Abstract
Vitamin A is essential for vision and the growth/differentiation of almost all human organs. Plasma retinol binding protein (RBP) is the principle and specific carrier of vitamin A in the blood. Here we describe an optimized technique to produce and purify holo-RBP and two real-time monitoring techniques to study the transport of vitamin A by the high-affinity RBP receptor STRA6. The first technique makes it possible to produce a large quantity of high quality holo-RBP (100%-loaded with retinol) for vitamin A transport assays. High quality RBP is essential for functional assays because misfolded RBP releases vitamin A readily and bacterial contamination in RBP preparation can cause artifacts. Real-time monitoring techniques like electrophysiology have made critical contributions to the studies of membrane transport. The RBP receptor-mediated retinol transport has not been analyzed in real time until recently. The second technique described here is the real-time analysis of STRA6-catalyzed retinol release or loading. The third technique is real-time analysis of STRA6-catalyzed retinol transport from holo-RBP to cellular retinol binding protein I (CRBP-I). These techniques provide high sensitivity and resolution in revealing RBP receptor's vitamin A uptake mechanism.
Keywords: Biochemistry, Issue 71, Molecular Biology, Genetics, Cellular Biology, Molecular Biology, Anatomy, Physiology, Ophthalmology, Proteomics, Proteins, Membrane Transport Proteins, Vitamin A, retinoid, RBP complex, membrane transport, membrane receptor, STRA6, retinol binding protein
Introduction
Vitamin A is an organic molecule that is essential for human survival and the proper functioning of almost all human organs. Vitamin A derivatives (retinoids) participate in diverse biochemical and cellular events including the sensing of light for vision 1,2 and the regulation of gene expression and protein translation during embryonic development and in adult tissues 3-6. Although retinol has the ability to diffuse systemically, evolution came up with plasma retinol binding protein, a specific carrier protein for vitamin A transport in the blood to achieve high efficiency and specificity and to avoid toxicity associated with random diffusion 7-10. A high-affinity receptor that binds to RBP and takes up vitamin A was hypothesized in the 1970s 11-13. Despite evidence accumulated in three decades on the existence of the RBP receptor 14-31, the receptor hypothesis was debated for many years due to the existence of an incorrect definition of holo-RBP. The correct definition of holo-RBP is that it is the high affinity 1:1 complex between retinol and RBP. Repeated extraction of holo-RBP by organic solvent is necessary to produce apo-RBP. This definition is used by almost all labs studying RBP 7,9,32-35 or the RBP receptor 14-31,36-42. The incorrect definition of holo-RBP that was used to disprove the existence of the RBP receptor is the acute mixture of free retinol with apo-RBP. Since the function of the RBP receptor in vitamin A uptake from holo-RBP is to release retinol from holo-RBP, the RBP receptor would play no role in retinol uptake if retinol is free to begin with (as proposed by the incorrect definition of holo-RBP).
The recent identification of RBP receptor as a multitransmembrane domain protein called STRA636 and its function in vitamin A uptake from holo-RBP 36-43 strongly argues against the hypothesis that RBP does not need a receptor to deliver vitamin A. Detailed analyses revealed that STRA6 has 9 transmembrane domains with the N-terminus located extracellularly and C-terminus located intracellularly 40. Located between transmembrane 6 and 7 is an essential RBP binding domain 39. STRA6 is coupled to both LRAT and CRBP-I in vitamin A uptake from holo-RBP, but neither LRAT nor CRBP-I is absolutely required for enhanced STRA6 activity 41. STRA6's ability to catalyze retinol release from holo-RBP is the key to its vitamin A uptake activity 41. By relying on STRA6 to release its retinol, vitamin A delivery by RBP can transport vitamin A to target cells in peripheral tissues with high specificity and efficiency.
The critical importance of holo-RBP definition and preparation is illustrated not only by the historical debate on the existence of the RBP receptor, but also by three related recent papers based on holo-RBP definitions different from the original and correct definition 44-46. The first paper used the holo-RBP definition that was used to disapprove the existence of the RBP receptor to study the RBP receptor 44. The second and third papers came up with a third definition of holo-RBP that made it even less likely for retinol to be studied to form a proper complex with RBP 45,46. These studies prepared 3H-retinol/RBP by mixing holo-RBP (not even apo-RBP) with 3H-retinol. Since this assay did not have 3H-retinol/RBP formed and did not remove excessive free 3H-retinol 45,46, it is not an assay for 3H-retinol uptake from 3H-retinol/RBP, but is a free 3H-retinol diffusion assay. It has been shown previously that STRA6 does not enhance cellular uptake of free retinol by LRAT 38 or CRBP-I 41. Virtually all retinol is bound to RBP in the blood and there is no detectable free retinol. A main function of the RBP receptor is to catalyze retinol release from holo-RBP during retinol uptake from holo-RBP 41. If the retinol is artificially released or is in the free form to begin with 45,46, the RBP receptor is not needed. The dramatically different results obtained from the free retinol diffusion assay as compared to assays based on correctly prepared holo-RBP illustrate that correct preparation of RBP is crucial for its functional assays.
