Abstract
High throughput phenotyping (phenomics) is a powerful tool for linking genes to their functions (see review1 and recent examples2-4). Leaves are the primary photosynthetic organ, and their size and shape vary developmentally and environmentally within a plant. For these reasons studies on leaf morphology require measurement of multiple parameters from numerous leaves, which is best done by semi-automated phenomics tools5,6. Canopy shade is an important environmental cue that affects plant architecture and life history; the suite of responses is collectively called the shade avoidance syndrome (SAS)7. Among SAS responses, shade induced leaf petiole elongation and changes in blade area are particularly useful as indices8. To date, leaf shape programs (e.g. SHAPE9, LAMINA10, LeafAnalyzer11, LEAFPROCESSOR12) can measure leaf outlines and categorize leaf shapes, but can not output petiole length. Lack of large-scale measurement systems of leaf petioles has inhibited phenomics approaches to SAS research. In this paper, we describe a newly developed ImageJ plugin, called LeafJ, which can rapidly measure petiole length and leaf blade parameters of the model plant Arabidopsis thaliana. For the occasional leaf that required manual correction of the petiole/leaf blade boundary we used a touch-screen tablet. Further, leaf cell shape and leaf cell numbers are important determinants of leaf size13. Separate from LeafJ we also present a protocol for using a touch-screen tablet for measuring cell shape, area, and size. Our leaf trait measurement system is not limited to shade-avoidance research and will accelerate leaf phenotyping of many mutants and screening plants by leaf phenotyping.
Keywords: Plant Biology, Issue 71, Cellular Biology, Molecular Biology, Physiology, Computer Science, Arabidopsis, Arabidopsis thaliana, leaf shape, shade avoidance, ImageJ, LeafJ, petiole, touch-screen tablet, phenotyping, phenomics
Protocol
1. Plant Materials
Note that this plant growth protocol is aimed for detecting shade avoidance response. You can grow plants under your favorite condition.
Sprinkle Arabidopsis thaliana seeds on water soaked filter papers in 9 cm Petri dishes and store (stratify) them at 4 °C for four days in the dark.
Transfer these Petri dishes to simulated sun conditions: 80-100 μE photosynthetically active radiation (PAR) and far-red supplement to bring the R:FR ratio to 1.86. Use long day conditions (16 hr light/8 hr dark) and constant temperature of 22 °C. Incubate in this condition for three days to allow the seeds to germinate.
Transfer germinated seed to soil and keep plants under sun condition. For large-scale experiments, we recommend preparing small tags for labeling each plants by using Data Merge Manager in Microsoft Word 2004 (or later) for making labels.
Eleven days after transfer to soil, move half of the plants to shade condition: same as sun but with supplemental far-red light to bring the R/FR ratio to 0.52.
After an additional twelve days, the plants are ready for leaf imaging. At this stage the older leaves have fully matured whereas younger leaves are still expanding, so that you capture a snapshot of development. You may want to choose a different developmental time depending on your needs.
2. Capturing Dissected Leaf Images
Prepare transparency sheets labeled with plant genotype and growth condition with five rectangular frames. One frame corresponds to leaves from one plant. Microsoft Excel can be used to print a consistent grid with labels.
Dissect leaves of twenty-six day old plants.
Scan leaves at 600 dpi on a flat-bed scanner. Note that leaves from one plant should be placed vertically within a black window in a sandwich of transparent sheets. Avoid touching leaves to a black window frame and overlapping leaves, which will give errors in following procedures.
3. Leaf Image Analysis by LeafJ
Download ImageJ from http://malooflab.openwetware.org/Resources.html#Software_Tools or http://bitbucket.org/jnmaloof/leafj. Drag the LeafJ.jar file into the plugins folder of ImageJ.
Open an image file in ImageJ 1.45s or later14.
Split the image into three-color channels (red, green, and blue) by "Image > Color > Split Channels" and apply threshold to the image in the blue channel.
Select all of the leaves from one plant by a rectangle tool (Figure 1A).
Select "LeafJ" from the plugin menu.
Select annotation information for this plant from the dialog box that appears. You can edit the default values that appear here by clicking "edit these options".
After running LeafJ plugin and before clicking "OK" button, edit traced lines from the region of interest (ROI) manager window (if necessary; Figure 1B). A touch-screen tablet (such as an iPad) is useful for this procedure. iPads can be connected to a computer as an external monitor using Air Display software.
