Abstract
Background
General cognitive ability is usually lower in individuals with schizophrenia, partly due to genetic influences. However, the specific genetic features related to general cognitive ability are poorly understood. Individual variation in a specific type of mutation, uncommon genetic deletions, has recently been linked with both general cognitive ability and risk for schizophrenia.
Methods
We derived measures of the aggregate number of “uncommon” deletions (i.e., those occurring in 3% or less of our combined samples) and the total number of base pairs affected by these deletions in individuals with schizophrenia (N = 79) and healthy controls (N = 110), and related each measure to the first principal component of a large battery of cognitive tests, a common technique for characterizing general cognitive ability. These two measures of mutation load were also evaluated for relationships with total brain gray matter, white matter, and lateral ventricle volume.
Results
The groups did not differ on genetic variables. Multivariate general linear models revealed a group (controls versus patients) by uncommon deletion number interaction, such that the latter variable was associated with lower general cognitive ability and larger ventricles in patients but not controls.
Conclusions
These data suggest that aggregate uncommon deletion burden moderates central features of the schizophrenia phenotype.
Keywords: copy number variations, schizophrenia, cognition, ventricles, mutations, intelligence
Introduction
Schizophrenia and general cognitive ability are highly heritable, correlated human phenotypes whose genetic foundations remain poorly understood despite decades of intensive study. Specific single nucleotide polymorphisms (SNPs) account for less than one percent of each type of phenotypic variation [1, 2]. The aggregate impact of many thousands of SNPs may be as high as 34% for schizophrenia [3] and 51% for general cognitive ability [4], though this remains controversial [5, 6]. Among the many ideas offered to account for the “missing heritability” [7] two seem especially promising. First, copy number variations (CNVs), a type of mutation, may represent another important source of genetic variation for schizophrenia and cognitive ability. Second, there is a growing realization that the effects of different genetic variations (SNPs or CNVs) may depend on “genetic background”. The current study examined a specific type of CNV, “uncommon” deletions, for its possible impact on general cognitive ability in two groups presumed to differ in genetic background, healthy controls and individuals with schizophrenia.
Several lines of evidence suggest that CNVs can impact brain development. First, rare, large CNVs are more common among patients with diverse neurodevelopmental disorders, including schizophrenia [8], autism [9], epilepsy [10], and ADHD [11]. The proportion of cases attributable to such anomalies remains uncertain. Second, CNVs capture much more nucleotide variation than do single nucleotide substitutions [12], and they have a higher de novo mutation rate [13, 14]. Their adverse impact is revealed by evidence of negative selection pressure [13]. Third, CNVs may be related to general cognitive ability, a trait with a distinctive developmental trajectory that relies upon specific, distributed cortical networks [15]. Consistent with this proposition, in a study of individuals with attention deficit hyperactivity disorder, a subsample with intellectual disability had an especially high number of CNVs [11]. Further, we have also recently reported that uncommon deletions may be related to intelligence in individuals with alcohol abuse [16]. In a relatively small sample of individuals originally recruited for a study designed to investigate genetic the correlates of alcohol dependence, we found that the total length of uncommon deletions (i.e., those occurring in less than 5% of our sample) correlated with the full scale intelligence quotient from the Wechsler Abbreviated Scale of Intelligence at r = −0.30 [16]. The association of intelligence with uncommon deletions did not appear to be influenced by alcohol dependence per se, as covarying a common measure of dependence had no effect on results.
These results linking CNVs with intelligence are broadly consistent with a body of research that has used an indirect marker of mutation load, fluctuating anatomic asymmetry, which refers to non-directional asymmetries in physical body characteristics that are symmetric at the population level, such as the length of the two ears. The idea that fluctuating asymmetry partly, even if imperfectly, reflects mutation load is supported by studies linking increased fluctuating asymmetry with factors that increase the number of mutations, such as exposure to radiation [17] or heat [18], and factors that increase their functional impact (inbreeding depression [19]). A recent meta-analysis demonstrated small but reliable associations between aggregate measures of fluctuating asymmetry and intelligence [20].
In contrast, a recent large-scale study of healthy controls found no relationship between uncommon deletions and general cognitive ability, raising concerns about sample-specific effects [21]. The relevance of “genetic background” has been highlighted in recent reports focusing on both schizophrenia and intelligence. Mitchell [22] notes that it is extremely common for mutations to be modified by additional variants in the genetic background (see also [23]). Johnson [24] has argued, based in part on the results of selective breeding studies, that the effects of specific genes depends upon other genetic variations such as mutations, as well as variable environmental triggers.