RBP can be purified from human serum 41, but the procedure is complex and the yield is low. An alternative approach is to produce RBP in E. coli. Because E. coli does not have the ability to correctly fold mammalian secreted proteins with more than one pair of disulfide bonds like RBP, it is essential to refold RBP and purify the correctly folded protein. Misfolded proteins not only behave differently from corrected folded RBP in various assays, but also cause protein aggregation during storage. For the same reason, apo-RBP is only produced from high-quality holo-RBP. We describe here an optimized protocol to produce high quality RBP 100% loaded with retinol through bacterial expression, refolding, and HPLC purification. HPLC purification not only removes incorrectly folded RBP but also significant bacterial contamination that can cause serious artifacts if RBP is used in signal transduction assays. We also describe two sensitive real-time monitoring techniques to study retinol transport by STRA6. Both techniques depend on high quality RBP. Due to space limitations, the classic techniques of radioactive retinoid-based and HPLC-based vitamin A uptake assays are not described here.
Protocol
1. Production, Refolding, and HPLC Purification of Holo-RBP
Transform BL-21cells with the pET3a vector harboring the cDNA for human RBP with 6x His tag on the N-terminus. Grow the transformed BL-21 cells in a shaker at 37 °C in 40 ml LB media with carbenicillin until O.D. at 600 nm reaches 0.5. Induce RBP protein expression by adding IPTG to 1mM. Grow the bacteria at 37 °C another 5 hr.
RBP produced in E.coli is mostly present in insoluble inclusion bodies. To enrich inclusion bodies, pellet cells at 10,000 x g for 20 min. Then sonicate the cells on ice in 5 ml PBS with protease inhibitors, followed by 2 cycles of freezing and thawing. Pellet down the inclusion bodies at 20,000 x g for 30 min at 4 °C. Remove supernatant and keep the pellet on ice.
Solubilize the inclusion body pellet by sonication in 10 ml 7.5 M guanidine hydrochloride. Add a half volume (5 ml) of 25 mM Tris buffer, pH 9.0 and DTT to a final concentration of 10 mM. Vigorously mix solution on a vortex mixer overnight at room temperature to fully reduce and denature proteins.
Centrifuge the protein solution to remove insoluble material at 18,000 x g for 20 min at 4 °C. After the spin, transfer the supernatant to a new tube and place the tube on ice.
Prepare four volumes (60 ml) of refolding buffer (25 mM Tris, pH 9.0, 0.3 mM cystine, 3.0 mM cysteine, 1 mM EDTA). Degas the refolding buffer. Also prepare 600 μl 10 mM retinol in ethanol under dim red light. Keep it in the dark on ice. Both the refolding buffer and retinol solution need to be freshly prepared.
Cool both the protein solution and the refolding buffer on ice for at least 30 min. Higher temperature likely reduces the quality of refolded RBP. Add 1 ml refolding buffer dropwise and 10 μl retinol solution in turn while protein solution is vigorously mixed in a beaker on ice. Repeat this step until all refolding buffer and retinol are added. Keep stirring the reaction on ice in the dark for 5 hr.
Spin down the refolding reaction at 24,000 x g for 30 min at 4 °C. It is essential to remove the aggregates right after the refolding reaction.
Concentrate the supernatant solution using Amicon Ultra 15 concentrator (MWCO 10 K) to about 10 ml. Do not concentrate the refolded solution more than this level. Concentrating too much will lead to protein aggregation and lowers the final yield and quality of correctly folded RBP. Dilute the concentrated sample with cold PBS to 100 ml. The purpose of this step is to remove the components in the refolding reaction that interfere with nickel purification.
Spin the solution one more time at 24,000 x g for 20 min at 4 °C. It is important to remove any precipitate. Transfer the supernatant to two 50 ml tubes, each containing 1 ml Ni-NTA slurry that has been washed with PBS. Rotate the tubes at 4 °C for one hour. Incubating diluted solution for 1 hr can cause the precipitation of misfolded proteins which need to be removed.
Spin down the tubes at 500 x g for 3 min. Remove supernatant carefully. Anything floating should be removed. Add cold PBS to 30 ml for each tube. Spin again and remove the supernatant. Repeat this process until you see nothing but beads in the tube.
Transfer the Ni-NTA resin to an empty column. Wash resin with 20 column volumes of 10 mM imidazole, 500 mM NaCl in PBS. Elute His-RBP in 5 column volumes of 100 mM imidazole in PBS. Do not store this solution for an extended period of time, and freezing reduces the final yield and quality. For the best result, purify RBP by HPLC immediately.
Dialyze His-RBP purified from the Ni-NTA resin against 25 mM Tris, pH 8.4, and 120 mM NaCl. A concentrator can also be used for buffer exchange. Clear samples for HPLC by centrifugation at 16,000 x g for 10 min at 4 °C right before each run.
Purify holo-RBP on HPLC using an ion exchange column AX-300 by a NaCl step gradient (220 mM 12 min, 360 mM 15 min, and 1,000 mM 15 min) using 25 mM Tris, pH 8.4 as mobile phase at 1 ml/min. As NaCl concentration rises, holo-His-RBP is released while apo-RBP and misfolded RBP stay bound to the column (Figure 1). If RBP refolding is not done optimally, misfolded RBP (e.g. misfolded RBP-I in Figure 1) can predominate the whole preparation.
