Abstract
The pathogen and parasite community that inhabits every free-living organism can control host vital rates including lifespan and reproductive output. To date, however, there have been few experiments examining pathogen community assembly replicated at large-enough spatial scales to inform our understanding of pathogen dynamics in natural systems. Pathogen community assembly may be driven by neutral stochastic colonization and extinction events or by niche differentiation that constrains pathogen distributions to particular environmental conditions, hosts, or vectors.
Here, we present results from a regionally-replicated experiment investigating the community of barley and cereal yellow dwarf viruses (B/CYDV's) in over 5000 experimentally planted individuals of six grass species along a 700 km latitudinal gradient along the Pacific coast of North America (USA) in response to experimentally manipulated nitrogen and phosphorus supplies. The composition of the virus community varied predictably among hosts and across nutrient-addition treatments, indicating niche differentiation among virus species. There were some concordant responses among the viral species. For example, the prevalence of most viral species increased consistently with perennial grass cover, leading to a 60% increase in the richness of the viral community within individual hosts (i.e., coinfection) in perennial-dominated plots. Furthermore, infection rates of the six host species in the field were highly correlated with vector preferences assessed in laboratory trials. Our results reveal the importance of niche differentiation in structuring virus assemblages. Virus species distributions reflected a combination of local host community composition, host species-specific vector preferences, and virus responses to host nutrition. In addition, our results suggest that heterogeneity among host species in their capacity to attract vectors or support pathogens between growing seasons can lead to positive covariation among virus species.
Introduction
Historically, the study of host-pathogen interactions has focused on three components essential to the completion of a pathogen lifecycle forming the ‘disease triangle’: an infectious microbe, a host, and a favorable environment [1], [2], [3]. While many studies have probed the interactions among the vertices of the disease triangle (e.g., [1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16]), investigations of the larger ecological context in which host-pathogen-environment interactions occur, including the interactions among multiple microbial species infecting the same host, multiple species in a host community, and the impact of multiple abiotic parameters have generally been observational or limited in spatial scope [17], [18], [19], [20], [21].
Nevertheless, the experimental investigation of interacting pathogen communities is important, because every free-living organism is host to a wide array of microbial symbionts that range in effects from pathogenic to mutualistic [22], [23], [24], [25], [26], [27], and the richness and composition of pathogen and parasite communities in particular can alter nearly every vital rate of a host including lifespan and reproductive output [27], [28], [29], [30], [31], [32], [33]. For example, coinfection by multiple pathogens can increase the severity of disease, as is the case for humans infected by multiple strains of human immunodeficiency virus (HIV), HIV and malaria, or the hepatitis C virus and the trematode Schistosoma mansonii [30], [31], [32], [33]. Infection by multiple viral or fungal pathogens can also increase mortality in plant hosts [28], [29]. Despite the importance of interactions among pathogens within a host, we know relatively little about the assembly of pathogen communities in natural systems, and we know of no field experiments that have been replicated at scales large enough to inform our understanding of how the biotic and abiotic context interact to mediate the natural assembly of pathogen communities in multi-host systems.
As in the community assembly of free-living species, variation in pathogen community structure can arise from stochastic infection events (i.e., neutral or dispersal assembled communities) or from niche differentiation that constrains the pathogens present in each host (i.e., niche assembled communities) [25], [34], [35]. Although neutral and niche processes are not mutually exclusive, their expectations can be used to assess the importance of species' traits in controlling community composition, beyond the variation expected from neutral processes [36], [37]. In particular, neutral theory predicts that species are functionally equivalent and should show no consistent changes in relative abundance across changing environments, including predictable composition among host species, sites, or along fertility gradients [36], [37]. In contrast, predictable variation in species composition across environmental gradients suggests trait-based sorting, supporting the importance of niches for understanding pathogen coexistence [36], [37]. Understanding the relative contribution of neutral and niche processes for coinfection ultimately could inform strategies for disease management and prevention.
Pathogen niches can be defined by processes that alter host, pathogen, and vector interactions [2], [4], [5], [6], [7], [8]. These processes can operate at a wide range of scales, from regional gradients in abiotic conditions such as climate or nutrient supply [17], [21], [25], [38], [39], [40] or biotic host and vector community composition [19], [20], [25], [40], [41], [42] down to pathogen and vector interactions with individual hosts [9], [10], [11], [43], [44], [45]. Although the abiotic and biotic environment has been shown to regulate many pathogens of global importance based on observational studies (e.g., malaria, dengue fever, Lyme disease, and West Nile virus) [2], [4], [5], [17], [19], [21], [39], [41], [43], experimental work has been largely confined to plant systems due the ethical and logistical difficulty of conducting large-scale manipulations of human and wildlife pathogens [40], [46], [47], [48].
Here we measure the prevalence and co-occurrence of five viral pathogens (barley and cereal yellow dwarf viruses; B/CYDV's) in over 5000 experimentally planted hosts of six different species at five sites spanning a 700 km latitudinal gradient along the Pacific coast of North America (USA). At each site, we experimentally manipulated supplies of nitrogen and phosphorus, creating within-site gradients in fertility and stoichiometry. Using data on regional viral occurrence and coinfection within hosts by these five viruses, we assess the extent to which the composition of viral species within hosts changed among host species, natural gradients in local plant composition, sites, and local gradients of two different nutrients. We use these data to quantify the processes and contingencies of pathogen community assembly in a natural environment.
Methods
Study System
B/CYDVs are aphid-vectored RNA viruses (family Luteoviridae) that infect the phloem of grasses and cause one of the most devastating of all diseases in cereal crops worldwide [49], [50]. In addition, B/CYDVs have been implicated as a causal agent in one of the most dramatic and persistent biological invasions worldwide, the conversion of nine million ha of grasslands in California (USA) from native perennial to exotic annual dominated [51], [52], [53]. B/CYDVs are known to infect over 150 grass species and are carried by over 25 aphid species [50], [54]. There is no vertical transmission of the viruses to the seeds (hosts only become infected through vector feeding after germination), and the viruses do not replicate within the aphid vectors [3], [55]. Hosts become infected through vector feeding after germination and the viruses spread systemically through the host phloem tissue. Once infected, perennial grasses are not known to clear themselves of viruses and thus generally accumulate viruses over time [56]. Because seeds do not carry B/CYDV infections, grasses with an annual life history are uninfected at the start of each growing season.
BYDVs and CYDVs belong to various viral genera [28].We track five common species (BYDV-MAV, BYDV-PAV, BYDV-SGV, BYDV-RMV, CYDV-RPV). The most common aphid vectors at our study sites are Rhopalosiphum padi, R. maidis, Sitobion avenae, Metopolophium dirhodum, and Schizaphis graminum (E. T. Borer, unpublished data), and these vector species differ strongly in their transmission efficiency of each virus species. Three of the aphids are generalist vectors that can transmit two viruses. R. padi is effective at transmitting BYDV-PAV and CYDV-RPV and S. avenae and M. dirhodum are effective at transmitting BYDV-MAV and BYDV-PAV [28]. Two aphid species, S. graminum and R. maidis, are specialists and can only transmit a single virus effectively (BYDV-SGV and BYDV-RMV, respectively) [28].
Experimental Design
Field nutrient-addition experiment
We conducted a full factorial experiment of nitrogen and phosphorus addition at five sites in California (McLaughlin Reserve and the Sierra Foothill and Hopland Research and Extension Centers) and Oregon (Baskett Slough and William Finley National Wildlife Refuges). Borer et al. [40] provide additional details on study sites and experimental design. The California sites were arrayed in longitudinal transect at approximately 39°N latitude and represent a gradient in mean rainfall from 939 mm yr−1 near the coast (Hopland) to 711 mm yr−1 further inland (Sierra Foothill). The two Oregon sites (Baskett and Finley) were at about 45°N latitude and have an average of 1198 and 1030 mm of rain per year, respectively. In addition to rainfall, the latitudinal gradient also corresponds to increasing cover of perennial grasses [25].
We established two experimental blocks at each site, with each block composed of four 40×40 m plots. These plots were randomly assigned to one of the four factorial combinations of phosphorus addition (Control or 4.3 g P m−2 yr−1 as triple super phosphate) or nitrogen addition (Control or 4 g N m−2 yr−1 as calcium nitrate). Fertilization treatments were applied quarterly starting in December 2006. In summary, the entire experiment was composed of 40 experimental units (40×40 m plots)−5 sites×2 blocks×2 levels of nitrogen×2 levels of phosphorus. Due to a field error, all samples from 5 plots at the Finley site could not be assayed for B/CYDVs.