In the current study we evaluated the relationship between the overall burden of uncommon deletions and general cognitive ability in two samples that likely differ in underlying genetics, healthy controls and individuals with schizophrenia. Our primary goals were (1) to test the hypothesis that the number and length of rare deletions would correlate negatively with the first principal component emerging from a large battery of neuropsychological tests, a common way to operationalize general cognitive ability, and (2) to determine if the hypothesized negative relationship between uncommon deletions and general intellectual ability differed in individuals with schizophrenia and healthy controls. A secondary goal was to explore relationships between overall measures of cranial volumes – gray matter, white matter, and ventricle volumes – and measures of uncommon deletions in both groups.
Methods
Participants
The participants were recruited through the Mind Clinical Imaging Consortium (MCIC). This consortium includes research teams at the Mind Research Network and University of New Mexico, Massachusetts General Hospital, the University of Minnesota, and the University of Iowa (see [25] for additional details). A total of 385 individuals were originally studied, though CNV, MRI and cognitive data were available only for 243 individuals. The current analysis is limited to the subset of these individuals who stated their racial background was “white”. CNVs, like single nucleotide polymorphisms, show population stratification, and use of a single reference genome may influence measurement of the total CNV burden. Hence, we could not accurately determine the “rareness” of CNVs in non-white groups using frequency data from a predominantly white sample and our numbers of minority participants were not sufficient to perform within-group analyses of CNV frequencies. Analysis of principal components reflecting population stratification from SNP data revealed that four self-described “white” individuals did not cluster with the rest of the white sample, so these were eliminated from further analysis. The final sample included 79 individuals with schizophrenia (57 males, 22 females) and 110 controls (67 males, 43 females). The number of participants recruited from each site were: Albuquerque, NM (19 patients/21 controls), Boston, MA (13/14), Minneapolis, MN (18/19), and Iowa City, IA (28/57).
A comprehensive clinical diagnostic assessment included either the Structured Clinical Interview for the DSM IV (SCID [26]) or the Comprehensive Assessment of Symptoms and History (CASH [27]). The mean length of illness for patients with schizophrenia was 12.02 years (SD = 11.07, range 0 – 42). Most patients were currently being treated with antipsychotics (77/79; 96%) and very few were antipsychotic naïve (2/79; 2.5%). Symptoms of schizophrenia were evaluated with the Scale for the Assessment of Positive Symptoms [28] and the Scale for the Assessment of Negative Symptoms [29]. Healthy controls were recruited from the general community through medical clinics and advertisements in local newspapers. Exclusionary criteria for the control group were presence of a physical or neurologic disorder affecting brain function, and lifetime history of any Axis I disorder, including substance abuse or dependence. Controls were not excluded if they had a first-degree relative with an Axis I psychiatric disorder. The Institutional Review Board at each site approved this study.
Intellectual assessment
All participants were administered a comprehensive battery of neuropsychological tests tapping these domains: reading ability, verbal fluency, working memory, verbal abstraction, nonverbal reasoning, attention, and visuomotor and executive skills. To facilitate accurate assessment of factor structure, the entire sample (N = 247) was used for a Principal Components Analysis (PCA), though subsequent analyses were limited to the white sample, as described above. [The first PC calculated for the subsample reported here with both MRI and genetic data correlated r = .96 with the first PC computed from the original, larger sample. All significant effects (see Results) were also significant with the first PC calculated on the smaller subset, but as the factor structure may be more accurately estimated in the larger, original sample, these results will be reported below]. A total of 25 test scores were entered for each participant for the PCA (see Supplementary Table S1 for a list of specific test variables and factor loadings). The first component was utilized in all analyses as a measure of general cognitive ability. Across the total number of expected data points (247 participants with 25 variables), a total of 2.4% were missing values. To avoid discarding participants through list-wise deletion, missing values of specific tests were replaced by the individual group mean. As the total proportion of total individual test scores imputed in this way was very small, this procedure likely had negligible effects on the results.
Genetic analyses
DNA extracted from blood samples was genotyped using Illumina HumanOmin1- quad chip, including 1,140,419 markers. The intensity values (LRR) and beta allele frequency (BAF) for the markers in autosomes were used for CNV detection. The details of CNV detection were described in [19, 30, 31]. Briefly, outlier correction was applied to replace the isolated large LRR values with median ± 2 standard deviation and principal component based correction was performed to eliminate variation induced by experimental or GC (guanine-cytosine) content factors. Two principal components were corrected here. Data from all participants passed the quality control on LRR (LRR d<0.28) [32]. The data were then segmented using a circular binary segmentation algorithm and a hidden Markov model algorithm (PennCNV) independently. Only segments (spanning at least 3 markers) detected by both algorithms (segments overlapping or apart by less than 3 markers) went through a final single to noise ratio check calculated by the ratio of the segment mean LRR over neighboring LRRs. CNV segments failing the signal-to-noise ratio check or overlapping with telomere or centromere larger than 50% were excluded, as well as small segments with less than 500 base pairs. Three subjects’ CNV data were excluded due to a large number of CNV calls (> 3 SD from mean).
