, siRNA delivery, Histamine, Endosomal escape, Transfection, Immunotoxicity
1. Introduction
In recent years, the use of small interfering RNAs (siRNAs) has gained much attention as therapeutics for the treatment of various diseases, due to their ability to selectively target and cleave complementary mRNA sequences for efficient gene silencing [1–4]. siRNA is composed of double-stranded RNA molecules that are ca. 20–25 nucleotides long with inherent negative charge and macromolecular size. The success of siRNA-based therapeutics is largely dependent on the efficient delivery of siRNA into the cytoplasm of the target cells while inducing minimal toxicity. Examples of the hurdles towards the proficient delivery of siRNA include extracellular and intracellular enzymatic degradation, limited cellular uptake, entrapment into endocytic vesicles that form upon the endocytosis of the siRNA, and limited release into the cytoplasm of the transfected cells [1,5,6]. Internalization of siRNA via endocytic pathways usually ends into acidified endosomes and later into lysosomes, where enzymatic degradation can occur, in addition to hindering the siRNA from reaching their target sites into the cytoplasm [7,8]. One way to circumvent these challenges is to complex the negatively-charged siRNA with positively-charged lipids/polymers, where electrostatic interactions between siRNA and cationic carriers result in the formation of positively-charged nanocomplexes, which partially protect the siRNA against degradation and enhance cellular uptake [9–12]. In addition, several types of membrane disruptive polymers and imidazole-containing polymers have been investigated to facilitate escape from the endosomes and achieve efficient transportation of genetic materials into the cytosol [13–18]. In particular, imidazole-containing polymers have gained much attention recently, as they exhibit pKa ~6 and can undergo protonation in the acidic endosomal environment leading, eventually, to efficient endosomal disruption [17,18].
An intricate system that can electrostatically bind to nucleic acids, mediate cellular entry of the nanocomplexes and trigger mechanisms for endosomal lysis, either by interaction with the endosomal membrane or via the proton sponge effect, partially fulfills the design requirements for the cytoplasmic delivery of nucleic acids [15,19,20]. Some of these systems depend on incorporating two functional groups of low and high pKas, for example, poly(l-Lysine) as a segment with high pKa and poly[(3-morpholinopropyl) aspartamide] as a low-pKa polymer. The high-pKa polymer interacts electrostatically with the nucleic acid at physiological pH, whereas the low-pKa polymer remains mostly non-ionized at this pH. Protonation of the low-pKa polymer can buffer the endosomes and aid in pumping protons concurrently with chloride ions (i.e. endosomal burst via the proton sponge effect) [21] or the additional positive charges (unbound to DNA/RNA) provide new sites for electrostatic interactions with the endosomal membrane. Both effects may enhance the ability of the vector to escape from the endosomes and release their cargoes into the cytoplasm [22,23].
Our group has a long standing interest in designing polymeric systems and chemistries to engineer synthetic nanostructures, known as shell crosslinked knedel-like nanoparticles (SCKs) that have exhibited promise for a diverse set of biomedical applications, including nucleic acids delivery, due to their versatility in size, shape, and morphology, along with a synthetic approach that allows for ease of chemical modification, orthogonal functionalization and multivalency [7,24–31]. SCKs are core–shell nanoparticles composed of amphiphilic block copolymers that self assemble in water, followed by selective crosslinking throughout adjacent chains in the hydrophilic shell layer, which allows SCKs to possess structural and kinetic stabilities. Cationic SCKs (cSCKs), with poly(acrylamidoethylamine) (PAEA) comprising the shells for electrostatic interaction with negatively-charged nucleic acids, have emerged as a promising class of nanomaterials for highly efficient transfection of nucleic acids of various structures [32–34]. In this study, a new class of cSCKs was designed by incorporating various amounts of histamines and primary amines (as low- and high-pKa functionalities) into their shells to achieve enhanced cell transfection with reduced toxicity, by absorbing protons at pH values between the physiological and endosomal pH range, resulting in disruption of endosomal membranes. These cSCKs were electrostatically complexed with siRNA and investigated for their cellular uptake, cytotoxicity, immunogenicity and endosomal escape in RAW 264.7 mouse macrophages and OVCAR-3 cells.
2. Materials and methods
2.1. Materials
Polymerizations were performed on a double manifold with glassware and syringes thatweredriedinanoven(100 °C) for at least 1 h, and with syringes that were washed with N2 (3×), prior to use. tert-Butyl acrylate (t-BA, 99%, Aldrich), styrene (99%, Aldrich), toluene (anhydrous, 99.8%, Aldrich), N,N,N′,N″,N″-pentamethyldiethylenetriamine (PMDETA, 99% Aldrich), copper (I) bromide (CuBr, 99.99%, Aldrich), ethyl 2-bromo propionate (99%, Aldrich), trifluoroacetic acid (TFA, 95%; Aldrich), 1-(3′-dimethylaminopropyl)-3-ethylcarbodiimide methiodide (EDCI, 98%, Aldrich), hydroxybenzotriazole (HOBT, Aldrich), 2–(1H–benzotriazol–1–yl)-1,1,3,3-tetramethyluronium hexafluoro phosphate (HBTU, Aldrich), N,N-diisopropylethylamine (DIPEA, 99.5%, Aldrich). The amphiphilic block copolymer of poly(acrylic acid)160-block-polystyrene30 (PAA160-b-PS30) was prepared using atom transfer radical polymerization (ATRP) of protected monomer precursors followed by deprotection, according to previously reported methods [35]. All other reagents were obtained from Sigma–Aldrich and used as received. Spectra/Por membrane tubes were purchased from Spectrum Medical Industries, Inc. (Waltham, MA), and were used for dialysis. Nanopure water (18 MΩ cm) was acquired by means of a Barnstead Nanopure ultrapure water purification system (Thermo Scientific, Asheville, NC).
2.2. Measurements
1H NMR and 13C NMR spectra were recorded on an Inova 300 or Mercury 300 MHz interfaced to a UNIX computer using VnmrJ software. Samples were prepared as solutions in CDCl3, CD2Cl2, or DMF-d7 and solvent protons were used as internal standard. Gel permeation chromatography (GPC) was conducted on a system equipped with a Waters Chromatography, Inc. (Milford, MA) model 1515 isocratic pump and a model 2414 differential refractometer with a three-column set of Polymer Laboratories, Inc. (Amherst, MA) Styragel columns (PLgel 5 μm Mixed C, 500 Å, and 104 Å, 300 × 7.5 mm columns) and a guard column (PLgel 5 μm, 50 × 7.5 mm). The system was equilibrated at 40 °C in tetrahydrofuran (THF), which served as the polymer solvent and eluent (flow rate set to 1.00 mL min−1). The differential refractometer was calibrated with Polymer Laboratories, Inc. polystyrene standards (300–467,000 Da). Polymer solutions were prepared at a concentration of ca. 3 mg/mL with 0.05% (v/v) toluene as flow rate marker and an injection volume of 200 μL was used. Data were analyzed using Empower Pro software from Waters Chromatography Inc. IR spectra were recorded on an IR Prestige 21 system (Shimadzu Corp., Japan). A small amount of sample was placed to cover the ATR crystal for IR measurements. Data were analyzed using IRsolution software. Thermogravimetric analysis was performed under N2 atmosphere using a Mettler Toledo model TGA/DSC1 with a heating rate of 10 °C min−1. Measurements were analyzed using Mettler Toledo STARe software v.10.00.
