Abstract
Background and Purpose
Prior studies demonstrated association between mitochondrial DNA variants and ischemic stroke (IS). We investigated whether variants within a larger set of oxidative phosphorylation (OXPHOS) genes encoded by both autosomal and mitochondrial DNA were associated with risk of IS and, based on our results, extended our investigation to intracerebral hemorrhage (ICH).
Methods
This association study employed a discovery cohort of 1643 individuals, a validation cohort of 2432 individuals for IS, and an extension cohort of 1476 individuals for ICH. Gene-set enrichment analysis (GSEA) was performed on all structural OXPHOS genes, as well as genes contributing to individual respiratory complexes. Gene-sets passing GSEA were tested by constructing genetic scores using common variants residing within each gene. Associations between each variant and IS that emerged in the discovery cohort were examined in validation and extension cohorts.
Results
IS was associated with genetic risk scores in OXPHOS as a whole (odds ratio (OR)=1.17, p=0.008) and Complex I (OR=1.06, p=0.050). Among IS subtypes, small vessel (SV) stroke showed association with OXPHOS (OR=1.16, p=0.007), Complex I (OR=1.13, p=0.027) and Complex IV (OR 1.14, p=0.018). To further explore this SV association, we extended our analysis to ICH, revealing association between deep hemispheric ICH and Complex IV (OR=1.08, p=0.008).
Conclusions
This pathway analysis demonstrates association between common genetic variants within OXPHOS genes and stroke. The associations for SV stroke and deep ICH suggest that genetic variation in OXPHOS influences small vessel pathobiology. Further studies are needed to identify culprit genetic variants and assess their functional consequences.
Keywords: OXPHOS, mitochondria, stroke, genes
INTRODUCTION
Despite remarkable advances in prevention, diagnosis, and treatment, stroke remains the world’s second-leading cause of death and a leading cause of disability. While many modifiable environmental factors contribute to stroke risk, there are ample data demonstrating a genetic risk component as well 1. Recent genome-wide association studies (GWAS) have demonstrated that common DNA variants influence risk of ischemic stroke 2-4.
A prior study demonstrated that common mitochondrial variants influence risk of ischemic stroke 4. The mitochondrial genome is vital to the assembly of the oxidative phosphorylation (OXPHOS) apparatus, but the majority of OXPHOS structural proteins are encoded within the autosomal genome 5. The OXPHOS apparatus consists of five complexes that are necessary to maintain aerobic homeostasis and preserve reduction/oxidation (redox) balance in the cellular environment. Multiple rare disorders are caused by mutations of OXPHOS genes, many of which result in neurodegenerative or stroke-like phenotypes, including seizures, metabolic infarcts, and encephalomyopathies 6. Additionally, OXPHOS fitness plays a role in the neuronal response to and recovery after oxidative stress 7.
We hypothesized that common genetic variants in OXPHOS genes, both within the autosomal and mitochondrial genome, influence the risk of stroke. To test this hypothesis, we performed a pathway-based genetic association analysis interrogating genetic variants within OXPHOS genes. We initially performed a cumulative test of all common genetic variation within OXPHOS loci by employing a gene set enrichment analysis (GSEA) technique. This allowed us to investigate whether the OXPHOS pathway was enriched for association with stroke risk. Based on this analysis, we sought to ascertain and quantify the role of these variants by calculating a genetic risk score from OXPHOS genes in the Massachusetts General Hospital (MGH) Ischemic Stroke GWAS. We then replicated this risk score association in a separate dataset comprised of individuals from the Ischemic Stroke Genetics Study (ISGS) and Siblings with Ischemic Stroke Study (SWISS). We then tested the same risk score in intracerebral hemorrhage (ICH) using individuals from the International Stroke Genetics Consortium ICH GWAS (ISGC ICH).
MATERIALS AND METHODS
Subjects
Genetic data and phenotypic information were contributed by the MGH Ischemic Stroke GWAS 8, ISGS 9, SWISS 10, and the ISGC ICH GWAS 11 [TABLE 1]. Additional control individuals for the MGH dataset were contributed by the MIGen Consortium, a case-control study of genetic risk for myocardial infarction 12. Hospital-based ischemic stroke case and control recruitment and phenotype ascertainment were performed according to protocols described previously, and stroke subtypes were assigned by TOAST criteria 8-10. In cases where ischemic stroke subtype data was unavailable, individuals were dropped from subtype analyses but were allowed to remain in all-cause ischemic stroke analyses (n=124 in MGH, n=387 in ISGS/SWISS). Multi-center hospital-based ICH case and control recruitment and phenotype ascertainment were performed according to protocols described previously 11. Location of ICH was assigned by stroke neurologists, based on standard criteria with central adjudication 11, 13. Institutional review boards from all participating centers approved the study, and all participants gave informed consent for data collection, genotyping, and analysis of genetic data.
Table 1.