Recover the correctly folded holo-RBP from the peak fractions at 360 mM NaCl. Carefully monitor the elution profile of correctly folded and misfolded His-RBP. At 1,000 mM NaCl, most of misfolded His-RBP should be released.
Fractions containing holo-RBP are pooled, concentrated, and dialyzed against PBS overnight at 4 °C. Check the final yield and quality (330 nm/280 nm) by spectrophotometer. Correctly folded product should have a 330 nm/280 nm ratio of slightly higher than 1 (Figure 1).
To produce apo-RBP, add an equal volume of heptane to the purified holo-RBP and mix gently by rotating overnight at 4 °C. Centrifuge at 16,000 g for 10 min at 4 °C and carefully transfer the bottom aqueous phase to a new tube (avoiding the precipitate at the interface). Repeat the process three times with a 3 hr incubation for each. Check absorption on a Nanodrop spectrophotometer to make sure the 330 nm peak has decreased to the background level.
2. Real-time Monitoring of STRA6-catalyzed Retinol Release and Retinol Loading
Block a black 96-well Microfluor-2 plate (Thermo Scientific) with 200 μl of Blocker Casein (Pierce) overnight at 4 °C to prevent nonspecific sticking of holo-RBP to the plastic. Blocker Casein gives the best blocking effect of the all the blocking solutions we tested.
Membranes freshly prepared from cells expressing STRA6 or control cells are washed using PBS and passed through a Hamilton syringe (Gastight #1710) 6 times to produce a uniform suspension in PBS. We typically use membranes derived from 1/100 to 1/20 of cells grown on a 100 mm dish per reaction.
Wash the coated wells of the Microfluor-2 plate once with 200 μl of PBS. Cool the plate on ice before addition of membrane suspension (50 μl per well).
Real time monitoring of retinol fluorescence is measured using fluorescence optics of the POLARstar Omega (BMG Labtech) with the excitation filter 320ex and the emission filter 460-10. The program is set to the Endpoint reading mode. The Plate Mode can also be used to measure a time course if the time intervals are not varied. The signal from each time point is the average of 10 measurements (orbital averaging with a diameter of 2 mm) and the gain is set at 1,800. The plate is shaken for 10 sec at 500 rpm using double-orbital shaking before each measurement.
To start the retinol release assay, the wells are read once using the Endpoint reading mode before holo-RBP (typically 1 μM final concentration) is added at 0 min. As soon as holo-RBP is added, the reaction is monitored continuously every 5-10 min for 1-3 hr. To start the retinol loading assay, all-trans retinol (typically 1 μM final concentration) is added to the wells under dim red light. The plate is read once before apo-RBP is added at 0 min. As soon as apo-RBP is added, the reaction is monitored continuously every 5-10 min for 1-3 hr.
After all measurements are done, download the raw data (example shown in Figure 2) from the Omega Data Analysis software (BMG Labtech) to Microsoft Excel for data analysis. Each time point generates a file that contains the readings of all experimental conditions. For example, if there are 20 time points, consolidate 20 files into one Excel file for data analysis.
For data analysis, the fluorescence signals before the addition of retinol or holo-RBP at 0 min are considered background signals and subtracted from the final fluorescence signals at all time points. Although the background signals are low compared to retinol fluorescence in holo-RBP, subtracting the background eliminates the nonspecific influence of light scattering by the membranes and makes it possible to focus on the retinol fluorescence of holo-RBP or retinol added at 0 min.
3. Real-time Monitoring of STRA6-catalyzed Transport of Retinol From Holo-RBP to CRBP-I
Retinol-EGFP is a new fluorescence resonance transfer (FRET) pair that has recently been established 41. STRA6-catalyzed retinol transport from holo-RBP to EGFP-CRBP-I is monitored in real time by measuring retinol-EGFP FRET. EGFP-CRBP-I fusion protein with 6XHis tag (EGFP-CRBP-I) is produced in mammalian cells and is purified on Ni-NTA resin before the experiment. EGFP-CRBP-I can be stored in PBS with protease inhibitors for a short period of time at 4 °C. Alternatively the fusion protein is frozen in -80 °C in small aliquots.
Before the reactions, prepare a blocked Microfluor-2 plate and membranes as described above. Wash the coated wells of the Microfluor-2 plate once with 200 μl PBS before addition of membrane suspension (50 μl per well).
Real-time retinol-EGFP FRET is measured using POLARstar Omega with the excitation filter 320ex and the emission filters 460-10 and 510-10 using simultaneous duel emission fluorescence optics. The program is set to the Endpoint reading mode. The Plate Mode can also be used to measure a time course if the time intervals are not varied. The signal from each time point is the average of 10 measurements (orbital averaging with a diameter of 2 mm) and the gain is set at 1,800 per channel. The plate is shaken for 10 sec at 500 rpm using double-orbital shaking before each measurement.
To start the reactions, EGFP-CRBP-I (typically 1 μM final concentration) is added to the wells. The plate is read once using the Endpoint reading mode before holo-RBP is added at 0 min. As soon as holo-RBP is added, the reaction is monitored continuously by measurement every 5-10 min for 1-2 hr.
After all measurements are done, download the raw data (example shown in Figure 3) from the Omega Data Analysis software to Microsoft Excel for data analysis as described above.