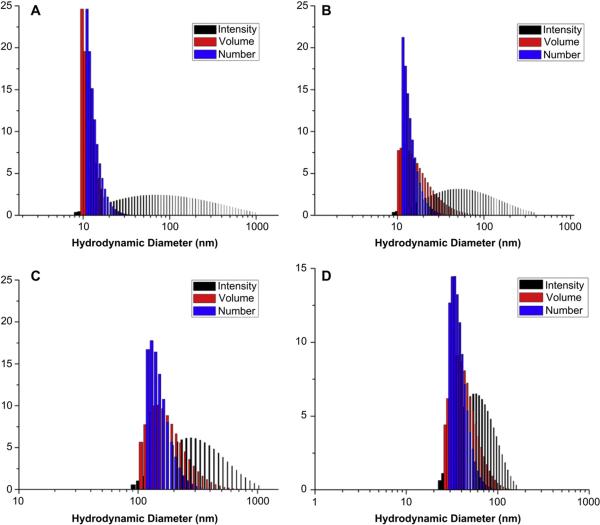
Samples for TEM were prepared by depositing 5 μL of sample to glow discharged carbon coated copper grids. Excess sample was wicked off using filter paper and the grids were allowed to dry in air for 1 min. The grids were then stained with 5 μL of 2% uranyl acetate and excess stain was wicked off using filter paper. Specimens were observed on a JEOL 1200EX transmission electron microscope operating at 100 kV and micrographs were recorded at calibrated magnifications using an SIA-15C CCD camera. The final pixel size was 0.42 nm/pixel. The number-average particle diameters (Dav) and standard deviations were generated from the analysis of particles from at least two different micrographs (n = 100). Dynamic light scattering (DLS) measurements were conducted using Delsa Nano C from Beckman Coulter, Inc. (Fullerton, CA) equipped with a laser diode operating at 658 nm. Size measurements were made in water (n = 1.3329, η = 0.890 cP at 25 ±1 °C; n = 1.3293, η = 0.547 cP at 50 ±1 °C; n = 1.3255, η = 0.404 cP at 70 ±1 °C). Scattered light was detected at 165° angle and analyzed using a log correlator over 70 accumulations for a 0.5 mL of sample in a glass size cell (0.9 mL capacity). The photomultiplier aperture and the attenuator were automatically adjusted to obtain a photon counting rate of ca. 10 kcps. The calculations of the particle size distribution and distribution averages were performed using CONTIN particle size distribution analysis routines. Prior to analysis, the samples were filtered through a 0.45 μm Whatman Nylon membrane filter (Whatman Inc., Piscataway, NJ). The samples in the glass size cell were equilibrated at the desired temperature for 5 min before measurements were made. The peak average of histograms from intensity, volume or number distributions out of ca. 70 accumulations was reported as the average diameter of the particles. Absorption measurements were made using a UV-2550 system (Shimadzu Corp., Japan) using PMMA cuvettes. Spectra were analyzed with UV-Probe 2.33 software.
2.3. Synthesis of poly(acrylamidoethylamine)160-boc-b-polystyrene30 (PAEA-Boc160-b-PS30)(1)
PAA160-b-PS30 (150 mg, 9.86 μmol) was allowed to dissolve in DMF (7.50 mL) for 30 min prior to the addition of HOBT (324 mg, 2.39 mmol) and HBTU (912 mg, 2.39 mmol) in a DMF solution. The reaction was allowed to stir for 30 min and diisopropylethylamine (180 mg, 1.34 mmol) was added along with N-boc-ethylenediamine (389 mg, 2.39 mmol). The reaction was allowed to stir overnight at room temperature for 24 h and dialyzed against 150 mm NaCl solution for 2 d, then against nanopure water for 5 d. After dialysis, the solution was lyophilized to obtain a white powder (380 mg, 95.0% yield). (Mn)NMR = 38.7 kDa. (Mn)GPC = 47.0 kDa. (Mw)GPC = 60.1 kDa. PDI = 1.27. IR: 3630–3120, 2960, 2910, 1650, 1620, 1540, 1450, 1380, 1260, 1180, 1030, 890, 830 cm−1. 1H NMR (DMSO-d6) δ: 1.0 – 2.0 (br, polymer backbone, Boc), 2.2 ((br,-CHCH2-), 2.8–3.4 (br,-NCH2CH2N-) 6.2–7.3 (br, ArH) ppm. 13C NMR (DMSO-d6) δ: 26.7, 31.6–41.0, 56.5, 80.2, 126.4–128.1, 145.5, 175.3 ppm. DSC: (Tg) = 28 °C, 106 °C. Tonset = 132 °C, Tdecomposition: [(132–239 °C) 29% mass loss; (239–475 °C) 47% mass loss; 24% mass remaining].
2.4. Synthesis of poly(acrylamidoethylamine-Boc)0.85-g-poly(acrylamidoethylimi dazole)0.15-b-polystyrene30 ((PAEA-Boc0.85-g-PAEI0.15)160-b-PS30)(2)
2.4.1. General procedure for incorporation of N-boc ethylenediamine and histamine onto the polymer backbone
PAA160-b-PS30 (150 mg, 9.9 μmol) was allowed to dissolve in DMF (7.50 mL) for 30 min prior to the addition of HOBT (324 mg, 2.39 mmol) and HBTU (912 mg, 2.39 mmol) in a DMF solution. The reaction was stirred for 30 min and diisopropylethylamine (180 mg, 1.34 mmol) was added along with N-boc-ethylenediamine (330 mg, 2.1 mmol) and histamine (40 mg, 0.2 mmol) from a stock solution in DMF. The reaction was allowed to stir overnight at room temperature for 24 h and dialyzed against 150 mm NaCl solution for 2 d, then against nanopure water for 5 d. After dialysis the solution was lyophilized to obtain a white powder (380 mg, 95.0% yield). (Mn)NMR = 37.2 kDa. IR: 3600–3000, 2965, 1700, 1650, 1538, 1498, 1380, 1288, 1160 cm−1. 1H NMR (DMSO-d6) δ: 1.0–2.0 (br, polymer backbone, Boc), 2.2 (br,-CHCH2-), 2.8–3.4 (br,-NCH2CH2N- and -NCH2CH2C-) 6.2–8.5 (br, ArH, styrene and histamine) ppm. 13C NMR (DMSO-d6) δ: 28.2, 31.6–41.0, 77.6, 117.9, 126.4–128.1, 134.7–136.4, 174.5 ppm. Tonset = 186 °C, Tdecomposition: [(186–255 °C) 22% mass loss; (255–449 °C) 60% mass loss; 18% mass remaining].
2.5. Synthesis of poly(acrylamidoethylamine)0.50-Boc-g-poly(acrylamidoethylimi dazole)0.50-b-polystyrene30 ((PAEA0.50-Boc-g-PAEI0.50)160-b-PS30)(3)
Polymer 3 was synthesized according to the general procedure for preparation of graft copolymer as mentioned above. (Mn)NMR = 34.4 kDa. IR: 3600–3000, 2950, 1700, 1640, 1545, 1460, 1375, 1250, 1160 cm−1. 1H NMR (DMSO-d6) δ:1.0–2.0 (br, polymer backbone, boc), 2.2 ((br,-CHCH2-), 2.8–3.4 (br,-NCH2CH2N- and -NCH2CH2C-) 6.2–8.5 (br, ArH, styrene and histamine) ppm. 13C NMR (DMSO-d6): δ: 28.2, 31.6–38.0, 77.7, 117, 126.4–128.1, 134.6, 174.3 ppm. Tonset = 183 °C, Tdecomposition: [(183–237 °C) 6% mass loss; (237–263 °C) 20% mass loss; 74% mass remaining].
2.6. Synthesis of poly(acrylamidoethylimidazole)160-b-polystyrene31 (PAEI160-b-PS30)(4)
PAA160-b-PS30 (150 mg, 9.86 μmol) was allowed to dissolve in DMF (7.50 mL) for 30 min prior to the addition of HOBT (324 mg, 2.39 mmol) and HBTU (912 mg, 2.39 mmol) in a DMF solution. The reaction was stirred for 30 min and diisopropylethylamine (180 mg, 1.34 mmol) was added along with histamine (270 mg, 2.43 mmol) from a stock solution in DMF. The reaction was allowed to stir overnight at room temperature for 24 h and dialyzed against 150 mm NaCl solution for 2 d, then against nanopure water for 5 d. After dialysis the solution was lyophilized to obtain a white powder (380 mg, 95.0% yield). (Mn)NMR = 30.5 kDa. IR: 3650-3000, 2940, 1630, 1540, 1440, 13750, 1250, 1125 cm−1. 1H NMR (DMSO-d6) δ:1.0–2.0 (br, polymer backbone), 2.2 ((br,–CHCH2−), 2.8–3.4 (br, −NCH2CH2C−) 6.2–8.5 (br, ArH, styrene and histamine) ppm. 13C NMR (DMSO-d6): δ:35–42.3, 117.9, 126.4–128.1, 134.7–136.4, 174.5 ppm. Tonset = 108 °C, Tdecomposition: [(108–283 °C) 11% mass loss; (283–500 °C) 48% mass loss; 41% mass remaining].