Study populations
| MGH/MIGen Cases |
MGH/MIGen Controls |
ISGS/SWISS Cases |
ISGS/SWISS Controls |
ISGC ICH Cases |
ISGC ICH Controls |
|
|---|---|---|---|---|---|---|
| n | 484 | 1159 | 1048 | 1384 | 928 | 909 |
| - Small vessel | 55 | --- | 197 | --- | --- | --- |
| - Large artery | 114 | --- | 223 | --- | --- | --- |
| - Cardioembolic | 191 | --- | 241 | --- | --- | --- |
| - Deep ICH | --- | --- | --- | --- | 430 | --- |
| - Lobar ICH | --- | --- | --- | --- | 409 | --- |
| Sex (% female) | 0.39 | 0.41 | 0.43 | 0.52 | 0.52 | 0.51 |
|
Age at enrollment
(mean, SD) |
66.5 (14.6) | 47.5 (8.5) | 64.8 (13.6) | 66.5 (12.6) | 72.4 (11.5) | 73.0 (8.4) |
| Hypertension (%) | 0.62 | 0.55 | 0.64 | 0.34 | 0.71 | 0.55 |
| DM 2 (%) | 0.21 | 0.18 | 0.19 | 0.11 | 0.20 | 0.09 |
| Atrial fibrillation (%) | 0.12 | 0.12 | 0.08 | 0.04 | 0.22 | 0.20 |
| Current smoker (%) | 0.20 | 0.18 | 0.16 | 0.06 | 0.17 | 0.20 |
| Warfarin use (%) | --- | --- | --- | --- | 0.08 | 0.03 |
ICH = intracerebral hemorrhage, n = number of cases/controls, SD = standard deviation, DM 2 = type 2 diabetes mellitus, --- = not applicable
Genotyping and Imputation
Blood samples from MGH/MIGen were processed and genotyped using the Affymetrix 6.0 platform, while ISGS and SWISS were assayed with the Illumina 660W and 1M platforms according to previously-published protocols 8-10. Blood samples for the ISGC ICH cases and controls were genotyped on Illumina 660W 11. For harmonization across platforms, all datasets had additional genotypes imputed using PLINK v1.07 (http://pngu.mgh.harvard.edu/~purcell/plink) and the International HapMap Project phase 3 reference dataset (http://www.hapmap.org). Captured mitochondrial variants from the genotyping arrays were extracted 14, and raw intensity files were inspected visually by C.D.A. and A.B. to confirm accuracy of genotype calls.
Genomic Quality Control
For all analyzed cohorts, quality control of genotyped individuals included gender-sex discordance, filtering for missingness by individual > 0.1, missingness by SNP > 0.05, and minor allele frequency (MAF) < 0.01. Individuals displaying cryptic relatedness (p-hat ≥ 0.125) and genotypes with significant departure from Hardy-Weinberg Equilibrium (p<10E-5) were excluded from analysis 15. Autosomal imputation was performed using PLINK v1.07 after quality control filtering. Mitochondrial imputation was performed using a haplotype-based approach with reference datasets from GenBank and Mitokor after additional mtDNA-specific quality control measures [Supplementary Appendix I] 16. Post-imputation, SNPs were excluded with MAF < 0.01 or RSQR quality index < 0.3.
OXPHOS SNP Selection
Genes encoding proteins directly involved in the OXPHOS respiratory chain were selected based on published criteria from a chemical dissection of mitochondrial function, yielding a total of 95 genes in the autosomal genome and 13 genes in the mitochondrial genome 5. SNPs falling within these genes +/− 100 kilobases and passing quality control filtering were extracted from the MGH and ISGS/SWISS datasets after imputation and included in the final analysis. Sub-analyses were performed for genes grouped according to each OXPHOS respiratory complex, classified according to annotation in the Ensembl Genome Browser (http://www.ensembl.org) [Supplementary Table S1 and S2].
Population Structure and Control
Only individuals of European ancestry were analyzed in the present study. Population structures for autosomal and mitochondrial variants were assessed independently due to their significantly different inheritance patterns, using principal component analysis 16. Autosomal principal components (PCs) 1-5 were extracted for each individual, and were added in association testing of autosomal SNPs until no additional reduction in genomic inflation factor (GIF) could be achieved (PC1-2 in all analyses). Mitochondrial PCs 1-10 were extracted for each individual, and were similarly added in association testing of mitochondrial SNPs until mitochondrial GIF was minimized (PC1-5 in all analyses).
Gene-set Enrichment Analysis
Testing for cumulative OXPHOS pathway associations with ischemic stroke risk was performed using the GSEA method 17 as implemented in the GenGen v.2010Apr29 software package 18. GSEA was implemented in this study as a preliminary screen of the OXPHOS pathway prior to generation of genetic scores, as a means to minimize the possibility of any false positive associations. The GSEA method determines whether variants within a pre-defined biological pathway contain more associations with the chosen phenotype than would be expected by chance alone. For ischemic stroke, GSEA testing was performed in the ISGS/SWISS cohort as a preliminary analysis prior to genetic score generation. GSEA was performed in the ISGS/SWISS replication cohort rather than the MGH/MIGen discovery cohort to prevent any chance enrichment of OXPHOS association in the MGH/MIGen cohort from influencing gene sets chosen for genetic score testing. A separate GSEA was performed in the ISGC ICH cohort. Results are reported as a permutation-derived empiric p-value (100,000 gene-set permutations) for gene set association with the ischemic stroke or ICH risk, with the null hypothesis derived from random sampling of an equal number of variants of similar MAFs chosen from genes not within the OXPHOS pathway. Using this technique, the significance threshold for our GSEA was set at empiric p < 0.05.