Deletions were termed “uncommon” if the occurred in 3% or less of the combined group of participants. In our prior [16] study we calculated the frequency of deletions and duplications as compared to the total number of both types of CNVs. However, as we found no relationship of IQ with duplications in our previous work, we now focus on deletions, in which case it makes sense to determine rareness with respect to deletions specifically. In the current study uncommon deletions were thus defined with reference only to the total number of deletions (approximately one-half of the total number of CNVs). Given our sample size, and the need to have a reasonable number of deletions for statistical analyses of aggregate totals, we summed all deletions occurring at a frequency of 3% or less in the entire sample to provide an aggregate total. In the analyses below, we focused on the total number of deletions meeting this criterion and the total number of base pairs lost in this aggregate number of deletions. For descriptive purposes, we provide data on number and length of these other types of CNV measures: uncommon duplications, common deletions, and common duplications.
Neuroimaging Acquisition and Analysis
Structural MRI scans were obtained on a 1.5T Siemens Sonata scanner at three sites (Mind research Network, University of Iowa, and Massachusetts General Hospital) and on a 3T Siemens Trio scanner at one site (University of Minnesota). The T1-weighted structural brain scans at each of the four sites were acquired with a coronal gradient echo sequence: TR=2530 ms for 3T, TR=12ms for 1.5T; TE=3.79 for 3T, TE=4.76ms for 1.5T; TI=1100 for 3T; Bandwidth=181 for 3T, Bandwidth=110 for 1.5T; 0.625°—0.625 voxel size; slice thickness 1.5mm; FOV, 256°—256°—128 cm matrix; FOV=16 cm; NEX=1 for the 3T, NEX=3 for the 1.5T. Freesurfer software (http://surfer.nmr.mgh.harvard.edu, Version 4.0.1) was used to align images, determine the gray-white tissue boundary, and estimate cortical gray matter and white matter volumes. Ventricle size was represented as the ratio of lateral ventricle volume to total intracranial volume. Additional details of image analysis and quality control procedures are provided in Ehrlich et al. [33].
Statistical analyses
Statistical analyses were conducted using SPSS v. 19.0.0. Primary analyses were hierarchical multiple regressions predicting general cognitive ability and brain volumes. In each of these, group (schizophrenia vs. controls), and sex, age, and ethnicity (Anglo vs. Hispanic) were entered in step one, then the uncommon deletion variable (number or length) and the interactions of the deletion variable with age and with group were entered in step two. We included ethnicity to control for possible impact on the general cognitive ability measure. No correction for multiple comparisons was made for the two regression models utilizing uncommon deletion measures, as these tests reflect specific a priori hypotheses based on prior studies. For the three analyses of brain volume compartments, the adjusted critical p value was 0.015. To evaluate the effect of using different scanners at each site we conducted a multivariate GLM analysis with three dependent variables (GM volume, WM volume, and ventricle volume, with site and group (and their interaction) as main variables and sex, age, and ethnicity as covariates. There was no significant effect of site (p = .18) or site X group interaction (p = .97). Hence, the effect of site was not modeled in analyses reported below.
Results
Demographic, cognitive, and brain volume variables from both groups are presented in Table 1, and clinical characteristics of the patient group are described in Table 2. As expected, individuals with schizophrenia had larger ventricle/intracranial volume ratios (t (114.77) = 4.06, p < 0.001), less gray matter volume (t(191) = 3.95, p < 0.001), and lower general intellectual functioning (t(113.88) = 10.69. p < 0.001). A trend was noted for reduced white matter volume (t(191) = 1.75, p = 0.082). Patients and controls did not differ in the aggregate number of uncommon deletions (Patients: = 11.34 (SD = 4.14; Controls = 11.87, SD = 4.75; ns) or the total number of base pairs affected by these deletions (Patients = 117,936, SD = 176,927; Controls = 118,552 (SD = 166,644; ns). The average length of the uncommon deletions was 10,400 base pairs in the patient group and 9,988 base pairs in controls.
Table 1.
Demographic, clinical, and cognitive characteristics of patients with schizophrenia and controls. Group comparisons were conducted with independent-sample t-tests and chi-square tests.
| Patients (N = 79) | Controls (N = 110) | p | |
|---|---|---|---|
| Age | 35.01 (11.43) | 31.68 (10.90) | 0.04 |
| Education | 13.65 (2.530 | 15.27 (1.99) | <0.001 |
| WRAT3RT | 47.54 (6.09) | 51.05 (3.98) | <0.001 |
| Percent Anglo | 83.8 | 79.8 | ns |
| Parental SES | 2.72 (.95) | 2.66 (.71) | ns |
| Participant SES | 3.46 (.99) | 2.65 (.53) | <0.001 |
| Gen. Cognitive Ability (z-score) | −.48 (.89) | .64 (.44) | <0.001 |
| White Matter Volume | 492.00 (66.34) | 508.21 (65.80) | ns |
| Gray Matter Volume | 520.13 (73.48) | 556.06 (60.15) | <0.001 |
| Ventricle/Intracranial Volume | .0099 (.0058) | .0073 (.00025) | <0.001 |
Note: WRAT3RT = Wide Range Achievement Test – 3rd Ed., Reading Test; SES categories reflect Hollingshead 5-level categories (1 = highest, 5 = lowest).
