Summary
HEN1-mediated 2′-O-methylation has been shown to be a key mechanism to protect plant microRNAs (miRNAs) and small interfering RNAs (siRNAs) as well as animal piwi-interacting RNAs (piRNAs) from degradation and 3′ terminal uridylation [1–8]. However, enzymes uridylating unmethylated miRNAs, siRNAs, or piRNAs in hen1 are unknown. In this study, a genetic screen identified a second-site mutation hen1 suppressor1-2 (heso1-2) that partially suppresses the morphological phenotypes of the hypomorphic hen1-2 allele and the null hen1-1 allele in Arabidopsis. HESO1 encodes a terminal nucleotidyl transferase that prefers to add untemplated uridine to the 3′ end of RNA, which is completely abolished by 2′-O-methylation. heso1-2 affects the profile of u-tailed miRNAs and siRNAs and increases the abundance of truncated and/or normal sized ones in hen1, which often results in increased total amount of miRNAs and siRNAs in hen1. In contrast, overexpressing HESO1 in hen1-2 causes more severe morphological defects and less accumulation of miRNAs. These results demonstrate that HESO1 is an enzyme uridylating unmethylated miRNAs and siRNAs in hen1. These observations also suggest that uridylation may destabilize unmethylated miRNAs through an unknown mechanism and compete with 3′-to-5′ exoribonuclease activities in hen1. This study shall have implications on piRNA uridylation in hen1 in animals.
Results
heso1-2 Partially Rescues the Morphological Phenotypes of hen1-2 and hen1-1
In Arabidopsis hen1-2, an asparagine to aspartic acid substitution impaired HEN1 activity, reduced miRNA abundance, and caused pleiotropic developmental defects such as delayed growth and reduced fertility (short siliques) [9]. An ethyl methanesulfonate (EMS) mutagenized population of hen1-2 was screened for second-site mutations that rescued the fertility defects of hen1-2 [10]. This genetic screen was expected to identify components destroying unmethylated miRNAs because lack of them might increase the abundance of miRNAs and therefore suppress the fertility defects of hen1-2. NRPD1 and NRPD2/NRPE2, two essential genes for siRNA biogenesis, were identified from this screen [10–13].
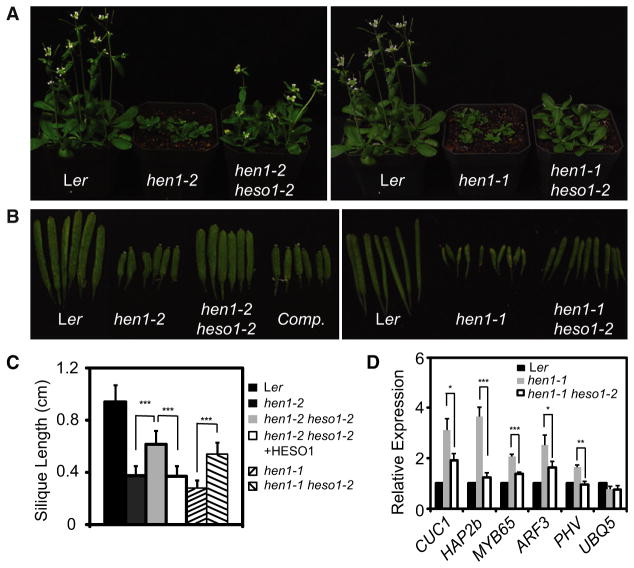
We further screened the EMS mutagenized hen1-2 population and isolated a suppressor with partially rescued vegetative phenotypes and longer siliques (Figures 1A and 1B). The average silique length of this suppressor was ~1.6- fold of that of hen1-2 (Figures 1B and 1C). Backcross analysis revealed that a single recessive mutation caused the phenotypic changes in this suppressor. Following the nomenclature of the companion paper [14], the mutation was named hen1 suppressor1-2 (heso1-2).
