Abstract
Altered RNA processing is an underlying mechanism of amyotrophic lateral sclerosis (ALS). Missense mutations in a number of genes involved in RNA function and metabolisms are associated with ALS. Among these genes is angiogenin (ANG), the fifth member of the vertebrate-specific, secreted ribonuclease superfamily. ANG is an angiogenic ribonuclease, and both its angiogenic and ribonucleolytic activities are important for motor neuron health. Ribonuclease 4 (RNASE4), the fourth member of this superfamily, shares the same promoters with ANG and is co-expressed with ANG. However, the biological role of RNASE4 is unknown. To determine whether RNASE4 is involved in ALS pathogenesis, we sequenced the coding region of RNASE4 in ALS and control subjects and characterized the angiogenic, neurogenic, and neuroprotective activities of RNASE4 protein. We identified an allelic association of SNP rs3748338 with ALS and demonstrated that RNASE4 protein is able to induce angiogenesis in in vitro, ex vivo, and in vivo assays. RNASE4 also induces neural differentiation of P19 mouse embryonal carcinoma cells and mouse embryonic stem (ES) cells. Moreover, RNASE4 not only stimulates the formation of neurofilaments from mouse embryonic cortical neurons, but also protects hypothermia-induced degeneration. Importantly, systemic treatment with RNASE4 protein slowed weight loss and enhanced neuromuscular function of SOD1G93A mice.
Keywords: Ribonuclease 4, Angiogenin, Angiogenesis, Neurogenesis, Neuroprotection, Amyotrophic lateral sclerosis (ALS)
Introduction
Secreted ribonucleases (RNASEs) are the only enzyme family that is vertebrate-specific [1]. Apparently, a new gene emerged after the separation of vertebrates from invertebrates [2]. This gene rapidly duplicated and evolved into a distinct family of 13 paralogs [3]. In addition to the universal ribonucleolytic activity, many members of this family have unique biologic activities. RNASE2 and RNASE3 are known as Eosinophils-Derived Neurotoxin (EDN) [4] and Eosinophils Cationic Protein (ECP) [5], respectively, as these proteins were originally isolated from eosinophils and are neurotoxic to cerebral Purkinje cells [6]. RNASE2/EDN is also able to activate human dendritic cells to produce cytokines and growth factors from the immune system in response to pathogen stimulation [7]. RNASE3/ECP has been shown to be anti-parasitic [8], anti-bacterial [9], and anti-viral [10]. RNASE5 was originally isolated from HT-29 human colon adenocarcinoma cell-conditioned medium and was identified as angiogenin (ANG) based on its angiogenic activity [11]. ANG is upregulated in human cancers and plays an important role in stimulating cell growth and in promoting cell survival [12]. The biological activities of ANG have been shown to be related to its ability in regulating RNA metabolism. For example, the growth-promoting activity of ANG is dependent on stimulation of ribosomal RNA (rRNA) transcription [13-15], while its pro-survival activity relies on the production of tRNA-derived, stress-induced RNAs (tiRNA) [16-18] that suppress global protein translation and save anabolic energy [19].
Among the 13 members of this superfamily, ribonuclease 4 (RNASE4) is the most conserved gene across the different vertebrate species [20]. The amino acid homology across the different vertebrate species is as high as 94% [20]. RNASE4 has a very strict substrate specificity–strongly preferring an uridine at the 3′-side of the cleavage site [21]. These unique properties of RNASE4 suggest that it may have an as yet undefined, more specific biological activity. Like ANG, RNASE4 was originally isolated from HT-29 cell-conditioned medium [21] but its angiogenic activity has not been clearly characterized.
At the protein level, human ANG and RNASE4 have 38.7% identity (Supplementary Fig. S1). ANG and RNASE4 genes are located in the same locus and share the same promoters. The locus of mouse Ang1 and Rnase4 genes have been reported to contain two non-coding exons followed by two distinct exons encoding Ang1 and Rnase4, respectively [22]. The two non-coding exons are preceded by two promoters that control liver-specific and tissue-specific expression [22]. We have found that human RNASE4 and ANG have a similar gene arrangement (Supplementary Fig. S2). As a consequence of this unique gene structure, RNASE4 and ANG are often co-expressed [23]. A search of expressed sequence tag (EST) database has revealed that RNASE4 and ANG mRNA have the same 5′-UTR region [24]. We have identified transcripts of both ANG and RNASE4 having the 5′UTR that contains either exon I or exon II, but not both (Supplementary Fig. S3). This unusual arrangement and regulation of RNASE4 and ANG genes suggest that they may have complementary or supplementary biological activities. The discovery that loss-of-function mutations in ANG gene are associated with amyotrophic lateral sclerosis (ALS) [25, 26] and that ANG protein is neuroprotective against stress-induced motor neurons degeneration [27, 28] prompted us to further characterize the biological activity of RNASE4 and to examine its possible role in ALS pathogenesis. We sequenced the coding exon (exon IV) of RNASE4 for potential ALS-associated mutations and SNPs, prepared the recombinant protein and characterize its angiogenic, neurogenic and neuroprotective activities. No mutations in the coding region of RNASE4 were associated with ALS. However, a previously reported SNP rs3748338 A/T(-39) (Table 1 and Supplementary Fig. S4), which results in an amino acid change from Thr to Ser at position −13 of the signal peptide, show association with ALS that is marginally significant (p=0.042). Consistent with a potential role of RNASE4 in motor neuron physiology, we found that WT RNASE4 protein is not only angiogenic, but also neurogenic and neuroprotective. RNASE4 induces angiogenesis in in vitro, ex vivo and in vivo assays. It also induces neurosphere formation and neuronal differentiation of mouse embryonic stem (ES) and embryonal carcinoma cells. It stimulates neurofilament formation of mouse embryonic cortical neurons and prevents stress-induced degeneration. Moreover, administration of RNASE4 protein in SOD1G93A mice slowed down their weight loss. There is also a trend that RNASE4-treated SOD1G93A mice regained some of their neuromuscular function. In light of the important role of RNA processing and metabolism in ALS [29-31], our results suggest that haploinsufficiency of RNASE4 might be a risk factor and that RNASE4 protein therapy, either alone or together with ANG, might be beneficial to ALS patients.
Table 1.
Relative frequencies of RNase4 SNPs in the SALS and control population
| ALS, n=1,575 | Control, n=658 | |||||
|---|---|---|---|---|---|---|
| Nucleotide 1 | Amino acid 1 | Known SNP | Cases | MAF | Cases | MAF |
| A(-39)CC → TCC |
T(-13) → S | rs3748338 | 413 (11)2 |
13.81 % | 152 | 11.55 % |
| C(28)GG → TGG |
R(10) → W | No | 9 | 0.29 % | 3 | 0.23 % |
| GAA(144) → GAC |
E(48) → D | No | 1 | 0.03 | 1 | 0.08 % |
| GAG(219) → GAA |
E (73) → E | rs11758275 8 |
1 | 0.03 % | 0 | 0 |
| G(223)TA → ATA |
V(75) → I | No | 2 | 0.06 % | 1 | 0.08 % |
| GC(293)G → GTG |
A (98) → V | No | 1 | 0.03 % | 1 | 0.08 % |
The numbering of amino acid and nucleotide started from the first amino acid in the mature protein and the first nucleotide of the corresponding codon.
the number in parenthesis is the cases of homozygous changes.
Materials and methods
Samples and mutation screening
Clinical specimens were obtained under a discarded tissue protocol approved by the institutional review board. Genomic DNA was extracted from peripheral leukocytes using standard protocol. The coding exon of RNASE4 was amplified by PCR with primers located in adjacent intron and non-coding regions, respectively, and the amplicons were sequenced bidirectionally. The sequences of the primers are as follows. Forward. 5′-ACCTTATTTCTCCTGCCCCTTG-3′; reverse: 5′-AAGCCCAGCCTCATTCATTACAG-3′.
Preparation of recombinant proteins
WT RNASE4 gene was cloned from genomic DNA of HeLa cells with the following PCR primers. Forward, 5′-GGAGATATCATATGCAGGATGGCATGTAC-3′; Reverse, 5′CCGGGATCCCTAACCGTCAAAGTGC-3′. PCR products were cloned into pET11 vector between the BamHI and NdeI sites. The resultant plasmid (pET11 -hRNASE4) was sequence confirmed and transformed into BL21 (DE3) cells for protein expression. K40A mutation was generated by QuickChange II Site-Directed Mutagenesis Kit from Stratagene using pET11 - hRNASE4 as the template and the following primers: Forward, 5′- CTTTGTATCACTGCgcGCGCTTCAACACCTT-3′; Reverse, 5′- AAGGTGTTGAAGCGCgcCCAGTGATACAAAG-3′. The resultant plasmid (pET11 - hRNASE4-K40A) was sequenced confirmed and cloned into BL21 (DE3) cells. Recombinant RNASE4 and RNASE4-K40A were induced and purified essentially following the protocol described for production of recombinant ANG [32] with the following modification. One liter of bacterial culture was pelleted and lysed by sonication in lysis buffer consisting of 50 mM Tris-HCI and 2 mM EDTA, pH 8.0, and centrifuged for 20 min at 20,000 × g. The pellet was re-suspended in the lysis buffer containing 1% (v/v) Triton X-100, sonicated, centrifuged, and washed one more time in the lysis buffer. The inclusion bodies were then dissolved in 10 ml of 0.1 M Tris-HCI, 2 mM EDTA, pH8.0, containing 7.0 M guanidine hydrochloride, 0.15 M reduced glutathione by stirring under nitrogen for 2 h. The solution was then added dropwise to 500 ml of 0.5 M L-arginine-HCI, pH 8.0, containing 0.6 mM oxidized glutathione. The diluted solution was kept at RT for 24 h to allow protein refolding to take place. It was centrifuged at 10,000 × g for 30 min, and the supernatant was diluted fivefold with water, and applied to an SP-sepharose column pre-equilibrated with 10 mM Tris-HCI, 0.2 M NaCI, pH 8.0. The proteins were eluted with a 0.2-0.8 M NaCl gradient in 10 mM Tris-HCl, pH8.0, and further purified by reversed-phase HPLC on a C4 (phenomenex) column. A 45 min linear gradient from 100% solvent A (0.1% TFA) to 100% solvent B (acetonitrile:isopropanol:water, 3:2:2, in 0.08%TFA) was used to elute RNASE4 and RNASE4-K40A.