2.7. Synthesis of poly(acrylamidoethylamine)-b-polystyrene30 (PAEA-160-b-PS30)(5)
2.7.1. General procedure for the removal of boc groups
A white powder of 1 (153 mg, 3.54 μmol) was allowed to dissolve in TFA (7.00 g, 61.4 mmol) and stirred at room temperature overnight. The following day, TFA was removed under a stream of N2. The polymer was then dissolved in DMSO and dialyzed against 150 mM NaCl solution for 1 d followed by dialysis against nanopure water for 3 d. The solution was then lyophilized to obtain the deprotected polymer. (Mn)NMR = 22.1 kDa. IR: 4000–2360, 1670, 1560, 1470, 1190, 1130, 855, 810, 750 cm−1. 1H NMR (DMSO-d6): δ 1.0–2.0 (br, polymer backbone), 2.2 (br,−CHCH2−), 2.8–3.4 (br, −NCH2CH2N−) 6.2–7.3 (br, ArH), 7.2–8.4 (br, NH) ppm. 13C NMR (DMSO-d6): δ: 26.7, 31.6–38.0, 40.5, 126.4–128.1, 145.5, 175.0 ppm. Tonset = 173 °C, Tdecomposition: [(173–296 °C) 14% mass loss; (296–484 °C) 48% mass loss; 38% mass remaining].
2.8. Synthesis of poly(acrylamidoethylamine)0.85-g-poly(acrylamidoethylimi dazole)0.15-b-polystyrene30 ((PAEA0.85-g-PAEI0.15)160-b-PS30)(6)
Polymer 6 was prepared according to the general procedure for removal of Boc groups as mentioned above. (Mn)NMR = 23.4 kDa. IR: 3600–3000, 2940, 1640, 1538, 1450, 1225 cm−1. 1H NMR (DMSO-d6) δ:1.0–2.0 (br, polymer backbone), 2.2 ((br, −CHCH2−), 2.8–3.4 (br, −NCH2CH2N− and −NCH2CH2C−) 6.2–9.5 (br, ArH, styrene and histamine, NH) ppm. 13C NMR (DMSO-d6): δ:31.6–41.0, 117.9, 126.4–128.1, 134.7–136.4, 174.8 ppm. Tonset = 193 °C, Tdecomposition: [(193–469 °C) 70% mass loss; 30% mass remaining].
2.9. Synthesis of poly(acrylamidoethylamine)0.50-g-poly(acrylamidoethylimi dazole)0.50-b-polystyrene30 ((PAEA0.50-g-PAEI0.50)160-b-PS30)(7)
Polymer 7 was synthesized according to the general procedure for removal of Boc groups as mentioned above. (Mn)NMR = 26.3 kDa. IR: 3600–3000, 2925, 1750, 1625, 1525, 1450, 1375, 1250, 1160 cm−1. 1H NMR (DMSO-d6) δ:1.0–2.0 (br, polymer backbone), 2.2 ((br, −CHCH2−), 2.8–3.4 (br, −NCH2CH2N− and −NCH2CH2C−) 6.2–9.5 (br, ArH, styrene and histamine, NH) ppm. 13C NMR (DMSO-d6): δ: 31.6–38.0, 117, 126.4–128.1, 134.6, 174.8 ppm. Tonset = 188 °C, Tdecomposition: [(188–294 °C) 27% mass loss; (294–497 °C) 62% mass loss; 11% mass remaining].
2.10. General procedure for micellization (8–11)
Each polymer (5.0 mg) was directly dissolved in nanopure water (5.0 mL) and sonicated using a water bath sonicator for 10 min to obtain clear micellar solution at a concentration of 1 mg/mL. The solution was then allowed to stir overnight at room temperature. DLS and zeta-potential measurements are listed in the Supporting information (S4–S7).
2.11. Preparation of cSCKs of PAEA160-b-PS30 (12)
2.11.1. General procedure for preparation of cSCKs
To 8 (5.00 mL), sodium carbonate (20 μL of 1.0 M solution) was added to adjust the pH to ca. 8.0. A diacid crosslinker, 4,15-dioxo-8, 11-dioxa-5,14-diazaoctadecane-1,18-dioic acid, (2.0 mg) was activated with HOBT (1.7 mg, 2.2 equiv. per COOH) and HBTU (4.8 mg, 2.2 equiv. per COOH) in 300 μL of DMF for 30 min. The activated crosslinker solution was slowly added to the micellar solution to crosslink ca. 5% of the amines. The reaction mixture was allowed to stir overnight, and then transferred to a dialysis tube (MWCO ca. 6000–8000 Da) and dialyzed against nanopure water for 3 d to obtain clear cSCK solution at 0.88 mg/mL. (Dh)number (DLS) = 21 ± 6nm; (Dh)volume (DLS) = 30 ± 18 nm; (Dh)intensity (DLS) = 106 ± 81 nm. Zeta potential = 26 ± 2 mV (in nanopure water pH 5.5).
2.12. Preparation of cSCKs of (PAEA0.85-g-PAEI0.15)160-b-PS30 (13)
cSCKs of 13 were prepared from the micelles following the general procedure for crosslinking as listed above. (Dh)number (DLS) = 14 ± 4; (Dh)volume (DLS) = 21 ± 4 nm; (Dh)intensity (DLS) = 147 ± 61 nm nm. Zeta potential = 35 ± 1 mV (pH 5.5).
2.13. Preparation of cSCKs of (PAEA0.50-g-PAEI0.50)160-b-PS30 (14)
cSCKs of 14 were prepared from the micelles following the general procedure for crosslinking as listed above. (Dh)number (DLS) = 19 ± 5 nm; (Dh)volume (DLS) = 29 ± 19 nm; (Dh)intensity (DLS) = 168 ± 163 nm. Zeta potential = 50 ± 2 mV (pH 5.5).
2.14. Potentiometric titration experiments
The buffering capacities of the nanoparticles were determined by acid-base titration assays over pH range of 12.0–2.0, as reported previously [19]. Briefly, cSCKs (9.0 μmoles of amine) were dissolved in 15 mL NaOH (0.01 N with 150 mm NaCl) and then titrated with HCl (0.01 N with 150 mm NaCl). The buffering capacity was measured as the volume of HCl needed to change pH from 5.1 to 7.4.
2.15. Cytotoxicity assays
RAW 264.7 (2 × 104 cells/well) mouse macrophages were plated in 96-well plate in Dulbecco's Modified Eagle Medium (DMEM) (10% fetal bovine serum and 1% penicillin/streptomycin). Cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2 for 24 h to adhere. Then, the medium was replaced with a fresh medium 1-h prior to the addition of 20 μL of the cSCKs and Lipofectamine to 100 μL of the medium (final concentrations ranged from 0–100 μg/mL). The cells were incubated with the formulations for 24 h and washed once with phosphate-buffered saline (PBS) and 100 μL of the complete media was added to the cells. MTS combined reagent (20 μL) was added to each well (Cell Titer 96® Aqueous Non-Radioactive Cell Proliferation Assay, Promega Co., Madison, WI). The cells were incubated with the reagent for 2 h at 37 °C in a humidified atmosphere containing 5% CO2 protected from light. Absorbance was measured at 490 nm using SpectraMax M5 (Molecular Devices Co., Sunnyvale, CA). The cell viability was calculated based on the relative absorbance to the control-untreated cells. The calculation of the IC50 values and the statistical analysis were performed using GraphPad Prism four-parameter fit, considering the 0% and 100% cell viabilities are for the control medium (no cells) and cells with no treatment, respectively.
