SUMMARY
ATP-dependent NuRD repressor complexes involve combinatorial assembly of its subunits. However, the mechanism of gene transcription by MTA1/NuRD remains enigmatic. Here we report that MTA1 methylation by G9a methytransferase and demethylation by LSD1 determines the nucleosome remodeling and transcriptional outcome. Contrary to the current static repressor model of the NuRD complex, we discovered that MTA1 association with nucleosomes and co-repressor/ co-activator complexes is dynamic. While methylated MTA1 is required for the NuRD repressor complex, demethylated MTA1 recognizes the bivalent histone H3K4-AcK9 mark and recruits co-activator NURF-trithorax remodeling complex in a signaling-dependent manner. MTA1’s lysine 532 methylation represents a molecular switch as methylated and de-methylated MTA1 nucleate NuRD or NURF complexes with opposite functions in a cyclical manner. In addition, MTA1 possesses an inherent histone amplifier activity with an instructive role in impacting the epigenetic landscape, providing a new perspective to the molecular governance of dual co-regulator functions of a master coregulator.
Introduction
Multicellular organisms must regulate the process of signaling-dependent transcription precisely to ensure a balanced gene expression. Gene stimulation or repression is intimately influenced by dynamic remodeling of chromatin (Perissi et al, 2010). Chromatin remodeling complexes (CRCs) are unique macromolecular machineries that utilize energy from ATP-hydrolysis to disrupt nucleosome-DNA interface for creating access points for coregulator proteins to influence the process of transcription. The nucleosome remodeling and histone deacetylase (NuRD) complex is one of the four major families of CRCs that comprise of evolutionarily conserved subunit members (Li et al., 2012). Metastatic tumor antigen 1 (MTA1), a core-subunit of the NuRD complex, is the only dual coregulator with an expected corepressor activity but unusual ability to stimulate transcription (Manavathi and Kumar, 2007; Li et al., 2012). Since MTA1 is a “hub” gene and a global genetic modifier (Lehner B et al., 2006; Ghanta et al., 2011) and the fact, genetic or siRNA-mediated depletion of MTA1 in mammalian cells is accompanied by a genome-wide stimulation of a large number of transcripts (Ghanta et al., 2011), underscoring the physiologic significance of a distinctive coactivator function of MTA1. These observations pose a continuing paradox in the field as MTA1/NuRD complex was originally thought to be static and corepressive in nature. Despite the remarkable growth of new information on the biochemistry and biology of MTAs since its discovery and the plethora of information about the role of MTA/NuRD complex in the mammalian cells (Li et al., 2012), we still do not know the precise mechanism that could explain this paradox and the underlying physiologic switch that results in a loss of MTA1’s corepressor function, a prerequisite to exert its coactivator activity. Here we have attempted to answer these outstanding questions in the field and provide novel molecular insights of the dual functionality of coregulators in general.
Results
MTA1 methylation directs orderly formation of the NuRD complex
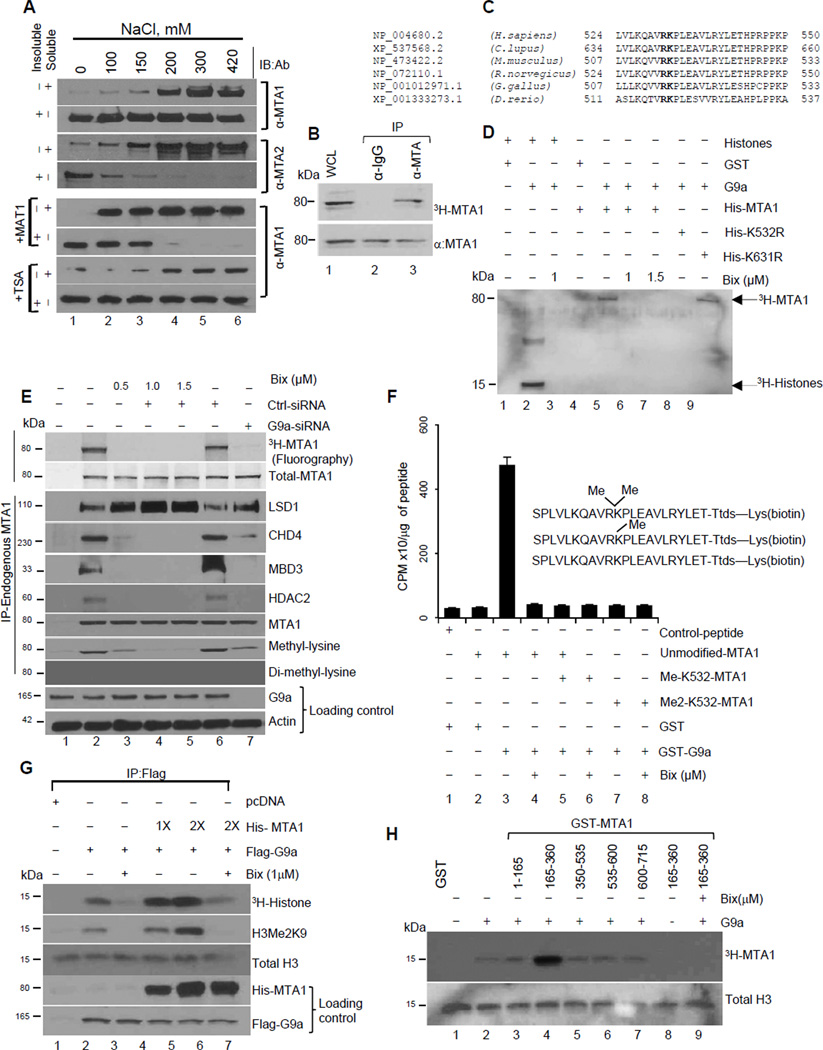