Ribonucleolytic assay
Ribonucleolytic activities of WT and K40A RNASE4 were examined using yeast tRNA as the substrate as previously described for ANG [33]. Reactions were initiated either by adding the enzyme. Varying amount RNASE were added to a final volume of 300 μl system including 600 ng yeast tRNA, 0.33 M Hepes, 0.33 M NaCl, pH 7.0, and 0.1 mg/ml RNASE-free BSA. After incubation at 37 °C for 120 min, 700 l of ice-cold 3.4% perchloric acid was added and incubated on ice for 10min, centrifuge at 14,000 × g for 10 min at 4 °C, and the absorbance at 260 nm of the supernatants was recorded. All buffers and water used above was filtered through Sep-Pak cartridge (Waters) to ensure that the system is RNASE-free. Experiments were done in triplicates.
Cells
P19 cells were maintained in DMEM plus 10% FBS and antibiotics (100 μg/ml streptomycin and 100 Units/ml penicillin). HUVEC were maintained in human endothelial basal growth medium plus 5 ng/ml bFGF. PA6 cells were cultured in α-MEM plus 10% FBS. Cell growth was measured either by MTT assay or with a Coulter counter. Mouse ES cells were maintained on mitomycin C-inactivated mouse embryonic fibroblasts (MEF) in ES culture medium in the presence of mLIF medium supplement (500 units/ml).
In vitro angiogenesis assays
Each well of the 48-well plate was coated with 50 l growth factor-reduced Matrigel. HUVEC, 1.5 × 104 per well, was seeded on the Matrigel and cultured in the presence or absence or testing materials for 4 h. Cells were fixed in 3.7% paraformaldehyde and photographed. The images were analyzed by ImageJ software to calculate the length or area of capillary-like structures as well as the numbers of circled tubular structure. Experiments were done in triplicates and were repeated at least three times.
Ex vivo angiogenesis assay
Thoracic aortic vessels from 2 month old C57BL/6J mice were dissected and transferred to a dish containing ice-cold MEM. The fibroadipose tissues were removed with microdissecting forceps and scissors. The aorta were cut into 0.5 mm pieces and washed extensively in cold PBS, and placed in Matrigel-coated 48-well plates. Each well was covered with another 50 l Matrigel and incubated in human endothelial SMF basal growth medium in the presence or absence of testing materials for 15 days. New angiogenic sprouts were stained with 20μl MTT (5 mg/ml in PBS) for 2 h, photographed and analyzed by ImageJ.
In vivo angiogenesis assay
A cold solution of 0.5 ml Matrigel with or without 0.5 μg/ml testing proteins was injected subcutaneously into C57BL/6 mice. The Matrigel plug was removed 4 days later and processed for immunohistochemical staining of blood vessels with anti-von Willebrand Factor (vWF) IgG.
Neurosphere formation and neurite outgrowth
P19 cell were seeded on PA6 monolayer and cultured in α-MEM plus 0.1% nonessential amino acids, 0.1% knockout serum replacement, 0.5 μM retinoic acid, and 200 ng/ml testing proteins for 24 h. Embryonal bodies were counted under phase light microscope at 10 × magnification. The numbers of embryonal bodies from the entire well were counted. Experiments were done in triplicates and were repeated three times.
For neurite outgrowth, P19 cells were cultures as described above but for 216 h. Cells were fixed successively in 4% paraformaldehyde for 15 min at RT and cold methanol for 5 min at −20 °C, blocked with 0.1% gelatin, 0.5% BSA and 0.1% Tween 20 in TBS or 1 h at RT. Neurofilaments were stained by a rabbit anti-Neurofilament medium chain at 1:500 dilution at 4°C overnight and Alexa 488-labeled goat anti-rabbit F(ab’) at 1:400 dilution for 1 h at RT.
Mouse ES cell differentiation
The protocol of Bibel et al [34] was followed. Briefly, mouse ES cells (J15) were dissociated from MEF feeders, and cultured in ES medium in petri dishes for 4 days. On day 5, the embryonal bodies were suspended, reseeded in petri dishes, and cultured in DMEM supplemented with 10% FBS, 2 mM L-glutamine, 1X non-essential amino acids, and 0.05 mM β-mercaptoethanol in the presence of 5 μM retinoic acid, 0.2 μg/ml ANG, or 0.2 μg/ml RNASE4 for another 4 days. On day 8, embryonal bodies were dissociated, plated on laminin-coated cell culture dishes, and cultured in B27 medium in the presence of various differentiation inducers for another 2 days and stained for glial fibrillary acidic protein (GFAP) by immunofluorescence.
Primary mouse embryonic cortical neuron culture
Brain cortical cells were isolated from C57/B6SJL embryos (E14) and seeded in 6 wells at a density of 1.1 × 106 per cm2 in neurobasal medium plus B27 and 0.5 mM L-glutamine for 36 h. The medium was changed and 10 μM Ara-c was added and cultured for another 6 days with medium change every two days. Ara-c was removed and the cells were cultured in neurobasal medium plus B27 in the presence or absence of testing material for additional 6 days. For hypothermia-induced degeneration, the cells were put at RT temperature for 40 min and returned to the cell culture incubator for additional 3 h. Neurofilaments were stained as described above.
Immunofluorescence
HUVE cells and P19 cells were cultured in their respective media on cover slips. Exogenous human RNASE4 protein, when present, was added at a final concentration of 0.5-1 μg/ml and incubated with the cells for 1 h. Cells were fixed with 4% formaldehyde at RT for 20 min, permeabilized with 1% Triton X-100, and incubated with affinity-purified RNASE4 polyclonal antibody (1 μg/ml) and Alexa 488-labeled goat anti-rabbit IgG.
In situ hybridization (ISH) of RNase4 mRNA
Human RNASE4 and mouse Rnase4 DNA were amplified by PCR from HeLa cell DNA and C57B6/SJL mouse tail genomic DNA as templates. The primers were as follows: Human RNASE4, forward, 5′-GGGTAATACGACTCACTATAGGGCGAaccttatttctcctgccccttg-3′ (up case: T7 sequence); reverse, 5′-aagcccagcctcattcattacag-3′. Mouse Rnase4, forward, 5′- GGGTAATACGACTCACTATAGGGCGAtccgggtccaggcactttcta-3′; reverse, 5′-gtg ctggttcttgccctgtatcta-3′. cRNA probes were produced by in vitro transcription with T7 RNA polymerase from 1 μg of the above purified PCR products as template and were labeled with digoxigenin (Roche) per manufacturer’s protocol. Control cRNA probes were generated from the control DNA template included in the kit. For in situ hybridization, the slides were deparaffinized with Xylene (2 × 10 min), washed in sequence with 100, 75, 50, and 25% ethanol and PBS each for 5min. The slides were then treated with 2 μg/ml of Proteinase K for 10 min, washed with 2 mg/ml of Lysine and PBS each for 5min, and incubated in acetylation buffer (0.25% acetic acid anhydride in 0.1 M Triethanolamine, pH 8.0) for 20 min. After two washes with 4 × SSC, the slides were prehybridized in Hybridization buffer (5 × SSC, 0.5 mg/ml heparin, 0.8 mg/ml salmon sperm DNA) at 45 °C for 60 min. cRNA probes were added onto the slides and incubated at 45 °C overnight. Hybridization signals were detected and visualized with the digoxigenin Nucleic Acid Detection Kit from Roche following manufacturer’s protocol.
Treatment of SOD1G93A mice with RNASE4 protein
Animal experiments were approved by Institutional Animal Care and Use Committee. A SOD1G93A mouse colony was established from a breeding pair of B6SJL-Tg(SOD1G93A)1Gur/J mice purchased from The Jackson Laboratory. Human SOD1G93A transgene copy number was determined by real time qPCR. Gender- and litter-matched mice at 10 weeks of age were separated into two groups and treated with weekly i.p. injection of PBS or RNASE4 at 10 μg/mouse. The mice were weighed once a week and were tested on a rotarod (20 rpm) for neuromuscular function weekly. Probability of survival was determined by Kapalan-Meyer analysis. The identities of the samples were blinded and were decoded only when the entire experiment and the analysis was finished.
Statistical analysis
The data obtained from independent experiments are presented as the mean ± SEM; they were analyzed using a paired Student’s t test with Mann-Whitney test, one-way analysis of variance (ANOVA) with Kruskal-Wallis test, or two-way ANOVA for multiple comparisons using SPSS software version 16. For rotarod experiments, repeated measures ANOVA was used. Genetic association analysis was done with Hardy-Weinberg equilibrium (http://ihg.gsf.de/cgibin/hw/hwa1.pl). Differences were considered significant at P < 0.05.
Results
Polymorphism of human RNASE4 gene
The coding region of RNASE4 gene from 1,575 sporadic ALS patients and 658 controls was sequenced for potential ALS-associated mutations and SNPs. We found a total of 6 SNPs in these 2,233 subjects (Table 1). The most abundant SNP is the nucleotide change from A to T at position −39, which corresponds to the known SNP rs37484338 in the data base. This change results in a change of amino acid from Thr to Ser in the signal peptide position −13. We observed a total of 11 and 413 homozygous and heterozygous T alleles in the 1,575 ALS subjects, representing a minor allele frequency of 13.81%. However, in the 658 control subjects, there were 152 heterozygous but no homozygous T alleles, representing a minor allele frequency of 11.55%. Hardy-Weinberg equilibrium and tests of association (http://ihg.gsf.de/cgibin/hw/hwa1.pl) showed that the p value for the allele frequency difference is 0.042, and the p value for the common odds ratio from Armitage’s trend test is 0.031. None of the other 5 SNPs reached a statistically significant difference between ALS and control subjects. These genetic data suggest that there is an association between the presence of a serine at position −13 in RNASE4 and ALS susceptibility.