We selected six target host species that are common throughout Pacific Coast grasslands and represent congeneric or contribal pairings of a native perennial and exotic annual grass. There are few native annual grasses in these systems [57]. The phylogenetic groupings were as follows (annual/perennial): Brome (Bromus hordeaceus/Bromus carinatus), Oat (Avena fatua/Koeleria macrantha), and Rye (Taeniatherum caput-medusae/Elymus glaucus). Note that K. macrantha is referred to by its synonym K. cristata by Borer et al. [40] in their related analysis of data from this experiment. Seeds of each species were collected at each site or from as nearby as possible. Hosts were pre-germinated in 25×25 mm soil plugs in greenhouses at Oregon State University, Corvallis, Oregon and transplanted to each site in January 2008, prior to the first aphid flight. One individual of each species was planted within a series of 20–30 quadrats (20×50 cm) arrayed along transects within each experimental plot. Except for being marked with twist-ties, the experimental hosts rapidly became indistinguishable from naturally recruiting individuals in the community.
We collected all aboveground tissue from the 5,095 surviving experimental hosts in late May and early June 2008, after about 5 months of growing in the field. These aboveground tissues were weighed to the nearest 0.01 g and assayed for five B/CYDV species (BYDV-MAV, BYDV-PAV, BYDV-SGV, BYDV-RMV, CYDV-RPV) using double antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) with antibodies supplied by Agdia, Elkhart, IN, USA. Individual hosts were randomly distributed between the Mitchell lab at UNC and the Power lab at Cornell University. In rare cases of putative infection by serologically related viruses that were associated nearly 1∶1 within hosts, we regarded the weaker of the two reactions as a cross-reaction to the more reactive virus rather than a coinfection. This conservative approach did not affect the overall prevalence estimates but may have somewhat reduced our estimate of coinfection by serologically related viruses. Note that we were only able to assay about half of the hosts for BYDV-SGV (2,320 hosts) due to a lack of antibody. We conducted analyses on both subsets of the data (5,095 hosts with 4 viruses and 2,320 hosts with 5 viruses). As results are qualitatively similar, we present the results with the full suite of viruses but fewer individual hosts.
We collected data on plant biomass and community composition at two locations along transects in each of the 40×40 m plots. We estimated total plant biomass in each plot by clipping all aboveground plant material in two 0.1×1 m strips, sorting it into categories (previous year's dead material and current year's grass, forb, legume, woody, bryophyte growth), drying it to constant mass at 60°C, and weighing it to the nearest 0.01 g. We estimated areal cover of all species in an adjacent 0.5×1 m quadrat. Cover of each species was estimated independently, so that total summed cover exceeds 100% in quadrats with multilayer canopies.
Laboratory aphid experiments
We use aphid fecundity and preference data from two experiments published by Borer et al. [58] to explain prevalence differences among host species: Preferential herbivory and Greenhouse multivector fecundity (Experiments III and IV in Borer et al. 2009a). In the Preferential-herbivory experiment 30 aphids (R. padi) were placed in the center of 3.78 l L pots in which one individual of 10 different host species (including our six focal species) were planted around the pot perimeter. Aphids were counted on all plants after 24 hours and, because plants of the same age differed in size, counts were calculated on a per gram of host tissue basis. For the Greenhouse multivector fecundity experiment, three species of aphids were placed in small mesh enclosures on replicate individuals of six our focal host species that were subjected to two levels of nitrogen (control or nitrogen addition). Our measure of fecundity was the number of offspring produced by each adult during a four day assay. See Borer et al. [58] for additional details of the aphid studies.
Statistical analyses
All analyses were conducted using R version 2.12 [59]. We tested for overall changes in the viral community using Permutational Multivariate Analysis of Variance (PerMANOVA) using the adonis function in the R vegan library. PerMANOVA is analysis of variance that compares distance matrices in which significance is determined using a permutation test. PerMANOVA is analogous to MANOVA and redundancy analyses [60], [61], [62]. Here we used 999 permutations and Jaccard's distance matrices. Only infected plants were included in the analyses, as uninfected plants all had a distance of zero and were uninformative. In the PerMANOVA tests, we use two approaches to control for spatial variability in our tests for the effects of the experimental treatments applied at the plot scale (nitrogen, phosphorus, and host species). First, we constrain the permutations to only occur among experimental units within each block (blocks are treated as strata in the adonis function). Second, we include key covariates (e.g., host community composition) while allowing permutations to be unconstrained. The first method of constrained permutations is the most powerful test of our experimental treatments, while the second method provides more biological insight into larger scale variability. Univariate analyses used mixed-effects regression models using the nlme R library. State, site and block within site were included as random effects in all mixed models. Fixed effects were determined using backwards selection as in Crawley [63].
Results
Overall infection prevalence across all 2320 hosts assayed for all five viruses was 25.5%, and 44% of the infected hosts were coinfected by multiple viruses. Infection patterns were similar in the larger data set (5095 hosts) assayed for four viruses; infection was 23.6% and coinfection of infected plants was 52.5%. There was a great deal of variation among the plots in terms of overall virus prevalence (0–68%), plant biomass (46–481 g m−2), annual grass cover (0–149%), perennial grass cover (0–66%), forb (non-host) cover (0–105%) and host (grass) species richness (1–7.2 species m−2). See supplement in Borer et al. [40] for a table of site-level means of total infection prevalence, soil chemistry, plant biomass, and plant species cover.
Host survival ranged from 79–88% among the host species, but was not statistically different [40]. Prevalence of each virus species across the study plants were as follows: BYDV-RMV (14.0%), BYDV-SGV (12.4%), BYDV-PAV (11.1%), CYDV-RPV (9.5%), and BYDV-MAV (9.4%). The mean number of viruses carried by infected hosts was 2.26±0.08 SEM. As has been found in other data sets [25], [56], viruses carried by the same vectors were strongly correlated with one another in coinfected hosts. Among the 10 pairwise combinations of virus species, the highest correlations were between the S. avenae vectored viruses, BYDV-MAV and BYDV-PAV, (r = 0.56, p<0.001) and the R. padi vectored viruses, BYDV-PAV and CYDV-RPV, (r = 0.43, p<0.001).
Among naturally infected hosts in control plots, the viral community varied at all spatial scales greater than individual plots (i.e. >1600 m2: block, site and state; Table S1). Furthermore, viral community composition differed among hosts in models that controlled for among-plot variability by constraining permutations to occur within plots (e.g., among quadrats within a block; Table S2; Figure 1). In particular, T. caput-medusae had consistently higher virus infection prevalence than other species, and A. fatua, generally considered a reservoir species for these pathogens [47], [52], had low to intermediate infection prevalence by all viral groups compared to other host species (Table S3; Figure 1).
Figure 1. Prevalence of five plant viruses in (A) six grass hosts: Avena fatua (AVEFAT), Bromus carinatus (BROCAR), Bromus hordeaceus (BROHOR), Elymus glaucus (ELYGLA), Koeleria macrantha (KOEMAC), and Taeniatherum caput-medusae (TAECAP), and (B) at five study sites in Oregon and California.
