Abstract
Micrometer and nanometer grooved surfaces have been determined to influence cellular orientation, morphology, and migration through contact guidance. Cells typically elongate along the direction of an underlying groove and often migrate with guidance provided by constraints of the pattern. This phenomenon has been studied primarily using linear grooves, post, or well patterns. We investigated the behavior of mouse embryonic fibroblasts on non-linear, sinusoidal wave grooves created via electron beam lithography on a polymethyl methacrylate (PMMA) substrate that was spin-coated onto a positively charged glass surface. Three different wave patterns, with varying wavelengths and amplitudes, and two different line patterns were created. Cell orientation and adhesion was examined after 4, 24, and 48 hours after cell seeding. Attachment strength was studied via subjecting cells on substrates to centrifugal force following a 24-hour incubation period. For all wave patterns studied, it was noted that cells did not reside within the groove, rather they were observed to cross over each groove, residing both inside and outside of each wave pattern, aligning linearly along the long axis of the pattern. For the linear patterns, we observed that cells tended to reside within the grooves, consistent with previous observations. The ability to add texture to a surface to manipulate cell adhesion strength and growth with only localized attachment, maintaining free space in curvilinear microtopography underlying the cell, may be a useful addition for tissue engineering and the fabrication of novel biomedical devices.
Keywords: contact guidance, fibroblasts (3T3), micro patterning, electron beam lithography, cell alignment, micro topography
Introduction
Mammalian cells in vivo exist in a complex environment with a multitude of signals that affect cellular behavior. Of these signals, there are naturally occurring micro- and nanotopographical cues within the extracellular matrix, which influence cell arrangement, migration, and orientation [1–4]. A universal goal of tissue engineering is to create materials capable of regulating and manipulating spatial cues, such that cell types may be spatially organized and directed, as they are in vivo. As new implant materials are developed, understanding cell-substrate interactions has become increasingly important. For many years, it has been recognized that cells respond to underlying substratum topographical features [5]. This phenomenon is termed contact guidance and is characterized by cellular response to micrometer and submicrometer surface features [6,7]. It has become clear that nearly all cell types will react to surface topography through adhesion, spreading, migration, and/or proliferation [8–14]. To date, this behavior has been studied predominantly on surfaces in which linear grooved patterns are created through micro-machining, photolithography, and electron beam lithography [1,2,6,14–31]. Linear groove patterns typically result in cell elongation, migration guidance along the direction of the grooves, and reorganization of the cytoskeleton [32]. Micro- and nano-pillars [16,19] and micro- and nano-wells have been found to affect cell adhesion to surface substrata by either increasing or decreasing cell attachment depending on spacing and feature size [33–35]. Hexagonal and random patterns have also been studied [34,36], though the majority of contact guidance has involved linear grooves or rectilinear arrays of posts and wells. In nature, curvilinear architecture exists in tissues and organs and is often vital for organ geometry and function. In this study, we investigate the behavior of mouse embryonic fibroblasts (3T3), on curvilinear, sinusoidal wave grooves created using electron-beam lithography (EBL) on a polymethyl methacrylate (PMMA) substrate.
Materials and Methods
Pattern Fabrication
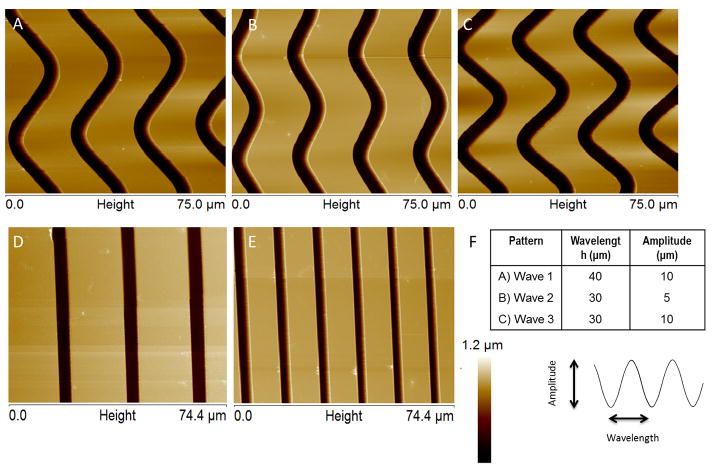
A diced 10 mm × 10 mm positively charged microscope slide (Globe Scientific Inc., Paramus, NJ, USA: charge density proprietary) was spin coated with a polymethyl methacrylate (PMMA) photoresist (950 PMMA C4, Microchem, Newton, MA, USA) at 2000 rpm for 45 seconds, resulting in a polymer thickness between 650 and 680 nm, measured by Veeco Dimension 3100 atomic force microscope (AFM; Bruker AXS, Santa Barbara, CA, USA). The coated chip was hot-baked for 1 minute to remove any excess residues and to facilitate PMMA resist adhesion. It was subsequently subjected to electron beam lithography, using a FEI Inspec scanning electron microscope (SEM; Hilsboro, OR, USA) equipped with nanopattern generation system (NPGS; JC Nabity, Bozeman, MT, USA) to etch line and wave patterns designed using DesignCAD Express (dimensions described in Figure 1). The etched polymer was developed for 1 minute with 1:3 methyl isobutyl ketone/isopropyl alcohol mixture (MIBK/IPA; Microchem Corp., Newton, MA, USA), transferred to isopropyl alcohol (IPA; Honeywell, Morristown NJ, USA) for 30 seconds, rinsed with deionized water, and dried with nitrogen gas. The patterns were measured by the Veeco Dimension 3100 AFM.