Figure 1. heso1-2 Mutation Partially Suppresses the Phenotypes of hen1-1 and hen1-2.
(A) Morphological phenotypes of the indicated genotypes.
(B) Siliques from plants of the indicated genotypes. Ler is WT plants. hen1-1 heso1-2 and hen1-2 heso1-2 are hen1-1 and hen1-2 harboring the heso1-2 mutation, respectively. Comp is hen1-2 heso1-2 harboring the HESO1 genomic DNA.
(C) Average silique length in various genotypes. Thirty siliques from six plants for each genotype were included in the analysis. ***p < 0.001.
(D) The transcript levels of miRNA and ta-siRNA targets in various genotypes. Target mRNA accumulation in various genotypes was quantified by qRT-PCR and compared with those of WT. Quantifications are normalized with ACTIN2 transcript. UBQ5: UBQUITIN5, which served as an internal control. The WT value is 1. *p < 0.05; **p <0.01; ***p < 0.001.
We next evaluated whether heso1-2 suppressed hen1-2 through restoring HEN1 activity. In this scenario, heso1-2 should not suppress the null hen1-1 allele. However, hen1-1 heso1-2 grew better, produced longer siliques (~2-fold increase), and flowered earlier than hen1-1 (Figures 1A–1C). To confirm that miRNA methylation status was not altered in hen1-1 heso1-2, we performed a periodate/β-elimination assay. Periodate/β-elimination treatment of unmethylated but not methylated miRNAs would result in faster migration during electrophoresis under denaturing condition, which could be detected by northern blot [15]. After chemical treatment, miR167 showed similar mobility alterations in hen1-1 heso1-2 as in hen1-1 (Figure S1). These results demonstrated that heso1-2-mediated phenotypic rescue of hen1 did not require HEN1 methylase activity.
heso1-2 Partially Restores miRNA and trans-acting siRNA Function in hen1-1
The rescue of developmental defects of hen1 by heso1-2 could be best explained by the recovery of small RNA-mediated gene regulation, because dysregulation of small RNA targets was thought to be the major reason for the morphological phenotypes of hen1 [16, 17]. We therefore compared the transcript levels of four miRNA targets, CUC, PHV, HAP2b, and MYB65 and a trans-acting siRNA (ta-siRNA) target, ARF3, in hen1-1 heso1-2 with those in hen1-1 by quantitative RT-PCR (qRT-PCR) [17, 18]. As previously reported, their steady-state transcript levels were increased in hen1-1 compared with those in Ler plants (wild-type [WT] plants) [17, 18]. Relative to hen1-1, the transcript levels of these targets but not the control UBQUITIN 5 were significantly reduced in hen1-1 heso1-2 (Figure 1D). These results indicated that heso1-2 might partially restore miRNA and ta-siRNA function in hen1-1.
heso1-2 Alters miRNA and siRNA Profiles in hen1-2 and hen1-1
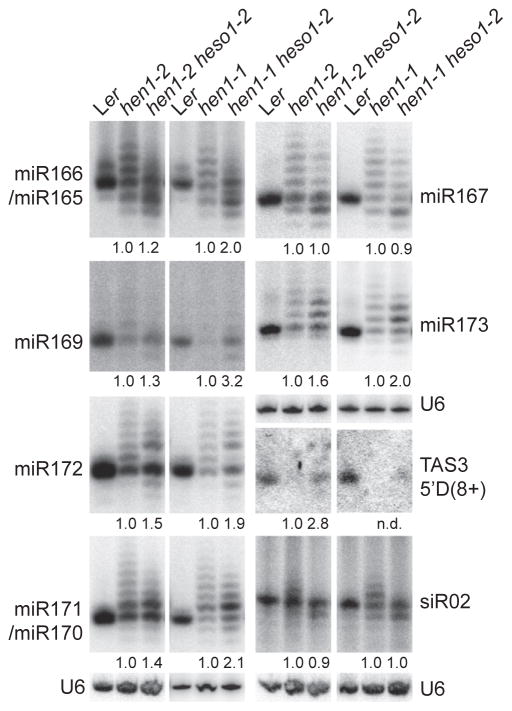
We next examined several miRNAs in hen1-2, hen1-1, hen-1-1 heso1-2, and hen1-2 heso1-2 by northern blot. Tailed miRNAs were still present in hen1-1 heso1-2 and hen1-2 heso1-2 (Figure 2). To determine whether the tail consisted of untemplated Us in hen1-1 heso1-2, we performed a primer-extension analysis of miRNA-specific RT-PCR products amplified from miRNAs ligated to adaptors from hen1-1 heso1-2 [2]. A ladder of primer-extension products was observed in both hen1-1 heso1-2 and hen1-1, indicating the presence of U-tailed miRNA in hen1-1 heso1-2 (Figure S2B). This result was consistent with the deep sequencing result in the companion paper [14].