Genetic Score Generation in Ischemic Stroke
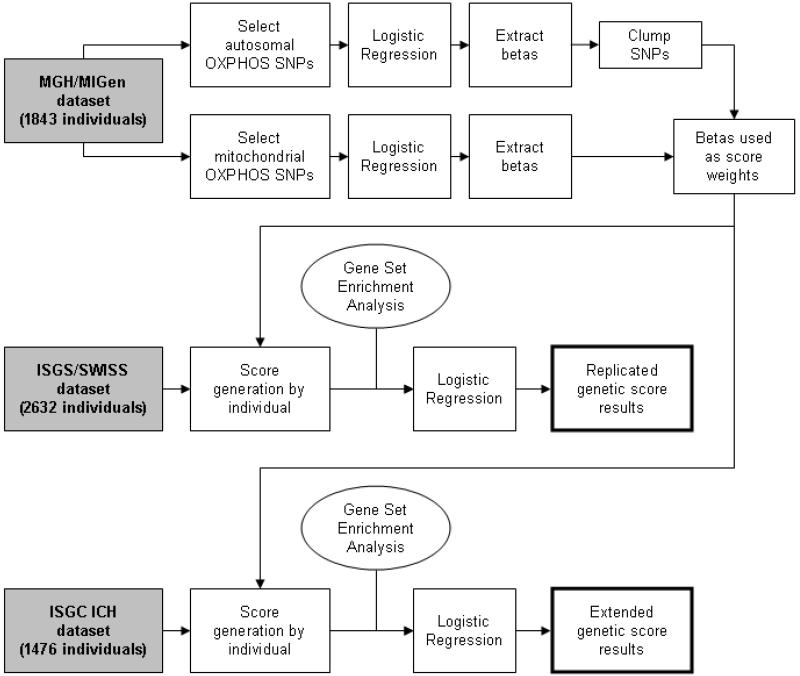
Combined effects of all autosomal and mitochondrial OXPHOS SNPs were evaluated using a score-based method previously described 4, 12 [FIGURE 1]. Briefly, each OXPHOS SNP was tested for association with all-cause ischemic stroke risk in the MGH/MIGen discovery case-control dataset. The results of this analysis, expressed as a beta coefficient for the risk allele at each SNP, were then clumped according to linkage disequilibrium (LD) using the “clump” function in PLINK v1.07. Only the SNP with the highest significance value was retained in regions where LD > 0.6 between SNPs. No additional pruning or thresholding was performed in an attempt to “optimize” the SNPs included in the genetic score. These beta coefficients were then applied to the corresponding OXPHOS SNPs in the ISGS/SWISS validation dataset. All subsequent analyses in replication and extension were based on this single set of beta coefficients from the MGH/MIGen discovery cohort in association with all-cause ischemic stroke [Supplementary Table S3]. Following beta coefficient extraction, the MGH/MIGen discovery cohort was not included in any further analyses. A risk score was generated for each individual by summing the beta coefficients associated with each risk allele present in the individual. Informed by GSEA results, scores were developed for all OXPHOS complexes, Complex I, and Complex IV. Because the risk score distributions failed testing for normality by Shapiro-Wilk, the score was divided into quintiles in an unsupervised fashion using the “cut” command in STATA v10.0 (www.stata.com) for association testing.
Figure 1.
Flowchart describing the MGH/MIGen discovery, ISGS/SWISS validation, and ISGC ICH extension cohorts. ICH = intracerebral hemorrhage, OXPHOS = all structural proteins directly contributing to oxidative phosphorylation complex function, SNP = single nucleotide polymorphism.
Genetic Score Association Testing in Ischemic Stroke
The ischemic stroke risk score quintiles were used as the independent variable in an ordinal logistic regression model for ischemic stroke risk, using age and sex as pre-specified covariates. Results reported represent risk (as expressed by odds ratio) per unit increase in score quintile. SNPs that were present in the MGH discovery cohort but were absent from the ISGS/SWISS validation cohort were dropped from the analysis (n=30). Additional covariates of hypertension, diabetes, and atrial fibrillation were also tested. Results of genetic score testing in the replication and extension datasets represent independent tests. Therefore p < 0.05 in the replication and extension datasets is considered statistically significant. All regression analyses were performed using STATA v10.0.
Extension of Genetic Score Analysis to Intracerebral Hemorrhage
The same beta coefficients of association with ischemic stroke from the MGH/MIGen discovery cohort were applied to individuals within the ISGC ICH cohort, again resulting in a risk score for each individual. Risk score quintiles were then used as the independent variable in a logistic regression for ICH risk, using age, sex, hypertension, and warfarin exposure as pre-specified covariates. For ICH, scores were developed only for all OXPHOS and Complex IV. Separate analyses were performed for deep and lobar ICH subtypes. Cerebellar and multi-compartment ICH were included in the all ICH analysis, but were dropped from deep and lobar analyses.
Post-hoc Power Calculation
Power for discovery of association between individual variants within OXPHOS genes and ischemic stroke was computed using the Genetic Power Calculator 19, with calculated ORs of 1.10, 1.20 and 1.40 and MAF of 0.10, 0.20 and 0.30. For this analysis, alpha was set at 6 × 10E-5 (842 independent tests for autosomal and mitochondrial variants within OXPHOS genes).