Table 2.
Clinical characteristics of patients with schizophrenia (N = 79).
| Clinical Measures | Patients with schizophrenia |
|---|---|
| Years of illness (mean, sd) | 12.02 (11.07) |
| Abnormal Involuntary Movement Scale | 0.38 (0.72) |
| SANS/SAPS scores | |
| Positive | 4.81 (2.86) |
| Negative | 7.99 (3.74) |
| Disorganized | 1.95 (1.95) |
| Diagnostic subtypes (N, %) | |
| Paranoid | 45 (56.9) |
| Disorganized | 3 (3.8) |
| Undifferentiated | 24 (30.4) |
| Residual | 4 (5.1) |
| Schizophreniform | 3 (3.8) |
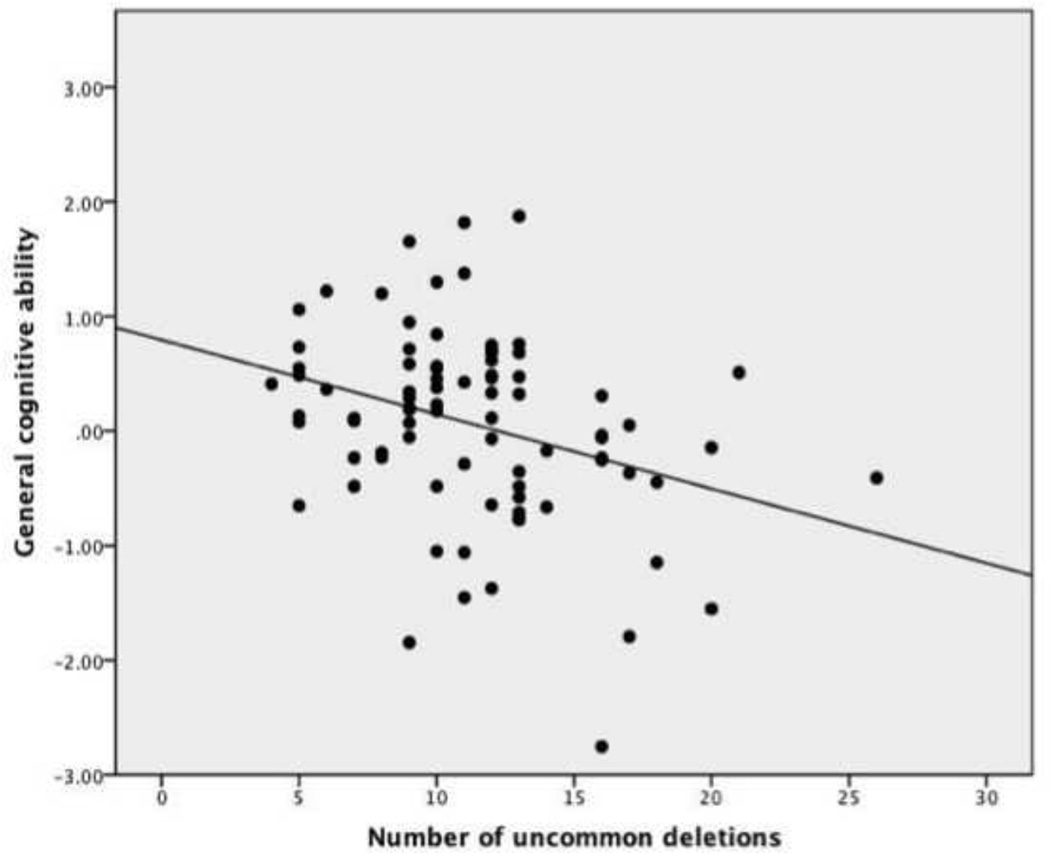
Separate regression analyses were performed for the total number of uncommon deletions and the total number of base pairs affected by these deletions. In each, the variables entered at step one (age, sex, group, and ethnicity) captured a highly significant amount of variance in general intellectual ability (F(4,189) =40.68, adjusted R square = 0.45, p < 0.001). For the number of uncommon deletions, the three variables entered at step two led to a significant R square change capturing 5.1 percent of the remaining variance (F(3, 186) = 6.54, p < 0.001). The effect of deletion number was significant (standardized beta = −0.19, p < 0.001), as was its interaction with group (standardized beta = −0.18, p = 0.001). The interaction of deletion number with age was not significant. In contrast, for the total length of uncommon deletions, the three variables entered at step two did not yield a significant R square change (F(3, 186) = 0.17, ns). Thus uncommon deletion number was related to general cognitive ability, but the total length of uncommon deletions was not. Further examination of the group by deletion number interaction revealed that the effect of deletions on general cognitive ability was limited to the patient group. Partial correlations (controlling for age, sex, and ethnicity) with general cognitive ability s were r = −0.36 (p = 0.001) for the patient group, and r = −0.02 (ns) for controls. Figure 1 shows the scatterplot relating uncommon deletion number and general cognitive ability in the patient group.