Figure 1.
AFM Images of patterns created on PMMA using electron beam lithography. Darker regions represent exposed positively charged glass. PMMA thickness of 650–680 nm, groove width of 4.5–5 μm, and groove spacing of 20 μm on all patterns except for pattern E, which has a groove spacing of 10 μm. Height scale is 0 to 1.2 μm for all patterns. The chart (F) shows the wavelength and amplitude of each of the wave patterns. A, B, and C represent wave 1, wave 2, and wave 3 respectively. D) 20 μm spaced line pattern E) 10 μm spaced line pattern.
Cell Culture
NIH 3T3 mouse fibroblasts (ATCC; Manassas, VA, USA) were grown in Dulbecco’s Modification of Eagle’s Medium (DMEM; Mediatech Inc., Manassas, VA, USA) supplemented with 10% Newborn Calf Serum, 1 M HEPES, 1% antibiotic, and 2% L-glutamine, all purchased from Lonza (Walkersville, MD, USA). Cells at 80–90% confluence were detached using trypsin (Lonza, Walkersville MD) and collected via centrifugation. Cells were reuspended to yield a final concentration of 200,000 cell/ml. 200 μl of cell suspension was placed on the surface of the chips and allowed to seed for 10 minutes (approximately 40,000 cells seeded per chip), after which an appropriate amount of media was added to the culture dish. Cells were then incubated i.n 95% air, 5% CO2 at 37°C for 4, 24, or 48 hours before staining.
Immunocytochemistry
Fibroblasts were stained for the visualization of actin filaments using TRITC-conjugated Phalloidin (Millipore, Billerica, MA, USA), and nuclei using fluoroshield with DAPI (Sigma-Aldrich). Some cells were stained for vinculin (focal adhesions) using mouse monoclonal antibody to vinculin (Millipore, Billerica, MA, USA) and FITC-conjugated mouse anti-IgG (Sigma-Aldrich, St. Louis, MO, USA). Briefly, cells were fixed with 4% para-formaldehyde for 15 minutes, washed with 0.05% Tween-20 (Fisher Scientific, USA) in 1x PBS, then permeated with 0.1% Triton-X (Sigma-Aldrich). Cells were then washed and blocked with 1% bovine serum albumin (BSA; Sigma Aldrich) in 1x PBS. If staining for vinculin, cells were incubated in mouse anti-vinculin (1 mg/ml, Millipore) diluted at 1:250 in 0.1% BSA in 1x PBS for 1 hour. Cells were then washed and incubated in FITC-conjugated mouse anti-IgG and TRITC-Phalloidin for 30 minutes. After washing, cells were mounted on cover slips using fluoroshield with DAPI.
Resistance to Detachment
After seeding fibroblasts as described above, cells were incubated in 95% air, 5% CO2 at 37°C for 24 hours. The patterned substrates were then removed from the incubator and stained with NucBlue live cell stain (Life Technologies, Carlsbad, CA, USA). The number of cells per pattern was counted using a fluorescent microscope (Zeiss Axiovert 135, New York, NY, USA) and the chip was placed in a 15 ml centrifuge tube filled with cell culture media. The surface was kept parallel to the tube walls using an insert designed and rapid prototyped specifically for our surfaces. The tubes were then centrifuged at 57 × g. The surfaces were immediately removed from the centrifuge tubes and the cells per pattern were counted. Percentage cells retained per pattern type was calculated.
Image Analysis
An inverted epi-fluorescence Nikon microscope at both 10× and 60× was used for gathering fluorescent images of stained cells. Cell number, length, and width data was collected using ImageJ (National Institutes of Health, Bethersda, VA, USA).