Power to detect an association between the genetic risk score and stroke phenotypes was computed using the expected proportion of variance explained, assuming that the overall information content of the score would account for 0.5%, 1% or 5% of variance between cases and controls. Power calculations were performed for ischemic stroke, ischemic stroke subtypes, ICH, and ICH deep and lobar subgroups.
RESULTS
Genotyping Quality Control and Imputation Results
Implementation of quality control and imputation methods left 1843 individuals and 707 autosomal SNPs in the MGH/MIGen cohort, 2632 individuals and 677 autosomal SNPs in the ISGS/SWISS cohort, and 1837 individuals and 707 autosomal SNPs in the ISGC ICH cohort [FIGURE 1]. 135 mitochondrial SNPs were retained in all three cohorts after extraction and haplotype-based imputation [Supplementary Appendix II].
Gene Set Enrichment Analysis in Ischemic Stroke
GSEA was performed using the ISGS/SWISS cohort, testing the OXPHOS gene-set 100,000 times against randomly-assigned gene sets of equal size [TABLE 2]. GSEA testing for the full OXPHOS gene-set and those within each respiratory complex demonstrated associations between ischemic stroke and the full OXPHOS gene-set (p=0.012) as well as OXPHOS Complexes I and IV (p=0.024 in both). Among ischemic stroke subtypes, GSEA revealed significant association between SV stroke and OXPHOS Complex I and IV (p = 0.008 and p = 0.005, respectively), although there was only a trend toward association between SV stroke and the full OXPHOS gene-set (p=0.091).
Table 2.
Gene set enrichment analysis (p-values)
| Cases/Controls | Full OXPHOS |
Complex I |
Complex II/III |
Complex IV |
Complex V |
|
|---|---|---|---|---|---|---|
|
All ischemic
stroke |
1048/1384 | 0.012 | 0.024 | > 0.20 | 0.024 | 0.11 |
| - CE | 241/1384 | > 0.20 | > 0.20 | > 0.20 | > 0.20 | > 0.20 |
| - LA | 223/1384 | > 0.20 | > 0.20 | > 0.20 | > 0.20 | > 0.20 |
| - SV | 197/1384 | 0.091 | 0.008 | > 0.20 | 0.005 | > 0.20 |
|
| ||||||
| All ICH | 928/548 | >0.20 | 0.12 | --- | 0.035 | --- |
| - Lobar ICH | 409/548 | >0.20 | >0.20 | --- | >0.20 | --- |
| - Deep ICH | 430/548 | >0.20 | 0.16 | --- | 0.009 | --- |
Association between gene sets and ischemic stroke (ISGS/SWISS) and ICH (ISGC ICH), with p-values reported from 100,000 permutations against the null.
CE = cardioembolic stroke, ICH = intracerebral hemorrhage, LA = large artery stroke, SV = small vessel stroke, OXPHOS = all structural proteins directly contributing to oxidative phosphorylation complex function, --- = analysis not performed.
Gene Set Enrichment Analysis in Intracerebral Hemorrhage
As with our ischemic stroke analysis, GSEA was performed in the ISGC ICH cohort in order to determine whether OXPHOS gene-sets were enriched for association with ICH risk [TABLE 2]. Based on our results from ischemic stroke GSEA, only the full OXPHOS, Complex I, and Complex IV gene-sets were carried over for testing in ICH. An association was found between all ICH and Complex IV (p=0.035). After restricting cases to deep and lobar subgroups, deep ICH retained association with Complex IV (p=0.008).
Genetic Score Analysis in Ischemic Stroke
Based on GSEA results, only SNPs in the full OXPHOS gene-set as well as Complexes I and IV were used to calculate genetic scores. Similarly, only all-cause ischemic stroke and the SV subtype were carried forward for genetic score analysis [TABLE 3]. Application of beta coefficient-based scores in the ISGS/SWISS cohort demonstrated associations between a score comprised of the full OXPHOS gene-set and ischemic stroke (odds ratio (OR)=1.17, 95% confidence interval (CI)=1.03–1.33). This full OXPHOS score was also associated with the SV stroke subtype (OR=1.16, 95% CI=1.04–1.29). Of note, these genetic score results were largely driven by autosomal variants, with results deviating less than 20% when mitochondrial variants were excluded [Supplementary Table S6].
Table 3.
Genetic score analysis
| Cases/Controls | Full OXPHOS | Complex I | Complex IV | ||||
|---|---|---|---|---|---|---|---|
| OR (95% CI) |
p | OR (95% CI) |
p | OR (95% CI) |
p | ||
| All ischemic stroke |
1048/1384 | 1.17 (1.03- 1.33) |
0.008 | 1.06 (1.00-1.12) |
0.050 | 1.05 (0.99-1.12) |
0.075 |
| SV only | 197/1384 | 1.16 (1.04- 1.29) |
0.007 | 1.13 (1.01-1.26) |
0.027 | 1.14 (1.02-1.27) |
0.018 |
|
| |||||||
| All ICH | 928/548 | --- | --- | --- | --- | 1.08 (1.01-1.17) |
0.039 |
| Deep only |
430/548 | --- | --- | --- | --- | 1.14 (1.03-1.25) |
0.008 |
Genetic score analysis results in the ISGS/SWISS validation cohort and ISGC ICH extension cohort, performed only on gene sets and subgroups with significant p-values in gene set enrichment analysis. Odds ratios represent risk per unit increase in risk score quintile.