Preparation and biochemical characterization of recombinant RNASE4 protein and K40A variant
The unique structure of RNASE4 and ANG genes, the fact that these proteins are co-expressed, and the possible association of SNP rs3748338 with ALS encouraged us to consider the possible neuroprotective activity of RNASE4. We therefore prepared the recombinant WT protein as well as the enzymatically attenuated K40A variant. K40 was chosen because it is an essential residue constituting the catalytic triad that is conserved across the members of this superfamily [33]. Mutations at K40 will diminish most of the catalytic activity of this family of proteins [35, 36]. The coding region of RNASE4 gene was amplified from HeLa genomic DNA and cloned into E. coli expression vector pET11a which has been successfully used to express ANG [32]. K40A mutation was generated by site-directed mutagenesis and was cloned into the same vector. We were able to obtain pure and active RNASE4 proteins as shown by SDS-PAGE (Fig. 1a) and yeast tRNA ribonucleolytic assay (Fig. 1b). As predicted, the K40A variant of RNASE4 has reduced but did not eliminate ribonucleolytic activity. It retains 5.4% of the activity of WT RNASE4 when yeast tRNA was used as a substrate (Fig. 1b). This finding is unusual as K40 is an essential residue for catalysis for this family of proteins; prior studies have shown that mutations at this position result in diminished enzymatic activity of other members of this family of enzymes. For example, replacing K40 in ANG by Q decreases ribonucleolytic activity by 2,000-fold [35], and K41C mutation in RNase A results in a 9,000-fold decrease in enzymatic activity [36]. It is unclear how K40A RNASE4 retains its ribonucleolytic activity, however, the findings here are consistent with this variant having enhanced angiogenic activity (see below).
Fig. 1.
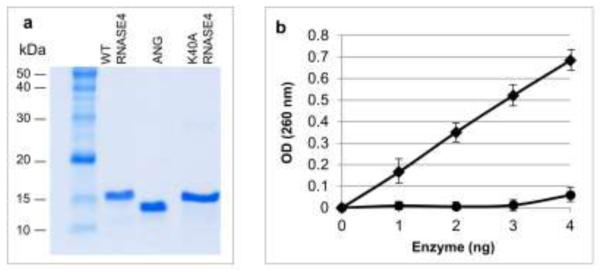
Ribonucleolytic activity of recombinant wide type RNASE4 and the K40A variant. a SDS-PAGE and Coomassie blue staining of WT and K40A RNASE4 from one of the four preparations. Recombinant ANG protein was included as a control. b Ribonuclolysis of yeast tRNA by WT RNASE4 (squares) and K40A RNASE4 (circles). Data shown are mean ± SEM of four independent preparations with triplicates in each enzyme concentration.
Characterization of the angiogenic activity of RNASE4
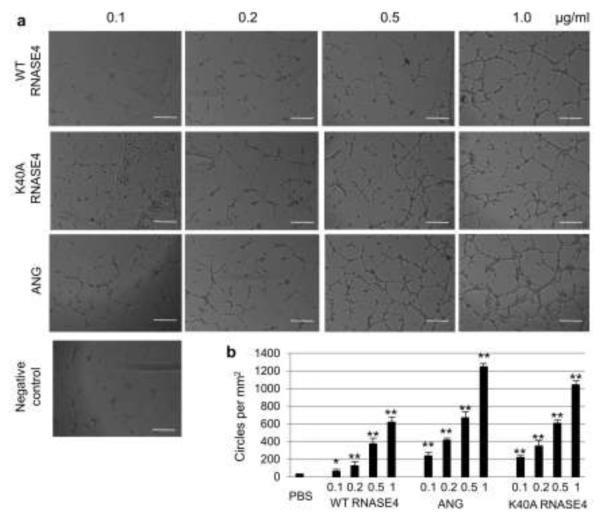
Among the 13 members of this family of protein, only RNASE5/ANG has been shown to be angiogenic. All the vertebrate ANG, from human to fish, are angiogenic and ribonucleolytic. Moreover, the ribonucleolytic activity is essential for angiogenesis. The ability of RNASE4 to induce angiogenesis has not been clearly demonstrated. RNASE4 was first co-isolated with ANG from HT-29 cell conditioned-medium and was originally given a name of HT-RNase and was shown to be inactive in chick chorioallantoic membrane (CAM) angiogenesis assay [11]. In the original study, two pools were obtained from a reversed-phase HPLC. Pool C, which contained mostly RNASE4, was active in the CAM assay only at >40 ng/egg, whereas Pool D that contained ANG was active at 0.5 ng/egg. It was concluded that the low angiogenic activity in Pool C was due to the ANG contaminant. However, a recent study showed that RNase4 isolated from bovine milk was able to induce endothelial tube formation [37], suggesting that RNASE4 is angiogenic. But in the same study, bovine RNase A, a clearly non-angiogenic protein, was also shown to induce the formation of endothelial cell tubes [37]. A clear understanding of the angiogenic activity of RNASE4 would be both scientifically and clinically significant as RNASE4 might be an ALS-associated gene. We therefore first examined the ability of WT and K40A RNASE4 to induce endothelial cell tube formation, a widely used angiogenesis assay [38]. As shown in Fig. 2a, both WT and K40A RNASE4 were able to induce tubule formation of HUVEC cultures in Matrigel. ANG was included as a positive control. RNASE4 had about 50% of the activity of ANG in this assay. For example, RNASE4 at 1 μg/ml and ANG at 0.5 μg/ml had an equivalent activity (Fig. 2b). It is interesting to note that the K40A variant actually has a higher angiogenic activity than the WT and is only slightly lower than ANG. No endotoxin was detected in the preparation. Heat inactivation or proteolysis abolished all the activity (data not shown) indicating that an intact peptide and structure is important. No angiogenic activity of RNase A/RNASE1, EDN/RNASE2, and ECP/RNASE3 were detected in the same assay system (Supplementary Fig. S5).
Fig. 2.
RNASE4 induces endothelial cell tube formation. a Microscopic images of endothelial cell tubes formed in various concentration of WT and K40A RNASE4. ANG and PBS were used as positive and negative controls, respectively. The images shown are representative of at least four sections from three independent experiments. Bar, 0.1 mm. (B) Number of circled tubular structures per mm2. Values are mean ± SEM of three independent experiments. Statistical analysis was performed by two-way ANOVA. *, p<0.05; **, p<0.01.
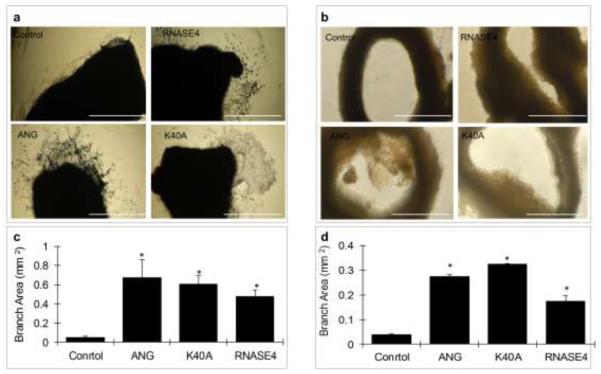
Next, we performed an ex vivo angiogenesis assay using mouse aortic ring explant cultures [39]. Both WT RNASE4 and K40A variant were able to stimulate endothelial sprouts from the inner side of the aorta so that the sprouts grew outward in the inside-out flipped aortic rings (Fig. 3a) and inward in the unflipped ones (Fig. 3b). Quantitative analysis with the ImageJ program indicated that the area covered by endothelial branch in the control, ANG, K40A and WT RNASE4 were 0.05 ± 0.01, 0.67 ± 0.19, 0.61 ± 0.09, and 0.47 ± 0.07 mm2, respectively, in inside-out flipped culture (Fig. 3c); and were 0.04 ± 0.01, 0.27 ± 0.01, 0.32 ± 0.01, and 0.17 ± 0.02 mm2, respectively, in unflipped culture (Fig. 3d). Thus, it is clear that RNASE4 elicits an angiogenic response in mouse aorta explant culture. Like the results with the in vitro endothelial cell tubule formation assay (Fig. 2), the K40A variant has a higher activity than the WT RNASE4, suggesting that if ribonucleolytic activity is necessary for RNASE4 to induce angiogenesis, the remaining 5% activity in R40A variant is sufficient for this purpose. It is noteworthy that while WT RNASE4 has a slightly lower angiogenic activity as compared with ANG, the activity of K40A RNASE4 variant and ANG are comparable.
Fig. 3.
RNASE4 stimulates endothelial sprouts from mouse aortic explants. a Outward growth of endothelial sprouts from aortic rings that have been flipped inside out. The images are representative of three rings from one of three repeats. Bar, 0.1 mm. ANG and PBS were used as positive and negative controls, respectively. b ImageJ analysis of the area covered by endothelial sprouts from flipped aortic rings. Data are presented as mean ± SEM from three independent experiments. c Inward growth of endothelial sprouts from unflipped mouse aortic rings. The images are representative of three rings from one of two repeats. d ImageJ analysis the area covered by endothelial sprouts from unflipped aortic rings. Data are presented as mean ± SEM from two independent experiments. Bar, 1.0 mm. Statistical analysis was performed by two-way ANOVA. *, p<0.001.
To confirm the in vitro and ex vivo finding that RNASE4 is angiogenic, we examined the ability of RNASE4 to induce blood vessel formation in the Matrigel plug in vivo angiogenesis assay [40]. As shown in Fig. 4a, a vigorous neovessel growth was observed in the Matrigel plugs in the presence of both WT RNASE4 and the K40A variant. ANG, used as a positive control, also elicited a strong angiogenic response. Quantitative analysis indicates that the vessel density per mm2 in control Matrigel and in those containing ANG, WT RNASE4 and K40A variant was 49 ± 7, 250 ± 10, 184 ± 39, and 189 ± 58, respectively (Fig. 4b). Thus, RNASE4 and its K40A variant are active in in vitro, ex vivo and in vivo angiogenesis assays. Taken together, these results clearly demonstrated that human RNASE4 is angiogenic.