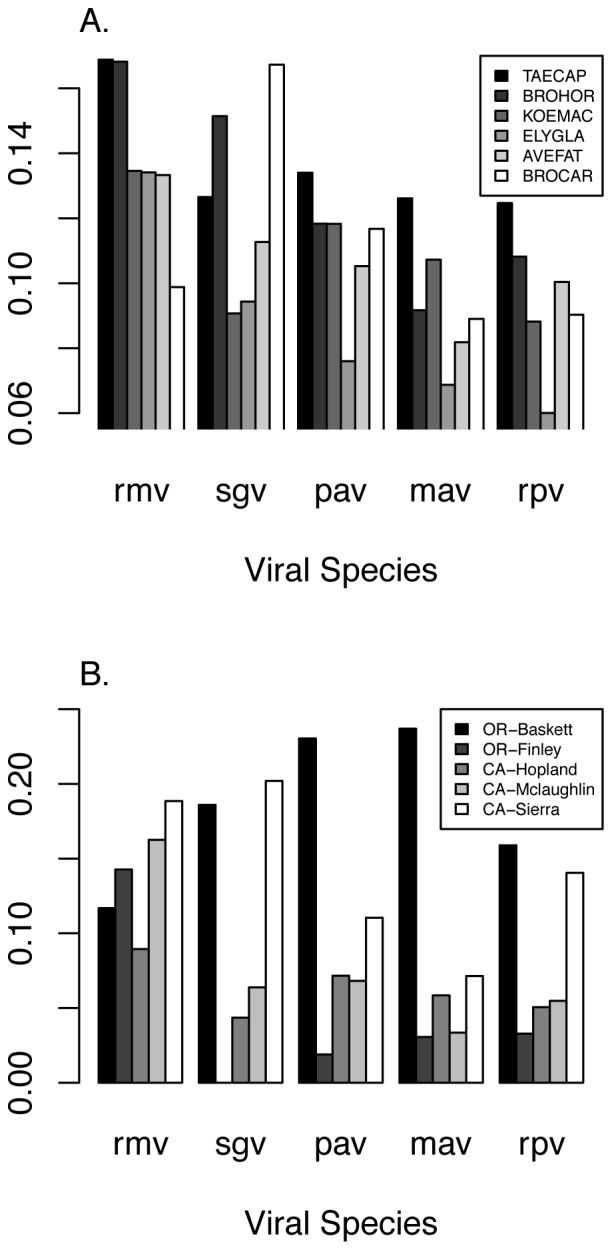
While viral community composition differed among host species, these among-host differences were not associated with host traits (p>0.05) including host lifespan (annual or perennial), provenance (native or exotic), or phylogenetic group (bromes, oats, or rye), and so we only present models here where species identity is treated as a single categorical variable. The strongest covariate explaining the relative prevalence rates in the different host species was the preference by the aphid vector, R. padi, for those host species in the laboratory Preferential-herbivory experiment) (Figure 2). There were no significant relationships between prevalence of the associated viruses in field hosts and aphid fecundity on the same host species in the laboratory Greenhouse multivector fecundity experiment (p>0.05).
Figure 2. Correlations of an aphid vector (Rhopalosiphum padi)'s feeding preference with the B/CYDV prevalence.
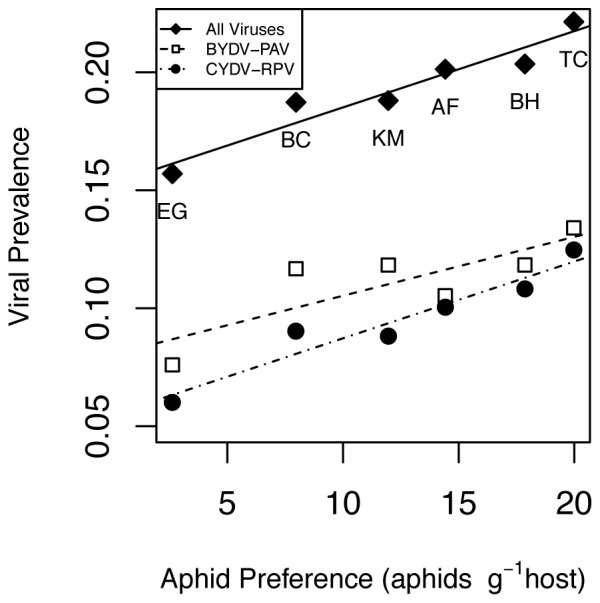
Plot show overall B/CYDV prevalence (infection by any virus) and the prevalences of two viruses (BYDV-PAV and CYDV-RPV) vectored by R. padi. The six grass hosts are as follows: Avena fatua (AF), Bromus carinatus (BC), Bromus hordeaceus (BH), Elymus glaucus (EG), Koeleria macrantha (KM), and Taeniatherum caput-medusae (TC). Correlations between feeding preference and overall prevalence of infection (r = 0.96), BYDV-PAV (r = 0.82), and CYDV-RPV (r = 0.96) were significantly greater than 0 (p<0.05).
Viral community composition also was correlated with host community composition (cover of perennial grasses, annual grasses and forbs), aboveground plant biomass, and phosphorus addition (Tables S4 and S5; Figures 1, 3, 4). Among-host differences in viral community composition were not significant indicating that they were obscured by large-scale environmental gradients (Table S4).
Figure 3. Prevalence of five viruses along gradients in plot host community composition.
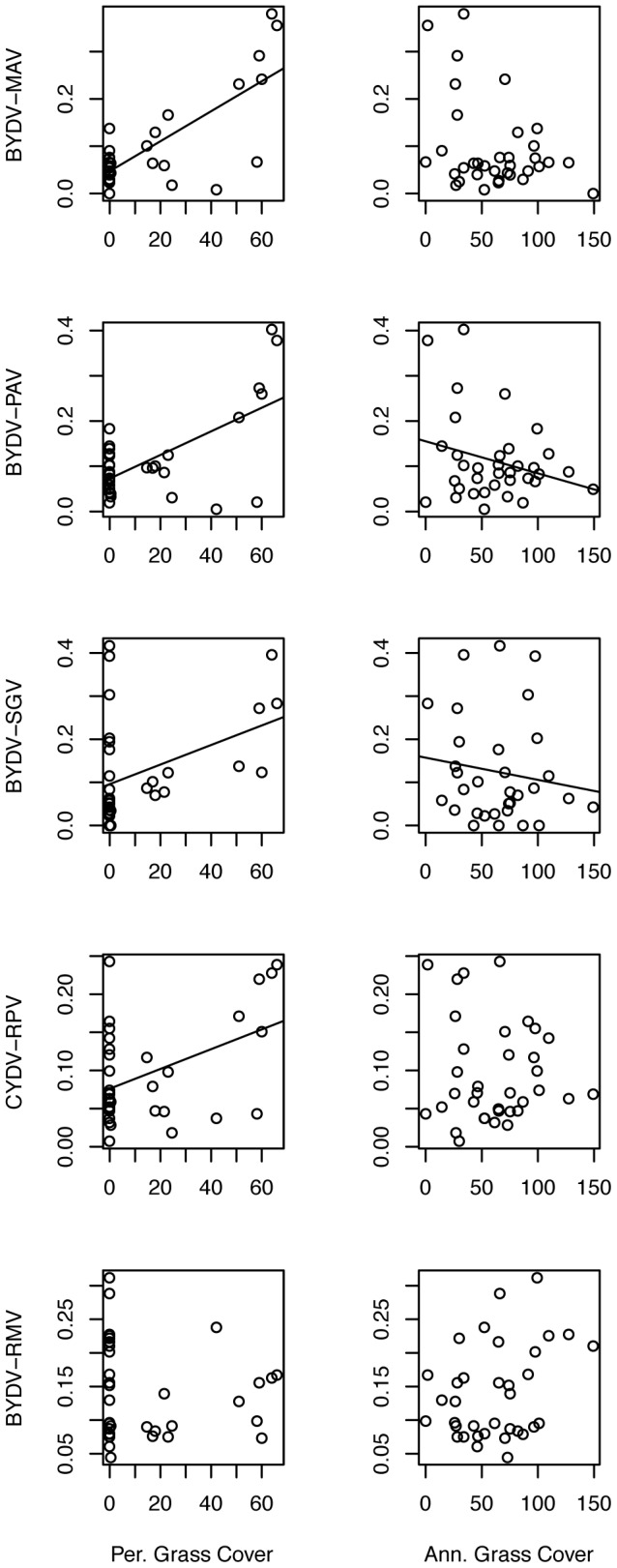
Lines indicate the slope of significant relationships.
Figure 4. Effects of phosphorus addition on the prevalence of five viral species.