2.16. Multi-plex assay
The RAW 264.7 cells were treated with medium (control), unmodified cSCKs and 15%-His-cSCKs (5 μg/mL) for 24 h. The supernatants were then collected and centrifuged for 10 min at 13,000 rpm. Serial dilutions of standards of cytokines were also prepared in the same diluent utilized for the samples (i.e. cell-culture medium). Control, standards and nanoparticle-treated samples (50 μL) were incubated with antibody-conjugated magnetic beads for 30 min in the dark. After washing, the detection antibody was added to the wells and incubated in the dark for 30 min under continuous shaking (300 rpm). After washing, streptavidin-phycoerythrin was added to every well and incubated while protected from light for 10 min under the same shaking conditions. Finally, after several washings and resuspension in the assay buffer and shaking, the expression of the mouse cytokines, interleukin (IL)-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12 (P40), IL-12 (P70), IL-13, IL-17, Eotaxin, Granulocyte-colony-stimulating factor (G-CSF), granulocyte macrophage-colony-stimulating factor (GM-CSF), Interferon-γ (IFN-γ), keratinocyte-derived chemokine (KC), monocyte chemotactic protein (MCP)-1, macrophage inflammatory protein (MIP)-1α, MIP-1β, regulated upon activation normal T-cell expressed and presumably secreted (RANTES) and tumor necrosis factor-α (TNF-α) was measured immediately using Bio-plex 200 system with HTF and Pro II Wash station (Bio-Rad Laboratories, Inc., Hercules, CA) and the data were analyzed using the Bio-plex Data Pro software.
2.17. Gel shift assay
Agarose gels (1%) were prepared in Tris-acetate-EDTA buffer (Bio-Rad Laboratories, Inc., Hercules, CA). The siRNA (5′-Cy3-(sense strand)-GGCCACAUCGGAUUUCACU, Mw = 13814 g/mol, Dharmacon, Chicago, IL), either free or complexed to the various nanoparticle formulations at nitrogen (total concentrations of the primary amines and histamines)-to-phosphate (N/P) ratios ranging from 0.25–6 (1.3 μg siRNA/25 μL/well), were mixed with glycerol (20% v/v) prior to the electrophoresis. Gel electrophoresis was carried out using a horizontal apparatus at 100 V for 30 min and fluorescence imaging of the separated siRNA bands was performed using a ChemiDoc XRS (Bio-Rad Laboratories, Inc.).
2.18. Death-siRNA transfection assays
RAW 264.7 mouse macrophages (2 × 104 cells/well) and human ovarian adenocarcinoma cells (OVCAR-3) (5 × 103 cells/well) were plated in a 96-well plate in DMEM and RPMI-1640 medium (10% and 20% fetal bovine serum, for the RAW 264.7 and OVCAR-3, respectively and 1% penicillin/streptomycin). Cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2 for 24 h to adhere. Then, the medium was replaced with a fresh medium 1-h prior to the addition of the siRNA-loaded cSCKs (100 nm final concentrations of AllStars death- or negative control-siRNA (Qiagen, Valencia, CA)) at N/P ratio of 5, unless otherwise indicated. The cells were incubated with the various formulations for 24 h and washed extensively with PBS and the cell viability was measured 24 h later by measuring the relative cell viability of the cells treated with death-siRNA to the negative control-loaded cSCKs. The cell viabilities were measured as described in the cytotoxicity section. The Lipofectamine-siRNA complexes were prepared according to the manufacturer instructions and the transfection efficiency was measured following the same procedures of the siRNA-cSCKs complexes. The effect of endosomal acidification on transfection efficiency was studied by incubating cells with 200 nm bafilomycin A1 (Sigma–Aldrich, St. Louis, MO) 30-min prior to the transfection and by adding the same concentration of bafilomycin to the transfection medium. The toxicity of siRNA complexes could be also measured by comparing the cell viability of the cells treated with the negative control-siRNA complexes to that of the control-untreated cells.
2.19. Laser scanning confocal microscopy (LSCM)
RAW 264.7 mouse macrophages (1 × 105 cells/well) were plated in glass-bottom six-well plate (MatTek Co., Ashland, MA) in DMEM medium. Cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2 for 24 h to adhere. Then, the medium was replaced with a fresh media 1-h prior to the addition of siRNA-loaded cSCKs or Lipofectamine (100 nm final concentration of the 5′-Cy3-siRNA). The cells were incubated with the formulations for 3 h and washed extensively with PBS. Then, DRAQ-5 (Biostatus Ltd., Shepshed, Leicestershire, UK) was utilized to stain the nucleus (30 min incubation, followed by extensive washing with PBS). Cells were then fixed with 1% formaldehyde for 20 min, washed once with PBS. The cells were then stored in 1 mL PBS in the refrigerator and analyzed by laser scanning confocal microscopy (LSM 510, Zeiss, Jena, Germany). The images were collected under the same conditions (laser power, detector gain, etc.) for consistency, and λexcitation of 543 and 633 nm were utilized for the Cy3 and DRAQ-5, respectively.
2.20. Statistical analysis
Values are presented as mean ± SD of at least three independent experiments. Significance of the differences between two groups was evaluated by Student's t test (unpaired) or between more than two groups by one-way ANOVA followed by Tukey's multiple comparison tests. Differences between different groups were considered significant for p values less than 0.05.
3. Results and discussion
3.1. Synthesis of polymers
A library of cationic micelles (8–11) and shell crosslinked nanoparticles (cSCKs, 12–14) was prepared from amphiphilic block copolymers (4–7) in a stepwise process, each of which was self-assembled into micelles followed by chemical crosslinking as illustrated in Schemes 1 and 2. Each of the amino- and/or histamine-functionalized amphiphilic block copolymers was derived from the same block copolymer precursor PAA160-b-PS30, which was prepared by a two-step ATRP process of tBA and styrene, followed by deprotection (Supporting information, S1–S3). Chemical modification reactions onto the PAA backbone and selective acidolysis then introduced the amino and histamine side chain groups. The acrylic acid repeat units were converted to acrylamides, each carrying a protected amino group (0%-His), imidazole (100%-His) or mixture of both at various ratios (85:15 (15%-His) and 50:50 (50%-His)), by using an excess of N-Boc ethylenediamine and/or histamine in the presence of HOBT, HBTU and DIPEA in DMF, yielding polymers 1, 2, 3 and 4. In the final step, the Boc protecting groups were removed by direct dissolution of 1, 2 and 3 in TFA overnight, followed by removal of the TFA under a N2 stream, to obtain the block copolymers 5, 6 and 7.
Scheme 1.
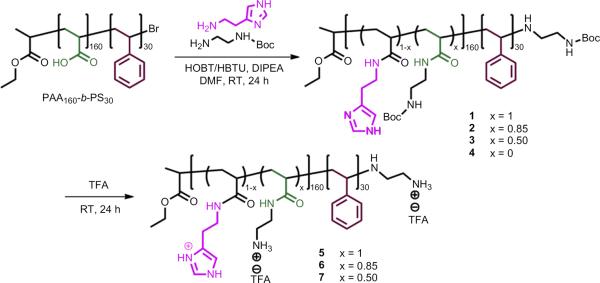
Incorporation of various amounts of primary amines and histamines onto a segment of an amphiphilic block copolymer backbone by a sequential two-step process of amidation followed by deprotection.
Scheme 2.
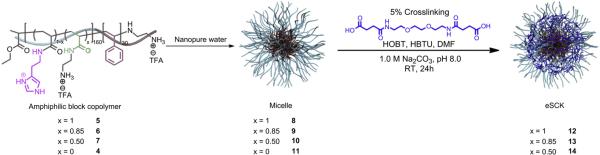
Self assembly of polymers with various amounts of primary amines and histamines (4–7) into micelles (8–11) followed by crosslinking selectively in the hydrophilic shell regions of (8–10) to obtain cSCKs (12–14).
3.2. Preparation and characterization of cSCKs
cSCKs were prepared by the solution-state assembly of amphiphilic block copolymers in water followed by crosslinking selectively throughout the hydrophilic shell layer (Scheme 2). Self assembly was induced by the direct dissolution of the amphiphilic block copolymer (5, 6, 7, and 4) at 1 mg mL−1 in molecular biology grade water to yield micelles 8, 9, 10 and 11, respectively (0%-His-, 15%-His-, 50%-His- and 100%-His-micelles, respectively). The series of cSCKs 12, 13 and 14 (0%-His-, 15%-His-, and 50%-His-cSCKs, respectively) was obtained by crosslinking approximately 5% of the amino groups along the hydrophilic segments of 8–10 by amidation with activated carboxylic acids (activated using 2.2 equiv. of both HOBT and HBTU relative to the COOH groups in the crosslinker in DMF) of the diacid crosslinker (4,15-dioxo-8,11-dioxa-5,14-diazaoctadecane-1,18-dioic acid) at pH ca. 8 (adjusted using 1.0 M Na2CO3 (aq.)). The crosslinking reaction was allowed to proceed overnight, followed by extensive dialysis (MWCO 6–8 kDa) to remove the unreacted small molecule starting materials and reaction by-products. The 100%-His-micelles did not have any cross-linking sites due to the complete incorporation of histamine groups onto the polymer backbone and, hence, were not converted to the corresponding cSCKs; further studies were continued with the micellar form.