Figure 1.
Scatterplot of the relationship between number of uncommon deletions (3% criterion) and general cognitive ability (corrected for age, sex, and ethnicity) in patients with schizophrenia (N = 79, partial r = −0.36).
In the combined sample, significant positive relationships were noted between general cognitive ability and gray matter volume (r = 0.22, p < 0.002) and white matter volume (r = 0.19, p < 0.008). However, hierarchical regression analyses (as above) revealed no significant relationships between measures of uncommon deletion burden and either gray or white tissue volume. However, in regressions predicting ventricle size with uncommon deletion number, the effects of variables entered at step one (age, sex, ethnicity, and group) was significant (F(4, 187) = 13.18, p < 0.001), and a significant R square change was seen for the set of variables entered at step two (F(3, 184) = 3.82, p = 0.018). Significant individual effects were noted for the effect of deletion burden (standardized beta = 0.18, p = 0.009) and the interaction of deletion burden with group (standardized beta = 0.18, p = 0.007). The partial correlations (controlling for age, sex, and ethnicity) with ventricle size was r = 0.30 (p = 0.008) for the patient group, and r = 0.00 (ns) for controls.
Prokosch et al. [34] suggested that one reason a large first component emerges from principal components analyses of batteries of cognitive tests may be the broad impact of mutation load across diverse tests. If so, a relatively greater mutation load should result in similar ability levels across tests, and hence, a relatively larger first component. We split our combined sample at the median for uncommon deletion number and determined the magnitude of the first component in each group. In the high mutation load group 38% of total variance was captured vs. 28% in the low mutation load group, consistent with Prokosch et al.’s perspective.
Finally, we examined correlations between symptom scales and deletion burden measures within the patient group. No significant relationships were observed between positive or negative symptoms and either deletion measure. However, disorganized symptoms were found to correlate significantly with the number of uncommon deletions (r = 0.25, p = 0.021). Disorganized symptoms also correlated with ventricle size (r = 0.27, p = 0.016), but not with general cognitive ability. As these correlations were not corrected for multiple comparisons, they should be considered as exploratory, awaiting replication in a larger sample.
Discussion
Our major observations are: (1) that a greater number of uncommon deletions is associated with reduced general cognitive ability in individuals with schizophrenia, but not healthy controls; and, (2) that a greater number of uncommon deletions is associated with increased ventricle size in individuals with schizophrenia, but not controls. As no group differences were evident in the total number of uncommon deletions, our basic findings likely reflect an interaction between deletion burden, probably best considered a nonspecific genetic effect, and the specific, unknown genetic abnormalities related to schizophrenia. In the context of a genetic propensity for schizophrenia, greater deletion burden leads to lower intellectual ability and larger lateral ventricles.
One possibility is that, among individuals with schizophrenia, the non-specific genetic component of uncommon deletion burden may lead to reduced resilience or canalization [35], rendering affected individuals (e.g., those with genetic liabilities specific to schizophrenia) more vulnerable to the adverse effects of the specific genetic component. This putative gene × gene interaction (i.e., epistasis) may complement, and act similarly to, important gene × environment interactions seen in studies of schizophrenia. For example, the neural effects of obstetric complications [36] and both cannabis and alcohol use [37, 38] are greater in individuals with a genetic liability for schizophrenia.
We detected no relationship between deletion burden and general intellectual ability in healthy controls, an observation that perhaps sheds some light on our prior observation of a significant relationship in individuals with alcohol use disorders [16]. Though we cannot assert the null hypothesis, the current results, along with those of MacLeod et al. [21], suggest that there is relatively little general, population-wide association between deletion burden and intellectual ability. As Johnson [24] suggested, the nature of genetic factors that contribute to phenotypic variation in intelligence may depend upon the “genetic background”, which reflects a host of different influences, including comorbid genetically-mediated psychopathology. The significant comorbidity of alcohol disorders and schizophrenia [39] has long been noted and perhaps reflects shared genetic background [40].