Results
Pattern Generation and Characterization
AFM imaging (Fig. 1) revealed that the electron beam lithography process created well-defined sinusoidal and linear grooves in PMMA substrates. PMMA thickness was between 650 and 680 nm and groove width was between 4.6 and 5 μm in all patterns. PMMA was completely etched in areas exposed to the electron beam, thus exposing the underlying positively charged glass, shown by the dark regions in the AFM images. Wave patterns were defined by their amplitude (A) and wavelength (λ) in μm: wave 1: A=40, λ=10; wave 2: A=30, λ=5; wave 3: A=30, λ=10; all wave patterns had groove spacing of 20 λm. Line patterns were defined by their line spacing, 20 μm or 10 μm. Unpatterned PMMA was used as a control surface.
Fibroblast Alignment
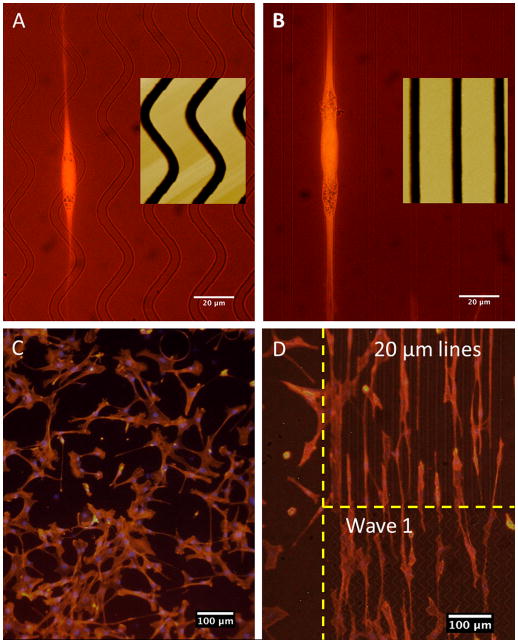
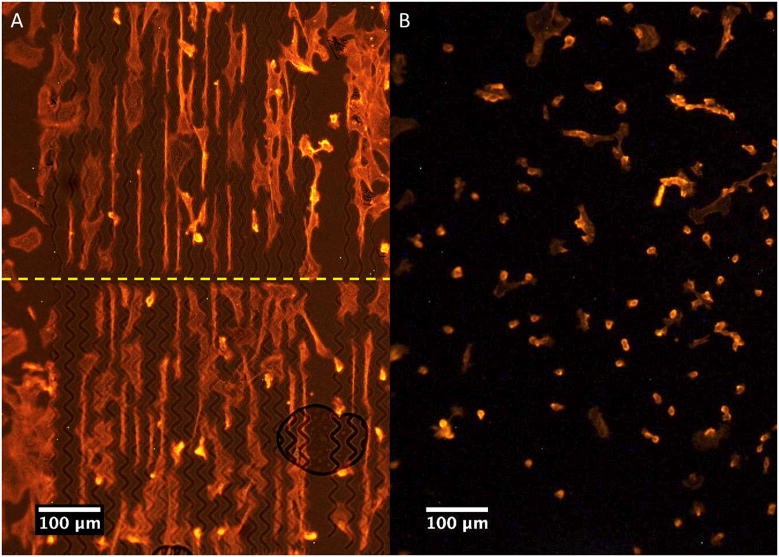
Fibroblast alignment along the long axis of the pattern was seen with cells grown on PMMA patterned surface with either wave or line patterns (Fig. 2). Interestingly, alignment and morphology between wave and line patterns appeared similar despite the distinct difference in pattern shape (Fig. 2A,B). In contrast to cells on patterned surface, fibroblasts on unpatterned control surface tended to be randomly oriented, spreading out in all directions (Fig. 2C), whereas cells on the patterned surfaces tended to be elongated in a single direction and oriented within the constraints of the patterns (Fig. 2A,B,C). Length and width of cells were measured after 24 and 48 hours and compared between pattern types. However, no significant differences were seen between data sets (data not shown).
Figure 2.
Fibroblast alignment on PMMA. A) Wave 1 pattern, 60x. B) 20 μm spaced lines, 60x. C) Control surface (unpatterned), 10x. D) Patterned surface of 10 μm spaced lines on top and wave 1 pattern on bottom, 10x.