To understand the physicochemical properties of the cationic micelles and cSCKs, their sizes and zeta-potential values were determined. Dynamic light scattering (DLS) analysis showed that the hydrodynamic diameters of the micelles and cSCKs decreased slightly with increasing amounts of histamines incorporated. The number-averaged diameters of the nanoparticles were less than 20 nm in all cases (Fig. 1). It was difficult to obtain high quality TEM images for all of the samples, but for those that gave adequate images, the core diameters were measured to be ca. 10 nm (Supp info Fig. S2), consistent with the hydrodynamic DLS-measured overall diameter values. Zeta potential values of the micelles and cSCKs, each in nanopure water at pH 5.5 were determined initially, and found to be positive for 8–10 and 12–14, but negative for 11 having only histamines and no primary amino groups. Given the pKa of histamine groups, typically 6–6.5, the observation of a negative zeta potential at pH 5.5 was hypothesized to be due to the use of nanopure water without high salt content and/or the presence of only histamine groups and, therefore, was then followed by a full study of each of the micellar solutions 8–11 at pH 5.0, 6.0 and 7.4 (PBS buffer, 10 mM NaCl). As the micelles 11 (100%-His) did not have any primary amines in the polymer backbone for crosslinking reaction, this study of zeta potential measurements at different pHs was performed only for the micellar solutions. As expected, at pH 7.4, all of the samples were found to be positively-charged except the 100%-His-micelles, which were negatively-charged (Fig. 2A). Generally, zeta-potential values were found to increase with the decrease in pH, which indicates higher protonation of polymers at acidic pH, corresponding to the endosomal environment. Additionally, nanoparticles with the highest amount of histamine content showed the least zeta potential values at all pHs, and were confirmed to be positively-charged at and below pH 6.0. At pH 7.4, the 50% His-micelles were insoluble and 100% His-micelles exhibited low zeta-potential values, whereas the 15%-His-micelles demonstrated similar behavior to the unmodified cSCKs.
Fig. 1.
DLS histograms from measurements of nanoparticles with various amounts of primary amines and histamines: (A) 0%-His-cSCK (12), (B) 15%-His-cSCK (13), (C) 50%-His-cSCK (14) and (D) 100%-His-micelles (11).
Fig. 2.
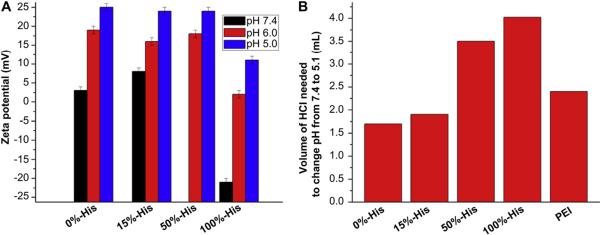
(A) Zeta-potential measurements of micelles 8–11 with increasing histamine content, each at pH 5.0, 6.0 and 7.4. (B) Relative buffering capacity of cSCK nanoparticles 12–14 and micelles 11 containing various amounts of histamine obtained from potentiometric titration experiments, and comparison with PEI. Each bar represents the amount of HCl (in mL) needed to change the pH of the nanoparticle solutions from pH 7.4 to 5.1.
To determine the buffering capacities of each type of cationic nanostructures, potentiometric titration studies were conducted over a pH range of 12–2, with focus over the range of 7.4 to 5.1 of relevance to extracellular to endosomal/lysosomal transitions (Fig. 2B). The nanoparticles were dissolved in NaOH(aq) and titrated by gradual addition of HCl. The particles were found to require increasing amounts of HCl to change the pH from 7.4 to 5.1 (17 μmol–41 μmol) with increasing histamine content (12 to 14 to 11, respectively). cSCK nanoparticles with 0% histamines (12) required the least amount (1.7 mL) of HCl to change pH from 7.4 to 5.1, while micellar nanoparticles with the highest histamine content (100% His, 11) needed the highest amount (4.1 mL) of HCl to induce the same change. In general, increasing histamine content directly correlated with the amount of acid needed, demonstrating the increased buffering capacities of nanoparticles with greater histamine content. In comparison to PEI, which contains combinations of primary, secondary and tertiary amines, the primary amino-containing cSCKs, 12, and the cSCKs with few histamines, 13, gave lower buffering capacity, whereas each of the cSCK and micelles with 50% and 100% histamines gave higher buffering capacities. Nanoparticles with higher buffering capacity are expected to buffer the endosomes to a greater extent and, hence, result in higher endosomal disruption [15,20,21].
3.3. Toxicity and immunogenicity of cSCKs
The cytotoxicity of nanoparticles containing various amounts of histamine and primary amine groups was evaluated in RAW 264.7 mouse macrophages, and compared to Lipofectamine. Increasing the histamine contents significantly increased the IC50 values of the nanoparticles from 16.3 μg/mL to 20.7 μg/mL (p < 0.05), for the cSCKs with 0% and 15% histamines, 12 and 13, respectively. The IC50 values could not be determined for nanoparticles that contained 50% and 100% histamine at the tested concentrations (0–100 μg/mL), due to the high cell viability measured over this concentration range. The IC50 value of the tested Lipofectamine was significantly higher than the cSCKs of 100% and 85% primary amines (12 and 13, respectively), but the cell viabilities were much lower than the cSCK (14) and micelles (11) with lower amine contents. These results reveal that increasing the histamine content reduced the toxicity of the nanoparticles, which is in accordance with previously published data from our group showing that reducing the amount of primary amines per polymer (by incorporating tertiary amines or carboxylic acids into the polymer structure), reduces the toxicity of the nanoparticles [7,33]. With the higher cytotoxicities observed for the cSCKs having low histamine content, and because the nanoparticles with histamine content greater than 15% demonstrated low cellular uptake and transfection efficiency (vide infra), we decided to compare the immunotoxicities of the unmodified- and 15%-His-modified cSCKs.
The release of cytokines from cells treated with nanomaterials is usually utilized as an indication of possible immunotoxicity of nanoparticles and their immunomodulatory effects [36–39]. The structure and composition of nanoparticles can greatly impact their ability to interact with the various components of the immune system, where unintentional immunomodulation can be serious and cause severe adverse reactions and complications [36,40,41]. Cationic liposomes, for instance, are known to induce the secretion of proinflammatory cytokines (e.g. TNF-α, IFN-γ and IL-12) [41]. Inorganic anionic zinc oxide nanoparticles also stimulated the release of IL-6 and TNF-α cytokines from dendritic cells treated with the nanoparticles [36].
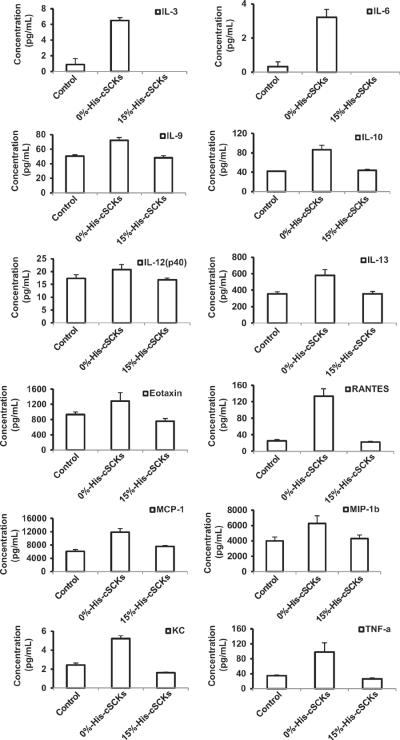
The immunotoxicities of the 0%- and 15%-His-cSCKs were studied by measuring the levels of 23 cytokines upon treatment of RAW 264.7 mouse macrophages with the nanoparticles for 24 h (Fig. 3). Generally, lower secretion of the cytokines was observed from cells treated with the histamine-modified cSCKs, 13, as compared to the cSCKs with 100% primary amines, 12. A similar trend was observed for most of the tested cytokines, which indicate the potential immunotoxicity of the 0%-His-cSCKs as compared to the histamine modified ones. The differences between the levels of the secreted cytokines were significant for 12 cytokines, IL-3, IL-6, IL-9, IL-10, IL-12(p40), IL-13, Eotaxin, RANTES, MCP-1, MIP-1β, KC and TNF-α (Fig. 3). The incorporation of histamine into the polymer precursor significantly reduced the toxicity of the nanoparticles and their ability to stimulate the cells and induce the secretions of cytokines, which might be due to the lower charge density, cell-binding affinity and cellular uptake, as will be discussed in the next sections.