Fig. 4.
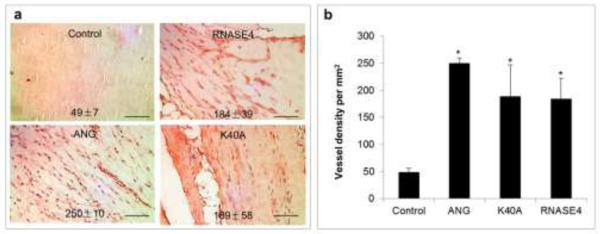
RNASE4 induces neovessel growth into Matrigel plug implanted under mouse skin. a IHC staining with vWF antibodies. The images shown are representative of at least four sections from three independent experiments. b ImageJ analysis of vWF-positive spots. Data are presented as mean ± SEM from three independent experiments. Bar, 100 μm. Statistical analysis was performed by two-way ANOVA. *, p<0.001.
The ribonucleolytic activity of RNASE4 is essential for angiogenesis
The finding that K40A variant retains 5.4% of the ribonucleolytic activity of RNASE4 was unexpected. It needs to point out that the enzymatic activity that K40A variant retains is still about 10 times higher than that of ANG. This may explain the enhanced angiogenic activity of K40A in all three angiogenesis assays. In order to know whether the ribonucleolytic activity of RNASE4 is essential for its angiogenic activity, we treated RNASE4 protein with diethylpyrocarbonate (DEC) that chemically modifies Lys residues and completely abolished the enzymatic activity of ribonucleases. DEPC-treated RNASE4 fails to cleave both yeast tRNA (Fig. 5a) and in HeLa rRNA even with prolonged incubation, indicating that DEPC treatment has completely abolished the enzymatic activity of RNASE4. Fig. 5c shows that DEPC treatment also completely abolished the ability of RNASE4 to induce endothelial tubule formation. These results indicate that the ribonucleolytic activity of RNASE4 is essential for angiogenesis and that the reason for an enhanced angiogenic activity of R40A RNASE4 variant is the remaining ribonucleolytic activity.
Fig. 5.
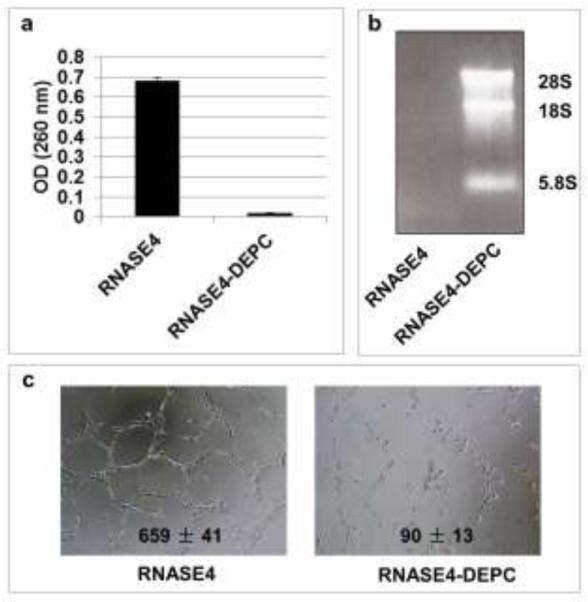
Ribonucleolytic activity of RNASE4 is essential for its angiogenic activity. a RNASE4 proteins was treated with 5 mM DEPC and its ribonucleolytic activity was examined by yeast tRNA assay. b DEPC-treated RNASE4 fails to cleave rRNA. c DEPC treatment abolishes angiogenic activity of RNASE4.
RNASE4 stimulates neuronal differentiation and neurite outgrowth
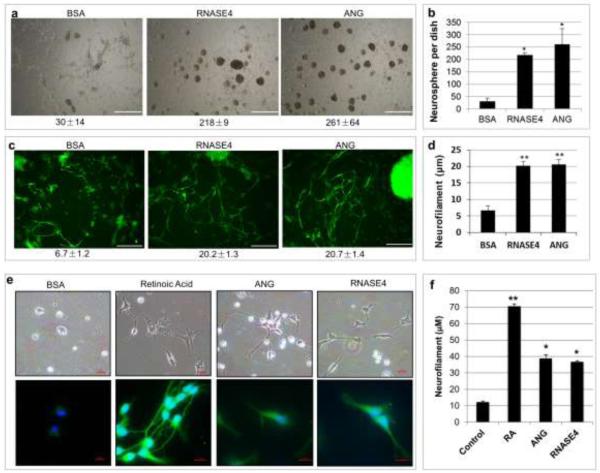
The neurogenic activity of RNASE4 was first examined in P19 mouse embryonal carcinoma cells. P19 cells are pluripotent and have stem cell-like property. These cells are able to both self-renew and differentiate into various types of neural cells [41, 42]. We found that both ANG and RNASE4 were able to induce the formation of neurospheres or embroynal bodies (Fig. 6a). The number of these neurospheres in a 35 mm dish after 24h incubation was 218 ± 9 and 261 ± 64, respectively, in the presence of RNASE4 and ANG, representing a 7.3- and 8.7-fold increase over that formed in the presence of control protein BSA (30 ± 14) (Fig. 6b). As the formation of neurospheres is a strong indication of neuronal differentiation [43, 44], these results indicate that both ANG and RNASE4 induce neurogenesis. Definite evidence for neuronal lineage commitment upon ANG and RNASE4 stimulation is unclear at present. But it is clear that ANG and RNASE4 were able to stimulate neurite outgrowth in a prolonged culture of P19 cell (Fig. 6c). After 9 days culture, the average length of neurofilaments in the presence of RNASE4 and ANG was 20.2 ± 0.8 and 20.7 ± 1.4 μm, respectively, which is 3.0- and 3.1-fold higher than that in the presence of BSA (6.7 ± 1.2 μm) (Fig. 6d). It has been previously reported that ANG stimulates neurite outgrowth and pathfinding of P19 cells [28]. Our results confirm this activity of ANG as well as demonstrate that RNASE4 has a similar activity both in angiogenesis and in neurogenesis.
Fig. 6.
Effect of RNASE4 on neuronal differentiation. a RNASE4 stimulates neurosphere formation of P19 cells. P19 cells were cultured on PA6 supporting cell layers in the presence of 0.2 μg/ml of BSA, RNASE4, or ANG for 24 h. The images are representative of at least four areas from three independent experiments. Bar, 0.5 mm. b Numbers of neurosphere counted from the entire 35-mm dish. Data shown are means ± SEM of three independent experiments in triplicates. Statistical analysis was performed by two-way ANOVA. *, p<0.001. c Cells were cultured as described in A for 216 h and stained for neurofilaments with an anti-neurofilament medium chain IgG. The images are representative of at least four areas from three independent experiments. Bar, 20 μm. d ImageJ analysis of neurofilament length from data shown in c. Data shown are means ± SEM of three independent experiments in triplicates. Statistical analysis was performed by two-way ANOVA. **, p<2×10−9. e RNASE4 stimulates mouse embryonic stem cell differentiation. Mouse ES cells were induced by 0.5 mM retinoic acid, 0.2 μg/ml ANG, or 0.2 μg/ml RNASE4 for 6 days. The top panel is the morphology of differentiated progenitor cells derived from the embryoid bodies. Bar, 15 μm. The bottom panel is GFAP-positive neuroprogenitor cells. Bar, 10 μm. f ImageJ analysis of the length GFP-positive neurofilaments. *, p<0.01. **, p<0.001.
In order to better understand neural differentiation activity of RNASE4, we examined its ability to induce differentiation of mouse ES cells. Fig. 6e shows that both RNASE4 and ANG induced differentiation of mouse ES cells into GFAP-positive progenitor neurons. The length of neurofilaments in the presence of ANG and RNASE4 was 38.8 ± 2.3 and 36.8 ± 0.7 μm, respectively, which is 3.1- and 3.0-fold higher than that in the negative control (BSA: 12.3 ± 0.7), and is 55% and 52% of that in the positive control (Retinoic acid: 70.5 ± 1.5). These results further demonstrate the neurogenesis activity of RNASE4 and angiogenin.
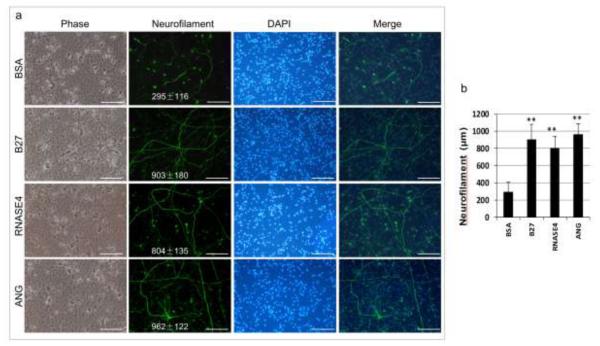
ANG and RNASE4 are also able to stimulate neurofilament growth of primary neurons. As shown in Fig. 7, mouse cortical neurons isolated from E14 embryos were able to survive for 12 days in neurobasal medium, but very little neurofilament growth was noted when only BSA was present (Fig. 7a). In the presence of B27, the serum-free supplements for growth and long-term viability of primary neurons [45], an extensive network of neurofilaments was formed. A similar network of neurofilaments was formed in the presence of RNASE4 and ANG. The length of neurofilaments formed in the presence of BSA, B27, RNASE4, and ANG were 295 ± 116, 903 ± 180, 804 ± 135, and 962 ± 122 μm, respectively (Fig. 7b). Thus, the activity of ANG and RNASE4 in stimulating neurofilament growth from the primary culture of the mouse embryonic cortical neuron is comparable to or greater than that of the B27 positive control. These results clearly indicate that both RNASE4 and ANG have neurogenic activity.
Fig. 7.