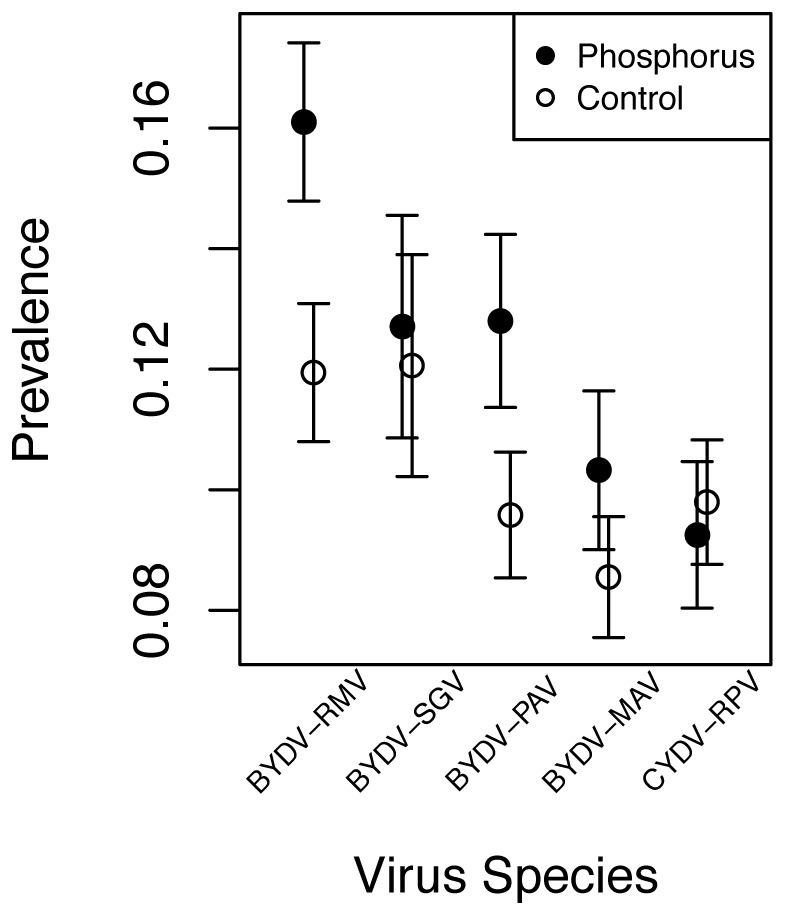
Error bars represent ± one SE from the mean.
To understand the nature of these strong effects of host-community composition, phosphorus, and host identity on viral community composition, we examined the univariate coefficients for the responses of individual viruses in the PerMANOVA models (Table S5). All viruses except BYDV-RMV increased with cover of perennial grasses, and BYDV-PAV and BYDV-SGV decreased with cover of annual grasses (Figure 3). As a result of this concordant response, viral richness within infected hosts increased 58% from 1.97 virus species per host in annual-only plots to 3.13 virus species per host in plots with more than 50% cover of perennial grasses (Figure 5; Table S6). Two viruses, BYDV-RMV and BYDV-PAV, increased in prevalence by approximately 30% in response to phosphorus addition, while the remaining viruses were unaffected (Figure 4).
Figure 5. Coinfection (viral species richness per infected host) of five viruses along a gradient of perennial grass cover.
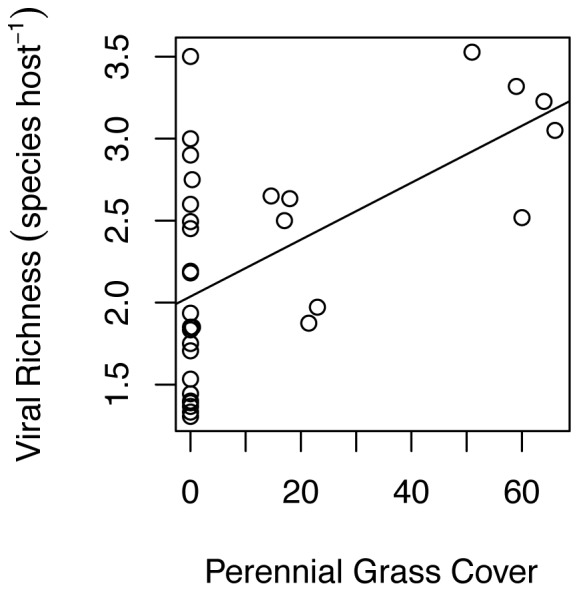
The line represents the slope of this relationship.
Discussion
Much of our understanding of host-pathogen interactions arises from detailed studies examining the coupled interactions between a single pathogen, one host species, and the environment [3], [6], [7], [8]. It is increasingly apparent that interactions in the disease triangle are mediated by a larger ecological context that includes multiple pathogens and host species, gradients of abiotic parameters, and complex landscapes [5], [19], [21], [41], [43], [64], [65]. Nevertheless, few studies have attempted to examine the drivers of multi-pathogen communities across a range of hosts and environmental conditions.
In a unique, regionally replicated, multi-host experiment, Borer et al. [40] found that that the infection of hosts by B/CYD viruses increased with additional phosphorus and the cover of long-lived, perennial hosts. While this host-centered analysis clarified the role of these factors for host infection, it did not clarify the effects of environmental factors on the composition and richness of the virus community within hosts. Here we show how changes in infection prevalence at the scale of hosts are determined by the niches of individual virus species. For example, our current analyses demonstrate that the increase in host infection prevalence with increasing perennial host abundance was caused by a concordant response of most viral species leading to a corresponding increase in viral richness within hosts. In contrast, the increase in overall prevalence arising from phosphorus addition [40] was primarily driven by an increase in two viruses (BYDV-PAV and BYDV-RMV). Thus, our current virus-centered analysis of environmental drivers of virus community diversity and composition within hosts provides novel insights into mechanisms leading to patterns of host infection.
Our current work demonstrates that the composition of the viral community within individual hosts varied across a wide range of spatial scales from hundreds of km to a few meters. Among control plots, representing natural biotic and abiotic gradients, the composition of the viral community differed among states (Oregon or California), sites, and blocks within sites. Specifically, the viral community varied with local host-community composition, phosphorus addition, host species identity, and vector foraging preferences among host species, suggesting the greater importance of niche differentiation compared to neutral processes in determining virus community assembly within hosts.
Host community composition was the most important determinant of spatial variability in viral community richness and composition, and these host-specific patterns of virus prevalence could be the result of interspecific variation in plant susceptibility to virus infection. Four of the five viruses increased in prevalence as perennial grass cover increased, resulting in a net 60% increase in the mean number of virus species in each host. Virus responses to host identity and nutrient supply were more complex, with the prevalence of each virus bearing a unique signal of response to phosphorus addition and host identity. For example, phosphorus addition had the strongest effects on the prevalence of BYDV-PAV and BYDV-RMV, and viruses differed in their ordering of the host species in terms of prevalence. Viruses depend on host cellular machinery to complete their replication cycle and require specific interactions between virus- and host-encoded factors [15]. A disruption of virus-host recognition, which could be caused by the presence of a tolerance or resistance gene in the host genome, can inhibit viral infection [66]. While some tolerance and resistance sources have been identified in wild relatives of cultivated grasses [67], [68], [69], these have been very limited in number. Our results suggest that their presence in the six host species selected in this study and impact on virus prevalence would be a fruitful avenue for future investigation.
In addition to host identity and phosphorus addition, virus community composition also was related strongly to differences in vector competence and host preference. As has been seen in observational studies and modeling work in this system [25], [56], viruses that share a vector coinfect hosts more frequently. In addition, virus distribution among hosts reflected vector-feeding preferences. Host specificity for the viruses carried by the widespread and common vector, R. padi, (BYDV-PAV and CYDV-RPV) were predicted extremely well by aphid preferences among host species in lab experiments (r = 0.82 and r = 0.96 respectively), and this correlation with vector preference was reflected in the overall prevalence of these viruses in the field, demonstrating that vector behavior and choices are important components of the non-random viral community assembly observed in this system.
Recent research in disease ecology has demonstrated that abiotic and biotic contexts are critical determinants of pathogen dynamics. A particular focus has been the prediction of human impacts, such as nutrient addition, habitat loss, biological invasions, and climate change, on pathogen epidemics [21], [38], [40], [41], [51], [70]. For example, the impacts of such globally critical human diseases as malaria and Lyme disease are mediated by both climate and the composition of the host community [39], [41], [43]. Recent work on plant pathogens has led to major advances by experimentally testing ecological drivers of pathogen distribution at scales or in ways that are logistically or ethically impossible in human and wildlife pathogen systems [40], [46], [47], [48], [71]. The work here extends our knowledge of the more complex dynamics of multi-host, multi-pathogen systems.
The importance of linking host and pathogen communities is highlighted by our finding that multiple viruses coinfected about half of infected hosts, and that coinfection rates arise from predictable differences in the environmental niches of the pathogens comprising the community. Specifically, we have demonstrated experimentally that the structure of multi-pathogen communities arises from niche differentiation among individual pathogens in response to the abiotic environment, host community composition, and the characteristics of individual hosts. For example, while phosphorus supplies increased overall infection rates [40], our current analyses demonstrate that the aggregate response predominantly reflects the large increases of a few virus species (BYDV-PAV and BYDV-RMV). Thus, the hosts in a phosphorus-rich system have both greater total infection prevalence and altered pathogen composition, contrasting strongly with the expectation that changing environments should not lead to alterations of species richness for neutrally-assembled communities [36], [37]. The altered community composition can be important, as infection by multiple viruses can reduce host fitness, especially if viruses are distantly related [28], [56].