Fig. 3.
The expression of IL-3, IL-6, IL-9, IL-10, IL-12(p40), IL-13, Eotaxin, RANTES, MCP-1, MIP-1β, KC and TNF-α were particularly enhanced upon the treatment of Raw 264.7 mouse macrophages with the unmodified-cSCKs (0%-His-cSCKs) for 24 h, as compared to the 15%-His-cSCKs and the control-untreated cells.
3.4. Nanocomplexes of siRNA-cSCKs for efficient gene silencing
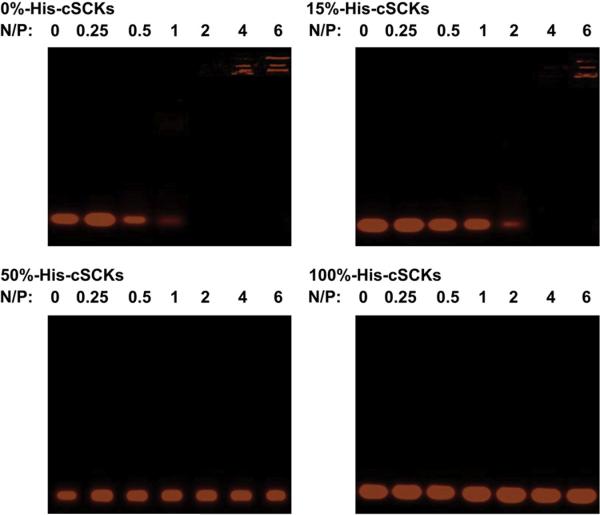
The binding affinity of Cy3-labeled siRNA to the micelle and cSCK nanoparticles of various compositions was studied using agarose gel electrophoresis (Fig. 4). Only the cSCKs with 0%- and 15%-histamine contents were able to efficiently bind siRNA at N/P ratios of 2 and 4, respectively. The incorporation of histamine reduced the binding affinity of cSCKs and micelles to the siRNA. In a previous study, the reduction of primary amine contents of cSCKs was also correlated with a reduced binding affinity to nucleic acids (plasmid DNA) [33]. Increasing the histamine contents reduces the amount of protonated primary amines available to electrostatically interact with the negatively-charged siRNA, and hence decreases the siRNA-binding affinity. The cellular uptake and transfection experiments were then performed at N/P ratio of 5 to ensure the complete siRNA-binding for both kinds of nanoparticles, 12 and 13.
Fig. 4.
Gel-shift assay of Cy3-labeled siRNA, either naked (N/P = 0) or complexed to micelles or cSCKs of various composition at increasing N/P ratios from 0.25–6. Complete binding is observed at N/P ratios of 2 and 4, for 0% and 15% histamine-modified cSCKs, whereas no binding was observed for cSCKs and micelles that contain 50% and 100% histaminemodified shells, respectively.
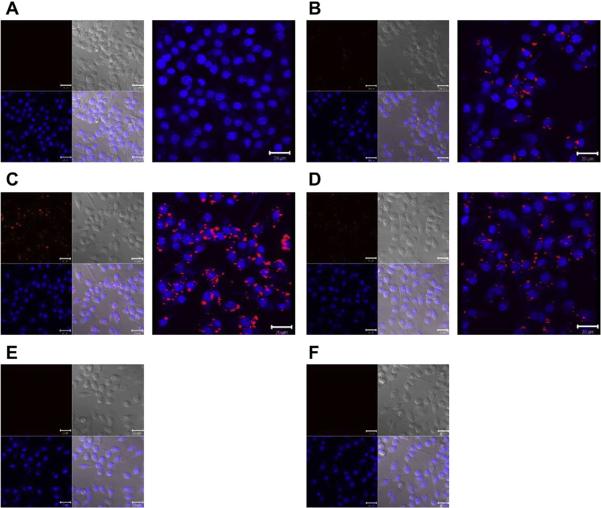
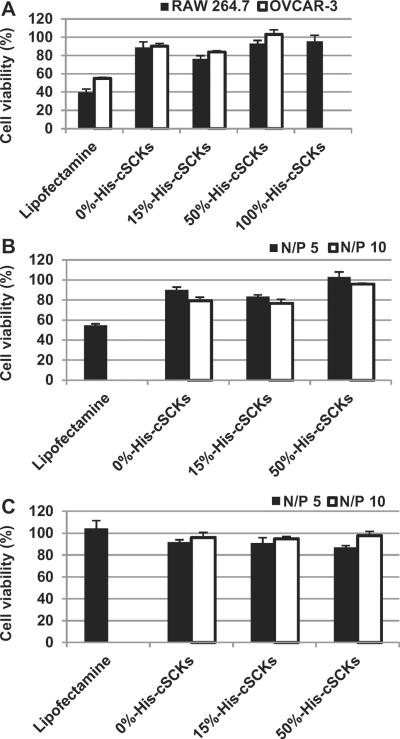
The cellular uptake and transfection efficiency of cSCKs and Lipofectamine were studied using laser scanning confocal microscopy and siRNA-cell death assay using RAW 264.7 mouse macrophages and OVCAR-3 cancer cells (Figs. 5 and 6). The siRNA-cell death assay was performed by measuring the relative viabilities of cells treated with nanoparticles-loaded with siRNA that induces cell death versus the cells-treated with the same nanoparticles but loaded with a negative control siRNA. No cellular uptake or transfection efficiency was observed in cells treated with the 50%-His-cSCKs and 100%-His-micelles. Higher cellular uptake was observed for the cells treated with the 0%-His-cSCKs than the cells treated with the 15%-His-cSCKs and Lipofectamine as indicated by the confocal microscopy images (Fig. 5), probably due to the higher binding affinity of the nanoparticles that are fully functionalized with primary amines to the cell membrane as compared to the other nanoparticles. However, the transfection efficiency of the 15%-histamine-modified cSCKs and the Lipofectamine was higher than the 0%-His-cSCKs. The incorporation of histamine increased the buffering capacities of the nanoparticles, as shown by the potentiometric titration experiments (Fig. 2B), which is expected to have facilitated greater endosomal release of siRNA/nanoparticles into the cytoplasm and thus resulted in higher intracellular bioavailability. The cellular uptake is not always correlated with transfection efficiency as it does not measure the ability of nanoparticles to reach their target sites into the cytoplasm/nucleus and additionally, does not differentiate between drug molecules that are being delivered into the cytoplasm or entrapped within endosomes/lysosomes. Elsabahy et al. [22] have observed higher transfection efficacy and Bcl-2 protein knockdown from antibody fragment-targeted poly(amidoamine)-based polyion complex micelles loaded with Bcl-2 siRNA than the corresponding non-targeted poly(amidoamine)/siRNA complexes, although the latter showed higher cellular uptake as measured by flow cytometric analysis and shown by confocal microscopy images.
Fig. 5.
Laser scanning confocal microscopy analysis of the control-untreated RAW 264.7 mouse macrophages (A), cells treated with Cy3-siRNA (100 nm) complexed with Lipofectamine (B), 0%-His- (C), 15%-His- (D), 50%-His- (E) and 100%-His-nanoparticles (F) at N/P ratio of 5. Two (left panel) and three (right panel)-dimensional images were collected for the cells (A–D), while only two-dimensional images were collected for E and F. On the two-dimensional images, the nucleus were stained with DRAQ5 nuclear stain (blue panel), whereas Cy3-labeled siRNA appears in red. The transmitted light-images and merged images are also indicated. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Transfection efficiency of death-siRNA complexed with Lipofectamine or cSCKs of varying primary amine and histamine ratios into OVCAR-3 cells and RAW 264.7 mouse macrophages at N/P ratios of 5 and 10. (A) The transfection of the death-siRNA/cSCK complexes in the two different cell lines at an N/P ratio of 5. (B) The effect of N/P ratio on the transfection efficiency in the OVCAR-3 cells, determined by comparison of death-siRNA/cSCKs vs. negative control-siRNA/cSCKs. (C) The effect of N/P ratio on the cytotoxicity of cSCKs in OVCAR-3 cells, determine by comparison of negative control-siRNA/cSCKs vs. control, untreated cell assays.
