Abstract
MoaA, a radical S-adenosylmethionine (SAM) enzyme, catalyzes the first step in molybdopterin biosynthesis. This reaction involves a complex rearrangement in which C8 of guanosine triphosphtate is inserted between the C2′ and the C3′ carbons of the ribose. This study identifies the site of initial hydrogen atom abstraction by the adenosyl radical and advances a mechanistic proposal for this unprecedented reaction.
Molybdenum is a required nutrient for plants, animals and microorganisms and is involved in many redox reactions implicated in the global carbon, sulfur, and nitrogen cycles1,2. With the exception of nitrogenase, all molybdenum-requiring enzymes use molybdopterin as the metal binding ligand. Nitrate reductase, sulfite oxidase, xanthine dehydrogenase and aldehyde oxidase are well known molybdopterin dependent enzymes. The biosynthesis of molybdopterin is similar in plants, animals and microorganisms where the first steps utilizing guanosine-5'-triphosphate (1; GTP) as the precursor are catalyzed by the enzymes MoaA and MoaC (Scheme 1). MoaA is a member of the radical-SAM superfamily of proteins and harbors two [4Fe-4S]2+,1+ clusters. The N-terminal [4Fe-4S] cluster is likely to be responsible for the reductive cleavage of SAM to the 5′-deoxyadenosyl (5′-dA) radical and L-methionine3,4 and the second cluster binds to N1 of GTP5. Upon generation of the 5′-dA radical, GTP is transformed into (2) via a complex rearrangement reaction where the C8 atom of the purine is inserted between the C2′ and the C3′ atoms of the ribose moiety (Scheme 1). This is an unprecedented reaction in ribose chemistry. MoaC then catalyzes the intra-molecular cyclization reaction of (2) to (3) which is oxidized to (4) prior to analysis because of the instability of (3).4 The goal of this study was to obtain mechanistic insights regarding the rearrangement reaction catalyzed by MoaA by identifying the initial site of hydrogen abstraction on GTP by the 5′-dA radical.
Scheme 1.
Reactions catalyzed by MoaA and MoaC. MoaA catalyzes the rearrangement of GTP (1) to (2) in an adenosyl radical dependent reaction involving the insertion of C8 of guanine between C2′ and C3′ of the ribose. MoaC catalyzes the formation of (3). Also shown in the figure is the oxidation of (3) to (4) by I2/KI. This compound is fluorescent and is more stable than (3).
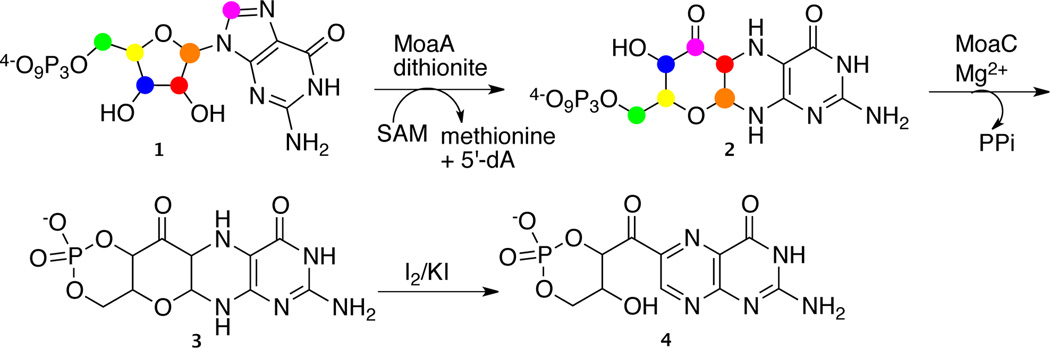
The 5′-dA (5) generated in the MoaA catalyzed reaction was first isolated and characterized. The HPLC chromatograms in Figure 1A were obtained from anaerobic reaction mixtures containing MoaA (125 mM), GTP (1mM), SAM (1.5 mM), and sodium dithionite (excess). The blue trace corresponds to the assay where all the components were present, the other traces are controls from which MoaA, SAM or sodium dithionite was omitted.
Figure 1.