Figure 2. heso1-2 Affects miRNA and siRNA Profiles in hen1-1 and hen1-2.
miRNAs and siRNAs in various genotypes were monitored by northern blot. U6 was used as a loading control. Ler served as a size control. miR166/165 and miR171/170, the paired miRNAs were recognized by the same probe because of sequence similarities.
The number below hen1-1 heso1-2 and hen1-2 heso1-2 indicated the fold changes of total amounts of miRNAs or siRNAs caused by heso1-2 in hen1-1 and hen1-2, respectively. n.d. represents not detected in hen1-1. To obtain the total amount of a miRNA and siRNA in each genotype, we quantified all individual bands in each sample and added them together. The total amount of a miRNA and siRNA in hen1-1 heso1-2 and hen1-2 heso1-2 was then normalized to U6 RNA and compared with that in hen1-1 and hen1-2, respectively.
Quantification analyses showed that heso1-2 increased the total amounts of miR166/165, miR169, miR171/170, miR172, and miR173 in hen1-1 and hen1-2, respectively (Figure 2), but not of miR167, whose overall levels had no obvious change (Figure 2). heso1-2 appeared to have a greater effect in hen1-1 than hen1-2, presumably due to the fact that hen1-1 is a null allele, whereas hen1-2 is a weak allele (Figure 2). For the examined miRNAs, truncated forms were increased in abundance in hen1-1 heso1-2 (2.2- to 4.3-fold of those in hen1-1) and hen1-2 heso1-2 (1.8- to 6.8-fold of those in hen1-2), compared with hen1-1 and hen1-2, respectively (Figure 2; Figure S2A). Sequencing analysis of miR167 demonstrated that truncations occurred at 3′ end (Table S1), in agreement with deep-sequencing analysis results in the companion paper [14]. The normal-sized miR169, miR171, miR172, and miR173 were increased in accumulation as well by heso1-2 in hen1-1 and hen1-2, respectively (Figure 2; Figure S2). heso1-2 also appeared to reduce the abundance of tailed miR167 and miR166/165 (>21 nt for miR167, >22 nt for miR166/165; Figure 2; Figure S2A). Although the levels of short-tailed miR171 (22–23 nt), miR172 (23–24 nt), and miR173 (23–24 nt) were increased by heso1-2 in hen1-1 and hen1-2, the abundance of longer ones (>24 nt for miR171, >25 nt for miR172 and miR173) was reduced (Figure 2; Figure S2A). These results suggested that heso1-2 increases the levels of normal-sized, 3′ truncated, and/or short-tailed miRNAs in hen1, which often led to an increase of the total amount of miRNAs.
We next examined the accumulation of a heterochromatic siRNA siR02 and a trans-acting siRNA TAS3-5′D8(+). heso1-2 increased the total amount of TAS3-5′D8(+), which lacks detectable U-tailed forms in hen1 and double mutants, but not siR02 (Figure 2). However, heso1-2 did increase the abundance of normal-sized and truncated siR02 and reduced accumulation of tailed isoforms in hen1-1 and hen1-2 (Figure 2).