CI = confidence interval, OR = odds ratio, OXPHOS = all structural proteins directly contributing to oxidative phosphorylation complex function, SV = small vessel stroke, --- = analysis not performed.
Nominal p-values are reported for association with ischemic stroke and ICH in the validation and extension cohorts.
In analysis of our Complex I score, ischemic stroke (OR=1.06, 95% CI=1.00–1.12) and the SV stroke subtype (OR=1.13, 95% CI=1.01–1.26) demonstrated significant association. For our Complex IV score, there was a trending association for all-cause stroke, and a significant association for SV stroke (OR=1.14, 95% CI=1.02-1.27).
Regression analyses for ischemic stroke were performed with and without the inclusion of vascular risk factors as covariates in logistic regression (hypertension, diabetes mellitus, and atrial fibrillation). These regressors did not demonstrate significant association with the genetic scores, and did not alter the results of the regression analysis (p = NS, data not shown).
Extension of Genetic Score Analysis to Intracerebral Hemorrhage
We constructed genetic scores in the ISGC ICH cohort based on beta coefficients from the MGH/MIGen ischemic stroke cohort [TABLE 3]. This analysis revealed association between a genetic score from Complex IV genes and all ICH (OR=1.08, 95% CI=1.01-1.17, p=0.039) as well as deep ICH (OR=1.14, 95% CI=1.03-1.25, p=0.008).
Post-hoc Power Calculation
Power calculations for discovery of individual OXPHOS genetic variants in association with ischemic stroke revealed a maximum power of 73% to detect variants conferring an OR of 1.4 in association with ischemic stroke risk, at an MAF of 0.30 [Supplementary Table S3]. SNPs conferring lower OR and lower MAF substantially limited study power to detect individual variants, as did restriction of samples to the SV subtype. We performed power calculations for our genetic score analyses based on percentage of variance explained by the genetic score, ranging from 0.5% to 5%. The genetic score analysis was powered at 21% to explain 0.5% of variance in ischemic stroke risk for all-cause strokes, and 3% to explain 0.5% of variance in the SV subtype. In application of this genetic score to ICH, power was 16% to explain 0.5% of variance in all ICH, and 8% to explain 0.5% of variance in deep ICH [Supplementary Table S4]. As a reference, percentages of variance explained in our logistic regression models incorporating genetic scores ranged from 0.5-1% in most analyses.
DISCUSSION
Our pathway-based analysis demonstrates that common genetic variants in OXPHOS genes are associated with risk of both ischemic stroke and ICH. These associations are robust, having passed GSEA and replication in independent cohorts. Stratifying ischemic stroke and ICH by subtype and OXPHOS genes by mitochondrial Complex, we reveal associations for Complex I and IV in SV stroke, and Complex IV in deep ICH. These sub-analyses retain significance despite a substantial restriction in sample size and SNP counts.
While ample evidence exists for rare mutations leading to severe OXPHOS dysfunction in a variety of familial mitochondrial syndromes with stroke phenotypes, our analysis provides evidence of a role for common genetic variants within OXPHOS in sporadic ischemic stroke and ICH. These results contribute to a growing body of evidence linking OXPHOS genetic and functional variation to common neurologic diseases, including Alzheimer Disease, Amyotrophic Lateral Sclerosis, and Parkinson Disease, to name a few 20-23.
Our sub-analyses restricted to variants within Complexes I and IV genes reveal additional parallels to rare mitochondrial syndromes. Mutations within Complex I account for up to 1/3 of the known respiratory chain diseases, and represent a major determinant of the redox state of the cell 24, 25. Complex IV is the final electron donor in the pathway, receiving electrons from Cytochrome C and passing them to oxygen. Complex IV dysfunction, in addition to causing early-life mitochondrial diseases such as Leigh’s Disease and encephalomyopathies, has also been implicated in neurodegenerative diseases 20. While Complex I is far larger than Complex IV (50 vs. 23 gene products), our demonstrated positive associations for both Complexes suggests that statistical power alone did not determine our results, and the correlations with existing knowledge of mitochondrial disease supports a possible role for these Complexes in sporadic human disease. Neither Complex I nor Complex IV dysfunction have effective treatments, although administration of cofactors has been reported to improve function in some instances 26.
Both SV stroke and deep ICH result from disease of cerebral small vessels, and share common risk factors such as diabetes and hypertension 27, 28. Our findings suggest a possible shared genetic contribution to small vessel pathobiology underlying SV stroke and deep ICH, which could be mediated through disruption in oxidative function at the tissue level, or through modification of upstream systemic or endothelial risk factors shared by the two diseases. We have previously reported an association between mitochondrial common variants and white matter hyperintensity volume (WMHV) 4, a phenotype to which SV stroke and deep ICH have been linked 29, 30. These new data provide additional support for the role of energy metabolism in small vessel disease. However, given that OXPHOS dysfunction can result in numerous physiologic derangements, including ATP depletion, reactive oxygen species generation, defects in cell signaling, and alteration in apoptotic thresholds, our demonstrated associations cannot directly inform the underlying pathobiology of this small vessel link. Functional studies to identify the mechanisms of bioenergetic dysfunction will be needed to build on these results.