Retinol-EGFP FRET is calculated by the dynamic change in the ratio of the acceptor/donor emission peaks47. The equation41 to calculate this ratio is [(510t-510b)/(460t-460b)], where 510t, 510b, 460t, and 460b represent emissions at 510 nm after initiation of the reaction (t=time point), at 510 nm before retinol or holo-RBP is added (b=background), at 460 nm after initiation of the reaction (t=time point), and at 460 nm before retinol or holo-RBP is added (b=background), respectively.
Representative Results
We present here representative results for holo-RBP production and purification by HPLC (Figure 1), real-time analysis of STRA6-catalyzed retinol release from holo-RBP and retinol loading into apo-RBP (Figure 2) and real-time analysis of STRA6-catalyzed retinol transport from holo-RBP to EGFP-CRBP-I (Figure 3).
Without refolding, RBP produced in bacteria is almost completely misfolded due to the presence of many incorrect disulfide bonds. Therefore, refolding in the presence of the ligand retinol is critically important in obtaining well-folded RBP. We found that if the refolding reaction is not done correctly, misfolded RBP species (e.g. misfolded RBP-I in Figure 1) can predominate the whole preparation and the yield of high quality holo-RBP can be very low after HPLC purification. Therefore, HPLC cannot correct the problem of poor refolding. As shown in Figure 1, even with the correct refolding reaction, RBP contains many species of proteins including apo-RBP, partially misfolded RBP, and correctly folded holo-RBP. Only HPLC purification can separate the perfectly folded RBP that is 100% loaded with retinol from misfolded RBP species. The most reliable criterion for correctly folded and 100% loaded holo-RBP is that the retinol peak of the purified protein is slightly higher than the protein peak. Partially misfolded RBP, even after HPLC purification, has a retinol peak that is significantly lower than the protein peak (Figure 1). Although partially misfolded RBP still binds retinol, the retinol/RBP complex is much less stable, and RBP more readily releases bound retinol. Misfolded RBP can lead to incorrect conclusions from RBP or vitamin A-related experiments. Another difference between correctly folded RBP and misfolded RBP is that correctly folded holo-RBP is stable for years at 4 °C in PBS. If the holo-RBP preparation has misfolded RBP contamination, insoluble materials will emerge after a period of storage. Insoluble materials can be observed after spinning down the solution at high speed at 4 °C. Apo-RBP is only produced from high-quality holo-RBP.
Once purified high-quality holo-RBP or apo-RBP is available, it can be used to study STRA6-catalyzed retinol transport. Both the real-time retinol release assay and retinol loading assay depend on monitoring retinol fluorescence. One source of artificial decline of retinol fluorescence is the nonspecific binding of RBP to the plastic dish (once RBP is bound to the plastic wall, it can no longer be measured in solution). We have tested many blocking conditions and found that Blocker Casein (Pierce) is most effective in reducing this nonspecific and receptor-independent decline of retinol fluorescence. Another source of artificial decline of retinol fluorescence is poor quality of the RBP, as discussed above.
STRA6-catalyzed retinol release, retinol loading and retinol transport are relatively slow events, as shown in the representative data in Figures 2 and 3. The reactions are relatively slow because each RBP only binds one molecule of vitamin A and all STRA6-catalyzed reactions depend on both RBP binding and RBP dissociation to continue. For STRA6-catalyzed retinol release reaction to continue, holo-RBP can only bind after apo-RBP dissociates. For STRA6-catalyzed retinol loading reaction to continue, apo-RBP can only bind after holo-RBP dissociates. For all the real-time monitoring techniques described here, we found that the ideal frequency of measurement is one measurement every 5 min or 10 min. Although more frequent measurements can create higher temporal resolution, the resulting data files will be extremely large because each time point creates an Excel file.
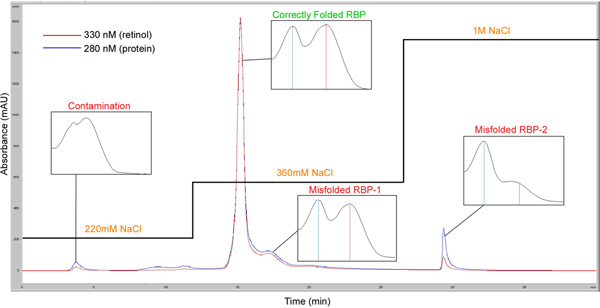 Figure 1. Purification of refolded RBP on HPLC to obtain holo-RBP 100% loaded with retinol. Nickel resin-purified refolded RBP is applied to ion exchange column AX-300 by a NaCl step gradient (220 mM 10 min, 360 mM 15 min, and 1,000 mM 15 min). The correctly folded holo-His-RBP is released and eluted at 360 nM NaCl. Absorption spectra (250 nm-400 nm) of the peaks corresponding to correctly folded holo-RBP, misfolded RBP-1, misfolded RBP-2, and contamination are also shown (protein peak is indicated by a blue vertical line and retinol peak is indicated by red vertical line). Click here to view larger figure.