We also evaluated the effect of heso1-2 mutation on miRNA accumulation in WT. However, northern blot showed that the levels of miR167, miR171, and miR173 in heso1-2 are comparable with those in WT (Figure S2C).
HESO1 Encodes a Cytoplasm and Nucleus- Localized Terminal Nucleotidyl Transferase
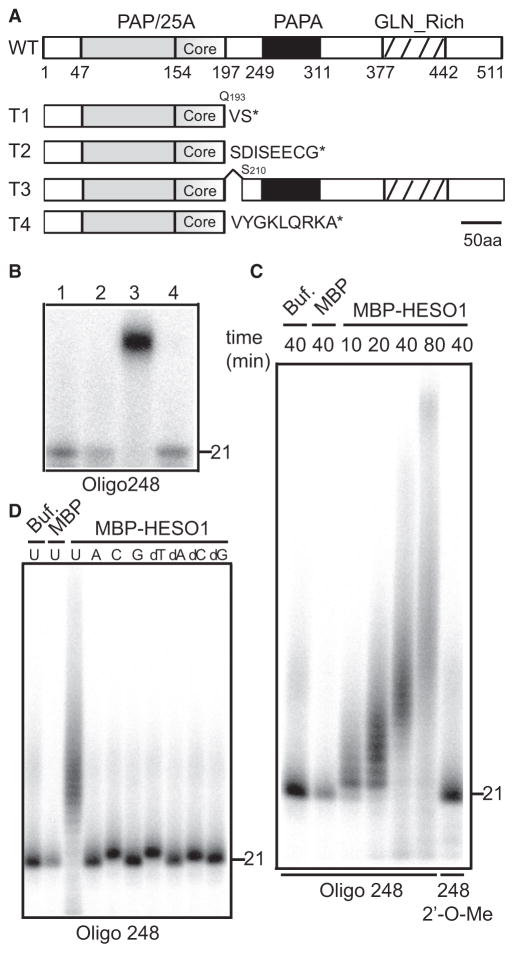
We next used a map-based cloning approach to determine the molecular nature of heso1-2 mutation. The mapping population was constructed by crossing hen1-2 heso1-2 with hen1-8, which carries the same mutation as hen1-2 but is in the Columbia-0 genetic background [10]. In the F2 segregating population, the plants with longer siliques were selected for marker analysis. The heso1-2 mutation was mapped to a ~70 kb region of chromosome 2 on the bacterial artificial chromosome (BAC) T5I7 (Figure S3A). Sequencing analysis of this region revealed a G-to-A change at the end of fifth intron of the gene At2g39740 (Figures S3B and S3C). Introduction of a WT genomic DNA covering At2g39740 promoter and coding regions into hen1-2 heso1-2 was able to restore miRNA and fertility phenotypes of hen1-2 (Figures 1B and 1C; Figure S3E). These results demonstrated that the G-to-A mutation in At2g39740 was responsible for the partial rescue of hen1-2. We named At2g39740 HEN1 SUPPRESSOR1 (HESO1). HESO1 encodes a putative nucleotidyl transferase that belongs to the DNA polymerase β-like superfamily [19]. It contains a poly A polymerase domain, (PAP/25A), a PAP-associated domain, and a glutamine rich region (Figure 3A). However HESO1 is distinct from canonical, eukaryotic PAPs because it lacks the RNA-binding motif (RRM) of classical PAP. HESO1 is thought to be a member of GLD-2-related family that adds nucleotides to the 3′ end of RNAs independent of templates [20].
Figure 3.
HESO1 Encodes a 3′ Terminal Uridylyl Transferase
(A) Schematic of HESO1 protein and its mutant forms generated by heso1-2. * represents protein stop codon. The amino acids introduced by heso1-2 following the truncated HESO1 are indicated. PAP/25A, poly A polymerase domain; PAPA, PAP associated domain; Core, core regions of PAP/25A; GLN_RICH, glutamine rich region.