Effect of RNASE4 on mouse embryonic cortical neurons. a Cortical neurons were isolated from E14 mouse embryos and cultured in neurobasal medium in the presence of 0.2 μg/ml RNASE4 or ANG for 12 days with a medium change on day 6. B27 was used as a positive control and BSA at 0.2 μg/ml was used as a negative control. The images are representative of at least four areas from three independent experiments. Bar, 0.5 mm. b ImageJ analysis of the length of the neurofilaments. Data shown are means ± SEM of three independent experiments in triplicates. Statistical analysis was performed by two-way ANOVA. **, p<0.005.
RNASE4 protects stress-induced neuronal degeneration
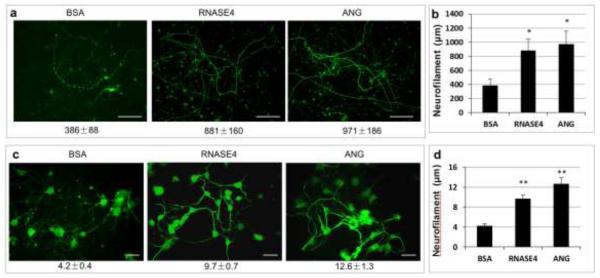
The neuroprotective activity of ANG has been demonstrated in various systems [27, 46-50]. To determine whether RNASE4 also possesses neuroprotective activity, we first examined its effect in preventing hypothermia-induced fragmentation of neurofilament derived from mouse embryonic cortical neuron culture. As have been shown in Fig. 7, a robust network of neurofilaments formed when mouse embryonic cortical neurons were cultured in the presence of B27. These neurofilaments fragmented into small pieces after being subjected to 25 °C for 40 min in the presence of control protein BSA (Fig. 8a). By contrast, both RNASE4 and ANG prevented hypothermia-induced neurofilament fragmentation, indicating that both are neuroprotective. The average length of the neurofilaments after being subjected to hypothermia was 386 ± 88, 881 ± 160, and 971 ± 186 μm in the cultures containing BSA, RNASE4, and ANG, respectively (Fig. 8b). Thus, RNASE4 and ANG have comparable neuroprotective activity. This is also the first report that ANG prevents stress-induced degeneration of primary neurons in culture.
Fig. 8.
RNASE4 protects stress-induced neuron degeneration. a Effect on hypothermia-induced neurofilament fragmentation of mouse cortical neurons. Mouse cortical neurons were cultured in the presence of B27 for 12 days. Cells were washed with neurobasal medium, incubated with 0.2 μg/ml RNASE4 or ANG at 37 °C for 1 hour, and then subjected to hypothermia treatment at 25 °C for 40 min. Cells were returned to incubator and continually cultured for 3 h and stained for neurofilaments. The images are representative of at least four areas from three independent experiments. Bar, 0.4 mm. b ImageJ analysis of the length of neurofilament. Data shown are means ± SEM of three independent experiments in triplicates. Statistical analysis was performed by two-way ANOVA. **, p<0.006. c Effect of serum starvation-induced degeneration of P19 neurofilaments. P19 cells were cultured on PA6 supporting cell layers in the presence of 0.5 μM retinoic acid for 216 h, subjected to serum starvation for 1 h in the absence or presence of 0.2 μg/ml RNASE4 or ANG, and stained with neurofilament antibody. The images are representative of at least four areas from three independent experiments. Bar, 10 μm d ImageJ analysis of the length of neurofilament. Data shown are means ± SEM of three independent experiments in triplicates. Statistical analysis was performed by two-way ANOVA.**, p<3×10−6.
To confirm the neuroprotective activity, we examined the effects of RNASE4 and ANG on serum withdrawal-induced neuronal degeneration of P19-derived neurofilaments. As shown in Fig. 8c, retinoic acid-induced P19 neurofilaments underwent fragmentation when subjected to serum starvation in the presence of BSA. However, no significant fragmentation was observed with the same treatment but in the presence of RNASE4 or ANG. ImageJ analyses indicate that the length of P19 cell-derived neurofilaments after 1 h serum starvation in the presence of BSA, RNASE4, and ANG was 4.2 ± 0.4, 9.7 ± 0.7, and 12.6 ± 1.3 μm, respectively. It is interesting to note that the protective activity of ANG seems to be significantly higher than that of RNASE4 (p<0.05), but it is clear that both RNASE4 and ANG protect serum withdrawal-induced fragmentation of P19-derived neurofilaments. RNASE4 had no effect on PA6 cell proliferation (Supplementary Fig. S6). Thus, the protective activity of RNASE4 is likely brought about by a direct action toward neurons.
RNASE4 undergoes nuclear translocation in HUVEC and P19 cells
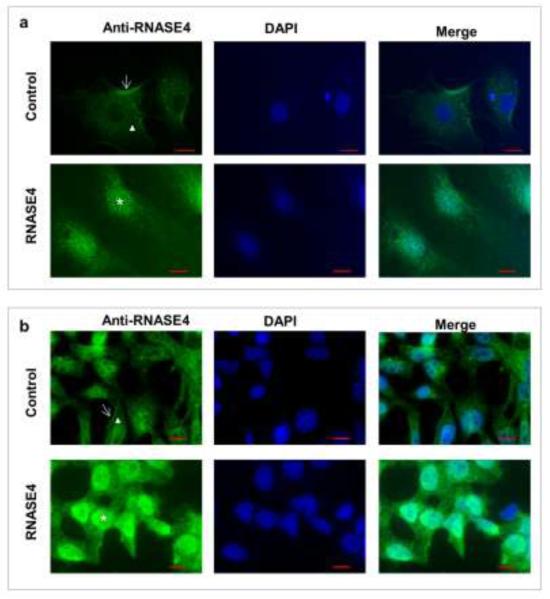
In order to gain mechanistic insights of the biological activity of RNASE4, we examined subcellular localization of RNASE4 with an affinity purified polyclonal antibody. Fig. 9 shows that endogenous RNASE4 is detectable on cell surface (indicated with arrows) and cytoplasm (indicated with triangles) of both HUVEC (Fig. 9a) and P19 cells (Fig. 9b). When exogenous RNASE4 was added to the cells, they were predominately accumulated in the nuclei (indicated with stars) of both cell lines. These results indicate that RNASE4 undergoes nuclear translocation in a similar manner as does ANG. Nuclear translocation of ANG is essential for its angiogenic [51], proliferative [52], and neuroprotective [26] activities. Therefore, although we do not know at present clearly whether nuclear translocation of RNASE4 is essential for its biological activity, it is certainly a plausible mechanism and a future subject of research.
Fig. 9.
Subcellular localization of RNASE4 in HUVE and P19 cells. a HUVE cells were incubated in the absence (top panel) or presence (bottom panel) of 0.5 μg/ml exogenous RNASE4 protein at 37 °C for 1 h. b P19 cells were incubated in the absence (top panel) or presence (bottom panel) of 1 μg/ml exogenous RNASE4 protein at 37 °C for 1 h. Immunofluorescence was carried out with affinity-purified RNASE4 polyclonal rabbit IgG and Alexa 488-labeld goat anti-rabbit IgG. Nuclei were stained with DAPI. Scale bar, 10 μm.
RNASE4 significantly slows down weight loss of SOD1G93A mice and slightly enhances their neuromuscular function
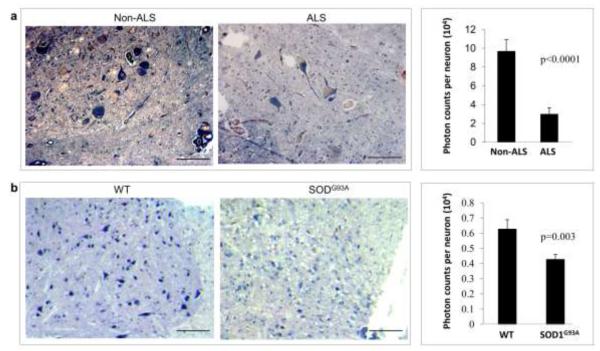
ANG has been shown to promote motor neuron survival and is beneficial to SOD1G93A mice [27]. These mice develop ALS like symptoms as a result of transgenic expression of human G93A mutated SOD1 gene [53]. Systemic delivery of ANG protein (1 g per mouse per day, ip) after disease onset enhanced muscle strength of SOD1G93A mice and prolonged their survival by 12 days [27]. The findings that a SNP on RNASE4 gene might be associated with ALS and that RNASE4 and ANG have similar angiogenic, neurogenic, and neuroprotective activities prompted us to examine the beneficial effect of exogenous RNASE4 on SOD1G93A mice. First, we used ISH to examine the mRNA level of RNASE4 in ALS and control spinal cords. Strong staining of RNASE4 mRNA in motor neurons was observed in non-ALS spinal cord, which was significantly decreased in ALS spinal cords (Fig. 10a). Quantitative analysis of the ISH images indicates that the total photon counts of RNASE4 mRNA staining per motor neurons of ALS and non-ALS spinal cord were 2.98 ± 0.71 × 104 and 9.67 ± 1.21 × 104, respectively, indicating that RNASE4 expression decreased by 69% in ALS. Similarly, the mRNA level of mouse Rnase4 is also decreased in the spinal cord motor neurons of SOD1G93A mice as compared to that of WT mice (Fig. 10b). The photon counts per spinal cord motor neuron in WT and SOD1G93A mice were 6.26 ± 0.62 × 103 and 4.27 ± 0.34 × 103, respectively, representing a 32% decrease in Rnase4 expression in the spinal code motor neurons of SOD1G93A mice.
Fig. 10.
Decreased RNASE4 mRNA level in the spinal cord motor neurons of ALS patients and in SOD1G93A mice. a In situ hybridization of human RNASE4 mRNA in the spinal cord of ALS patients and non-ALS control subject. Left panel, representative ISH images from one of the six patients. Bar, 10 μm. Right panel, ImageJ analysis of photon counts per motor neuron. Data shown are means ± SEM from six patients. Statistical analysis was performed by two-way ANOVA. b In situ hybridization of mouse Rnase4 mRNA in the spinal cord of WT and SOD1G93A mice. Left panel, representative ISH images from one of the six mice. Bar, 10 μm. Right panel, ImageJ analysis of photon counts per motor neuron. Data shown are means ± SEM from six patients. Statistical analysis was performed by two-way ANOVA.