Determination of the regiochemistry of the MoaA catalyzed hydrogen atom abstraction from GTP. A) HPLC analysis of the MoaA reaction mixture. The blue trace is from the reaction containing MoaA, SAM, GTP and dithionite. The red, green and violet traces are controls in which MoaA, SAM or sodium dithionite was omitted respectively. The peak at 14.5 min is 5′-dA. All the reactions were performed under anaerobic conditions. B) ESI-MS analysis of the 14.5 min peak indicates an m/z of 252.1 Da corresponding to the [M+H] for 5′-dA. D) MoaA assays were performed with GTP isotopomers and the resulting 5′-dA was analyzed by LCMS for deuterium incorporation. U-2H10-GTP is universally deuteriated GTP. The data show that a deuterium atom at C3′ of GTP is incorporated into 5′-dA demonstrating that MoaA catalyzes the generation of a radical at C3′ of GTP.
The product 5′-dA, eluting at 14.5 min, is formed only in the reaction mixture where all components are present. ESI-MS analysis (Figure 1B) and NMR analysis (Figure S9) confirmed the identity of the 14.5 min eluting compound.
Several (Figure 1D) site specifically deuterium labeled GMP isotopomers were synthesized and converted to GTP in situ using guanylate kinase, nucleoside diphosphate kinase and ATP/Mg2+ (Figures S2–S7). Deuterium transfer from labeled GTP to the 5′-dA was monitored by LCMS (Figure S1). Analysis of the 5′-dA generated using universally labeled GTP (i.e. deuterium on all non-exchangeable sites) reveled an increase of a single mass unit ([M+H] - 253.1Da) demonstrating that only one of the deuterium atoms was incorporated into 5′-dA. No deuterium incorporation was observed with 8-2H-GTP demonstrating that the deuterium was derived from the ribose. No deuterium incorporation was observed with 2′-2H-GTP and a single deuterium incorporation was observed with 3′,4′,5′,5′-2H4-GTP, further localizing the site of deuterium incorporation to C3′, C4′ or C5′ of the ribose. Final localization of the hydrogen atom transferred was achieved by demonstrating a single deuterium transfer from 3′-2H-GTP to 5′-dA.
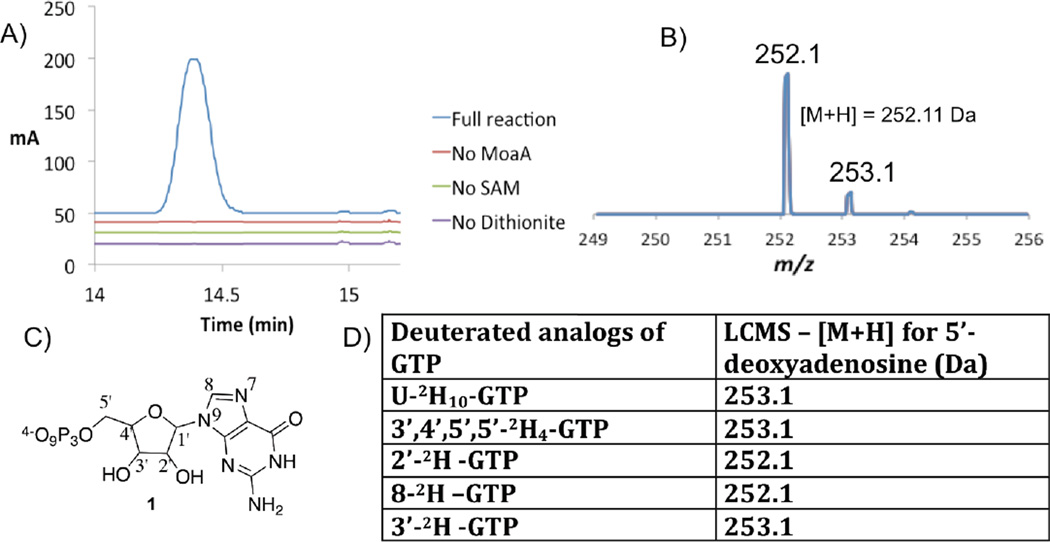
The pterin product (2) of the MoaA reaction was also analyzed. This eluted after 18.6 min (Figure 2A) and was present only in the reaction mixture containing GTP, SAM and dithionite. Since this product was oxygen sensitive, all the LCMS buffers were degassed and the LCMS samples were prepared in an anaerobic chamber. ESI-MS analysis revealed that the [M+H] for the signal at retention time 18.6 min is 524.00 Da (Figure 2B). This is consistent with structure (2).