Nutrient addition can act on pathogens through multiple pathways such as alteration of host community composition, host quality for vectors, pathogen reproduction, and host tolerance of infection. For example, nutrient additions often lead to declines in plant species diversity [79], [80], and declines in species diversity have been shown to lead to increased pathogen prevalence via the dilution effect [19], [20], [41], [48]. In this experiment, phosphorus addition increased total host biomass and total plant biomass, however it did not alter species richness or the relative abundance of perennial grasses [40]. We controlled for changes in total productivity and community composition in our analyses, and still found strong effects of phosphorus on the prevalence of some virus species (BYDV-PAV and BYDV-RMV). Thus, it is unlikely that the phosphorus effects on infection were driven solely by changes in vector or plant community composition, diversity, or productivity. While fertilization can alter vector performance or preference, aphid vector responses are primarily driven by nitrogen and not phosphorus [58]. Further, in experimental inoculations of our six focal host species, hosts physiologically adapted to greater nutrient supplies, e.g. with greater leaf nitrogen concentrations, were more susceptible to infection by BYDV-PAV [81]. Phosphorus has been shown to control viral reproduction rates [76], but the role of nutrient limitation of growth for each virus in this system remains unresolved and is a fruitful avenue for future research. Similarly, we do not have direct measurements of whether nutrient supplies differentially alter host tolerance or resistance to each of the B/CYDV's, as is the case for some other pathogens [77], [78]. Despite the complexity of these interactions, the consistently higher pathogen loads of hosts in eutrophic systems have important implications for disease management. Human activity has led to an eightfold increase in phosphorus and fourfold increase in nitrogen in the earth's ecosystems [72], [73], [74], [75], and nutrient additions have been shown to alter pathogen prevalence or reproduction in a wide array of human, animal, and plant systems [21], [40], [76], [77], [78].
Infection rates by each virus species generally increased with phosphorus addition, but the magnitude of this increase differed among host species. In particular, Avena fatua, an exotic annual grass that is known as a reservoir host species in this and other grass systems [47], [53], was associated with the greatest increase in prevalence of 4 of the 5 viral species in response to additional nutrients. In contrast, virus species responses to fertilization in Elymus glaucus, a widespread native perennial, consistently were less pronounced compared to A. fatua. More generally, these results highlight the important role of host species identity for determining virus infection and coinfection rates. Variation in host species traits is important for determining transmission rates and patterns of infection in many generalist pathogens including sudden oak death, Lyme disease, and West Nile virus [41], [81], [82], [83], and our current results suggest that host nutrition and physiology also may interact to control coinfection rates by related pathogen species.
Recent research in disease ecology has been building linkages between host community composition, host diversity and infection risk for single pathogens or specialist pathogens (e.g., pathogen spillover and the dilution effect) [19], [20], [41], [47], [48]. While there were no effects of host species richness in our system, we did find that infection risk by most viruses increased with the presence of long-lived perennial hosts as has been shown for aggregate infection risk [40]. Along a gradient from annual-dominated to perennial-dominated host communities, we found that the aggregated response of the individual viruses led to a 60% increase in the number of viruses carried by individual hosts. Given that coinfection rates can have strong effects on pathogen transmission and virulence [84], [85], [86], [87], it is important to expand the investigation of host community effects of pathogens to include coinfection.
Finally, we found that host-vector interactions were critical determinants of infection risk, as has been found in other systems such as mosquito-vectored pathogens [11], demonstrating that vector preference can provide a general key to predicting host infection rates in both animals and plants. Foraging preference of aphid vectors in the lab was a surprisingly strong predictor of infection among hosts in the field. The role of vector behavior is particularly important, as vector transmitted pathogens are the dominant form of emerging plant and animal diseases [88], [89]. There have been attempts to exploit vector feeding preferences to benefit human health by maintaining preferred hosts in close proximity to humans (i.e., zooprophylaxis) [43], [90], [91], however preferred hosts may also increase vector densities so it remains unclear if host preference will alter infection risk over longer periods of time [43]. Our results suggest that vector foraging preferences led to the observed strong and consistent differences in infection risk among hosts grown in close proximity. This linkage between laboratory measurements of vector behavior and infection risk across large geographic gradients suggests that disease control strategies may be able to effectively exploit vector preference to control disease risk.
While there has been growing evidence that host community and ecosystem context are critical drivers of host-pathogen interactions [19], [20], [21], [40], [41], [47], [48], [76], [77], [78], most of the existing evidence is based on observational studies, is not replicated at regional scales, or has focused on single pathogens. Here we experimentally demonstrate pathogen niche differentiation arising from differential responses of each pathogen species to host identity, the suite of hosts in the surrounding community, and the supply of nutrient resources. We also found concordant responses across these coexisting pathogens; most viral species were more common in the presence of long-lived hosts. This concordant response resulted in increases in both aggregate viral prevalence and coinfection within hosts. The net result was a mosaic of coinfection patterns in which the pathogen load of each host bore the imprint of its own identity and its biotic and abiotic context. Additional regionally-replicated, experimental studies of coinfection among other pathogen groups will provide the more general framework necessary to forecast the effects of human-induced changes to climate and nutrient supplies on global patterns of disease [46].
Supporting Information
Results of permutational multivariate analysis of variance (PERMANOVA) testing similarity in the viral community (BYDV-MAV, BYDV-PAV, BYDV-SGV, BYDV-RMV, CYDV-RPV) in infected individuals of six grass species (Avena fatua, Bromus carinatus, Bromus hordeaceus, Elymus glaucus, Koeleria macrantha, and Taeniatherum caput-medusae among states (Oregon or California), sites within states, blocks within sites, quadrats within plots, and host species within a quadrat. All terms not in the reduced model presented here were not significant (p>0.05). Sum of squares are sequential and so represent the nested spatial structure. Note these data are only from control plots and so represent background variability in the viral community. In addition, there is only a single plot per block, so there is not estimate of variability among plots within blocks.
(DOCX)
Results of permutational multivariate analysis of variance (PERMANOVA) testing the effect of factorial additions of nitrogen and phosphorus on the prevalence of five different viruses (BYDV-MAV, BYDV-PAV, BYDV-SGV, BYDV-RMV, CYDV-RPV) in infected individuals of six grass hosts (Avena fatua, Bromus carinatus, Bromus hordeaceus, Elymus glaucus, Koeleria macrantha, and Taeniatherum caput-medusae. Permutations were constrained within unique block by site combinations testing for the effects of nitrogen, phosphorus, and host species after controlling for variation among blocks, sites, and states. Full model contained all two-way interactions between nitrogen, phosphorus, and host species. Note that among-host differences were not associated with host lifespan (annual or perennial), provenance (native or exotic), or phylogenetic group (bromes, oats, or rye).
(DOCX)
Results of permutational multivariate analysis of variance (PERMANOVA) testng the effect of perennial grass cover, annual grass cover, forb cover, and factorial additions of nitrogen and phosphorus on the prevalence of five different viruses (BYDV-MAV, BYDV-PAV, BYDV-SGV, BYDV-RMV, CYDV-RPV) in infected individuals of six grass species (Avena fatua, Bromus carinatus, Bromus hordeaceus, Elymus glaucus, Koeleria macrantha, and Taeniatherum caput-medusae). Full model contained total live biomass, host species richness, perennial grass cover, annual grass cover, forb cover and all two-way interactions between.
(DOCX)
Linear coefficients for individual viral species from PERMANOVA (Table S2) testing the effect of factorial additions of nitrogen and phosphorus on the prevalence of five different viruses (BYDV-MAV, BYDV-PAV, BYDV-SGV, BYDV-RMV, CYDV-RPV) in infected individuals of six grass species (Avena fatua, Bromus carinatus, Bromus hordeaceus, Elymus glaucus, Koeleria macrantha, and Taeniatherum caput-medusae).
(DOCX)
Linear coefficients for individual viral species from PERMANOVA (Table S4) testing the effect of perennial grass cover, annual grass cover, forb cover, and factorial additions of nitrogen and phosphorus on the prevalence of five different viruses (BYDV-MAV, BYDV-PAV, BYDV-SGV, BYDV-RMV, CYDV-RPV) in infected individuals of six grass species (Avena fatua, Bromus carinatus, Bromus hordeaceus, Elymus glaucus, Koeleria macrantha, and Taeniatherum caput-medusae).