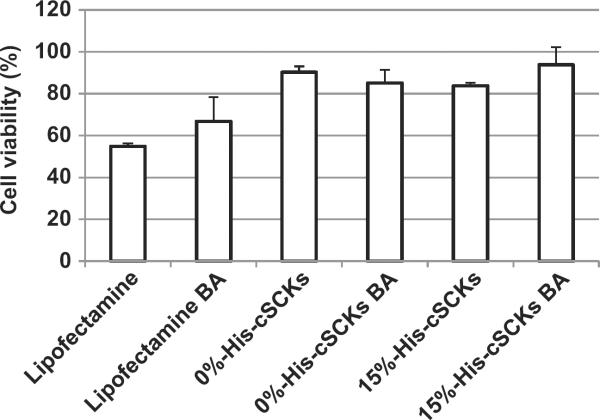
The effect of N/P ratio on the transfection efficiency was also measured using the same siRNA-cell death assay and showed a slight increase in transfection efficiency of the cSCKs by increasing the N/P ratio from 5 to 10 (Fig. 6B). However, no observable transfection was observed for the 50%-His-cSCKs at both N/P ratios. No toxicities were observed for the siRNA/cSCKs complexes at both N/P ratios (Fig. 6C). The role of endosomal acidification on the transfection of cSCKs was investigated by measuring the effect of bafilomycin A1 (an inhibitor of endosomal acidification) (Fig. 7) [21]. The pre-incubation of cells with bafilomycin had a slight effect on the transfection of Lipofectamine and 0%-His-cSCKs, but completely inhibited the transfection of the 15%-His-cSCKs. The non-acidified endosomes partially prevent the protonation of the histamine functionalities, which reduced their ability to destabilize the endosomes and to release a considerable amount of siRNA into the cytoplasm [22,42,43].
Fig. 7.
Transfection efficiency of death-siRNA (100 nm) complexed with Lipofectamine or cSCKs of varying compositions into OVCAR-3 cells, with and without pre-treatment with bafilomycin A1 (BA, 200 nm for 30 min before the transfection and continued during the transfection).
4. Conclusions
In this study, a library of block copolymer micelles and cSCKs has been created with different percentages of primary amines and histamines incorporated into the nanoparticle shells. The 50% and 100%-histamine-modified nanoparticles induced lower cytotoxicity as compared to Lipofectamine, whereas the cSCKs with 85% and 100% primary amines (15% and 0% histamines) were more toxic than Lipofectamine. Treatment of mouse macrophages with the unmodified cSCKs resulted in the secretion of large amounts of several cytokines, as compared to the control-untreated cells and the cells-treated with histamine-modified nanoparticles. Increasing the amount of histamines increased the buffering capacities of the nanoparticles at the expense of the siRNA-binding affinity, due to the reduced contents of primary amines. Intermediate primary amine and histamine levels resulted in higher intracellular concentrations of the complexed siRNA and higher gene silencing activity. Histamine incorporation of 15% appeared to be a threshold for the series of samples tested, with cSCKs having higher histamine contents completely lost the ability to bind siRNA and to interact with cell membrane and induce gene silencing. These nanoparticles are among the initial steps involved in the development of cSCKs for effective delivery of nucleic acids for clinical applications. However, a further probing of histamine contents between 15% and 50% may better identify the optimum balance of siRNA-binding, cellular uptake and buffering capacity. Expansion of the chemical compositional heterogeneity and synthesis of degradable analogs of these cSCKs is currently underway for extended in vitro and in vivo studies toward development of polymeric materials for nucleic acids delivery.
Supplementary Material
Acknowledgments
We acknowledge financial support from the National Heart Lung and Blood Institute of the National Institutes of Health as a Program of Excellence in Nanotechnology (HHSN268201000046C) and the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health (R01-DK082546). The Welch Foundation is also gratefully acknowledged for partial support through the W. T. Doherty-Welch Chair in Chemistry, Grant No. A-0001. The authors would like to thank Adriana Pavia for creating the Autodesk 3ds Max images and Prof. John-Stephen A. Taylor of Washington University in Saint Louis for creative inspiration.
Footnotes
Appendix A. Supplementary material Supplementary material associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.biomaterials.2012.07.054.
References
- [1].Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–38. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bumcrot D, Manoharan M, Koteliansky V, Sah DWY. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–9. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–70. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ferrari M. Experimental therapies: vectoring siRNA therapeutics into the clinic. Nat Rev Clin Oncol. 2010;7:485–6. doi: 10.1038/nrclinonc.2010.131. [DOI] [PubMed] [Google Scholar]
- [5].Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4:581–93. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- [6].Nishikawa M, Huang L. Nonviral vectors in the new millennium: delivery barriers in gene transfer. Hum Gene Ther. 2001;12:861–70. doi: 10.1089/104303401750195836. [DOI] [PubMed] [Google Scholar]
- [7].Elsabahy M, Wooley KL. Design of polymeric nanoparticles for biomedical delivery applications. Chem Soc Rev. 2012;41:2545–61. doi: 10.1039/c2cs15327k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Miyata K, Nishiyama N, Kataoka K. Rational design of smart supramolecular assemblies for gene delivery: chemical challenges in the creation of artificial viruses. Chem Soc Rev. 2012;41:2562–74. doi: 10.1039/c1cs15258k. [DOI] [PubMed] [Google Scholar]
- [9].Mastrobattista E, Hennink WE. Polymers for gene delivery: charged for success. Nat Mater. 2012;11:10–2. doi: 10.1038/nmat3209. [DOI] [PubMed] [Google Scholar]
- [10].Suma T, Miyata K, Ishii T, Uchida S, Uchida H, Itaka K, et al. Enhanced stability and gene silencing ability of siRNA-loaded polyion complexes formulated from polyaspartamide derivatives with a repetitive array of amino groups in the side chain. Biomaterials. 2012;33:2770–9. doi: 10.1016/j.biomaterials.2011.12.022. [DOI] [PubMed] [Google Scholar]
- [11].Takemoto H, Ishii A, Miyata K, Nakanishi M, Oba M, Ishii T, et al. Polyion complex stability and gene silencing efficiency with a siRNA-grafted polymer delivery system. Biomaterials. 2010;31:8097–105. doi: 10.1016/j.biomaterials.2010.07.015. [DOI] [PubMed] [Google Scholar]
- [12].Li WJ, Szoka FC. Lipid-based nanoparticles for nucleic acid delivery. Pharmaceut Res. 2007;24:438–49. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- [13].Murthy N, Robichaud JR, Tirrell DA, Stayton PS, Hoffman AS. The design and synthesis of polymers for eukaryotic membrane disruption. J Control Release. 1999;61:137–43. doi: 10.1016/s0168-3659(99)00114-5. [DOI] [PubMed] [Google Scholar]
- [14].Chen T, McIntosh D, He Y, Kim J, Tirrell DA, Scherrer P, et al. Alkylated derivatives of poly(ethylacrylic acid) can be inserted into preformed liposomes and trigger pH-dependent intracellular delivery of liposomal contents. Mol Membr Biol. 2004;21:385–93. doi: 10.1080/09687860400010516. [DOI] [PubMed] [Google Scholar]
- [15].Itaka K, Kanayama N, Nishiyama N, Jang WD, Yamasaki Y, Nakamura K, et al. Supramolecular nanocarrier of siRNA from PEG-based block catiomer carrying diamine side chain with distinctive pKa directed to enhance intracellular gene silencing. J Am Chem Soc. 2004;126:13612–3. doi: 10.1021/ja047174r. [DOI] [PubMed] [Google Scholar]
- [16].Cho YW, Kim JD, Park K. Polycation gene delivery systems: escape from endosomes to cytosol. J Pharm Pharmacol. 2003;55:721–34. doi: 10.1211/002235703765951311. [DOI] [PubMed] [Google Scholar]