Figure 1. Purification of refolded RBP on HPLC to obtain holo-RBP 100% loaded with retinol. Nickel resin-purified refolded RBP is applied to ion exchange column AX-300 by a NaCl step gradient (220 mM 10 min, 360 mM 15 min, and 1,000 mM 15 min). The correctly folded holo-His-RBP is released and eluted at 360 nM NaCl. Absorption spectra (250 nm-400 nm) of the peaks corresponding to correctly folded holo-RBP, misfolded RBP-1, misfolded RBP-2, and contamination are also shown (protein peak is indicated by a blue vertical line and retinol peak is indicated by red vertical line). Click here to view larger figure.
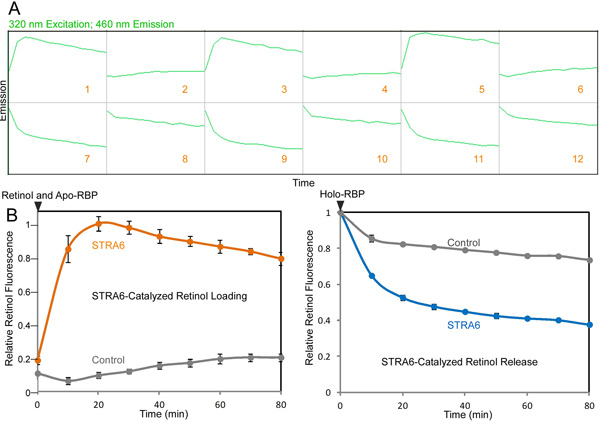 Figure 2. Real-time monitoring of STRA6-catalyzed retinol loading into apo-RBP and retinol release from holo-RBP. This assay played an important role in revealing what STRA6 can do by itself without LRAT or CRBP-I. A. An example of the raw data file for the STRA6-catalzyed retinol loading and retinol release as shown on the Omega Data Analysis software. To initiate the retinol release reaction, holo-RBP is added to STRA6 membrane or control membrane at 0 min. To initiate the retinol loading reaction, retinol is added to STRA6 membrane or control membrane premixed with apo-RBP at 0 min. Fluorescence measurements for 320 nm excitation and 460 nm emission revealed the time course of the reactions. Reactions 1 to 6 monitor retinol loading into apo-RBP (1, 3, and 5 are STRA6 reaction; 2, 4, and 6 are control reactions). Reactions 7 to 12 monitor retinol loading into apo-RBP (7, 9, and 11 are STRA6 reaction; 8, 10, and 12 are control reactions). B. The final calculated signals for reactions shown in A. The left graph was calculated based on reactions 1 to 6. The right graph was calculated based on reactions 7 to 12. The highest fluorescence signal is defined as 1 in the left graph. The fluorescence of holo-RBP added at 0 min is defined as 1 in the right graph. Click here to view larger figure.
Figure 2. Real-time monitoring of STRA6-catalyzed retinol loading into apo-RBP and retinol release from holo-RBP. This assay played an important role in revealing what STRA6 can do by itself without LRAT or CRBP-I. A. An example of the raw data file for the STRA6-catalzyed retinol loading and retinol release as shown on the Omega Data Analysis software. To initiate the retinol release reaction, holo-RBP is added to STRA6 membrane or control membrane at 0 min. To initiate the retinol loading reaction, retinol is added to STRA6 membrane or control membrane premixed with apo-RBP at 0 min. Fluorescence measurements for 320 nm excitation and 460 nm emission revealed the time course of the reactions. Reactions 1 to 6 monitor retinol loading into apo-RBP (1, 3, and 5 are STRA6 reaction; 2, 4, and 6 are control reactions). Reactions 7 to 12 monitor retinol loading into apo-RBP (7, 9, and 11 are STRA6 reaction; 8, 10, and 12 are control reactions). B. The final calculated signals for reactions shown in A. The left graph was calculated based on reactions 1 to 6. The right graph was calculated based on reactions 7 to 12. The highest fluorescence signal is defined as 1 in the left graph. The fluorescence of holo-RBP added at 0 min is defined as 1 in the right graph. Click here to view larger figure.
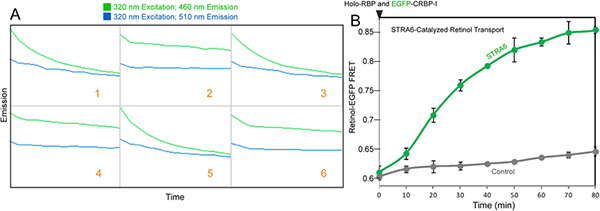 Figure 3. Real-time monitoring of STRA6-catalyzed retinol transport from holo-RBP to CRBP-I. A. An example of the raw data file that monitors the FRET between retinol and EGFP as shown on the Omega Data Analysis software. At 0 min, holo-RBP is added to the reactions containing STRA6 membrane (reactions 1, 3, and 5) or control membrane with EGFP-CRBP-I to initiate the reactions (reactions 2, 4, and 6). Simultaneous duel emission measurements for excitation at 320 nm revealed the green trace as the 460-10 emission time course and the blue trace as the 510-10 emission time course. B. The final calculated FRET signals for reactions shown in A. The FRET signals were calculated according to the methods in step 3.6. Click here to view larger figure.