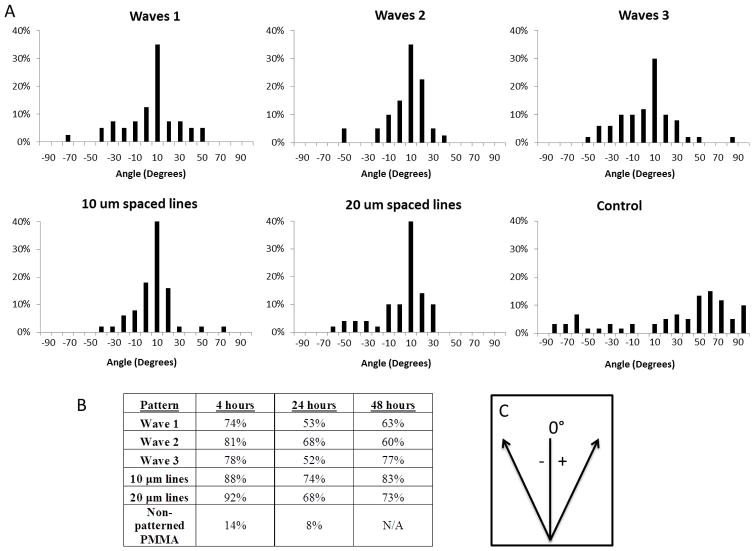
Cell alignment was defined by cells oriented within ±15 degrees of the pattern direction, angle measured using ImageJ. Cell alignment by the majority of cells was observed on all patterns types (Fig. 3). Plotted histograms show distribution of cell orientation on each of the patterns 24 hours after cell seeding (Fig. 3A). The distributions show clear cell preference for cell alignment along the pattern direction, 0°. For unpatterned PMMA control surfaces, the cells seemed to be randomly oriented showing no trend in cell alignment. Cell alignment was also observed 4 and 48 hours after cell seeding (Fig. 3B). The most cell alignment was seen 4 hours after cell seeding across all pattern types with the highest alignment seen on the line patterns: 10 μm with 88% alignment and 20 μm lines with 92% alignment, compared to 74%, 81%, and 78% alignment seen from wave patterns 1, 2, and 3, respectively. For all time frames, the line patterns had a higher percentage of cells that aligned to the pattern surface, except for after 48 hours, wave 2 had 77% alignment and 20 μm lines had 73% alignment.
Figure 3.
A) Fibroblast alignment 24 hours after seeding on patterned surface. Frequency on the histograms is plotted as % of total cells counted, 40–60 cells per pattern type. B) Percent cell alignment per pattern, cell alignment was considered to be ±15 degrees. C) Convention used during measuring orientation, where 0° represents pattern orientation.
It was noted that cells began to spread, elongate, and orient along the pattern direction just 4 hours after initial seeding. Fig. 3 shows cell adhesion 4 hours after seeding on A) the patterned surface and B) the unpatterned PMMA surface. Cells on the patterned surfaces were elongated and anchored across the grooves for both line and wave patterns. In contrast, cells on the control surface remained much smaller and rounded.
Intimacy of Cell Attachment to a Surface
In order to further understand the nature of cell adhesion to the patterned surface, z-stacked images were taken of fibroblasts lying on both wave and line patterns (Fig. 5). It was noted that on line patterns, the majority of each fibroblast resides within the groove of the pattern. Whereas, on the wave patterns, most of the cell lay on the top, flat portion of the PMMA and as the cell crossed a groove, it dipped into it. Vinculin focal points were seen both inside and outside of the grooves for both the wave and the line patterns, indicating that cells have adhesion points on both the glass and the PMMA surfaces.
Figure 5.
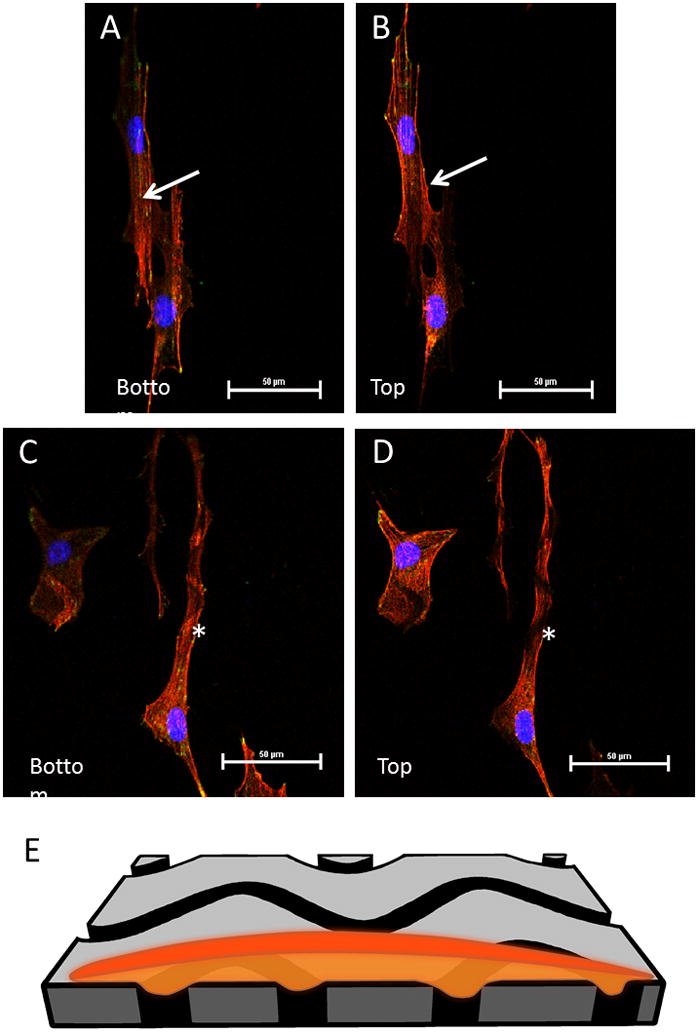