The reduction of RNASE4 expression in the spinal cord of human ALS patients and SOD1G93A mice offered an additional rationale for testing the potential therapeutic activity of RNASE4 protein in ALS treatment. SOD1G93A mice, at 11 weeks of age, were treated with weekly i.p. injection of PBS or RNASE4 protein at 10 μg per mouse. The neuromuscular function of the mice was examined by their performance in a rota-rod assay. As shown in Fig. 11a, weekly treatment of RNASE4 increased the time the mice could stay on a rotarod (20 rpm) from week 14 to week 18 to various degrees. The greatest difference was recorded at week 15, when the RNASE4-treated mice were able to stay on the rotarod for 333 ± 106 sec, whereas PBS-treated animals were able to stay on it for only 181 ± 68 sec. Although the difference did not reach statistical significance due to large inter-animal variance, the trend that RNASE4 treatment enhanced rotarod performance appears clear. RNASE4 treatment also decreased the rate of body weight decrease (Fig. 11b). In the early phase of the treatment (week 12 to 14), a slight increase in body weight in both control and treatment groups was observed. The average body weight of the PBS and RNASE4 groups at week 12 was essentially the same at 23.45 ± 0.70 and 23.53 ± 0.59 g, respectively. At week 15, when no weight loss was observed, the difference in body weight was already obvious at 23.87 ± 0.73 g for the PBS group and 24.24 ± 0.63 g for RNASE4 group, representing a 1.5% difference. This difference increased as the disease progressed and the animals started to lose weight. At week 17, the body weight of PBS and RNASE4 groups was 21.87 ± 0.76 and 22.93 ± 0.69 g, respectively, representing a difference of 4.6%. At weeks 20, and 21, the difference in body weight in the control and treatment group was 14.3 and 16.8%, with the p values of both being 0.007. These findings demonstrate that RNASE4 treatment has a clear benefit in slowing down the loss of body weight. By contrast, however, RNASE4 provided no statistically significant benefit in survival. The average survival of the PBS and RNASE4 group was 136.5 ± 1.9 and 139.3 ± 1.9 days, respectively (Supplementary Fig. S7) with a p value of 0.288. It should be pointed out that SOD1G93A mice develop very aggressive disease and progress rapidly. A 3 day life extension in this animal model should not be neglected even though the p value is not statistically significant. For example, Riluzole [54], the only FDA-approved drug for ALS, and Dexpramipexole [55], an experimental ALS drug currently in Phase III clinical trial, only extended survival of these mice for 9 and 7 days, respectively.
Fig. 11.
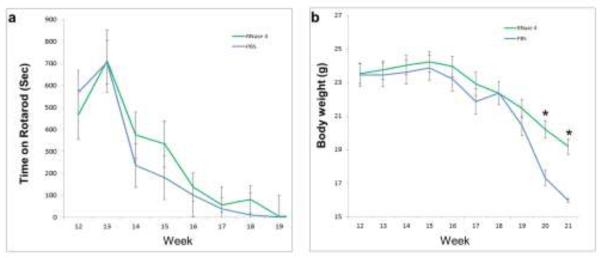
Effect of RNASE4 on SOD1G93A mice. Starting from 11 weeks of age, mice were treated with weekly i.p. injection of WT RNASE4 protein at 10 μg per mouse. Three independent experiments were performed with a total of 34 and 31 mice in the RNASE4 treatment and PBS control group, respectively. a Effect on rotarod performance at 20 rpm without revolving. Two successive measurements were recorded. An upper limit of 1,000 second was used. b Effect on body weight. Data shown are means ± SEM of all survived animals at each data point. Statistical analysis was performed by two-way ANOVA.*, p<0.01.
Discussion
The finding that RNSASE4 possesses angiogenic and neurogenic activity is significant and may have profound implications for neurodegenerative diseases, especially in ALS. Angiogenesis and neurogenesis are closely associated both anatomically and morphologically [56]. The nervous and vascular systems are developed in parallel and there is a high degree of cross-talk between them. Blood vessels and nerves are guided to their target and are often track alongside each other [57]. Vessels and nerves share many common principles and signals for pattern formation and survival. Molecules and signals that regulate neurogenesis have been found also regulate angiogenesis. For example, the semaphorin-plexin system has an essential role in both angiogenesis and neurogenesis [58]. Angiogenic factors, such as VEGF and ANG, have also been shown to play an important role in neurogenesis and in protecting stress-induced neuronal degeneration [59]. A defect in the expression and/or activity of angiogenic factors, such as loss of hypoxia regulation of VEGF expression [60] and missense mutations in the coding region of ANG [25], have been shown to be a causative or risk factor of ALS. Protein therapy with either recombinant VEGF [61] or ANG [27] was beneficial to SOD1G93A mice. Our results suggest that RNASE4 might be another angiogenic factor involved in ALS pathogenesis.
Our data show unequivocally that RNASE4 is angiogenic as shown by three different angiogenesis assays (Fig. 2-4). This finding can also be readily reproduced by different operators in different laboratories. Although this is an unexpected finding, as a prior study suggested RNASE4 is not angiogenic based on a CAM assay [11], this finding is not unreasonable as both RNASE4 and ANG share the same promoters and are often co-expressed and co-regulated (Supplementary Fig. S2 and S3). One possible explanation for absence of angiogenic activity of RNASE4 in the early CAM-based report could be that chicken do not have an RNase4 [2] so that mammalian RNASE4 does not induce neovessel growth. To demonstrate that the observed angiogenic activity of RNASE4 is not a general property of RNASEs, we examined the angiogenic activity of the other members of this superfamily and found that RNase A/RNASE1, EDN/RNASE2, and ECP/RNASE3 are not angiogenic in the in vitro endothelial cell tubule formation assay (Supplementary Fig. S5). The angiogenic activity of RNASES 6-13 is unknown at present.
Another unexpected finding is that the K40A mutation, which decreased the catalytic RNASE activity by 20-fold (Fig. 1), did not inhibit the angiogenic activity of RNASE4. By contrast, the angiogenic activity of the K40A variant is higher than the WT RNASE4 in all three angiogenesis assays employed in this study. It is notable that the ribonucleolytic activity of K40A RNASE4 variant, albeit 20-fold lower than that of WT RNASE4, is still higher than that of ANG toward the yeast tRNA substrate. The low ribonucleolytic activity of ANG is known to be essential for its biological activity in both angiogenesis [35, 62] and neurogenesis [50]. DEPC-treatment completely abolished both ribonucleolytic and angiogenic activity of RNASE4. Taken together, these results suggest that RNASE4 does not require a full enzymatic activity to induce angiogenesis but a certain degree of the enzymatic activity is essential.
The mechanism by which RNASE4 induces angiogenesis is not yet characterized. However, we found that RNASE4, like ANG, undergoes nuclear translocation in both HUVEC and P19 cells. The angiogenic activity of ANG is related to its ability to undergo nuclear translocation [51] and to stimulate rRNA transcription [15]. It is therefore plausible that RNASE4 may exert its biological activity also via a yet to be determined nuclear action. But RNASE4 did not stimulate rRNA transcription [13] and tiRNA production [17] under the same conditions under which ANG did. The identity of ANG receptor on the cell surface has not been fully characterized yet [63]. However, RNASE4 and ANG seem to bind at different sites or to different molecules although both undergo nuclear translocation (Fig. 9). RNASE4, at 10-fold higher concentration, did not inhibit nuclear translocation of ANG (data not shown), suggesting that binding of ANG to its cell surface receptor was not inhibited by RNASE4. Consistently, the dominant negative ANG variant H13A that inhibits WT ANG [62] did not inhibit the angiogenic activity of RNASE4 (data not shown). These results suggest that RNASE4 and ANG stimulate angiogenesis by different mechanisms. It is conceivable that the two ribonucleases, which are co-regulated and co-expressed from the same promoters, both have angiogenic activity but act through a different mechanism so that the activity is not entirely redundant so as to ensure adequate angiogenesis when needed.
Formation of embryonal bodies, or neurospheres, is an important step in neurogenesis [44]. After forming a neurosphere, neural progenitor cells differentiate and spread out into a specific lineage [64]. The finding that RNASE4 and ANG induce neural differentiation of both P19 and ES cells (Fig. 6) indicates that these two proteins are neurogenic. The mode of the neurogenic action of both RNASE4 and ANG is unknown at present. It is also unknown whether RNASE4 and ANG commit the neurosphere into a lineage-specific differentiation. However, we have found that, similar as ANG, RNASE4 induces neurite outgrowth of P19 cells (Fig. 6c and 6d), which are positive for motor neuron markers Islet1 and Peripherin [50]. ANG and RNASE4 also induce mouse ES cells to differentiate into GFAP-positive neuroprogenitor cells (Fig 5e and 6f). The effect of RNASE4 and ANG on neurite outgrowth appears to be a direct action on neurons rather than to be an indirect effect through the supporter cells. PA6 supporter cells are bone marrow-derived stromal cells that have biochemically-uncharacterized activities to induce ES cell differentiation [65]. Both RNASE4 and ANG did not stimulate PA6 cell proliferation (Supplementary Fig. S6). Consistent with the hypothesis that RNASE4 and ANG act on neurons directly, we found that both proteins stimulated neurofilament outgrowth of mouse embryonic cortical neurons cultured in a PA6-free system (Fig. 7). Notable, this is also the first report that ANG is able to stimulate P19 cells to form embryonal bodies and to stimulate neurofilament growth of mouse embryonic cortical neurons.