(B) Terminal uridylyl transferase activity of HESO1. Oligo 248 was 5′-end labeled with P32, incubated with buffer alone (1), purified MBP (2), MBP-HESO1 (3) or MBP-HESO1Δ16 (4) under the presence of UTP for 120 min, and resolved on a denaturing polyacrylamide gel.
(C) Time-course reaction of HESO1 activity and the effect of 2′-O-methylation on HESO1 activity. 5′ end–labeled oligo 248 or 2′-O-methyl oligo 248 was incubated with MBP-HESO1. The reaction was stopped at the indicated time.
(D) Preference of HESO1 to ribonucleotides and deoxyribonucleotides. 5′ end labeled oligo 248 was incubated with MBP-HESO1 under the presence of UTP, ATP, CTP, GTP, dATP, dTTP, dCTP, or dGTP for 40 min.
RT-PCR and subsequent sequencing analyses detected four abnormal but no WT HESO1 transcripts in hen1-2 heso1-2 (Figure S3D), indicating that the G-to-A mutation caused splicing defects. Either intron 5 was fully retained in the messenger RNA (mRNA) (transcript 1) or exon 6 was partially (transcript 2 and 3) or completely spliced out (transcript 4; Figure S3D). Transcripts 1, 2, and 4 contained in-frame stop codons, which should lead to the production of truncated proteins lacking the C-terminal region starting from amino acid 194 (Figure 3A; Figure S3D). Transcript 3 was predicted to produce a mutant protein, in which 16 amino acids (194–209) including the last 4 amino acids of PAP/5A core region were deleted (Figure 3A; Figure S3D). This deletion completely abolished nucleotidyl transferase activity of HESO1 (Figure 3B, described below). Thus, heso1-2 is most likely a null allele.
The alteration of U-tailing profile of small RNAs in hen1 heso1-2 together with the protein annotation suggested that HESO1 might be an enzyme responsible for the untemplated uridine addition to the 3′ end of miRNAs and siRNAs in hen1. Therefore, we tested whether HESO1 possessed terminal nucleotidyl transferase activity using a recombinant maltose-binding protein (MBP)-HESO1 protein. The recombinant MBP-HESO1 was expressed in E. coli and purified with maltose resin (Figure S3F). As controls, a MBP protein and a MBP-HESO1 protein lacking amino acid from 194 to 209 (MBP-HESO1Δ16 encoded by the transcript 3 in hen1-2 heso1-2, Figure S3F) were also expressed. We incubated the proteins with an in vitro synthesized 21 nt oligo RNA (oligo 248) [21], which was 5′ P32-labeled, under the presence of uridine triphosphate (UTP). After the reaction was stopped, the RNA was resolved on a polyacrylamide gel. MBP-HESO1 but not MBP and MBP-HESO1Δ16 lengthened the substrate RNA (Figure 3B), indicating that HESO1 added untemplated uridines to the 3′ end of oligo 248. A time-course experiment revealed that the RNA substrate was elongated during the reaction (Figure 3C), demonstrating that HESO1 had template-independent polymerase activity. A 2′-O-methyl group at the 3′ end of oligo 248 completely abolished HESO1 activity (Figure 3C), which is consistent with the notion that methylation prevents 3′ uridylation of plant miRNAs and siRNAs and animal piRNAs [1, 2, 8]. MBP-HESO1 was able to add one cytidine (C), deoxythymidine (dT), or deoxycytidine (dC) to the 3′ end of oligo 248 (Figure 3D). The results demonstrated that HESO1 has preference to add Us to the 3′ end of RNA under our assay conditions.