Export measurement results and associated information (file names, flowering time, dissected by, measured by, etc) to Microsoft Excel or equivalent software.
4. Leaf Cell Image Analysis in ImageJ
Fix dissected leaves as described in reference15 after scanning (step 2). FAA fixed leaves can be kept in 4 °C for at least 6 months.
Clear the leaves by changing FAA fixative to chloral hydrate solution and incubate leaves for 1~2 hr before microscopic observation15.
Mount leaves on microscope slides with trichomes facing up. Using 40x magnification on a compound microscope, image the mesophyll layer of the center of each leaf on either side of the main vein, avoiding cells near trichomes or veins.
Trace leaf cell outlines by ImageJ ROI manager tool with aid of the touch-screen tablet and a stylus (as described in step 3). Cell image analysis uses the built-in features of ImageJ but does not require LeafJ.
Representative Results
1. Leaf Images Showing Estimates of the Petiole and Leaf Blade Boundary, and Their Measurement Window
One of the most useful features of LeafJ is automated detection of leaf blade/petiole boundary (Figure 1). The LeafJ algorithm works as follows: the built-in ImageJ ParticleAnalyzer functionality is used to find and determine the orientation of the leaves inside of the user selection. For each leaf the width of the leaf is determined along the leaf's entire axis. Then the change in width at each position along the axis is determined using a running window (the mean width for the seven positions proximal to the focal position is subtracted from the mean width for the seven positions distal to the focal position). The petiole/blade boundary is defined as the first position beyond the leaf base where the change in width is greater than 90% of all calculated width differences. LeafJ does additional checks to increase reliability of this call; specifically LeafJ also requires that 1) the width exceeds the narrowest 5% of positions in the length (this prevents aberrant calls at the petiole base); 2) the region proximal to the focal position does not have a major change in width; and 3) the width of the leaf 20% distal to the focal position is at least 150% wider than at the focal position (true if the focal position is the boundary because 20% proximal to the boundary should be blade and therefore much wider).
Once LeafJ has defined the petiole/blade boundary, built-in ImageJ classes and methods are used to determine blade area, perimeter, and circularity. Built-in ImageJ methods are also used to fit an ellipse to the blade and to calculate the major and minor axes of that ellipse (then used as blade length and width). Petiole length is determined by a line that traces along the center of the petiole area.
2. Detection of Shade-induced Petiole Elongation
To ask whether LeafJ measurements were useful in assaying shade avoidance, we used a mixed effects model with treatment and leaf number as fixed effects and replicate as a random effect. We found that petiole length, blade area, blade length, blade width, and the petiole length/blade length ratio were significantly affected by shade treatment, whereas blade circularity and the blade length/blade width ratio were not (p<0.05). Our data showed that LeafJ plugin is useful for studies on leaf shade avoidance responses (Figure 2).
3. Accuracy and Speed of the LeafJ Plugin
To determine the performance of the LeafJ plugin in larger data sets we compared operation time and accuracy between manual and plugin measurements. For manual measurements we defined the petiole/ blade boundary as the place where the leaf width appeared to rapidly increase. It took an experienced researcher an average of 19 min 3 sec to measure one transparency with five plants (about 50 leaves) by manual measurement, while with LeafJ it took only 3 min 20 sec. Averaged across 5 transparencies, measuring with the plugin was 5.7 times faster than manual measurement. The manual measurement was done by a researcher with experience in making many manual measurements; a beginner would be considerably slower at manual measurement, resulting in an even greater advantage of LeafJ. We assessed accuracy by comparing data from the two methods; the data were highly correlated for all leaf parameters (Figure 3). Of the 3,532 data points there were 172 (4.9%) that showed strong differences between the methods (evidenced by being outliers on the correlation plots). We analyzed the cause of these outliers. Of 172 outliers, 29 were due to errors during LeafJ plugin measurement and 143 were due to errors in manual measurement. This error analysis also showed accuracy of plugin measurement.