Figure 3. Real-time monitoring of STRA6-catalyzed retinol transport from holo-RBP to CRBP-I. A. An example of the raw data file that monitors the FRET between retinol and EGFP as shown on the Omega Data Analysis software. At 0 min, holo-RBP is added to the reactions containing STRA6 membrane (reactions 1, 3, and 5) or control membrane with EGFP-CRBP-I to initiate the reactions (reactions 2, 4, and 6). Simultaneous duel emission measurements for excitation at 320 nm revealed the green trace as the 460-10 emission time course and the blue trace as the 510-10 emission time course. B. The final calculated FRET signals for reactions shown in A. The FRET signals were calculated according to the methods in step 3.6. Click here to view larger figure.
Discussion
We share here an optimized RBP production protocol because RBP production and purification procedures are critical to generating correctly folded RBP. Given the possibility of misfolded RBP species and the presence of trace amounts of bacterial proteins even in HPLC purified bacteria-produced RBP, it is helpful to use native RBP from serum to confirm a conclusion related to RBP. Urine RBP, which is commercially available, is a complex mixture of many species of RBP including apo-RBP and holo-RBP 48,49.
The real-time monitoring techniques described here are based on the intrinsic fluorescence of retinol. The original impetus to develop these assays was that we were limited by the information that can be revealed by the classic radioactive assays and HPLC-based assays. For example, we found that STRA6 has little vitamin A uptake activity by itself in vitamin A uptake assays, but we cannot easily explain the low activity of STRA6 in vitamin A uptake without LRAT or CRBP-I41. In retinyl ester-based assays, it is easy to understand that STRA6 by itself does not make retinyl ester because STRA6 is not an enzyme that has LRAT activity. However, even in retinol-based assays, STRA6 still has little activity by itself. Does STRA6 have any vitamin A uptake activity at all? If it does not, how can CRBP-I or LRAT accelerate its activity? If it does, why does it need CRBP-I or LRAT? Although radioactive assays and HPLC-based assays did not answer these questions, we reasoned that STRA6 should have the ability to release retinol from RBP. We also reasoned that a retinol fluorescence assay might reveal this activity. Retinol fluorescence measurement has made it possible to study the movement of free retinol in vertebrate photoreceptor cells after light bleaching of visual pigments in real time 50,51. How can RBP receptor activity be monitored by measuring retinol fluorescence? It was discovered in early 1970s by two research groups that retinol has dramatically enhanced fluorescence when bound to RBP 52,53. We found that STRA6-catalyzed retinol release from holo-RBP causes a decrease in retinol fluorescence, while STRA6-catalyzed retinol loading into apo-RBP causes an increase in retinol fluorescence. Although radioactive assays and HPLC-based assays have been used extensively to study vitamin A uptake, fluorescence assays had not been used to study the RBP receptor activity until recently, likely because the endogenous membranes used as the source of the receptor have very high background fluorescence and have a complex mixture of retinoid binding proteins/enzymes that makes it difficult to interpret the contribution of each protein. The currently available sensitive instruments are also what make these techniques more feasible. In addition to direct fluorescence measurement, the new retinol-EGFP FRET pair41 coupled with simultaneous duel emission optics makes it possible to study retinol transport to retinol binding proteins in real time and in multiple reactions simultaneously.
Disclosures
No conflicts of interest declared.
Acknowledgments
Supported by National Institutes of Health grant R01EY018144.
References
- Crouch RK, Chader GJ, Wiggert B, Pepperberg DR. Retinoids and the visual process. Photochem. Photobiol. 1996;64:613–621. doi: 10.1111/j.1751-1097.1996.tb03114.x. [DOI] [PubMed] [Google Scholar]
- Travis GH, Golczak M, Moise AR, Palczewski K. Diseases caused by defects in the visual cycle: retinoids as potential therapeutic agents. Annu. Rev. Pharmacol. Toxicol. 2007;47:469–512. doi: 10.1146/annurev.pharmtox.47.120505.105225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli JL. Biochemical pathways of retinoid transport, metabolism, and signal transduction. Clin. Immunol. Immunopathol. 1996;80:52–62. doi: 10.1006/clin.1996.0142. [DOI] [PubMed] [Google Scholar]
- Drager UC. Retinoic acid signaling in the functioning brain. Sci STKE. 2006;2006:pe10. doi: 10.1126/stke.3242006pe10. [DOI] [PubMed] [Google Scholar]
- Maden M. Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat. Rev. Neurosci. 2007;8:755–765. doi: 10.1038/nrn2212. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Dolle P. Retinoic acid in development: towards an integrated view. Nat. Rev. Genet. 2008;9:541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- Goodman DS. In: The Retinoids. Sporn MB, Boberts AB, Goodman DS, editors. Vol. 2. Academic Press; 1984. pp. 41–88. [Google Scholar]