In order to examine the localization of HESO1 in cells, we transiently expressed HESO1 under the control of a Cauliflower mosaic virus (CaMV) 35S promoter in Nicotiana benthamiana [22]. The transgene was able to produce functional protein because it caused more severe defects of hen1-2 when expressed (described below). The yellow florescence signal could be observed in both cytoplasm and nucleus in the leaf epidermal cells of N. benthamiana harboring 35S::HESO1-YFP (Figure S3G), suggesting that HESO1 might be localized in both cytoplasm and nucleus, consistent with the observation that both miRNAs and siRNAs were affected by heso1-2 in hen1-1 and hen1-2.
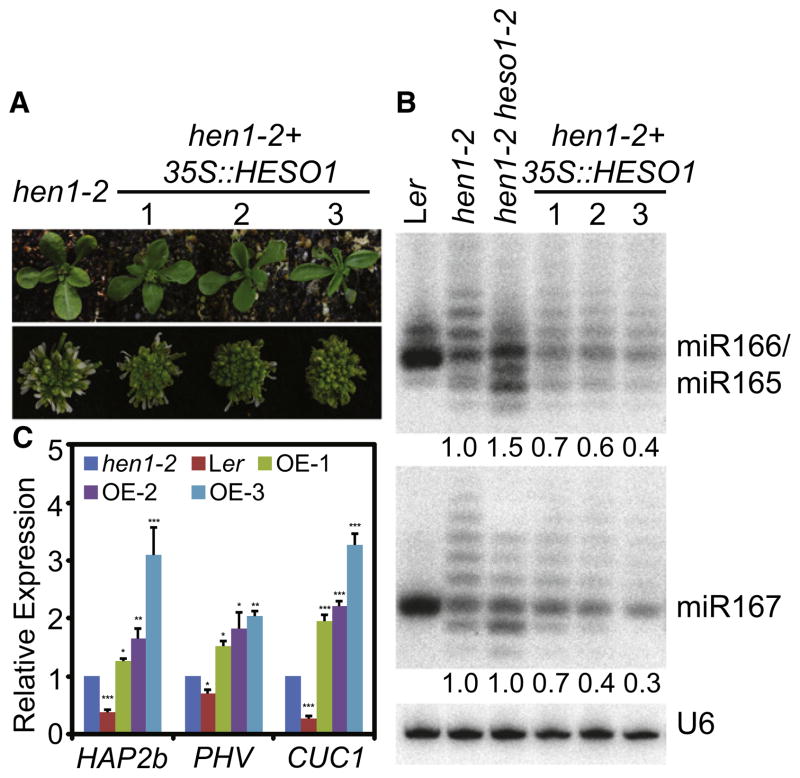
Overexpression of HESO1 in hen1-2 Causes More Severe Morphological Defects and Less miRNA Accumulation
The fact that lack of HESO1 in hen1 increased the accumulation of miRNAs suggested that HESO1-mediated uridylation might destabilize unmethylated miRNAs. If so, we would expect that overexpression of HESO1 in hen1 could further reduce the abundance of miRNAs and cause more severe developmental defects. We overexpressed HESO1 under the control of 35S promoter in hen1-2. In T1 transgenic plants, three transgenic hen1-2 heso1-2 with 4- to 12-fold increased transcript levels of HESO1 relative to hen1-2 were selected by qRT-PCR analysis (Figure S4). All of these three transgenic lines showed more severe leaf and flower morphological defects than hen1-2 (Figure 4A). Northern blot analysis revealed that the amount of miR166/165 and miR167 was reduced in three transgenic hen1-2 lines overexpressing HESO1 compared with hen1-2 (Figure 4B). This result supported that HESO1 might trigger degradation of unmethylated miRNAs. Furthermore, we detected increased miRNA target transcript levels in overexpression lines relative to hen1-2 (Figure 4C), indicating that the reduction of miRNA abundance may be the cause for severe morphological defects of overexpression lines.
Figure 4. Overexpression of HESO1 in hen1-2 Causes More Severe Phenotypes.
(A) Morphological phenotypes of various genotypes. Ler is WT plants. hen1-2 + 35S::HESO1 is hen1-2 overexpressing HESO1.