The deletion burden-general cognitive ability relationship in individuals with schizophrenia was not mediated by gray matter volume, consistent with our prior study utilizing an indirect measure of mutation load [41]. As individual variation in general cognitive ability has been related to many different characteristics of the brain, including more efficient white matter connections [42] and formal network properties [43], there are several other candidate mechanisms awaiting future study. A particularly relevant characteristic may be greater plasticity [44], which is linked with glutamatergic activity[45], and we have found reduced glutamate/glutamine concentrations in individuals with greater deletion burden [46].
Though preliminary, the observations regarding disorganized symptoms are of interest. This was the only symptom scale significantly related to either deletion burden or ventricle size. A recent report based on the Maudsley Twin Register found that “the heritability for the disorganized dimension remained significant when influences acting through liability to psychosis were set to zero, suggesting that some influences on disorganization are modifying factors independent of psychosis liability” (p.89) [47]. Deletion burden may function as just this kind of modifying factor.
In contrast to our prior study, we did not observe any effects for total uncommon deletion length. The reasons for the different results may reflect population-specific effects, or perhaps more likely, methodological issues. The current study, using a different genetic analysis platform, yielded a mean deletion length just over half the length observed in our prior study, despite an equivalent number of uncommon deletions.
Several limitations of the current study suggest cautious interpretation of results. Foremost among these, our sample was small for a genetic study, limiting statistical power. Further, the statistical analysis of rare genetic variants is especially affected by small sample size (see [48] for a comprehensive discussion). This may have limited our ability to detect group differences in deletion number or a deletion-ability relationship in controls. We also do not know if our results generalize across ethnic groups. Most importantly, there remains much work to be done in refining and clarifying CNV measures. Technical improvements in CNV call rate accuracy [49] will facilitate development of more valid measures of deletion burden. However, as noncoding variants are found in regulatory DNA, with demonstrated effects on disease [50], both types of deletions could exert important effects. Clearly, deletions of different types or in different locations in the genome could vary greatly in the impact on general cognitive ability, though large samples may be required to detect such effects. In future studies it will be important to evaluate the significance of deletions specifically affecting genes, the relative impact of single (heterozygous) vs. double (homozygous) deletions, and the effects of deletions near chromosomal hot-spots vs. those in other locations [51], as there may be important functional distinctions related to these characteristics of copy number deletions.
Supplementary Material
Acknowledgements
This research was supported by grants from the Department of Energy under Award Number DE-FG02-08ER64581, and the National Institute Health (grants 5P20RR021938, 1RC1MH089257 and R01EB005846. The authors wish to thank Marilee Morgan and the Mind Research Network Neurogenetics Core Laboratory for their assistance. Portions of this work were presented at the 2011 meeting of the Behavior Genetic Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosure: Dr. Schulz reports grants from NIMH, Astra-Zeneca, Otsuka, Myrida/RBM and consultations for Eli-Lilly and Genetech. All other authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Deary IJ, Johnson W, Houlihan LM. Genetic foundations of human intelligence. Human Genetics. 2009;126(1):215–232. doi: 10.1007/s00439-009-0655-4. [DOI] [PubMed] [Google Scholar]
- 2.Visscher PM, Goddard ME, Derks EM, Wray NR. Evidence-based psychiatric genetics, AKA the false dichotomy between common and rare variant hypotheses. Molecular Psychiatry. 2012;17(5):474–485. doi: 10.1038/mp.2011.65. [DOI] [PubMed] [Google Scholar]
- 3.Consortium TIS. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature Perspectives. 2009:460748–460752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, et al. Genomewide association studies establish that human intelligence is highly heritable and polygenic. Molecular Psychiatry. 2011;16(10):996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell KJ, Porteous DJ. Rethinking the genetic architecture of schizophrenia. Psychological Medicine. 2011;41(1):19–32. doi: 10.1017/S003329171000070X. [DOI] [PubMed] [Google Scholar]
- 6.Wahlsten D. The hunt for gene effects pertinent to behavioral traits and psychiatric disorders: From mouse to human. Developmental Psychobiology. 2012;54(5):475–492. doi: 10.1002/dev.21043. [DOI] [PubMed] [Google Scholar]