The neuroprotective activity of RNASE4 is similar to that of ANG in preventing hypothermia-induced fragmentation of neurofilaments derived from mouse embryonic cortical neurons (Fig. 8). RNASE4 also has a significant protective activity against serum starvation-induced degeneration of P19 cell-derived neurites, even though its activity is not as robust as ANG (Fig. 8). ANG has been shown to protect serum withdrawal-induced apoptosis of P19 cells through upregulation of Bcl-2 [47] and inhibition of nuclear translocation of apoptosis-inducing factor [66]. At this time, we do not know whether RNASE4 is also anti-apoptotic and whether the neuroprotective activity of RNASE4 and ANG is connected to a potential antiapoptotic activity, and if it is, whether these mechanisms are also Bcl-2-dependent.
There was a strong rationale to test the therapeutic activity of RNASE4 in ALS. First, the T allele of SNP rs3748338 is enriched in ALS patients (Table 1). The A and T allele encodes for Thr and Ser, respectively, at position −13 at the signal peptide region (Table 1 and Supplementary Fig. S1 and S4). We have not yet determined whether Thr to Ser change will affect the processing of the signal peptide or the secretion of the mature protein. A similar polymorphism on SNP rs11701 of ANG gene was reported [67] and was the origin of many subsequent studies that finally demonstrated that ANG is an ALS gene and that ANG protein therapy is beneficial to SOD1G93A mice [25-28, 48, 50]. Therefore, there is genetic evidence that RNASE4 may be relevant to ALS. A similar angiogenic, neurogenic, and neuroprotective activity of RNASE4 to that of ANG provide biochemical rationale that RNASE4 may be beneficial for motor neuron survival. Moreover, there is pathological evidence that RNASE4 expression is decreased in ALS spinal code of both human (Fig. 10a) and mice (Fig. 10b). As predicted, treatment of RNASE4 did enhance the neuromuscular function of SOD1G93A mice as assessed by their performance on a rotarod (Fig. 11a). RNASE4 treatment also significantly slowed down the rate of weight loss (Fig. 11b). We recognized that despite the beneficial influence of RNASE4 on weight loss and rotarod function, it only improve survival in the SOD1G93A mice for 3 days under the current treatment regimen of weekly i.p. injections at 10 μg per mouse. It would be of interest to see whether the therapeutic efficacy could be improved by increasing the frequency of injection or by administering the protein directly into the spinal cord by an osmotic pump. It will also be of interest to examine the potential effect of K40A variant that has an enhanced angiogenic activity. The therapeutic activity of a mixture of RNASE4 and ANG should also be considered and tested in future studies.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Health grant R01 NS 065237 (to GFH). GFH receives fund from NIH, ALS Therapy Alliance, and Harvard University Technology Development Accelerator Fund, Massachusetts Alzheimer Disease Research Center, and Tufts University CTSI program. RHB receives support from the National Institute for Neurological Disease and Stroke, the Angel Fund, the ALS Association, Project ALS, P2ALS. Pierre L. de Bourgknecht ALS Research Foundation, the Al-Athel ALS Foundation, and the ALS Therapy Alliance. We thank Robert Shapiro (Harvard Medical School) for helpful discussions on biochemical characterization of RNASE4, Helene F. Rosenberg (NIH/NIAID) for providing mouse Rnase4 expression plasmid, Jan Hofsteenge (Friedrich Miescher Institute, Switzerland) for providing pig RNASE4 expression plasmid.
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Cho S, Beintema JJ, Zhang J. The ribonuclease A superfamily of mammals and birds: identifying new members and tracing evolutionary histories. Genomics. 2005;85:208–220. doi: 10.1016/j.ygeno.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Cho S, Zhang J. Zebrafish ribonucleases are bactericidal: implications for the origin of the vertebrate RNase A superfamily. Mol Biol Evol. 2007;24:1259–1268. doi: 10.1093/molbev/msm047. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg HF, Tenen DG, Ackerman SJ. Molecular cloning of the human eosinophil-derived neurotoxin: a member of the ribonuclease gene family. Proc Natl Acad Sci U S A. 1989;86:4460–4464. doi: 10.1073/pnas.86.12.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker RL, Loegering DA, Ten RM, Hamann KJ, Pease LR, Gleich GJ. Eosinophil cationic protein cDNA. Comparison with other toxic cationic proteins and ribonucleases. J Immunol. 1989;143:952–955. [PubMed] [Google Scholar]
- 6.Fredens K, Dahl R, Venge P. The Gordon phenomenon induced by the eosinophil cationic protein and eosinophil protein X. J Allergy Clin Immunol. 1982;70:361–366. doi: 10.1016/0091-6749(82)90025-2. [DOI] [PubMed] [Google Scholar]
- 7.Yang D, Chen Q, Rosenberg HF, Rybak SM, Newton DL, Wang ZY, Fu Q, Tchernev VT, Wang M, Schweitzer B, et al. Human ribonuclease A superfamily members, eosinophil-derived neurotoxin and pancreatic ribonuclease, induce dendritic cell maturation and activation. J Immunol. 2004;173:6134–6142. doi: 10.4049/jimmunol.173.10.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamann KJ, Barker RL, Loegering DA, Gleich GJ. Comparative toxicity of purified human eosinophil granule proteins for newborn larvae of Trichinella spiralis. J Parasitol. 1987;73:523–529. [PubMed] [Google Scholar]
- 9.Lehrer RI, Szklarek D, Barton A, Ganz T, Hamann KJ, Gleich GJ. Antibacterial properties of eosinophil major basic protein and eosinophil cationic protein. J Immunol. 1989;142:4428–4434. [PubMed] [Google Scholar]
- 10.Rosenberg HF, Domachowske JB. Eosinophils, eosinophil ribonucleases, and their role in host defense against respiratory virus pathogens. J Leukoc Biol. 2001;70:691–698. [PubMed] [Google Scholar]
- 11.Fett JW, Strydom DJ, Lobb RR, Alderman EM, Bethune JL, Riordan JF, Vallee BL. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry. 1985;24:5480–5486. doi: 10.1021/bi00341a030. [DOI] [PubMed] [Google Scholar]
- 12.Li S, Hu GF. Angiogenin-mediated rRNA transcription in cancer and neurodegeneration. Int J Biochem Mol Biol. 2010;1:26–35. [PMC free article] [PubMed] [Google Scholar]
- 13.Xu ZP, Tsuji T, Riordan JF, Hu GF. The nuclear function of angiogenin in endothelial cells is related to rRNA production. Biochem Biophys Res Commun. 2002;294:287–292. doi: 10.1016/S0006-291X(02)00479-5. [DOI] [PubMed] [Google Scholar]
- 14.Xu ZP, Tsuji T, Riordan JF, Hu GF. Identification and characterization of an angiogenin-binding DNA sequence that stimulates luciferase reporter gene expression. Biochemistry. 2003;42:121–128. doi: 10.1021/bi020465x. [DOI] [PubMed] [Google Scholar]
- 15.Yoshioka N, Wang L, Kishimoto K, Tsuji T, Hu GF. A therapeutic target for prostate cancer based on angiogenin-stimulated angiogenesis and cancer cell proliferation. Proc Natl Acad Sci USA. 2006;103:14519–14524. doi: 10.1073/pnas.0606708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-Induced tRNA Fragments Inhibit Translation Initiation. Mol Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofsteenge J, Vicentini A, Zelenko O. Ribonuclease 4, an evolutionarily highly conserved member of the superfamily. Cell Mol Life Sci. 1998;54:804–810. doi: 10.1007/s000180050209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shapiro R, Fett JW, Strydom DJ, Vallee BL. Isolation and characterization of a human colon carcinoma-secreted enzyme with pancreatic ribonuclease-like activity. Biochemistry. 1986;25:7255–7264. doi: 10.1021/bi00371a002. [DOI] [PubMed] [Google Scholar]
- 22.Dyer KD, Rosenberg HF. The mouse RNase 4 and RNase 5/ang 1 locus utilizes dual promoters for tissue-specific expression. Nucleic Acids Res. 2005;33:1077–1086. doi: 10.1093/nar/gki250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Futami J, Tsushima Y, Murato Y, Tada H, Sasaki J, Seno M, Yamada H. Tissue-specific expression of pancreatic-type RNases and RNase inhibitor in humans. DNA Cell Biol. 1997;16:413–419. doi: 10.1089/dna.1997.16.413. [DOI] [PubMed] [Google Scholar]
- 24.Strydom DJ. The angiogenins. Cell Mol Life Sci. 1998;54:811–824. doi: 10.1007/s000180050210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, Patterson V, Swingler R, Kieran D, Prehn J, et al. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- 26.Wu D, Yu W, Kishikawa H, Folkerth RD, Iafrate AJ, Shen Y, Xin W, Sims K, Hu GF. Angiogenin loss-of-function mutations in amyotrophic lateral sclerosis. Ann Neurol. 2007;62:609–617. doi: 10.1002/ana.21221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kieran D, Sebastia J, Greenway MJ, King MA, Connaughton D, Concannon CG, Fenner B, Hardiman O, Prehn JH. Control of motoneuron survival by angiogenin. J Neurosci. 2008;28:14056–14061. doi: 10.1523/JNEUROSCI.3399-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian V, Feng Y. A new role for angiogenin in neurite growth and pathfinding: implications for amyotrophic lateral sclerosis. Hum Mol Genet. 2007;16:1445–1453. doi: 10.1093/hmg/ddm095. [DOI] [PubMed] [Google Scholar]