4. Cell Size and Cell Number Measurement
Independent of LeafJ we also developed an efficient work flow for measuring cell number and size. Theoretically cell number and size can be used to classify mutant plants into nine categories when compared with wild type; (1) smaller cells size with decreased cell numbers, (2) smaller cells with normal cell numbers, (3) smaller cells with increased cell numbers, (4) normal cell size with decreased cell numbers, (5) normal cell size with normal cell numbers, (6) normal cell size with decreased cell numbers, (7) larger cell size with decreased cell numbers, (8) larger cell size with normal cell numbers, (9) larger cell size with increased cell numbers15,16. We measured leaf cell parameters in 67 genotypes of Arabidopsis thaliana using our tablet-based cell size and cell number measurement method. We measured cell size of 8,629 cells from 877 leaves of 224 plants. Multiplying cell density by leaf area (measured by LeafJ), we estimated total leaf cell number data of 438 leaves from 219 plants. Our analysis placed these genotypes into six of the possible nine categories (Fig. 4). The largest category was (5): normal cell size and number; second largest was (8): larger cell size with normal cell number. Although follow-up work is needed, this suggests that our tablet-based method can be used to categorize mutants based on leaf cell size and number.
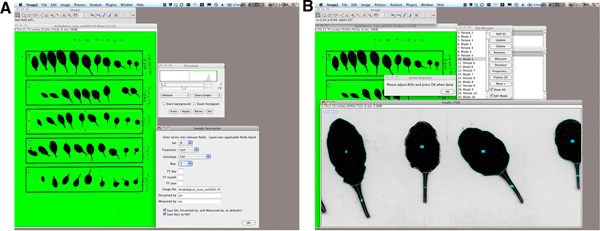 Figure 1. An example of petiole/leaf blade boundary detection and user interface. Note that LeafJ is able to define petiole/leaf blade boundary automatically (Figure 1B). Click here to view larger figure.
Figure 1. An example of petiole/leaf blade boundary detection and user interface. Note that LeafJ is able to define petiole/leaf blade boundary automatically (Figure 1B). Click here to view larger figure.
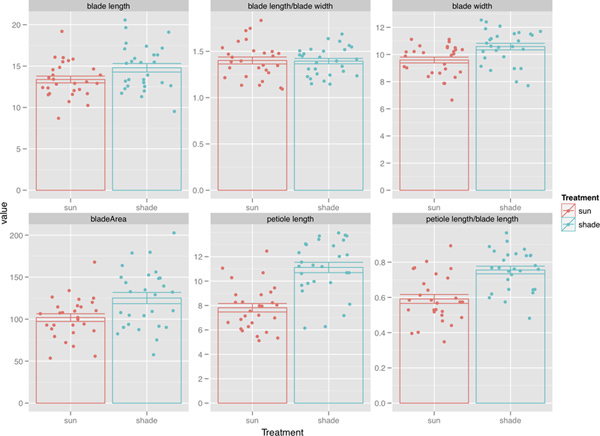 Figure 2. LeafJ plugin could detect shade avoidance responses in various leaf parameters. Leaf 3 to leaf 6 from seven wild type plants (Arabidopsis thaliana Columbia ecotype) under each condition (sun and shade) were examined. From top left to bottom right, the y-axis units are mm, mm, mm, mm2, ratio of petiole length to blade length, and ratio of blade length to blade width. Click here to view larger figure.
Figure 2. LeafJ plugin could detect shade avoidance responses in various leaf parameters. Leaf 3 to leaf 6 from seven wild type plants (Arabidopsis thaliana Columbia ecotype) under each condition (sun and shade) were examined. From top left to bottom right, the y-axis units are mm, mm, mm, mm2, ratio of petiole length to blade length, and ratio of blade length to blade width. Click here to view larger figure.
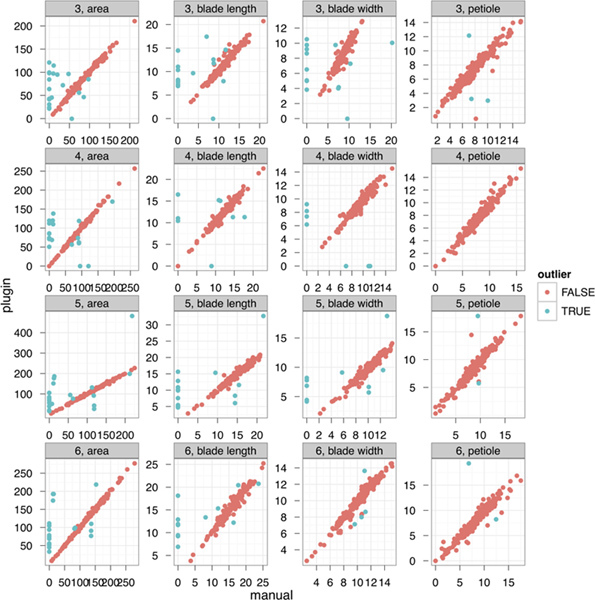 Figure 3. LeafJ plugin is highly accurate. Correlation of data from manual measurement and LeafJ plugin measurement from 3,532 data points. Each dot represents one leaf. Green dots indicate the 170 outliers in this graph. Axes of leaf area is mm2 all others are mm. Numbers in front of each parameter represent leaf position (i.e. "3" is the third leaf).