(DOCX)
Results of mixed-effects model testing the effect of perennial grass cover, annual grass cover, forb cover, and factorial additions of nitrogen and phosphorus on viral species richness (coinfection) by five different viruses (BYDV-MAV, BYDV-PAV, BYDV-SGV, BYDV-RMV, CYDV-RPV) in infected individuals of six grass hosts (Avena fatua, Bromus carinatus, Bromus hordeaceus, Elymus glaucus, Koeleria macrantha, and Taeniatherum caput-medusae. Full model contained total live biomass, host species richness, perennial grass cover, annual grass cover, forb cover and all two-way interactions between nitrogen, phosphorus, and host species. State, Site, Block, and Plot were treated as nested random effects.
(DOCX)
Acknowledgments
The work presented here would not have been possible without the support of many people and organizations. Marty Dekkers, Burl Martin, Emily Orling, and Jasmine Peters were responsible for much of the data collection. Vincent Adams performed the aphid feeding preference trials. We also acknowledge the generous support of the staff of the University of California Reserve System (McLaughlin Natural Reserve), University of California Research and Extension Centers (Hopland REC and Sierra Foothill REC), and the U.S. Fish and Wildlife Service (Baskett Slough and William L. Finley National Wildlife Refuges).
Funding Statement
Support for this project was provided, in part, by National Science Foundation (NSF) grants EF-0525666 and DEB-1015805 to E.T. Borer and E.W. Seabloom, EF-05-25641 and DEB-10-15909 to C.E. Mitchell, and EF-0525669 and DEB-1015903 to A.G. Power as part of the joint NSF-National Institutes of Health Ecology of Infectious Disease program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jones LR (1924) The Relation of Environment to Disease in Plants. American Journal of Botany 11: 601–609. [Google Scholar]
- 2. Colhoun J (1973) Effects of environmental factors on plant disease. Annual Review of Phytopathology 11: 343–364. [Google Scholar]
- 3.Agrios GN (1978) Plant Pathology. Orlando, Floriday, USA: Academic Press.
- 4. Kniskern JM, Rausher MD (2006) Environmental variation mediates the deleterious effects of Coleosporium ipomoeae on Ipomoea purpurea. Ecology 87: 675–685. [DOI] [PubMed] [Google Scholar]
- 5. Springer YP, Hardcastle BA, Gilbert GS (2007) Soil calcium and plant disease in serpentine ecosystems: a test of the pathogen refuge hypothesis. Oecologia 151: 10–21. [DOI] [PubMed] [Google Scholar]
- 6. Al-Naimi FA, Garrett KA, Bockus WW (2005) Competition, facilitation, and niche differentiation in two foliar pathogens. Oecologia 143: 449–457. [DOI] [PubMed] [Google Scholar]
- 7. Burdon JJ, Chilvers GA (1982) HOST DENSITY AS A FACTOR IN PLANT-DISEASE ECOLOGY. Annual Review of Phytopathology 20: 143–166. [Google Scholar]
- 8. Thrall PH, Godfree R, Burdon JJ (2003) Influence of spatial structure on pathogen colonization and extinction: a test using an experimental metapopulation. Plant Pathology 52: 350–361. [Google Scholar]
- 9. Carlsson-Graner U, Thrall PH (2006) The impact of host longevity on disease transmission: host-pathogen dynamics and the evolution of resistance. Evolutionary Ecology Research 8: 659–675. [Google Scholar]
- 10. Gilbert GS, Webb CO (2007) Phylogenetic signal in plant pathogen-host range. Proceedings of the National Academy of Sciences of the United States of America 104: 4979–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lyimo IN, Ferguson HM (2009) Ecological and evolutionary determinants of host species choice in mosquito vectors. Trends in Parasitology 25: 189–196. [DOI] [PubMed] [Google Scholar]
- 12. Dodds P, Thrall P (2009) Recognition events and host-pathogen co-evolution in gene-for-gene resistance to flax rust. Functional Plant Biology 36: 395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Flor H (1955) Host-parasite interactions in flax - its genetic and other implications. Phytopathology 45: 680–685. [Google Scholar]
- 14. Garcia-Arenal F, Fraile A, Malpica JM (2003) Variation and evolution of plant virus populations. International Microbiology 6: 225–232. [DOI] [PubMed] [Google Scholar]
- 15. Ahlquist P, Noueiry AO, Lee WM, Kushner DB, Dye BT (2003) Host factors in positive-strand RNA virus genome replication. Journal of Virology 77: 8181–8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fereres A, Moreno A (2009) Behavioural aspects influencing plant virus transmission by homopteran insects. Virus Research 141: 158–168. [DOI] [PubMed] [Google Scholar]
- 17. Minakawa N, Omukunda E, Zhou GF, Githeko A, Yan GY (2006) Malaria vector productivity in relation to the highland environment in Kenya. American Journal of Tropical Medicine and Hygiene 75: 448–453. [PubMed] [Google Scholar]
- 18. Lello J, Boag B, Fenton A, Stevenson IR, Hudson PJ (2004) Competition and mutualism among the gut helminths of a mammalian host. Nature 428: 840–844. [DOI] [PubMed] [Google Scholar]
- 19. Ezenwa VO, Godsey MS, King RJ, Guptill SC (2006) Avian diversity and West Nile virus: testing associations between biodiversity and infectious disease risk. Proceedings of the Royal Society B-Biological Sciences 273: 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Keesing F, Holt RD, Ostfeld RS (2006) Effects of species diversity on disease risk. Ecology Letters 9: 485–498. [DOI] [PubMed] [Google Scholar]
- 21. Pope K, Masuoka P, Rejmankova E, Grieco J, Johnson S, et al. (2005) Mosquito habitats, land use, and malaria risk in Belize from satellite imagery. Ecological Applications 15: 1223–1232. [Google Scholar]
- 22. Dobson A, Lafferty KD, Kuris AM, Hechinger RF, Jetz W (2008) Homage to Linnaeus: How many parasites? How many hosts? Proceedings of the National Academy of Sciences of the United States of America 105: 11482–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lindenfors P, Nunn CL, Jones KE, Cunningham AA, Sechrest W, et al. (2007) Parasite species richness in carnivores: effects of host body mass, latitude, geographical range and population density. Global Ecology and Biogeography 16: 496–509. [Google Scholar]
- 24. Strong DR, Levin DA (1975) Species richness of parasitic fungi of British trees. Proceedings of the National Academy of Sciences of the United States of America 72: 2116–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seabloom EW, Borer ET, Mitchell CE, Power AG (2010) Viral diversity and prevalence gradients in North American Pacific Coast grasslands. Ecology 91: 721–732. [DOI] [PubMed] [Google Scholar]
- 26. Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, et al. (2010) A Catalog of Reference Genomes from the Human Microbiome. Science 328: 994–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ (2010) Changes in the Composition of the Human Fecal Microbiome After Bacteriotherapy for Recurrent Clostridium difficile-associated Diarrhea. Journal of Clinical Gastroenterology 44: 354–360. [DOI] [PubMed] [Google Scholar]