Slices from a z-stacked image of fibroblasts on PMMA patterned surface. Red is phalloidin stained actin filaments, blue is DAPI (nuclei), and green is vinculin focal adhesions. A) Bottom portion of the z-stack, closest to the charged glass surface, most of the cell lies within the groove. B) Top portion of the z-stack, notice the absence of actin where the groove lies indicating that the cell is dipping into the groove, lying within it. The arrows indicate one of the grooves the cell is lying within. C) and D) show bottom and top, respectively, slice of a cell sitting across wave pattern 3. The majority of the cell lies on the PMMA surface, but the cell dips slightly into each groove as it crosses it. The star is placed just under one of the wave grooves. E) shows an illustration of how a cell goes into one of the grooved surfaces as it passes over the wave pattern.
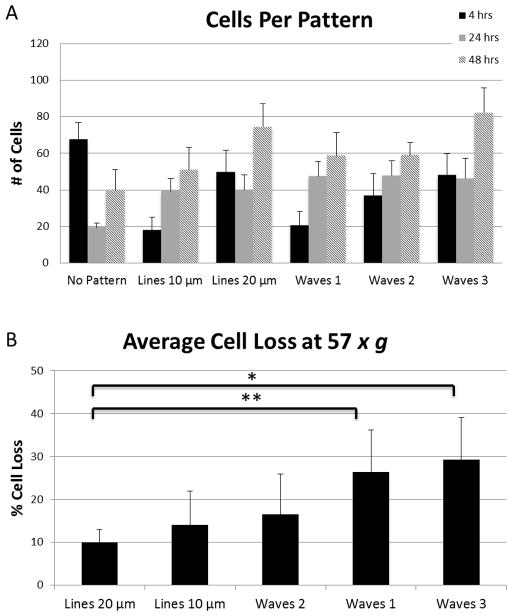
Cell residence on each of the patterns was found by counting the number of cells on each pattern 4, 24, and 48 hours after cell seeding (Fig. 6A). Wave pattern 3 and the 20 μm spaced line pattern had the most cell attachment after 48 hours. No significant differences were noted between pattern types.
Figure 6.
A) Cell adhesion on patterns 4, 24, and 48 hours after seeding. B) Average cell loss after 15 minutes exposure to 57 × g force from centrifuge. Error bars represent standard error. (*) p-value 0.059. (**) p-value 0.078.
As a measure of adhesion strength, we subjected the cells a centrifugal force of 57 × g for 15 minutes. Cells on the surface were counted before and after centrifugation so that percentage cell loss could be calculated. Cell loss ranged between 9 and 29% depending on pattern type (Fig. 6B). Differences between 20 μm spaced line pattern and wave 1 (p-value = 0.059) and 20 μm spaced line pattern and wave 3 (p-value = 0.078) were the most significant.
Discussion
The ability to control cell behavior on artificial materials is important in both tissue engineering and biomaterials. This work was designed to investigate cell behavior on non-linear, sinusoidal groove micropatterns. The use of electron beam lithography allowed for the precise control of pattern dimensions and the creation of three well-defined wave groove patterns and two linear patterns. The findings clearly demonstrate that fibroblasts will align themselves to non-linear wave grooved micropatterns as well as linear groove patterns but will randomly orient on unpatterned surface, with no general trend in orientation angle. We also noted that the patterned surfaces allow for faster cell adhesion and elongation compared to unpatterned PMMA. This may be a result of increased surface area on the patterned surfaces compared to the unpatterned surface. Additionally, the positive charge of the underlying glass surface in the patterned areas attracted the negative charge of the cell membrane.
Fibroblasts had the greatest alignment 4 hours after their initial seeding compared to alignment data 24 and 48 hours after seeding. We believe this is due to the fact that cells begin to migrate around the surface at 24 and 48 hour time points. As the cells move around the surface, alignment decreased slightly as not all cells travelled directly along the patterns.