Figure 2.
Characterization of the pterin formed in the MoaA catalyzed reaction. A) HPLC (320nm) chromatogram showing the second product (blue trace, 18.6 min) formed in the MoaA catalyzed reaction. This product was observed only in reactions where MoaA, GTP, SAM and sodium dithionite were present. Chromatograms from control reactions lacking MoaA, GTP, SAM or dithionite are also shown (red, green, purple and black traces respectively). B) ESI-MS of the compound eluting at 18.6 min using GTP as the substrate shows [M+H] = 524.00 Da consistent with structure (2). C) ESI-MS of the compound eluting at 18.6 min using 3′,4′,5′,5′-2H4-GTP as the substrate shows [M+H] = 527.02 Da consistent with the loss of deuterium from C3′ and retention of the three deuteria at C4′ and C5′. D) Mechanistic proposal for the reaction catalyzed by MoaA.
It was not possible to obtain an NMR spectrum of (2) due to its instability. The identity of this compound was therefore confirmed, as previously described,6 by treatment with MoaC followed by oxidation with I2/KI to give an air-stable product with structure (4) as shown by NMR analysis (Figures 1 and S8). It was not possible to analyze the pterin produced using deuteriated GTP by NMR because of the small quantities of the labeled samples available. However, when 3′,4′,5′,5′-2H4-GTP was used as a substrate, a 3 Da increase was observed to give [M+H] of 527.02 Da (Figure 2C). This is consistent with abstraction of the C3′ hydrogen by the 5′-dA radical and retention of the C4′ and C5′ hydrogens in the product.
A mechanistic proposal for the unprecedented carbon insertion reaction catalyzed by MoaA that is consistent with the labeling studies described here and the previously reported ENDOR and structural studies is shown in Figure 2D3,4,5. The 5′-dA radical abstracts the 3′ hydrogen atom from GTP bound to the second [4Fe-4S] cluster (6) resulting in radical (7) which then adds to C8 of the purine to give (8). Reduction of this radical by the liganded cluster would give (9). Hydrolysis of this strained system would give (10). A benzilic like rearrangement would give (11) completing the insertion of the purine carbon into the ribose.
Dehydration of (11) to (12) followed by conjugate addition gives (13). A final tautomerization completes the formation of (2). This mechanistic proposal has several testable features and is currently under investigation.
Supplementary Material
Acknowledgment
This research was supported by a grant from the National Institutes of Health (DK44083) and by the Robert A. Welch Foundation (A-0034) and by the Deutsche Forschungsgemeinschaft (Rudolf Virchow Center for Experimental Biomedicine, FZ 82).
Footnotes
ASSOCIATED CONTENT
Supporting Information Available: Detailed procedures for the synthesis of labeled GMP and GTP, HPLC methods, NMR data for (4), and procedures for the enzymatic reactions are provided in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Hille R. Trends Biochem. Sci. 2002;27(7):360–367. doi: 10.1016/s0968-0004(02)02107-2. [DOI] [PubMed] [Google Scholar]
- 2.Schwarz G, Mendel RR. Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu. Rev. Plant Biol. 2006;57:623–647. doi: 10.1146/annurev.arplant.57.032905.105437. [DOI] [PubMed] [Google Scholar]
- 3.Hänzelmann P, Schindelin H. Proc. Natl. Acad. Sci. U. S. A. 2004;101(35):12870–12875. doi: 10.1073/pnas.0404624101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hänzelmann P, Schindelin H. Proc. Natl. Acad. Sci. U. S. A. 2006;103(18):6829–6834. doi: 10.1073/pnas.0510711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lees NS, Häzelmann P, Hernandez HL, Subramanian S, Schindelin H, Johnson MK, Hoffman BM. JACS. 2009;131(26) doi: 10.1021/ja903978u. 9184-91850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson JL, Wuebbens MM, Rajagopalan KV. J. Biol. Chem. 1989;264(23):13440–13447. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.