- Blomhoff R, Green MH, Berg T, Norum KR. Transport and storage of vitamin A. Science. 1990;250:399–404. doi: 10.1126/science.2218545. [DOI] [PubMed] [Google Scholar]
- Newcomer ME, Ong DE. Plasma retinol binding protein: structure and function of the prototypic lipocalin. Biochim. Biophys. Acta. 2000;1482:57–64. doi: 10.1016/s0167-4838(00)00150-3. [DOI] [PubMed] [Google Scholar]
- Quadro L, Hamberger L, Colantuoni V, Gottesman ME, Blaner WS. Understanding the physiological role of retinol-binding protein in vitamin A metabolism using transgenic and knockout mouse models. Mol. Aspects Med. 2003;24:421–430. doi: 10.1016/s0098-2997(03)00038-4. [DOI] [PubMed] [Google Scholar]
- Heller J. Interactions of plasma retinol-binding protein with its receptor. Specific binding of bovine and human retinol-binding protein to pigment epithelium cells from bovine eyes. J. Biol. Chem. 1975;250:3613–3619. [PubMed] [Google Scholar]
- Bok D, Heller J. Transport of retinol from the blood to the retina: an autoradiographic study of the pigment epithelial cell surface receptor for plasma retinol-binding protein. Exp. Eye Res. 1976;22:395–402. doi: 10.1016/0014-4835(76)90177-9. [DOI] [PubMed] [Google Scholar]
- Rask L, Peterson PA. In vitro uptake of vitamin A from the retinol-binding plasma protein to mucosal epithelial cells from the monkey's small intestine. J. Biol. Chem. 1976;251:6360–6366. [PubMed] [Google Scholar]
- Heller M, Bok D. A specific receptor for retinol binding protein as detected by the binding of human and bovine retinol binding protein to pigment epithelial cells. Am. J. Ophthalmol. 1976;81:93–97. doi: 10.1016/0002-9394(76)90198-7. [DOI] [PubMed] [Google Scholar]
- Chen CC, Heller J. Uptake of retinol and retinoic acid from serum retinol-binding protein by retinal pigment epithelial cells. J. Biol. Chem. 1977;252:5216–5221. [PubMed] [Google Scholar]
- Bhat MK, Cama HR. Gonadal cell surface receptor for plasma retinol-binding protein. A method for its radioassay and studies on its level during spermatogenesis. Biochim. Biophys. Acta. 1979;587:273–281. doi: 10.1016/0304-4165(79)90360-x. [DOI] [PubMed] [Google Scholar]
- Rask L, Geijer C, Bill A, Peterson PA. Vitamin A supply of the cornea. Exp. Eye Res. 1980;31:201–211. doi: 10.1016/0014-4835(80)90078-0. [DOI] [PubMed] [Google Scholar]
- Torma H, Vahlquist A. Vitamin A uptake by human skin in. 1984;276:390–395. doi: 10.1007/BF00413360. [DOI] [PubMed] [Google Scholar]
- Torma H, Vahlquist A. Uptake of vitamin A and retinol-binding protein by human placenta in vitro. Placenta. 1986;7:295–305. doi: 10.1016/s0143-4004(86)80147-3. [DOI] [PubMed] [Google Scholar]
- Pfeffer BA, Clark VM, Flannery JG, Bok D. Membrane receptors for retinol-binding protein in cultured human retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 1986;27:1031–1040. [PubMed] [Google Scholar]
- Eriksson U, et al. Increased levels of several retinoid binding proteins resulting from retinoic acid-induced differentiation of F9 cells. Cancer Res. 1986;46:717–722. [PubMed] [Google Scholar]
- Ottonello S, Petrucco S, Maraini G. Vitamin A uptake from retinol-binding protein in a cell-free system from pigment epithelial cells of bovine retina. Retinol transfer from plasma retinol-binding protein to cytoplasmic retinol-binding protein with retinyl-ester formation as the intermediate step. J. Biol. Chem. 1987;262:3975–3981. [PubMed] [Google Scholar]
- Sivaprasadarao A, Findlay JB. The interaction of retinol-binding protein with its plasma-membrane receptor. Biochem. J. 1988;255:561–569. [PMC free article] [PubMed] [Google Scholar]
- Sivaprasadarao A, Findlay JB. The mechanism of uptake of retinol by plasma-membrane vesicles. Biochem. J. 1988;255:571–579. [PMC free article] [PubMed] [Google Scholar]
- Shingleton JL, Skinner MK, Ong DE. Characteristics of retinol accumulation from serum retinol-binding protein by cultured Sertoli cells. Biochemistry. 1989;28:9641–9647. doi: 10.1021/bi00451a015. [DOI] [PubMed] [Google Scholar]
- Sivaprasadarao A, Findlay JB. Structure-function studies on human retinol-binding protein using site-directed mutagenesis. Biochem. J. 1994;300(Pt. 2):437–442. doi: 10.1042/bj3000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melhus H, Bavik CO, Rask L, Peterson PA, Eriksson U. Epitope mapping of a monoclonal antibody that blocks the binding of retinol-binding protein to its receptor. Biochem. Biophys. Res. Commun. 1995;210:105–112. doi: 10.1006/bbrc.1995.1633. [DOI] [PubMed] [Google Scholar]
- Smeland S, et al. Tissue distribution of the receptor for plasma retinol-binding protein. Biochem. J. 1995;305(Pt. 2):419–424. doi: 10.1042/bj3050419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M, Sivaprasadarao A, DeSousa MM, Findlay JB. The transfer of retinol from serum retinol-binding protein to cellular retinol-binding protein is mediated by a membrane receptor. J. Biol. Chem. 1998;273:3336–3342. doi: 10.1074/jbc.273.6.3336. [DOI] [PubMed] [Google Scholar]