- 28. Miller WA, Rasochova L (1997) Barley yellow dwarf viruses. Annual Review of Phytopathology 35: 167–190. [DOI] [PubMed] [Google Scholar]
- 29. Hood ME (2003) Dynamics of multiple infection and within-host competition by the anther-smut pathogen. American Naturalist 162: 122–133. [DOI] [PubMed] [Google Scholar]
- 30. Lal RB, Rudolph D, Alpers MP, Sulzer AJ, Shi YP, et al. (1994) Immunologic cross-reactivity between structural proteins of human T-cell lymphotropic virus type I and the blood stage of Plasmodium falciparum. Clinical and Diagnostic Laboratory Immunology 1: 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kamal SM, Bianchi L, Al Tawil A, Koziel M, Khalifa KE, et al. (2001) Specific cellular immune response and cytokine patterns in patients coinfected with hepatitis C virus and Schistosoma mansoni. Journal of Infectious Diseases 184: 972–982. [DOI] [PubMed] [Google Scholar]
- 32. Kamal SM, Rasenack JW, Bianchi L, Al Tawil A, Khalifa KES, et al. (2001) Acute hepatitis C without and with schistosomiasis: Correlation with hepatitis C-specific CD4(+) T-cell and cytokine response. Gastroenterology 121: 646–656. [DOI] [PubMed] [Google Scholar]
- 33. Tirado SMC, Yoon KJ (2003) Antibody-dependent enhancement of virus infection and disease. Viral Immunology 16: 69–86. [DOI] [PubMed] [Google Scholar]
- 34. Power AG (1996) Competition between viruses in a complex plant-pathogen. Ecology 77: 1004–1010. [Google Scholar]
- 35. Dove ADM, Cribb TH (2006) Species accumulation curves and their applications in parasite ecology. Trends in Parasitology 22: 568–574. [DOI] [PubMed] [Google Scholar]
- 36. Chase JM (2007) Drought mediates the importance of stochastic community assembly. Proceedings of the National Academy of Sciences of the United States of America 104: 17430–17434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harpole WS, Tilman D (2006) Non-neutral patterns of species abundance in grassland communities. Ecology Letters 9: 15–23. [DOI] [PubMed] [Google Scholar]
- 38. Harvell CD, Mitchell CE, Ward JR, Altizer S, Dobson AP, et al. (2002) Ecology - Climate warming and disease risks for terrestrial and marine biota. Science 296: 2158–2162. [DOI] [PubMed] [Google Scholar]
- 39. Laneri K, Bhadra A, Ionides EL, Bouma M, Dhiman RC, et al. (2010) Forcing Versus Feedback: Epidemic Malaria and Monsoon Rains in Northwest India. Plos Computational Biology 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Borer ET, Seabloom EW, Mitchell CE, Power AG (2010) Local context drives infection of grasses by vector-borne generalist viruses. Ecology Letters 13: 810–818. [DOI] [PubMed] [Google Scholar]
- 41. LoGiudice K, Ostfeld RS, Schmidt KA, Keesing F (2003) The ecology of infectious disease: Effects of host diversity and community composition on Lyme disease risk. Proceedings of the National Academy of Sciences of the United States of America 100: 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cumming GS, Guegan JF (2006) Food webs and disease: Is pathogen diversity limited by vector diversity? Ecohealth 3: 163–170. [Google Scholar]
- 43. Dobson A, Cattadori I, Holt RD, Ostfeld RS, Keesing F, et al. (2006) Sacred Cows and Sympathetic Squirrels: The Importance of Biological Diversity to Human Health. Plos Medicine 3: 714–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Poulin R (1995) PHYLOGENY, ECOLOGY, AND THE RICHNESS OF PARASITE COMMUNITIES IN VERTEBRATES. Ecological Monographs 65: 283–302. [Google Scholar]
- 45. Hall GS, Peters JS, Little DP, Power AG (2010) Plant community diversity influences vector behaviour and Barley yellow dwarf virus population structure. Plant Pathology 59: 1152–1158. [Google Scholar]
- 46. Borer ET, Antonovics J, Kinkel LL, Hudson PJ, Daszak P, et al. (2011) Bridging taxonomic and disciplinary divides in infectious disease. Ecohealth [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Power AG, Mitchell CE (2004) Pathogen spillover in disease epidemics. American Naturalist 164: S79–S89. [DOI] [PubMed] [Google Scholar]
- 48. Mitchell CE, Tilman D, Groth JV (2002) Effects of grassland plant species diversity, abundance, and composition on foliar fungal disease. Ecology 83: 1713–1726. [Google Scholar]
- 49. Irwin ME, Thresh JM (1990) Epidemiology of Barley Yellow Dwarf - a study in ecological complexity. Annual Review of Phytopathology 28: 393–424. [Google Scholar]
- 50.D'Arcy C (1995) Symptomology and host range of Barley Yellow Dwarf. In: D'Arcy CJ, Burnett PA, editors. Barley Yellow Dwarf: 40 Years of Progress. St. Paul, Minnesota: The American Phytopathological Society. pp. 9–28.
- 51. Borer ET, Hosseini PR, Seabloom EW, Dobson AP (2007) Pathogen-induced reversal of native dominance in a grassland community. Proceedings of the National Academy of Sciences of the United States of America 104: 5473–5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Malmstrom CM, Hughes CC, Newton LA, Stoner CJ (2005) Virus infection in remnant native bunchgrasses from invaded California grasslands. New Phytologist 168: 217–230. [DOI] [PubMed] [Google Scholar]
- 53. Malmstrom CM, McCullough AJ, Johnson HA, Newton LA, Borer ET (2005) Invasive annual grasses indirectly increase virus incidence in California native perennial bunchgrasses. Oecologia 145: 153–164. [DOI] [PubMed] [Google Scholar]
- 54.Halbert S, Voegtlin D (1995) Biology and taxonomy of vectors of barley yellow dwarf viruses. In: D'Arcy CJ, Burnett PA, editors. Barley Yellow Dwarf: 40 Years of Progress. St. Paul, Minnesota: The American Phytopathological Society. pp. 217–258.
- 55. Rochow WF (1970) Barley Yellow Dwarf Virus: Phenotypic Mixing and Vector Specificity. Science 167: 875–878. [DOI] [PubMed] [Google Scholar]
- 56. Seabloom EW, Hosseini PR, Power AG, Borer ET (2009) Diversity and Composition of Viral Communities: Coinfection of Barley and Cereal Yellow Dwarf Viruses in California Grasslands. The American Naturalist 173: E79–E98. [DOI] [PubMed] [Google Scholar]
- 57. Seabloom EW, Williams JW, Slayback D, Stoms DM, Viers JH, et al. (2006) Human impacts, plant invasion, and imperiled, plant species in California. Ecological Applications 16: 1338–1350. [DOI] [PubMed] [Google Scholar]
- 58. Borer ET, Adams VT, Engler GA, Adams AL, Schumann CB, et al. (2009) Aphid fecundity and grassland invasion: invader life history is the key. Ecological Applications 19: 1187–1196. [DOI] [PubMed] [Google Scholar]
- 59.R Development Core Team (2010) R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria.
- 60. Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecology 26: 32–46. [Google Scholar]
- 61. Legendre P, Anderson MJ (1999) Distance-based redundancy analysis: Testing multispecies responses in multifactorial ecological experiments. Ecological Monographs 69: 1–24. [Google Scholar]
- 62. McArdle BH, Anderson MJ (2001) Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology 82: 290–297. [Google Scholar]
- 63.Crawley MJ (2007) The R Book. West Sussex, England: John Wiley & Sons Ltd. 942 p.