- 7.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stone JL, O'Donovan MC, Gurling H, Kirov GK, Blackwood DHR, Corvin A, et al. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455(7210):237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316(5823):445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mefford HC, Muhle H, Ostertag P, von Spiczak S, Buysse K, Baker C, et al. Genome-wide copy number variation in epilepsy: Novel susceptibility loci in idiopathic generalized and focal epilepsies. Plos Genetics. 2010;6(5) doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams NM, Zaharieva I, Martin A, Langley K, Mantripragada K, Fossdal R, et al. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet. 2010;376(9750):1401–1416. doi: 10.1016/S0140-6736(10)61109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beckmann JS, Estivill X, Antonarakis SE. Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability. Nat Rev Genet. 2007;8(8):639–646. doi: 10.1038/nrg2149. [DOI] [PubMed] [Google Scholar]
- 13.Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, et al. Population analysis of large copy number variants and hotspots of human genetic disease (vol 84, pg 148, 2009) American Journal of Human Genetics. 2009;84(4):550–551. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang F, Gu WL, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annual Review of Genomics and Human Genetics. 2009:10451–10481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deary IJ. Intelligence. Annual Review of Psychology. 2011:63453–63482. doi: 10.1146/annurev-psych-120710-100353. [DOI] [PubMed] [Google Scholar]
- 16.Yeo RA, Gangestad SW, Liu J, Calhoun VD, Hutchison KE. Rare copy number deletions predict individual variation in intelligence. Public Library of Science One. 2011;6(1):e16339. doi: 10.1371/journal.pone.0016339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moller AP. Developmental instability and sexual selection in stag beetles from Chernobyl and a control area. Ethology. 2002;108(3):193–204. [Google Scholar]
- 18.Debat V, Debelle A, Dworkin I. Plasticity, canalization, and developmental stability of the drosophila wing: joint effects of mutations and developmental temperature. Evolution. 2009;63(11):2864–2876. doi: 10.1111/j.1558-5646.2009.00774.x. [DOI] [PubMed] [Google Scholar]
- 19.Carter AJR, Weier TM, Houle D. The effect of inbreeding on fluctuating asymmetry of wing veins in two laboratory strains of Drosophila melanogaster. Heredity. 2009;102(6):563–572. doi: 10.1038/hdy.2009.13. [DOI] [PubMed] [Google Scholar]
- 20.Van Dongen S, Gangestad SW. Human fluctuating asymmetry in relation to health and quality: a meta-analysis. Evolution and Human Behavior. 2011;32(6):380–398. [Google Scholar]
- 21.MacLeod AK, Davies G, Payton A, Tenesa A, Harris SE, Liewald D, Ke X, Luciano M, Lopez LM, Gow AJ, Corley J, Redmond P, McNeill G, Pickles A, Ollier W, Horan M, Starr JM, Pendleton N, Thomson PA, Porteous DJ, Deary IJ. Genetic copy number variation and general cognitive ability. Public Library of Science One. doi: 10.1371/journal.pone.0037385. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell KJ. What is complex about complex disorders? Genome Biol. 2012;13(1) doi: 10.1186/gb-2012-13-1-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girirajan S, Brkanac Z, Coe BP, Baker C, Vives L, Vu TH, et al. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. Plos Genetics. 2011;7(11) doi: 10.1371/journal.pgen.1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson W. Understanding the genetics of intelligence: Can height help? Can corn oil? Current Directions in Psychological Science. 2010;19(3):177–182. [Google Scholar]
- 25.White T, Magnotta VA, Bockholt HJ, Williams S, Wallace S, Ehrlich S, et al. Global white matter abnormalities in schizophrenia: A multisite diffusion tensor imaging study. Schizophrenia Bulletin. 2011;37(1):222–232. doi: 10.1093/schbul/sbp088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.First M, Spitzer RL, Gibbon M, Williams JB. Structured Clinical Interview for DSM-IV-TR Axis I Disorders. Washington, DC: American Psychiatric Press, Inc; 1997. [Google Scholar]
- 27.Andreasen NC, Flaum M, Arndt S. The Comprehensive Assessment of Symptoms and History (Cash) - an instrument for assessing diagnosis and psychopathology. Archives of General Psychiatry. 1992;49(8):615–623. doi: 10.1001/archpsyc.1992.01820080023004. [DOI] [PubMed] [Google Scholar]
- 28.Andreasen NC. The Scale for the Assessment of Positive Symptoms. Iowa City: The University of Iowa; 1984. [Google Scholar]
- 29.Andreasen NC. The Scale for the Assessment of Positive Symptoms. Iowa City: The University of Iowa; 1983. [Google Scholar]