- [17].Putnam D, Gentry CA, Pack DW, Langer R. Polymer-based gene delivery with low cytotoxicity by a unique balance of side-chain termini. Proc Natl Acad Sci U S A. 2001;98:1200–5. doi: 10.1073/pnas.031577698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Luo SB, Cheng R, Meng FH, Park TG, Zhong ZY. Water soluble poly(histamine acrylamide) with superior buffer capacity mediates efficient and nontoxic in vitro gene transfection. J Polym Sci Polym Chem. 2011;49:3366–73. [Google Scholar]
- [19].Kanayama N, Fukushima S, Nishiyama N, Itaka K, Jang WD, Miyata K, et al. A PEG-based biocompatible block catiomer with high buffering capacity for the construction of polyplex micelles showing efficient gene transfer toward primary cells. Chem Med Chem. 2006;1:439–44. doi: 10.1002/cmdc.200600008. [DOI] [PubMed] [Google Scholar]
- [20].Fukushima S, Miyata K, Nishiyama N, Kanayama N, Yamasaki Y, Kataoka K. PEGylated polyplex micelles from triblock catiomers with spatially ordered layering of condensed pDNA and buffering units for enhanced intracellular gene delivery. J Am Chem Soc. 2005;127:2810–1. doi: 10.1021/ja0440506. [DOI] [PubMed] [Google Scholar]
- [21].Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7:657–63. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- [22].Elsabahy M, Wazen N, Bayo-Puxan N, Deleavey G, Servant M, Damha MJ, et al. Delivery of nucleic acids through the controlled disassembly of multifunctional nanocomplexes. Adv Funct Mater. 2009;19:3862–7. [Google Scholar]
- [23].Elsabahy M, Dufresne M-H, Leroux J-C. Polymeric micelles as versatile carriers for drugs and nucleic acids delivery. In: Torchilin V, Amiji M, editors. Handbook of materials for nanomedicine. Pan Stanford Publishing; Hackensack: 2010. pp. 169–234. [Google Scholar]
- [24].Elsabahy M, Wooley KL. Strategies toward well-defined polymer nanoparticles inspired by nature: chemistry versus versatility. J Polym Sci Polym Chem. 2012;50:1869–80. doi: 10.1002/pola.25955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Samarajeewa S, Shrestha R, Li YL, Wooley KL. Degradability of poly(lactic acid)-containing nanoparticles: enzymatic a through a cross-linked shell barrier. J Am Chem Soc. 2012;134:1235–42. doi: 10.1021/ja2095602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shrestha R, Shen YF, Pollack KA, Taylor JSA, Wooley KL. Dual peptide nucleic acid- and peptide-functionalized shell cross-linked nanoparticles designed to target mRNA toward the diagnosis and treatment of acute lung injury. Bioconjug Chem. 2012;23:574–85. doi: 10.1021/bc200629f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sorrells JL, Shrestha R, Neumann WL, Wooley KL. Porphyrin-crosslinked block copolymer assemblies as photophysically-active nanoscopic devices. J Mater Chem. 2011;21:8983–6. [Google Scholar]
- [28].Nyström AM, Wooley KL. The importance of chemistry in creating well-defined nanoscopic embedded therapeutics: devices capable of the dual functions of imaging and therapy. Acc Chem Res. 2011;44:969–78. doi: 10.1021/ar200097k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Welch MJ, Pressly ED, Rossin R, Hagooly A, Fukukawa KI, Messmore BW, et al. Structural effects on the biodistribution and positron emission tomography (PET) imaging of well-defined Cu-64-labeled nanoparticles comprised of amphiphilic block graft copolymers. Biomacromolecules. 2007;8:3126–34. doi: 10.1021/bm700541e. [DOI] [PubMed] [Google Scholar]
- [30].Sun G, Xu J, Hagooly A, Rossin R, Li Z, Moore DA, et al. Strategies for optimized radiolabeling of nanoparticles for in vivo PET imaging. Adv Mater. 2007;19:3157–62. [Google Scholar]
- [31].Thurmond KB, Huang H, Clark CG, Jr, Kowalewski T, Wooley KL. Shell cross-linked polymer micelles: stabilized assemblies with great versatility and potential. Colloid Surf B. 1999;16:45–54. [Google Scholar]
- [32].Zhang K, Fang H, Shen G, Taylor JS, Wooley KL. Well-defined cationic shell crosslinked nanoparticles for efficient delivery of DNA or peptide nucleic acids. Proc Am Thorac Soc. 2009;6:450–7. doi: 10.1513/pats.200902-010AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang K, Fang H, Wang Z, Li Z, Taylor JS, Wooley KL. Structure-activity relationships of cationic shell-crosslinked knedel-like nanoparticles: shell composition and transfection efficiency/cytotoxicity. Biomaterials. 2010;31:1805–13. doi: 10.1016/j.biomaterials.2009.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhang K, Fang H, Wang Z, Taylor J-SA, Wooley KL. Cationic shell-crosslinked knedel-like nanoparticles for highly efficient gene and oligonucleotide transfection of mammalian cells. Biomaterials. 2009;30:968–77. doi: 10.1016/j.biomaterials.2008.10.057. [DOI] [PubMed] [Google Scholar]
- [35].Ma QG, Wooley KL. The preparation of t-butyl acrylate, methyl acrylate, and styrene block copolymers by atom transfer radical polymerization: precursors to amphiphilic and hydrophilic block copolymers and conversion to complex nanostructured materials. J Polym Sci Polym Chem. 2000;38:4805–20. [Google Scholar]
- [36].Heng BC, Zhao X, Tan EC, Khamis N, Assodani A, Xiong S, et al. Evaluation of the cytotoxic and inflammatory potential of differentially shaped zinc oxide nanoparticles. Arch Toxicol. 2011;85:1517–28. doi: 10.1007/s00204-011-0722-1. [DOI] [PubMed] [Google Scholar]
- [37].Hussain S, Boland S, Baeza-Squiban A, Hamel R, Thomassen LC, Martens JA, et al. Oxidative stress and proinflammatory effects of carbon black and titanium dioxide nanoparticles: role of particle surface area and internalized amount. Toxicology. 2009;260:142–9. doi: 10.1016/j.tox.2009.04.001. [DOI] [PubMed] [Google Scholar]
- [38].Rettig L, Haen SP, Bittermann AG, von Boehmer L, Curioni A, Kramer SD, et al. Particle size and activation threshold: a new dimension of danger signaling. Blood. 2010;115:4533–41. doi: 10.1182/blood-2009-11-247817. [DOI] [PubMed] [Google Scholar]
- [39].Poland CA, Duffin R, Kinloch I, Maynard A, Wallace WAH, Seaton A, et al. Carbon nanotubes introduced into the abdominal cavity of mice show asbestos-like pathogenicity in a pilot study. Nat Nanotechnol. 2008;3:423–8. doi: 10.1038/nnano.2008.111. [DOI] [PubMed] [Google Scholar]
- [40].Tao WK, Mao XZ, Davide JP, Ng B, Cai MM, Burke PA, et al. Mechanistically probing lipid-siRNA nanoparticle-associated toxicities identifies jak inhibitors effective in mitigating multifaceted toxic responses. Mol Ther. 2011;19:567–75. doi: 10.1038/mt.2010.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cui ZR, Han SJ, Vangasseri DP, Huang L. Immunostimulation mechanism of LPD nanoparticle as a vaccine carrier. Mol Pharm. 2005;2:22–8. doi: 10.1021/mp049907k. [DOI] [PubMed] [Google Scholar]
- [42].Dufes C, Uchegbu IF, Schatzlein AG. Dendrimers in gene delivery. Adv Drug Deliv Rev. 2005;57:2177–202. doi: 10.1016/j.addr.2005.09.017. [DOI] [PubMed] [Google Scholar]
- [43].Hong SP, Leroueil PR, Janus EK, Peters JL, Kober MM, Islam MT, et al. Interaction of polycationic polymers with supported lipid bilayers and cells: nanoscale hole formation and enhanced membrane permeability. Bioconjug Chem. 2006;17:728–34. doi: 10.1021/bc060077y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.