Interestingly, we observed similar cell adhesion orientation between the wave and line patterns (Fig. 2A,B), despite the obvious difference in pattern shape between line and wave patterns. However, we noted a difference in how cells orient themselves on each pattern type. For linear patterns, cells tended to reside within each groove, which has been seen in other work [37]. However, on the wave micropatterns the cells tend to sit primarily on the PMMA surface and orient themselves across a single wave. This is a particularly interesting way of cell alignment in comparison to cell behavior on the linear grooved patterns as it seems that upon initial interaction with the wave pattern, cells do not differentiate a line from a wave. Yet, they orient themselves on the wave patterns in a way that would almost be equivalent to orthogonal alignment on a line pattern, which we do not see on any of the line patterns.
It was observed on all patterned surfaces that cell spreading was clearly greater 4 hours after cell seeding compared to the unpatterned control PMMA surface (Fig. 4). At this time point, fibroblasts on the unpatterened surface remained round and small, whereas cells on the patterned surface were elongated and spread across the patterned surface. At this time point, our data shows more cells typically reside on the unpatterened surface (Fig. 6). However, they may not be as tightly bound as they are on the textured surface because they are not spread out and elongated as they are on the patterned surface.
Figure 4.
Cellular morphology 4 hours after seeding on A) patterned surface, wave 2 (top) and wave 3 (bottom), and B) unpatterned control surface.
The actin staining helped to further understand how the cells were adhering to the wave and groove patterns. The z-stacked images (Fig. 5), verified that cells were indeed lying within each groove on the line patterns and also showed that the cells on the wave patterns dipped into the grooves as they crossed over them. The vinculin staining showed focal adhesions both inside and outside of each groove for the wave patterns. As far as adhesion strength, we observed a higher rate of cell retention during our centrifuge experiment for the linear patterns compared to any of the wave patterns (Fig. 6B). The highest rate of retention was seen on the 20 μm spaced line patterns, with significant differences seen between this group and wave patterns 1 and 3. Because fibroblasts tend to sit within the grooves on the line patterns, we can assume that 100% of the bottom surface of the cell is interacting with the underlying positively charged glass surface. For the wave patterned surfaces, typically about 50% of the cell was contacting PMMA and the other 50% interacting with the underlying positively charged substrate (calculated using ImageJ). Because the underlying positively charged glass surface attracted the negative charge of the cell membrane, we saw a difference in adhesion strength based on the percentage of cell area that came in contact with the glass surface. However, adhesion strength was not completely lost and the wave patterns were still able to retain cells. This may allow tissue-engineers to control the amount of cell adhesion or cell interaction that is seen on an implant surface, while still allowing for guidance and control of cellular orientation and migration. Additionally, as fibroblasts on the wave patterns reside atop the PMMA surface, it may be possible to deliver drugs, nutrients, or other solutes to cells via the underlying wave channels underneath them, i.e. a form of cellular irrigation.
Conclusion
We have shown that fibroblasts do not need a linear groove pattern to induce linear cell alignment through contact guidance. We have demonstrated fibroblast alignment using a sinusoidal wave micropattern, which is comparable to alignment using linear groove patterns. We observed that cells on a wave pattern will cross over grooves and reside both inside and outside of the groove as the cell aligns to the pattern, dipping into each groove as it is crossed. Creation of a pattern on PMMA surface with an underlying positively charged surface allows for faster adhesion compared to an unpatterned PMMA surface. The ability to add texture to a surface to manipulate cell adhesion strength and growth with only localized attachment and to maintain free space in curvilinear microtopography underlying the cell may be a useful addition for tissue engineering strategies and the fabrication of novel biomedical devices.
Highlights.
Fibroblasts may be aligned linearly on sinusoidal wave micropatterns, residing both within and atop of each wave pattern.
Adhesion strength is not completely limited on wave micropatterns despite decrease in cell-surface contact area.
Cell surface interaction/adhesion on implant surfaces can be modulated based on cell-surface contact area.
Underlying free spaces in curvilinear micropatterns may be useful for exposure of cells to fluids (cellular irrigation).