APOE allele status has been demonstrated to affect the risk and severity of lobar intracerebral hemorrhage, presumably due to a strong relationship between cerebral amyloid angiopathy and the lobar ICH subtype 11, 13. The present study suggests a relationship between OXPHOS variants and deep ICH only, contributing to growing evidence that deep and lobar ICH represent genetically distinct entities. Genetic approaches appears to be useful tools to explore the differences between these ICH subtypes, and hopefully can lead to a more comprehensive understanding of the pathogenesis of these similar but unique disease subtypes.
Limitations render our results preliminary. The magnitude of effect sizes for OXPHOS genetic scores in stroke risk in our analyses are small, but are in-line with the results from other GWAS efforts in ischemic and hemorrhagic stroke 2-4, 8, 11, 13. Our GSEA did not find associations for the large artery or cardioembolic stroke subtypes in ischemic stroke, or the lobar subtype in ICH. Given our power calculations, it is possible that the restriction in sample size for subtype-stratified analyses led to a false negative for these subtypes. We cannot, therefore, definitively demonstrate that the effect of OXPHOS genetic variants on ischemic or hemorrhagic stroke is isolated to SV ischemic or deep ICH subtypes. Many subjects in the MGH/MIGen and ISGS/SWISS datasets were previously used in prior study of mitochondrial variants in ischemic stroke, although the method of analysis differed between these studies 4. These datasets theoretically could be particularly enriched with OXPHOS associations, although the positive extension to the ISGC ICH cohort would not be predicted if this were the case.
Genetic score analysis, although effective in aggregating signals to detect association, cannot identify individual causative variants. As a result, we are unable to determine the particular genetic loci conferring risk in the present study. GWAS platform-based SNPs were used in this analysis, which are not highly enriched for functional variants likely to cause missense, nonsense, or splice-site mutations. It is possible that other common or rare genetic variants in the OXPHOS pathway lie in linkage disequilibrium with our included variants, exerting a more substantial effect in affected individuals. Given the small aggregate effect sizes of the genetic scores in our analysis, prohibitively large sample sizes would be required to achieve sufficient power to detect individual OXPHOS variants. The significance thresholds in the current study were set according to established techniques in GSEA and genetic score analysis, and can be considered robust due to the use of permutation in the case of GSEA, and the use of separate discovery and replication cohorts in the case of the genetic score analysis. Because the majority of genes encoding OXPHOS proteins are autosomal, we cannot determine whether the low risk contribution (≤20%) of mitochondrial variants to the risk scores for ischemic stroke and ICH in our analysis is due to an imbalance in SNP contributions to the genetic score or a true difference in risk proportion. Finally, we cannot determine whether the involvement of the OXPHOS pathway in ischemic stroke and ICH is mediated at the brain tissue level, or possibly through modification of systemic vascular or metabolic risk factors. Follow-up analyses will be required to address OXPHOS function in different tissue types.
CONCLUSION
Through a pathway-based analysis, we have demonstrated that genetic variation within genes involved in the OXPHOS apparatus associates with ischemic stroke risk, particularly the SV stroke subtype. Extension to ICH reveals retained association with OXPHOS Complex IV in deep ICH. Further studies will be necessary to clarify the functional impact of these variants on OXPHOS function.
Supplementary Material
ACKNOWLEDGEMENTS
This work used samples and clinical data from the National Institutes of Neurological Disorders and Stroke Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds). This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov). The project described was supported in part by a grant from the National Institute of Neurological Disorders and Stroke and National Institute on Minority Health and Health Disparities (NS U54NS057405). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NINDS or the NIH.
FUNDING AND SUPPORT
MGH/MIGen: These studies were funded by the American Heart Association/Bugher Foundation Centers for Stroke Prevention Research (0775010N), the NIH-National Institute for Neurologic Disorders and Stroke (NINDS) (R01 NS059727, U01 NS069208), The Keane Genetics Fund, and the Deane Institute for Integrative Research in Atrial Fibrillation and Stroke. The MIGen study was funded by the U.S. National Institutes of Health and National Heart, Lung, and Blood Institute’s STAMPEED genomics research program (R01 HL087676) and a grant from the National Center for Research Resources. The Broad Institute Center for Genotyping and Analysis is supported by grant U54 RR020278 from the National Center for Research resources. C.D.A., A.B., and N.S.R. were supported in part by the American Heart Association/Bugher Foundation Centers for Stroke Prevention Research, and C.D.A. is supported by the American Brain Foundation.
ISGS/SWISS: These studies were funded by the NIH-NINDS (R01 NS42733, R01 NS39987), the Intramural Research Program of the NIH-National Institute on Aging (NIA) (Z01 AG000954-06), and by the Marriott Disease Risk and Regenerative Medicine Initiative Award in Individualized Medicine and the Marriott Mitochondrial Fund. The inclusion of BLSA samples was supported in part by the Intramural Research Program of the NIH-NIA (Z01 AG000015-50). O.A.R. is supported by the American Heart Association, James and Esther King Biomedical Research Program, the Florida Department of Health, and the Myron and Jane Hanley Award in Stroke Research.