- 29.Baumer D, Ansorge O, Almeida M, Talbot K. The role of RNA processing in the pathogenesis of motor neuron degeneration. Expert Rev Mol Med. 2010;12:e21. doi: 10.1017/S1462399410001523. [DOI] [PubMed] [Google Scholar]
- 30.van Blitterswijk M, Landers JE. RNA processing pathways in amyotrophic lateral sclerosis. Neurogenetics. 2010;11:275–290. doi: 10.1007/s10048-010-0239-4. [DOI] [PubMed] [Google Scholar]
- 31.Strong MJ. The evidence for altered RNA metabolism in amyotrophic lateral sclerosis (ALS) J Neurol Sci. 2010;288:1–12. doi: 10.1016/j.jns.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 32.Holloway DE, Hares MC, Shapiro R, Subramanian V, Acharya KR. High-level expression of three members of the murine angiogenin family in Escherichia coli and purification of the recombinant proteins. Protein Expr Purif. 2001;22:307–317. doi: 10.1006/prep.2001.1434. [DOI] [PubMed] [Google Scholar]
- 33.Shapiro R, Weremowicz S, Riordan JF, Vallee BL. Ribonucleolytic activity of angiogenin: essential histidine, lysine, and arginine residues. Proc Natl Acad Sci U S A. 1987;84:8783–8787. doi: 10.1073/pnas.84.24.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bibel M, Richter J, Lacroix E, Barde YA. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat Protoc. 2007;2:1034–1043. doi: 10.1038/nprot.2007.147. [DOI] [PubMed] [Google Scholar]
- 35.Shapiro R, Fox EA, Riordan JF. Role of lysines in human angiogenin: chemical modification and site-directed mutagenesis. Biochemistry. 1989;28:1726–1732. doi: 10.1021/bi00430a045. [DOI] [PubMed] [Google Scholar]
- 36.Messmore JM, Fuchs DN, Raines RT. Ribonuclease a: revealing structure-function relationships with semisynthesis. J Am Chem Soc. 1995;117:8057–8060. doi: 10.1021/ja00136a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Liddo R, Dalzoppo D, Baiguera S, Conconi MT, Dettin M, Parnigotto PP, Grandi C. In vitro biological activity of bovine milk ribonuclease-4. Mol Med Report. 2010;3:127–132. doi: 10.3892/mmr_00000229. [DOI] [PubMed] [Google Scholar]
- 38.Donovan D, Brown NJ, Bishop ET, Lewis CE. Comparison of three in vitro human ‘angiogenesis’ assays with capillaries formed in vivo. Angiogenesis. 2001;4:113–121. doi: 10.1023/a:1012218401036. [DOI] [PubMed] [Google Scholar]
- 39.Masson VV, Devy L, Grignet-Debrus C, Bernt S, Bajou K, Blacher S, Roland G, Chang Y, Fong T, Carmeliet P, et al. Mouse Aortic Ring Assay: A New Approach of the Molecular Genetics of Angiogenesis. Biol Proced Online. 2002;4:24–31. doi: 10.1251/bpo30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akhtar N, Dickerson EB, Auerbach R. The sponge/Matrigel angiogenesis assay. Angiogenesis. 2002;5:75–80. doi: 10.1023/a:1021507031486. [DOI] [PubMed] [Google Scholar]
- 41.Bain G, Ray WJ, Yao M, Gottlieb DI. From embryonal carcinoma cells to neurons: the P19 pathway. Bioessays. 1994;16:343–348. doi: 10.1002/bies.950160509. [DOI] [PubMed] [Google Scholar]
- 42.McBurney MW, Rogers BJ. Isolation of male embryonal carcinoma cells and their chromosome replication patterns. Dev Biol. 1982;89:503–508. doi: 10.1016/0012-1606(82)90338-4. [DOI] [PubMed] [Google Scholar]
- 43.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 44.Kurosawa H. Methods for inducing embryoid body formation: in vitro differentiation system of embryonic stem cells. J Biosci Bioeng. 2007;103:389–398. doi: 10.1263/jbb.103.389. [DOI] [PubMed] [Google Scholar]
- 45.Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- 46.Cho GW, Kang BY, Kim SH. Human angiogenin presents neuroprotective and migration effects in neuroblastoma cells. Mol Cell Biochem. 2010;340:133–141. doi: 10.1007/s11010-010-0410-0. [DOI] [PubMed] [Google Scholar]
- 47.Li S, Yu W, Kishikawa H, Hu GF. Angiogenin prevents serum withdrawal-induced apoptosis of P19 embryonal carcinoma cells. FEBS J. 2010;277:3575–3587. doi: 10.1111/j.1742-4658.2010.07766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sebastia J, Kieran D, Breen B, King MA, Netteland DF, Joyce D, Fitzpatrick SF, Taylor CT, Prehn JH. Angiogenin protects motoneurons against hypoxic injury. Cell Death Differ. 2009;16:1238–1247. doi: 10.1038/cdd.2009.52. [DOI] [PubMed] [Google Scholar]
- 49.Steidinger TU, Standaert DG, Yacoubian TA. A neuroprotective role for angiogenin in models of Parkinson’s disease. J Neurochem. 2011;116:334–341. doi: 10.1111/j.1471-4159.2010.07112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subramanian V, Crabtree B, Acharya KR. Human angiogenin is a neuroprotective factor and amyotrophic lateral sclerosis associated angiogenin variants affect neurite extension/pathfinding and survival of motor neurons. Hum Mol Genet. 2008;17:130–149. doi: 10.1093/hmg/ddm290. [DOI] [PubMed] [Google Scholar]
- 51.Moroianu J, Riordan JF. Nuclear translocation of angiogenin in proliferating endothelial cells is essential to its angiogenic activity. Proc Natl Acad Sci U S A. 1994;91:1677–1681. doi: 10.1073/pnas.91.5.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsuji T, Sun Y, Kishimoto K, Olson KA, Liu S, Hirukawa S, Hu GF. Angiogenin is translocated to the nucleus of HeLa cells and is involved in ribosomal RNA transcription and cell proliferation. Cancer Res. 2005;65:1352–1360. doi: 10.1158/0008-5472.CAN-04-2058. [DOI] [PubMed] [Google Scholar]
- 53.Tu PH, Raju P, Robinson KA, Gurney ME, Trojanowski JQ, Lee VM. Transgenic mice carrying a human mutant superoxide dismutase transgene develop neuronal cytoskeletal pathology resembling human amyotrophic lateral sclerosis lesions. Proc Natl Acad Sci U S A. 1996;93:3155–3160. doi: 10.1073/pnas.93.7.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gurney ME, Cutting FB, Zhai P, Doble A, Taylor CP, Andrus PK, Hall ED. Benefit of vitamin E, riluzole, and gabapentin in a transgenic model of familial amyotrophic lateral sclerosis. Ann Neurol. 1996;39:147–157. doi: 10.1002/ana.410390203. [DOI] [PubMed] [Google Scholar]
- 55.Danzeisen R, Schwalenstoecker B, Gillardon F, Buerger E, Krzykalla V, Klinder K, Schild L, Hengerer B, Ludolph AC, Dorner-Ciossek C, et al. Targeted antioxidative and neuroprotective properties of the dopamine agonist pramipexole and its nondopaminergic enantiomer SND919CL2x [(+)2-amino-4,5,6,7-tetrahydro-6-Lpropylamino-benzathiazole dihydrochloride] J Pharmacol Exp Ther. 2006;316:189–199. doi: 10.1124/jpet.105.092312. [DOI] [PubMed] [Google Scholar]
- 56.Carmeliet P. Neuro-vascular link: from genetic insights to therapeutic perspectives. Bull Mem Acad R Med Belg. 2008;163:445–451. discussion 451-442. [PubMed] [Google Scholar]
- 57.Segura I, De Smet F, Hohensinner PJ, Ruiz de Almodovar C, Carmeliet P. The neurovascular link in health and disease: an update. Trends Mol Med. 2009;15:439–451. doi: 10.1016/j.molmed.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 58.Tran TS, Kolodkin AL, Bharadwaj R. Semaphorin regulation of cellular morphology. Annu Rev Cell Dev Biol. 2007;23:263–292. doi: 10.1146/annurev.cellbio.22.010605.093554. [DOI] [PubMed] [Google Scholar]
- 59.Lambrechts D, Lafuste P, Carmeliet P, Conway EM. Another angiogenic gene linked to amyotrophic lateral sclerosis. Trends Mol Med. 2006;12:345–347. doi: 10.1016/j.molmed.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 60.Oosthuyse B, Moons L, Storkebaum E, Beck H, Nuyens D, Brusselmans K, Van Dorpe J, Hellings P, Gorselink M, Heymans S, et al. Deletion of the hypoxia-response element in the vascular endothelial growth factor promoter causes motor neuron degeneration. Nat Genet. 2001;28:131–138. doi: 10.1038/88842. [DOI] [PubMed] [Google Scholar]
- 61.Zheng C, Nennesmo I, Fadeel B, Henter JI. Vascular endothelial growth factor prolongs survival in a transgenic mouse model of ALS. Ann Neurol. 2004;56:564–567. doi: 10.1002/ana.20223. [DOI] [PubMed] [Google Scholar]
- 62.Shapiro R, Vallee BL. Site-directed mutagenesis of histidine-13 and histidine-114 of human angiogenin. Alanine derivatives inhibit angiogenin-induced angiogenesis. Biochemistry. 1989;28:7401–7408. doi: 10.1021/bi00444a038. [DOI] [PubMed] [Google Scholar]
- 63.Hu GF, Riordan JF, Vallee BL. A putative angiogenin receptor in angiogenin-responsive human endothelial cells. Proc Natl Acad Sci U S A. 1997;94:2204–2209. doi: 10.1073/pnas.94.6.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bratt-Leal AM, Carpenedo RL, McDevitt TC. Engineering the embryoid body microenvironment to direct embryonic stem cell differentiation. Biotechnol Prog. 2009;25:43–51. doi: 10.1002/btpr.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kawasaki H, Mizuseki K, Nishikawa S, Kaneko S, Kuwana Y, Nakanishi S, Nishikawa SI, Sasai Y. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron. 2000;28:31–40. doi: 10.1016/s0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 66.Li S, Yu W, Hu GF. Angiogenin inhibits nuclear translocation of apoptosis inducing factor in a Bcl-2-dependent manner. J Cell Physiol. 2011 doi: 10.1002/jcp.22881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greenway MJ, Alexander MD, Ennis S, Traynor BJ, Corr B, Frost E, Green A, Hardiman O. A novel candidate region for ALS on chromosome 14q11.2. Neurology. 2004;63:1936–1938. doi: 10.1212/01.wnl.0000144344.39103.f6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.