Acknowledgments
Funding for this research was provided by the Pilot Grant from BIO5 Institute, The University of Arizona, and the Cardiovascular Biomedical Engineering Training Grant, U.S. National Institutes of Health T32HL007955.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bettinger CJ, Langer R, Borenstein JT. Engineering substrate topography at the micro- and nanoscale to control cell function. Angew Chem Int Ed. 2009;48:5406–5415. doi: 10.1002/anie.200805179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flemming RG, Murphy CJ, Abrams GA, Goodman SL, Nealey PF. Effects of synthetic micro- and nano-structured surfaces on cell behavior. Biomaterials. 1999;20:573–588. doi: 10.1016/s0142-9612(98)00209-9. [DOI] [PubMed] [Google Scholar]
- 3.Clark P, Connolly P, Curtis AS, Dow JA, Wilkinson CD. Topographical control of cell behaviour: II. multiple grooved substrata. Development. 1990;108:635–644. doi: 10.1242/dev.108.4.635. [DOI] [PubMed] [Google Scholar]
- 4.Dunn GA, Brown AF. Alignment of fibroblasts on grooved surfaces described by a simple geometric transformation. J Cell Sci. 1986;83:313–340. doi: 10.1242/jcs.83.1.313. [DOI] [PubMed] [Google Scholar]
- 5.Harrison R. The reaction of embryonic cells to solid stuctures. J Exp Zool. 1914;17:521–544. [Google Scholar]
- 6.Dalby MJ, Riehle MO, Yarwood SJ, Wilkinson CD, Curtis AS. Nucleus alignment and cell signaling in fibroblasts: response to a micro-grooved topography. Exp Cell Res. 2003;284:272–280. doi: 10.1016/s0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- 7.Weiss P, Garber B. Shape and movement of mesenchyme cells as functions of the physical structure of the medium: contributions to a quantitative morphology. Proc Nat Acad Sci USA. 1952;38:26–280. doi: 10.1073/pnas.38.3.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oakley C, Brunette DM. The sequence of alignment of microtubules, focal contacts and actin filaments in fibroblasts spreading on smooth and grooved titanium substrata. J Cell Sci. 1993;106:343–354. doi: 10.1242/jcs.106.1.343. [DOI] [PubMed] [Google Scholar]
- 9.Oakley C, Brunette DM. Topographic compensation: guidance and directed locomotion of fibroblasts on grooved micromachined substrata in the absence of microtubules. Cell Motility Cytoskeleton. 1995;31:45–58. doi: 10.1002/cm.970310106. [DOI] [PubMed] [Google Scholar]
- 10.Dunn G, Heath J. A new hypothesis of contact guidance in tissue cells. Exp Cell Res. 1976;101:1–14. doi: 10.1016/0014-4827(76)90405-5. [DOI] [PubMed] [Google Scholar]
- 11.Clark P, Connolly P, Curtis AS, Dow JA, Wilkinson CD. Topographical control of cell behaviour. I. simple step cues. Development. 1987;99:439–448. doi: 10.1242/dev.99.3.439. [DOI] [PubMed] [Google Scholar]
- 12.Brunette DM. Spreading and orientation of epithelial cells on grooved substrata. Exp Cell Res. 1986;167:203–217. doi: 10.1016/0014-4827(86)90217-x. [DOI] [PubMed] [Google Scholar]
- 13.Dalby MJ, Yarwood SJ, Johnstone HJH, Affrossman S, Riehle MO. Fibroblast signaling events in response to nanotopography: a gene array study. IEEE Trans Nanobiosci. 2002;1:12–17. doi: 10.1109/tnb.2002.806930. [DOI] [PubMed] [Google Scholar]
- 14.Curtis A, Wilkinson C. Topographical control of cells. Biomaterials. 1997;18:1573–1583. doi: 10.1016/s0142-9612(97)00144-0. [DOI] [PubMed] [Google Scholar]
- 15.Andersson A. The effects of continuous and discontinuous groove edges on cell shape and alignment. Exp Cell Res. 2003;288:177–188. doi: 10.1016/s0014-4827(03)00159-9. [DOI] [PubMed] [Google Scholar]
- 16.Baker D, Liu X, Weng H, Luo C. Fibroblast/fibrocyte: surface interaction dictates tissue reactions to micropillar implants. Biomacromolecules. 2011;12:997–1005. doi: 10.1021/bm1013487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou L, Firth JD, Uitto VJ, Brunette DM. Substratum surface topography alters cell shape and regulates fibronectin mRNA level, mRNA stability, secretion and assembly in human fibroblasts. J Cell Sci. 1995;108:1563–1573. doi: 10.1242/jcs.108.4.1563. [DOI] [PubMed] [Google Scholar]
- 18.Craighead H, James C, Turner AM. Chemical and topographical patterning for directed cell attachment. Curr Opin Solid State Mater Sci. 2001;5:177–184. [Google Scholar]