ISGC ICH: The Differences in the Imaging of Primary Hemorrhage based on Ethnicity or Race (DECIPHER) project was supported by Award Number U54NS057405 from the NIH-NINDS and National Institute on Minority Health and Health Disparities (NIMHD) (U54NS057405). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Neurological Disorders and Stroke or the National Institutes of Health (DECIPHER). The Genetic and Environmental Risk Factors for Hemorrhagic Stroke (GERFHS) study was supported by NIH-NINDS (NS36695 and NS30678), and by the Greater Cincinnati Foundation Grant (Cincinnati Control Cohort). The MGH ICH GWAS study was funded by NIH-NINDS (K23NS042695, 5K23NS059774, R01NS059727, and 5R01NS042147), the Keane Stroke Genetics Research Fund, the Edward and Maybeth Sonn Research Fund, by the University of Michigan General Clinical Research Center (M01RR000042), and by a grant from the National Center for Research Resources. The Hospital del Mar ICH (HM-ICH) study was funded by the Instituto de Salud Carlos III, Spanish Research Networks “Red HERACLES” (RD06/009) FEDER. The Jagiellonian University Hemorrhagic Stroke Study (JUHSS) was supported by a grant funded by the Polish Ministry of Education (NN402083934). The Lund Stroke Register (LSR) was funded by Lund University, Region Skåne, King Gustaf V’s and Queen Victoria’s Foundation, and the Swedish Medical Research Council (K2010-61X-20378-04-3). Biobank services and genotyping were done at Region Skåne Competence Centre (RSKC Malmö), Skåne University Hospital, Malmö, Sweden. Controls from the Medical University of Graz ICH (MUG-ICH) study were from the Austrian Stroke Prevention Study, which is a population-based study funded by the Austrian Science Fond grant numbers P20545-P05 and P13180; The Medical University of Graz supports the databank of the Austrian Stroke Prevention Study.
Footnotes
DISCLOSURES
None
Subject Codes: [55], [47], [44], [43]
STATEMENT OF CONTRIBUTION
Manuscript Preparation: Christopher Anderson (co-leader), Alessandro Biffi (co-leader), Jonathan Rosand. Data Acquisition: Christopher Anderson, Natalia Rost, Alison Ayres, Kristin Schwab, Anand Viswanathan, Michael Nalls, William Devan, Valerie Valant, Bjorn Hansen, Alessandro Biffi. Manuscript Revision: Natalia Rost, Michael Nalls, Owen Ross, Richa Saxena, James Meschia, William Devan, Valerie Valant, Jonathan Rosand, Bradford Worrall, Thomas Brott, Devin Brown, Bjorn Hansen, Joseph Broderick, Bo Norrving, Anand Viswanathan, Scott Silliman, David Tirschwell, Arne Lindgren, Agnieszka Slowik, Reinhold Schmidt, Magdy Selim, Jaume Roquer, Joan Montaner, Andrew Singleton, Steven Greenberg, Chelsea Kidwell, Daniel Woo. Data Analysis: Christopher Anderson, Alessandro Biffi, Michael Nalls. Study Management: Jonathan Rosand, Karen Furie, Joshua Goldstein, James Meschia, Andrew Singleton, Thomas Brott, Bradford Worrall, Owen Ross, Devin Brown, Joseph Broderick, Bo Norrving, Anand Viswanathan, Scott Silliman, David Tirschwell, Arne Lindgren, Agnieszka Slowik, Reinhold Schmidt, Magdy Selim, Jaume Roquer, Joan Montaner, Andrew Singleton, Steven Greenberg, Chelsea Kidwell, Daniel Woo.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
BIBLIOGRAPHY
- 1.Flossmann E, Schulz UG, Rothwell PM. Systematic review of methods and results of studies of the genetic epidemiology of ischemic stroke. Stroke. 2004;35:212–227. doi: 10.1161/01.STR.0000107187.84390.AA. [DOI] [PubMed] [Google Scholar]
- 2.Meschia JF. Advances in genetics 2010. Stroke. 2011;42:285–287. doi: 10.1161/STROKEAHA.110.605089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellenguez C, Bevan S, Gschwendtner A, Spencer CC, Burgess AI, Pirinen M, et al. Genome-wide association study identifies a variant in hdac9 associated with large vessel ischemic stroke. Nature genetics. 2012;44:328–333. doi: 10.1038/ng.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson CD, Biffi A, Rahman R, Ross OA, Jagiella JM, Kissela B, et al. Common mitochondrial sequence variants in ischemic stroke. Annals of neurology. 2011;69:471–480. doi: 10.1002/ana.22108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner BK, Kitami T, Gilbert TJ, Peck D, Ramanathan A, Schreiber SL, et al. Large-scale chemical dissection of mitochondrial function. Nature biotechnology. 2008;26:343–351. doi: 10.1038/nbt1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiMauro S, Schon EA. Mitochondrial disorders in the nervous system. Annual review of neuroscience. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- 7.Nicholls DG. Oxidative stress and energy crises in neuronal dysfunction. Annals of the New York Academy of Sciences. 2008;1147:53–60. doi: 10.1196/annals.1427.002. [DOI] [PubMed] [Google Scholar]