- 30.Chen J, Liu J, Calhoun V. IEEE International Conference on Bioinformatics and Biomedicine. Hong Kong: 2010. Correction off copy number data using principal component analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J, Liu J, Calhoun VD. A pipeline for copy number variation detection based on principal components analysis. Proceedings IEEE Bioinformatics and Biomedicine. 2011 doi: 10.1109/IEMBS.2011.6091763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Need AC, Ge DL, Weale ME, Maia J, Feng S, Heinzen EL, et al. A Genomewide investigation of SNPs and CNVs in schizophrenia. Plos Genetics. 2009;5(2) doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ehrlich S, Brauns S, Yendiki A, Ho B-C, Calhoun V, Schulz SC, Gollub RL, Sponheim SR. Associations of cortical thickness and cognition in patients with schizophrenia and healthy controls. Schizophrenia Bulletin. doi: 10.1093/schbul/sbr018. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prokosch MD, Yeo RA, Miller GF. Intelligence tests with higher g-loadings show higher correlations with body symmetry: Evidence for a general fitness factor mediated by developmental stability. Intelligence. 2005;33(2):203–213. [Google Scholar]
- 35.Euler M, Thoma RJ, Gangestad SW, Canive JM, Yeo RA. The Impact of developmental instability on voxel-based morphometry analyses of neuroanatomical abnormalities in Schizophrenia. Schizophrenia Research. 2009;115(1):1–7. doi: 10.1016/j.schres.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNeil TF, Cantor-Graae E, Weinberger DR. Relationship of obstetric complications and differences in size of brain structures in monozygotic twin pairs discordant for schizophrenia. American Journal of Psychiatry. 2000;157(2):203–212. doi: 10.1176/appi.ajp.157.2.203. [DOI] [PubMed] [Google Scholar]
- 37.Welch KA, McIntosh AM, Job DE, Whalley HC, Moorhead TW, Hall J, et al. The impact of substance use on brain structure in people at high risk of developing schizophrenia. Schizophrenia Bulletin. 2011;37(5):1066–1076. doi: 10.1093/schbul/sbq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Habets P, Marcelis M, Gronenschild E, Drukker M, van Os J Group. Reduced cortical thickness as an outcome of differential sensitivity to environmental risks in schizophrenia. Biological Psychiatry. 2011;69(5):487–494. doi: 10.1016/j.biopsych.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 39.Batel P. Addiction and schizophrenia. European Psychiatry. 2000;15(2):115–122. doi: 10.1016/s0924-9338(00)00203-0. [DOI] [PubMed] [Google Scholar]
- 40.Leung YJ, Sannesy S, Lyons MJ, Tsuang MT. Familial vulnerability to schizophrenia increases the risk of substance use: A twin study. American Journal of Medical Genetics Part B-Neuropsychiatric Genetics. 2004;130B(1):133–133. [Google Scholar]
- 41.Thoma RJ, Yeo RA, Gangestad SW, Halgren E, Sanchez NM, Lewine JD. Cortical volume and developmental instability are independent predictors of general intellectual ability. Intelligence. 2005;33(1):27–38. [Google Scholar]
- 42.Chiang MC, Barysheva M, Shattuck DW, Lee AD, Madsen SK, Avedissian C, et al. Genetics of brain fiber architecture and intellectual performance. Journal of Neuroscience. 2009;29(7):2212–2224. doi: 10.1523/JNEUROSCI.4184-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van den Heuvel MP, Stam CJ, Kahn RS, Pol HEH. Efficiency of functional brain networks and intellectual performance. Journal of Neuroscience. 2009;29(23):7619–7624. doi: 10.1523/JNEUROSCI.1443-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brans RGH, Kahn RS, Schnack HG, van Baal GCM, Posthuma D, van Haren NEM, et al. Brain plasticity and intellectual ability are influenced by shared genes. Journal of Neuroscience. 2010;30(16):5519–5524. doi: 10.1523/JNEUROSCI.5841-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng S, Zhang Y, Zhang JN, Wang H, Ren BX. Glutamate receptors and signal transduction in learning and memory. Molecular Biology Reports. 2011;38(1):453–460. doi: 10.1007/s11033-010-0128-9. [DOI] [PubMed] [Google Scholar]
- 46.Yeo RA, Gangestad SW, Gasparovic C, Liu J, Calhoun VD, Thoma RJ, Mayer AR, Kalyanam R, Hutchison KE. Rare copy number deletions predict individual variation in human brain neurometabolite concentrations in individuals with alcohol use disorders. Biological Psychiatry. 2011:70537–70544. doi: 10.1016/j.biopsych.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rijsdijk FV, Gottesman II, McGuffin P, Cardno AG. Heritability estimates for psychotic symptom dimensions in twins with psychotic disorders. American Journal of Medical Genetics Part B-Neuropsychiatric Genetics. 2011;156B(1):89–98. doi: 10.1002/ajmg.b.31145. [DOI] [PubMed] [Google Scholar]
- 48.Bansal V, Libiger O, Torkamani A, Schork NJ. Statistical analysis strategies for association studies involving rare variants. Nat Rev Genet. 2010;11(11):773–785. doi: 10.1038/nrg2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eckel-Passow JE, Atkinson EJ, Maharjan S, Kardia SLR, de Andrade M. Software comparison for evaluating genomic copy number variation for Affymetrix 6.0 SNP array platform. Bmc Bioinformatics. 2011;12 doi: 10.1186/1471-2105-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gangestad SW, Yeo RA, Liu J, et al. Rare deletions predict general cognitive ability, brain neurometabolite concentrations, and schizophrenia phenotype; presented at Deletions and cognitive ability 20 the annual meeting of the Human Behavior and Evolution Society; Montpellier, France. 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.