(B) miR167 and miR166/165 in various genotypes were monitored by northern blot. U6 was used as a loading control. Ler served as a miRNA size control; hen1-2 + 35S::HESO1, hen1-2 overexpressing HESO1. The number below overexpression lines indicated the fold changes of total amounts of miRNAs or siRNAs caused by overexpression of HESO1 in hen1-2, respectively. To obtain the total amount of a miRNA and siRNA in each genotype, we quantified all individual bands in each sample and added them together. The total amount of a miRNA and siRNA in HESO1 overexpression lines 2 was then normalized to U6 RNA and compared with that in hen1-2.
(C) The transcript levels of three miRNA targets in Ler, hen1-2, and three HESO1 overexpression lines. Target mRNA accumulation in various genotypes was quantified by qRT-PCR and compared with those of WT. Quantifications are normalized with ACTIN2 transcript. The hen1-2 value is 1. *p < 0.05; **p < 0.01; ***p < 0.001.
Discussion
The in vitro uridylyl transferase activity of HESO1 and the effect of heso1-2 on the profile of uridylated small RNAs in hen1 demonstrate that HESO1 is an enzyme targeting unmethylated miRNAs and siRNAs in hen1. The facts that heso1-2 increases the levels of miRNAs and overexpression of HESO1 reduces the abundance of miRNAs in hen1 demonstrate the hypothesis that uridylation triggers degradation of plant miRNAs and siRNAs or animal piRNAs in hen1 [1–4, 8, 21, 23]. It has been observed that uridylation triggers the degradation of miRNAs from 3′ end by exosome component RRP6 in vitro in Chlamydomonas [21]. However, the increased accumulation of 3′ truncated miRNAs and siRNAs in hen1 heso1-2 indicates that mechanisms other than 3′-to-5′ degradation might be employed by Arabidopsis to degrade uridylated miRNA and siRNAs. It is possible that uridylation may trigger highly progressive 3′-to-5′ degradation so that the 3′ truncated products was less accumulated in hen1. The presence of 3′ truncated miRNAs and siRNAs in hen1 also indicates that 3′-to-5′ degradation activities such as SDN1, exosome or homologs of Nibbler might act on miRNAs and siRNAs in hen1 as well [21, 24–26]. It is tempting to speculate that HESO1 may compete with 3′-to-5′ exoribonuclease activities or their products as substrates in Arabidopsis.
Additional terminal uridylyl transferases must act in miRNA and siRNA uridylation processes in hen1 because uridylated miRNAs and siRNAs are still present in hen1 heso1-2 and heso1-2 is most likely a null allele. Arabidopsis encodes nine additional HESO1 homologs (Figure S3H). One or more of them may add U tails to the unmethylated small RNAs in hen1 heso1-2. The increased accumulation of some miRNAs with short tails is observed in hen1 heso1-2. A possible explanation is that other nucleotidyl transferases may add a short tail to miRNA, which then can be used by HESO1 as substrates to elongate. However, other possibilities are also present. This observation also suggests that the short-tailed miRNAs may have a very slow degradation rate, or a certain U-tail length threshold may be required to trigger degradation in Arabidopsis. The reduced abundance of U-tailed miR166/165 and miR167 may reflect substrate preference of HESO1 homologs. Consistent with this, terminal nucleotidyl transferases GLD-2 and ZCCHC11 have been shown to specifically modify miR122 and miR26 in animals, respectively [27, 28]. Several putative terminal nucleotidyl transferases are also proposed to modify miRNAs in a miRNA-specific manner in human [29].
heso1-2 has no obvious effect on several examined miRNAs in WT, indicating that 2′-O-methylation prevents HESO1 activity. However, we cannot rule out the possibility that HESO1 has subtle effects on small RNA levels. In fact, tailed miRNAs and siRNAs are also present in WT plants [2]. In addition to miRNAs and siRNAs, HESO1 may have other substrates in vivo such as 5′ fragments of RNA-induced silencing complex cleavage products, U6 small nuclear RNA and ribosomal 5S RNAs. Uridylation of these RNAs has been established [30–32]. In animals, uridylation of pre-let7 is also observed [33–35]. Clearly, these possibilities need to be examined in the future.