- 64. Gaidet N, Caron A, Cappelle J, Cumming GS, Balanca G, et al. (2012) Understanding the ecological drivers of avian influenza virus infection in wildfowl: a continental-scale study across Africa. Proceedings of the Royal Society B-Biological Sciences 279: 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yoshida-Takashima Y, Nunoura T, Kazama H, Noguchi T, Inoue K, et al. (2012) Spatial Distribution of Viruses Associated with Planktonic and Attached Microbial Communities in Hydrothermal Environments. Applied and Environmental Microbiology 78: 1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sacristan S, Garcia-Arenal F (2008) The evolution of virulence and pathogenicity in plant pathogen populations. Molecular Plant Pathology 9: 369–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang ZY, Lin ZS, Xin ZY (2009) Research progress in BYDV resistance genes derived from wheat and its wild relatives. Journal of Genetics and Genomics 36: 567–573. [DOI] [PubMed] [Google Scholar]
- 68. Kosova K, Chrpova J, Sip V (2008) Recent advances in breeding of cereals for resistance to barley yellow dwarf virus - A review. Czech Journal of Genetics and Plant Breeding 44: 1–10. [Google Scholar]
- 69. Scholz M, Ruge-Wehling B, Habekuss A, Schrader O, Pendinen G, et al. (2009) Ryd4 (Hb): a novel resistance gene introgressed from Hordeum bulbosum into barley and conferring complete and dominant resistance to the barley yellow dwarf virus. Theoretical and Applied Genetics 119: 837–849. [DOI] [PubMed] [Google Scholar]
- 70. Tompkins DM, White AR, Boots M (2003) Ecological replacement of native red squirrels by invasive greys driven by disease. Ecology Letters 6: 189–196. [Google Scholar]
- 71. Borer ET, Mitchell CE, Power AG, Seabloom EW (2009) Consumers indirectly increase infection risk in grassland food webs. Proceedings of the National Academy of Sciences of the United States of America 106: 503–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tilman D, Fargione J, Wolff B, D'Antonio C, Dobson A, et al. (2001) Forecasting agriculturally driven global environmental change. Science 292: 281–284. [DOI] [PubMed] [Google Scholar]
- 73. Vitousek PM, Aber JD, Howarth RW, Likens GE, Matson PA, et al. (1997) Human alteration of the global nitrogen cycle: Sources and consequences. Ecological Applications 7: 737–750. [Google Scholar]
- 74. Vitousek PM, Mooney HA, Lubchenco J, Melillo JM (1997) Human domination of Earth's ecosystems. Science 277: 494–499. [Google Scholar]
- 75. Rockstrom J, Steffen W, Noone K, Persson A, Chapin FS, et al. (2009) A safe operating space for humanity. Nature 461: 472–475. [DOI] [PubMed] [Google Scholar]
- 76. Clasen JL, Elser JJ (2007) The effect of host Chlorella NC64A carbon : phosphorus ratio on the production of Paramecium bursaria Chlorella Virus-1. Freshwater Biology 52: 112–122. [Google Scholar]
- 77. Beck MA, Levander OA (2000) Host nutritional status and its effect on a viral pathogen. Journal of Infectious Diseases 182: S93–S96. [DOI] [PubMed] [Google Scholar]
- 78. Smith VH, Jones TP, Smith MS (2005) Host nutrition and infectious disease: an ecological view. Frontiers in Ecology and the Environment 3: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hillebrand H, Gruner DS, Borer ET, Bracken MES, Cleland EE, et al. (2007) Consumer versus resource control of producer diversity depends on ecosystem type and producer community structure. Proceedings of the National Academy of Sciences of the United States of America 104: 10904–10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Harpole WS, Tilman D (2007) Grassland species loss resulting from reduced niche dimension. Nature 446: 791–793. [DOI] [PubMed] [Google Scholar]
- 81. Cronin JP, Welsh ME, Dekkers MG, Abercrombie ST, Mitchell CE (2010) Host physiological phenotype explains pathogen reservoir potential. Ecology Letters 13: 1221–1232. [DOI] [PubMed] [Google Scholar]
- 82. Allan BF, Langerhans RB, Ryberg WA, Landesman WJ, Griffin NW, et al. (2009) Ecological correlates of risk and incidence of West Nile virus in the United States. Oecologia 158: 699–708. [DOI] [PubMed] [Google Scholar]
- 83. Rizzo DM, Garbelotto M (2003) Sudden oak death: endangering California and Oregon forest ecosystems. Frontiers in Ecology and the Environment 1: 197–204. [Google Scholar]
- 84. Seabloom EW, Borer ET, Jolles A, Mitchell CE (2009) Direct and indirect effects of viral pathogens and the environment on invasive grass fecundity in Pacific Coast grasslands. Journal of Ecology 97: 1264–1273. [Google Scholar]
- 85. Wen F, Lister RM, Fattouh FA (1991) Cross-protection among strains of barley yellow dwarf virus. Journal of General Virology 72: 791–799. [DOI] [PubMed] [Google Scholar]
- 86. Bentwich Z, Kalinkovich A, Weisman Z, Borkow G, Beyers N, et al. (1999) Can eradication of helminthic infections change the face of AIDS and tuberculosis? Immunology Today 20: 485–487. [DOI] [PubMed] [Google Scholar]
- 87. Jolles AE, Ezenwa VO, Etienne RS, Turner WC, Olff H (2008) Interactions between macroparasites and microparasites drive infection patterns in free-ranging African buffalo. Ecology 89: 2239–2250. [DOI] [PubMed] [Google Scholar]
- 88. Taylor LH, Latham SM, Woolhouse MEJ (2001) Risk factors for human disease emergence. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences 356: 983–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Power AG, Flecker AS (2003) Virus specificity in disease systems: are species redundant? In: Kareiva P, Levin SA, editors. The Importance of Species: Perspectives on Expendability and Triage. Princeton, NJ: Princeton University Press. pp. 330–346.
- 90. Service MW (1991) AGRICULTURAL-DEVELOPMENT AND ARTHROPOD-BORNE DISEASES - A REVIEW. Revista De Saude Publica 25: 165–178. [DOI] [PubMed] [Google Scholar]
- 91. Sota T, Mogi M (1989) EFFECTIVENESS OF ZOOPROPHYLAXIS IN MALARIA CONTROL - A THEORETICAL INQUIRY, WITH A MODEL FOR MOSQUITO POPULATIONS WITH 2 BLOODMEAL HOSTS. Medical and Veterinary Entomology 3: 337–345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results of permutational multivariate analysis of variance (PERMANOVA) testing similarity in the viral community (BYDV-MAV, BYDV-PAV, BYDV-SGV, BYDV-RMV, CYDV-RPV) in infected individuals of six grass species (Avena fatua, Bromus carinatus, Bromus hordeaceus, Elymus glaucus, Koeleria macrantha, and Taeniatherum caput-medusae among states (Oregon or California), sites within states, blocks within sites, quadrats within plots, and host species within a quadrat. All terms not in the reduced model presented here were not significant (p>0.05). Sum of squares are sequential and so represent the nested spatial structure. Note these data are only from control plots and so represent background variability in the viral community. In addition, there is only a single plot per block, so there is not estimate of variability among plots within blocks.
(DOCX)
Results of permutational multivariate analysis of variance (PERMANOVA) testing the effect of factorial additions of nitrogen and phosphorus on the prevalence of five different viruses (BYDV-MAV, BYDV-PAV, BYDV-SGV, BYDV-RMV, CYDV-RPV) in infected individuals of six grass hosts (Avena fatua, Bromus carinatus, Bromus hordeaceus, Elymus glaucus, Koeleria macrantha, and Taeniatherum caput-medusae. Permutations were constrained within unique block by site combinations testing for the effects of nitrogen, phosphorus, and host species after controlling for variation among blocks, sites, and states. Full model contained all two-way interactions between nitrogen, phosphorus, and host species. Note that among-host differences were not associated with host lifespan (annual or perennial), provenance (native or exotic), or phylogenetic group (bromes, oats, or rye).
(DOCX)
Results of permutational multivariate analysis of variance (PERMANOVA) testng the effect of perennial grass cover, annual grass cover, forb cover, and factorial additions of nitrogen and phosphorus on the prevalence of five different viruses (BYDV-MAV, BYDV-PAV, BYDV-SGV, BYDV-RMV, CYDV-RPV) in infected individuals of six grass species (Avena fatua, Bromus carinatus, Bromus hordeaceus, Elymus glaucus, Koeleria macrantha, and Taeniatherum caput-medusae). Full model contained total live biomass, host species richness, perennial grass cover, annual grass cover, forb cover and all two-way interactions between.
(DOCX)
Linear coefficients for individual viral species from PERMANOVA (Table S2) testing the effect of factorial additions of nitrogen and phosphorus on the prevalence of five different viruses (BYDV-MAV, BYDV-PAV, BYDV-SGV, BYDV-RMV, CYDV-RPV) in infected individuals of six grass species (Avena fatua, Bromus carinatus, Bromus hordeaceus, Elymus glaucus, Koeleria macrantha, and Taeniatherum caput-medusae).
(DOCX)
Linear coefficients for individual viral species from PERMANOVA (Table S4) testing the effect of perennial grass cover, annual grass cover, forb cover, and factorial additions of nitrogen and phosphorus on the prevalence of five different viruses (BYDV-MAV, BYDV-PAV, BYDV-SGV, BYDV-RMV, CYDV-RPV) in infected individuals of six grass species (Avena fatua, Bromus carinatus, Bromus hordeaceus, Elymus glaucus, Koeleria macrantha, and Taeniatherum caput-medusae).
(DOCX)
Results of mixed-effects model testing the effect of perennial grass cover, annual grass cover, forb cover, and factorial additions of nitrogen and phosphorus on viral species richness (coinfection) by five different viruses (BYDV-MAV, BYDV-PAV, BYDV-SGV, BYDV-RMV, CYDV-RPV) in infected individuals of six grass hosts (Avena fatua, Bromus carinatus, Bromus hordeaceus, Elymus glaucus, Koeleria macrantha, and Taeniatherum caput-medusae. Full model contained total live biomass, host species richness, perennial grass cover, annual grass cover, forb cover and all two-way interactions between nitrogen, phosphorus, and host species. State, Site, Block, and Plot were treated as nested random effects.
(DOCX)