Figure 3. LeafJ plugin is highly accurate. Correlation of data from manual measurement and LeafJ plugin measurement from 3,532 data points. Each dot represents one leaf. Green dots indicate the 170 outliers in this graph. Axes of leaf area is mm2 all others are mm. Numbers in front of each parameter represent leaf position (i.e. "3" is the third leaf).
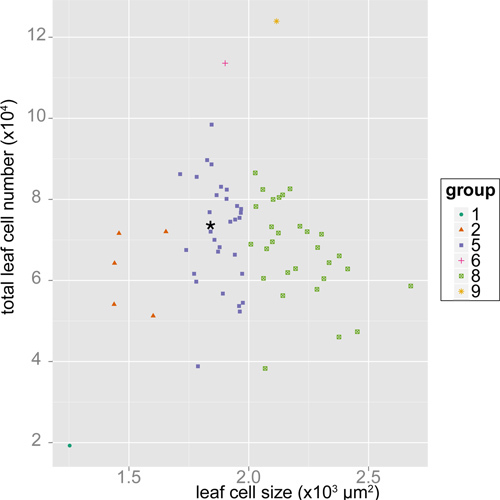 Figure 4. Scatter plot of leaf cell area and leaf cell number among 67 genotypes of Arabidopsis thaliana. Each point represents the phenotypes of each genotype grown under shade condition. Plants were classified into nine categories based on their difference from wild type (Col) as determined by a linear mixed-effects model and multiple-testing corrected p-values: (1) smaller cell size with decreased cell numbers, (2) smaller cell size with normal cell number, (3) smaller cell size with increased cell number, (4) normal cell size with decreased cell number, (5) normal cell size with normal cell number, (6) normal cell size with decreased cell number, (7) larger cell size with decreased cell number, (8) larger cell size with normal cell number, (9) larger cell size with increased cell number. "*" indicates the phenotype of wild type (Col).
Figure 4. Scatter plot of leaf cell area and leaf cell number among 67 genotypes of Arabidopsis thaliana. Each point represents the phenotypes of each genotype grown under shade condition. Plants were classified into nine categories based on their difference from wild type (Col) as determined by a linear mixed-effects model and multiple-testing corrected p-values: (1) smaller cell size with decreased cell numbers, (2) smaller cell size with normal cell number, (3) smaller cell size with increased cell number, (4) normal cell size with decreased cell number, (5) normal cell size with normal cell number, (6) normal cell size with decreased cell number, (7) larger cell size with decreased cell number, (8) larger cell size with normal cell number, (9) larger cell size with increased cell number. "*" indicates the phenotype of wild type (Col).
Discussion
Our "LeafJ" plugin enables measurement of petiole length semi-automatically, increasing throughput nearly 6 times over manual measurement. Petiole length is an important index of SAS and is also a landmark of other phenomena, such as submergence resistance and hyponastic growth17. Therefore this plugin may be useful to a wide range of plant researchers.
Our plugin is implemented in a well-established java-based free software, ImageJ. This enables easy cross-platform installation. Ease of modification of the program is also an advantage of the LeafJ plugin because ImageJ already has a large library of plugins that were written by Java and ImageJ macro languages (http://imagejdocu.tudor.lu/doku.php?id=tutorial:start). Currently we only tested Arabidopsis leaves, but our algorithm of petiole/leaf blade boundary detection could apply to other dicotyledonous leaves after some modification of the plugin.