Supplementary Material
Acknowledgments
We thank Xiang Liu, Shuxin Zhang, Meng Xie, and Heriberto Cerutti from the University of Nebraska Lincoln for critical reading of the manuscript and Heriberto Cerutti for providing RNA oligo 248 and 2′-O-methyalted oligo 248. This work was supported by National Science Foundation (NSF) MCB-1121193 (to B.Y.), a faculty seed grant and an Enhancing Interdisciplinary Teams Grant from the University of Nebraska Lincoln (to B.Y.), and NSF MCB-1021465 (to X.C.).
Footnotes
Supplemental Information includes four figures, two tables, and Supplemental Experimental Procedures and can be found with this article online at doi:10.1016/j.cub.2012.02.052.
References
- 1.Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJ, Roovers EF, Ladurner P, Berezikov E, Ketting RF. Hen1 is required for oocyte development and piRNA stability in zebra-fish. EMBO J. 2010;29:3688–3700. doi: 10.1038/emboj.2010.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 4.Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603– 1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA. 2007;13:1397–1401. doi: 10.1261/rna.659307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- 7.Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 8.Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Liu J, Cheng Y, Jia D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development. 2002;129:1085–1094. doi: 10.1242/dev.129.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu B, Bi L, Zhai J, Agarwal M, Li S, Wu Q, Ding SW, Meyers BC, Vaucheret H, Chen X. siRNAs compete with miRNAs for methylation by HEN1 in Arabidopsis. Nucleic Acids Res. 2010;38:5844–5850. doi: 10.1093/nar/gkq348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 12.Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, Mo B, Chen X. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol. 2012;22:689–694. doi: 10.1016/j.cub.2012.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boutet S, Vazquez F, Liu J, Béclin C, Fagard M, Gratias A, Morel JB, Crété P, Chen X, Vaucheret H. Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr Biol. 2003;13:843–848. doi: 10.1016/s0960-9822(03)00293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaucheret H, Vazquez F, Crété P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Aravind L, Koonin EV. DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwak JE, Wickens M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci USA. 2010;107:3906–3911. doi: 10.1073/pnas.0912632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapila J, de Rycke R, van Montagu M, Angenon G. An Agrobacterium-mediated transient gene expression system or intact leaves. Plant Sci. 1997;122:101–108. [Google Scholar]
- 23.van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 24.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han BW, Hung JH, Weng Z, Zamore PD, Ameres SL. The 3′-to-5′ exoribonuclease Nibbler shapes the 3′ ends of microRNAs bound to Drosophila Argonaute1. Curr Biol. 2011;21:1878–1887. doi: 10.1016/j.cub.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu N, Abe M, Sabin LR, Hendriks GJ, Naqvi AS, Yu Z, Cherry S, Bonini NM. The exoribonuclease Nibbler controls 3′ end processing of microRNAs in Drosophila. Curr Biol. 2011;21:1888–1893. doi: 10.1016/j.cub.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu S, Ivanov AR, Wolf DA, Mizgerd JP. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol. 2009;11:1157– 1163. doi: 10.1038/ncb1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res. 2011;21:1450–1461. doi: 10.1101/gr.118059.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y, Sinha K, Perumal K, Reddy R. Effect of 3′ terminal adenylic acid residue on the uridylation of human small RNAs in vitro and in frog oocytes. RNA. 2000;6:1277–1288. doi: 10.1017/s1355838200000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibrahim F, Rohr J, Jeong WJ, Hesson J, Cerutti H. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science. 2006;314:1893. doi: 10.1126/science.1135268. [DOI] [PubMed] [Google Scholar]
- 33.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.