- 8.Anderson CD, Biffi A, Rost NS, Cortellini L, Furie KL, Rosand J. Chromosome 9p21 in ischemic stroke: Population structure and meta-analysis. Stroke. 2010;41:1123–1131. doi: 10.1161/STROKEAHA.110.580589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meschia JF, Brott TG, Brown RD, Jr., Crook RJ, Frankel M, Hardy J, et al. The ischemic stroke genetics study (isgs) protocol. BMC neurology. 2003;3:4. doi: 10.1186/1471-2377-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meschia JF, Brown RD, Jr., Brott TG, Chukwudelunzu FE, Hardy J, Rich SS. The siblings with ischemic stroke study (swiss) protocol. BMC medical genetics. 2002;3:1. doi: 10.1186/1471-2350-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biffi A, Anderson CD, Jagiella JM, Schmidt H, Kissela B, Hansen BM, et al. Apoe genotype and extent of bleeding and outcome in lobar intracerebral haemorrhage: A genetic association study. Lancet neurology. 2011;10:702–709. doi: 10.1016/S1474-4422(11)70148-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nature genetics. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biffi A, Sonni A, Anderson CD, Kissela B, Jagiella JM, Schmidt H, et al. Variants at apoe influence risk of deep and lobar intracerebral hemorrhage. Annals of neurology. 2010;68:934–943. doi: 10.1002/ana.22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saxena R, de Bakker PI, Singer K, Mootha V, Burtt N, Hirschhorn JN, et al. Comprehensive association testing of common mitochondrial DNA variation in metabolic disease. American journal of human genetics. 2006;79:54–61. doi: 10.1086/504926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner S, Armstrong LL, Bradford Y, Carlson CS, Crawford DC, Crenshaw AT, et al. Quality control procedures for genome-wide association studies. Current protocols in human genetics / editorial board, Jonathan L. Haines … [et al.] 2011 doi: 10.1002/0471142905.hg0119s68. Chapter 1:Unit 1,19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biffi A, Anderson CD, Nalls MA, Rahman R, Sonni A, Cortellini L, et al. Principal-component analysis for assessment of population stratification in mitochondrial medical genetics. American journal of human genetics. 2010;86:904–917. doi: 10.1016/j.ajhg.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. American journal of human genetics. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S, Cherny SS, Sham PC. Genetic power calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 20.Muller WE, Eckert A, Kurz C, Eckert GP, Leuner K. Mitochondrial dysfunction: Common final pathway in brain aging and alzheimer’s disease--therapeutic aspects. Molecular neurobiology. 2010;41:159–171. doi: 10.1007/s12035-010-8141-5. [DOI] [PubMed] [Google Scholar]
- 21.Duan W, Li X, Shi J, Guo Y, Li Z, Li C. Mutant tar DNA-binding protein-43 induces oxidative injury in motor neuron-like cell. Neuroscience. 2010;169:1621–1629. doi: 10.1016/j.neuroscience.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Gallardo E, Iceta R, Iglesias E, Montoya J, Ruiz-Pesini E. Oxphos toxicogenomics and parkinson’s disease. Mutation research. 2011;728:98–106. doi: 10.1016/j.mrrev.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Coskun P, Wyrembak J, Schriner SE, Chen HW, Marciniack C, Laferla F, Wallace DC. A mitochondrial etiology of alzheimer and parkinson disease. Biochimica et biophysica acta. 2012;1820:553–564. doi: 10.1016/j.bbagen.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rich PR, Marechal A. The mitochondrial respiratory chain. Essays in biochemistry. 2010;47:1–23. doi: 10.1042/bse0470001. [DOI] [PubMed] [Google Scholar]
- 25.Stefanatos R, Sanz A. Mitochondrial complex I: A central regulator of the aging process. Cell Cycle. 2011;10:1528–1532. doi: 10.4161/cc.10.10.15496. [DOI] [PubMed] [Google Scholar]
- 26.Lagoa R, Graziani I, Lopez-Sanchez C, Garcia-Martinez V, Gutierrez-Merino C. Complex I and cytochrome c are molecular targets of flavonoids that inhibit hydrogen peroxide production by mitochondria. Biochimica et biophysica acta. 2011;1807:1562–1572. doi: 10.1016/j.bbabio.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 27.Putaala J, Liebkind R, Gordin D, Thorn LM, Haapaniemi E, Forsblom C, et al. Diabetes mellitus and ischemic stroke in the young: Clinical features and long-term prognosis. Neurology. 2011;76:1831–1837. doi: 10.1212/WNL.0b013e31821cccc2. [DOI] [PubMed] [Google Scholar]
- 28.Bokura H, Saika R, Yamaguchi T, Nagai A, Oguro H, Kobayashi S, Yamaguchi S. Microbleeds are associated with subsequent hemorrhagic and ischemic stroke in healthy elderly individuals. Stroke. 2011;42:1867–1871. doi: 10.1161/STROKEAHA.110.601922. [DOI] [PubMed] [Google Scholar]
- 29.Rost NS, Rahman RM, Biffi A, Smith EE, Kanakis A, Fitzpatrick K, et al. White matter hyperintensity volume is increased in small vessel stroke subtypes. Neurology. 2010;75:1670–1677. doi: 10.1212/WNL.0b013e3181fc279a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamada S, Saiki M, Satow T, Fukuda A, Ito M, Minami S, Miyamoto S. Periventricular and deep white matter leukoaraiosis have a closer association with cerebral microbleeds than age. European journal of neurology. 2012;19:98–104. doi: 10.1111/j.1468-1331.2011.03451.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.