- 19.Dalby MJ, Riehle MO, Sutherland DS, Agheli H, Curtis ASG. Use of nanotopography to study mechanotransduction in fibroblasts - methods and perspectives. Eur J Cell Biol. 2004;83:159–169. doi: 10.1078/0171-9335-00369. [DOI] [PubMed] [Google Scholar]
- 20.den Braber ET, de Ruijter JE, Smits HT, Ginsel LA, von Recum AF, Jansen JA. Quantitative analysis of cell proliferation and orientation on substrata with uniform parallel surface micro-grooves. Biomaterials. 1996;17:1093–1099. doi: 10.1016/0142-9612(96)85910-2. [DOI] [PubMed] [Google Scholar]
- 21.den Braber ET, de Ruijter JE, Ginsel LA, von Recum AF, Jansen JA. Quantitative analysis of fibroblast morphology on microgrooved surfaces with various groove and ridge dimensions. Biomaterials. 1996;17:2037–2044. doi: 10.1016/0142-9612(96)00032-4. [DOI] [PubMed] [Google Scholar]
- 22.den Braber ET, de Ruijter JE, Smits HT, Ginsel LA, von Recum AF, Jansen JA. Effect of parallel surface microgrooves and surface energy on cell growth. J Biomed Mater Res. 1995;29:511–518. doi: 10.1002/jbm.820290411. [DOI] [PubMed] [Google Scholar]
- 23.Wilkinson C. Nanostructures in biology. Microelectron Eng. 1995;27:61–65. [Google Scholar]
- 24.Gadegaard N, Martines E, Riehle MO, Seunarine K, Wilkinson CDW. Applications of nano-patterning to tissue engineering. Microelectron Eng. 2006;83:1577–1581. [Google Scholar]
- 25.Kim D-H, Han K, Gupta K, Kwon KW, Suh K-Y, Levchenko A. Mechanosensitivity of fibroblast cell shape and movement to anisotropic substratum topography gradients. Biomaterials. 2009;30:5433–5444. doi: 10.1016/j.biomaterials.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loesberg WA, te Riet J, van Delft FCMJM, Schön P, Figdor CG, Speller S, et al. The threshold at which substrate nanogroove dimensions may influence fibroblast alignment and adhesion. Biomaterials. 2007;28:3944–3951. doi: 10.1016/j.biomaterials.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 27.Meyle J, Wolburg H, von Recum AF. Surface micromorphology and cellular interactions. J Biomater Appl. 1993;7:362–374. doi: 10.1177/088532829300700404. [DOI] [PubMed] [Google Scholar]
- 28.Peterbauer T, Yakunin S, Siegel J, Hering S, Fahrner M, Romanin C, et al. Dynamics of spreading and alignment of cells cultured in vitro on a grooved polymer surface. J Nanomater. 2011;2011:1–10. [Google Scholar]
- 29.Su W-T, Liao Y-F, Chu I-M. Observation of fibroblast motility on a micro-grooved hydrophobic elastomer substrate with different geometric characteristics. Micron. 2007;38:278–285. doi: 10.1016/j.micron.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 30.Walboomers XF, Jansen JA. Cell and tissue behavior on micro-grooved surfaces. Odontology/Soc Nippon Dental Univ. 2001;89:2–11. doi: 10.1007/s10266-001-8178-z. [DOI] [PubMed] [Google Scholar]
- 31.Yoshinari M, Matsuzaka K, Inoue T, Oda Y, Shimono M. Effects of multigrooved surfaces on fibroblast behavior. J Biomed Mater Res A. 2003;65:359–368. doi: 10.1002/jbm.a.10521. [DOI] [PubMed] [Google Scholar]
- 32.Martínez E, Engel E, Planell JA, Samitier J. Effects of artificial micro- and nano-structured surfaces on cell behaviour. Ann Anatomy. 2009;191:126–135. doi: 10.1016/j.aanat.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Curtis ASG, Gadegaard N, Dalby MJ, Riehle MO, Wilkinson CDW, Aitchison G. Cells react to nanoscale order and symmetry in their surroundings. IEEE Trans Nanobiosci. 2004;3:61–65. doi: 10.1109/tnb.2004.824276. [DOI] [PubMed] [Google Scholar]
- 34.Dalby MJ, Gadegaard N, Wilkinson CDW. The response of fibroblasts to hexagonal nanotopography fabricated by electron beam lithography. J Biomed Mater Res A. 2008;84:973–979. doi: 10.1002/jbm.a.31409. [DOI] [PubMed] [Google Scholar]
- 35.Berry CC, Campbell G, Spadiccino A, Robertson M, Curtis ASG. The influence of microscale topography on fibroblast attachment and motility. Biomaterials. 2004;25:5781–5788. doi: 10.1016/j.biomaterials.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 36.Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, et al. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nature Mater. 2007;6:997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 37.Lee SW, Kim SY, Rhyu IC, Chung WY, Leesungbok R, Lee KW. Influence of microgroove dimension on cell behavior of human gingival fibroblasts cultured on titanium substrata. Clin Oral Implants Res. 2009;20:56–66. doi: 10.1111/j.1600-0501.2008.01597.x. [DOI] [PubMed] [Google Scholar]