During testing LeafJ plugin, we found most of the 14 errors came from human mistakes such as misplacing copied results on datasheets and/or mislabeling of plant genotypes. In rare cases the petiole/leaf blade boundary was called incorrectly, necessitating manual correction and creating additional risk of copy and paste mistakes. We could detect such errors after looking at data (a) by thresholding values (e.g. petioles longer than leaf length) and (b) by finding duplicated sample condition (e.g. "sun" or "shade"), genotype, or position of leaves.
Our touch-screen tablet method facilitated accuracy and speed of measurement. Limitation of our method is that communication between main computer and the touch screen tablet relies on the speed of the wireless local area network (LAN).
Disclosures
No conflicts of interest declared.
Acknowledgments
LeafJ was written by JNM while he was on sabbatical in Dr. Katherine Pollard's lab at the Gladstone Institutes.
This work was supported by a grant from the National Science Foundation (grant number IOS-0923752).
References
- Furbank RT, Tester M. Phenomics--technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011;16:635–644. doi: 10.1016/j.tplants.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Berger B, Parent B, Tester M. High-throughput shoot imaging to study drought responses. J. Exp. Bot. 2010;61:3519–3528. doi: 10.1093/jxb/erq201. [DOI] [PubMed] [Google Scholar]
- Borevitz JO. Natural genetic variation for growth and development revealed by high-throughput phenotyping in Arabidopsis thaliana. G3 (Bethesda) 2012;2:29–34. doi: 10.1534/g3.111.001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht DR, Bargmann CI. High-content behavioral analysis of Caenorhabditis elegans in precise spatiotemporal chemical environments. Nat. Methods. 2011;8:599–605. doi: 10.1038/nmeth.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, et al. Native environment modulates leaf size and response to simulated foliar shade across wild tomato species. PLoS ONE. 2012;7:e29570. doi: 10.1371/journal.pone.0029570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, et al. The developmental trajectory of leaflet morphology in wild tomato species. Plant Physiol. 2012;158:1230–1240. doi: 10.1104/pp.111.192518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. Shade Avoidance. The Arabidopsis Book. 2012. p. e0157. [DOI] [PMC free article] [PubMed]
- Smith H. In: Photomorphogenesis in Plants. Kendrick RE, Kronenberg GHM, editors. Kluwer Academic Publishers; 1994. pp. 377–416. [Google Scholar]
- Iwata H, Ukai Y. SHAPE: a computer program package for quantitative evaluation of biological shapes based on elliptic Fourier descriptors. J. Hered. 2002;93:384–385. doi: 10.1093/jhered/93.5.384. [DOI] [PubMed] [Google Scholar]
- Bylesjo M, et al. LAMINA: a tool for rapid quantification of leaf size and shape parameters. BMC Plant Biol. 2008;8:82. doi: 10.1186/1471-2229-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weight C, Parnham D, Waites R. LeafAnalyser: a computational method for rapid and large-scale analyses of leaf shape variation. Plant J. 2008;53:578–586. doi: 10.1111/j.1365-313X.2007.03330.x. [DOI] [PubMed] [Google Scholar]
- Backhaus A, et al. LEAFPROCESSOR: a new leaf phenotyping tool using contour bending energy and shape cluster analysis. New Phytol. 2010;187:251–261. doi: 10.1111/j.1469-8137.2010.03266.x. [DOI] [PubMed] [Google Scholar]
- Tsukaya H. Mechanisms of Leaf-shape determination. Annual Review of Plant Biology. 2006;57:477–496. doi: 10.1146/annurev.arplant.57.032905.105320. [DOI] [PubMed] [Google Scholar]
- Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Horiguchi G, Fujikura U, Ferjani A, Ishikawa N, Tsukaya H. Large-scale histological analysis of leaf mutants using two simple leaf observation methods: identification of novel genetic pathways governing the size and shape of leaves. Plant. J. 2006;48:638–644. doi: 10.1111/j.1365-313X.2006.02896.x. [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Ferjani A, Fujikura U, Tsukaya H. Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant. Res. 2006;119:37–42. doi: 10.1007/s10265-005-0232-4. [DOI] [PubMed] [Google Scholar]
- Pierik R, de Wit M, Voesenek LACJ. Growth-mediated stress escape: convergence of signal transduction pathways activated upon exposure to two different environmental stresses. New. Phytol. 2011;189:122–134. doi: 10.1111/j.1469-8137.2010.03458.x. [DOI] [PubMed] [Google Scholar]