- Vogel S, et al. Retinol-binding protein-deficient mice: biochemical basis for impaired vision. Biochemistry. 2002;41:15360–15368. doi: 10.1021/bi0268551. [DOI] [PubMed] [Google Scholar]
- Liden M, Eriksson U. Development of a versatile reporter assay for studies of retinol uptake and metabolism in vivo. Exp. Cell Res. 2005;310:401–408. doi: 10.1016/j.yexcr.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Kanai M, Raz A, Goodman DS. Retinol-binding protein: the transport protein for vitamin A in human plasma. J. Clin. Invest. 1968;47:2025–2044. doi: 10.1172/JCI105889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask L, et al. The retinol-binding protein. Scand. J. Clin. Lab Invest. Suppl. 1980;154:45–61. [PubMed] [Google Scholar]
- Monaco HL, Rizzi M, Coda A. Structure of a complex of two plasma proteins: transthyretin and retinol-binding protein. Science. 1995;268:1039–1041. doi: 10.1126/science.7754382. [DOI] [PubMed] [Google Scholar]
- Zanotti G, Berni R. Plasma retinol-binding protein: structure and interactions with retinol, retinoids, and transthyretin. Vitam. Horm. 2004;69:271–295. doi: 10.1016/S0083-6729(04)69010-8. [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, et al. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–825. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- Isken A, et al. RBP4 Disrupts Vitamin A Uptake Homeostasis in a STRA6-Deficient Animal Model for Matthew-Wood Syndrome. Cell Metab. 2008;7:258–268. doi: 10.1016/j.cmet.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golczak M, et al. Metabolic basis of visual cycle inhibition by retinoid and nonretinoid compounds in the vertebrate retina. J. Biol. Chem. 2008;283:9543–9554. doi: 10.1074/jbc.M708982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Yu J, Wiita P, Honda J, Sun H. An essential ligand-binding domain in the membrane receptor for retinol-binding protein revealed by large-scale mutagenesis and a human polymorphism. J. Biol. Chem. 2008;283:15160–15168. doi: 10.1074/jbc.M801060200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Yu J, Wiita P, Ter-Stepanian M, Sun H. Mapping the membrane topology and extracellular ligand binding domains of the retinol binding protein receptor. Biochemistry. 2008;47:5387–5395. doi: 10.1021/bi8002082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, et al. Receptor-mediated cellular uptake mechanism that couples to intracellular storage. ACS Chem. Biol. 2011;6:1041–1051. doi: 10.1021/cb200178w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Zhong M, Kassai M, Ter-Stepanian M, Sun H. STRA6-Catalyzed Vitamin A Influx, Efflux and Exchange. J. Membr. Biol. 2012;245:731–745. doi: 10.1007/s00232-012-9463-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, et al. Retinoid content, visual responses and ocular morphology are compromised in the retinas of mice lacking the retinol-binding protein receptor, STRA6. Invest. Ophthalmol. Vis. Sci. 2012;53:3027–3039. doi: 10.1167/iovs.11-8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DC, Jin H, Majumdar A, Noy N. Signaling by vitamin A and retinol-binding protein regulates gene expression to inhibit insulin responses. Proc. Natl. Acad. Sci. U.S.A. 2011;108:4340–4345. doi: 10.1073/pnas.1011115108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DC, O'Byrne SM, Vreeland AC, Blaner WS, Noy N. Cross Talk between Signaling and Vitamin A Transport by the Retinol-Binding Protein Receptor STRA6. Mol. Cell Biol. 2012;32:3164–3175. doi: 10.1128/MCB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DC, Croniger CM, Ghyselinck NB, Noy N. Mol. Cell Biol. In Press; 2012. Transthyretin blocks retinol uptake and cell signalling by the holo-retinol-binding protein receptor STRA6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- Peterson PA, Berggard I. Isolation and properties of a human retinol-transporting protein. J. Biol. Chem. 1971;246:25–33. [PubMed] [Google Scholar]
- Rask L, Vahlquist A, Peterson PA. Studies on two physiological forms of the human retinol-binding protein differing in vitamin A and arginine content. J. Biol. Chem. 1971;246:6638–6646. [PubMed] [Google Scholar]
- Koutalos Y. Measurement of the mobility of all-trans-retinol with two-photon fluorescence recovery after photobleaching. Methods Mol. Biol. 2010;652:115–127. doi: 10.1007/978-1-60327-325-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutalos Y, Cornwall MC. Microfluorometric measurement of the formation of all-trans-retinol in the outer segments of single isolated vertebrate photoreceptors. Methods Mol. Biol. 2010;652:129–147. doi: 10.1007/978-1-60327-325-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson PA, Rask L. Studies on the fluorescence of the human vitamin A-transporting plasma protein complex and its individual components. J. Biol. Chem. 1971;246:7544–7550. [PubMed] [Google Scholar]
- Futterman S, Heller J. The enhancement of fluorescence and the decreased susceptibility to enzymatic oxidation of retinol complexed with bovine serum albumin, beta-lactoglobulin, and the retinol-binding protein of human plasma. J. Biol. Chem. 1972;247:5168–5172. [PubMed] [Google Scholar]