While evaluating the affinities of the NuRD subunits to interact with chromatin in the HeLa cells, we discovered that a large fraction of MTA1 remains bound to chromatin even under high salt extraction buffer (420 mM NaCl). In contrast, other components were extractable at much lower concentration of NaCl (Figures 1A and S1A). However, inclusion of the methylation inhibitor 5′-deoxy-5′-methylthioadenosine (MAT1) but not HDAC inhibitor Trichostatin-A (TSA) facilitated MTA1 solubilization from chromatin (Figure 1A), suggesting that the MTA1 subunit of the NuRD complex interacts with chromatin in a methylation-dependent manner. Inhouse, bioinformatics analysis predicted that MTA1 may be subject to methylation. Accordingly, pulse-chase methylation assay confirmed that endogenous MTA1 is methylated under physiological conditions (Figure 1B). Results from site-directed mutagenesis of potential lysine residues (K61, K71, K182, K532, K554, K626, and K631) to arginine indicated that lysine 532 is the only functional conserved lysine in MTA1 subjected to methylation (Figure S1B). Sequence similarity of lysine 532 residue with known recognized targets of eukaryotic methyltransferases (Rathert et al., 2008) revealed G9a as a potential methyltransferase for MTA1 (Figure 1C). In an in vitro methyltransferase assay, we confirmed that recombinant G9a efficiently methylates His-MTA1 but not His-MTA1-K532R or His-MTA1-K532A in a G9a specific inhibitor BIX01294-dependent manner (Figures 1D and S1C). Furthermore, analysis of the endogenous MTA1 methylation status confirmed that MTA1 methylation is regulated in vivo by G9a methyltransferase as treatment of the HeLa cells with G9a inhibitor Bix-01294 or G9a-specific siRNA effectively inhibited MTA1 methylation as detected by anti-methyl lysine or MTA1 methylation site specific antibody (MTA1- K532-Me) that recognizes mono-methylated MTA1. (Figure 1E and S1D&E). MTA1 being a member of the multisubunit NuRD complex, prompted us to investigate the impact of MTA1 methylation on its ability to interact with associated proteins by analyzing the status of proteins bound to methylated vs non-methylated MTA1. We observed the enrichment of the NuRD components (CHD4, HDAC2, MBD3) in complexes with methylated MTA1, while LSD1 specifically associates with demethylated MTA1 (Figure 1E). We found that MTA1 is mono-methylated by G9a (Figure 1F). Further inclusion of the G9a inhibitor as well as methylation deficient MTA1-K532R mutant significantly decreases the amount of salt-resistant MTA1 (Figure S1F), revealing that MTA1-K532 methylation by G9a is an important determinant of MTA1 chromatin affinity. In brief, MTA1 binds to chromatin in a methylation-dependent manner and represents a non-histone substrate of G9a.
Figure 1. MTA1 is a methylated component of the NuRD complex.
(A) Salt solubilization assay using nuclei from untreated, MAT1 or TSA treated the HeLa cells. Salt soluble and resistant fractions were analyzed using anti-MTA1 and MTA2 antibodies (see also Figure S1A for analysis using anti-CHD3, -CHD4, -MBD and -Rbp antibodies). See also Figure S1A
(B) HeLa cells were labeled with 50uCi of (L-Methyl-3H) methionine in presence of protein synthesis inhibitor and incubated for 5h at 37°C. Cell lysates (~1mg) was used for immunoprecipitation using IgG or anti-MTA1 antibody. Immunoprecipitated proteins were electrophoresed in 10% SDS-PAGE, transferred to nitrocellulose membrane and developed by fluorography. Western blot for Anti-MTA1 serves as loading control. See also Figure S1B
(C) Sequence alignment of MTA1 highlighting a conserved G9a methylation site in bold.
(D) In vitro methylation assay using GST-G9a and His-MTA1, -K532R, and –K631R and bulk histones, S-Adenosyl-L-(methyl-3H)-methionine. Reaction products were analyzed by SDSPAGE and fluorography. See also Figure S1C.
(E) Untreated (control), Bix-treated (0.5–1.5µM), control-siRNA and G9a- siRNA treated HeLa cells were labeled with L-Methyl-3H) methionine in presence of protein synthesis inhibitor and incubated for 5 hr at 37°C. Cell lysates (~1mg) was used for immunoprecipitation using IgG or anti-MTA1 antibody. Immunoprecipitated proteins were electrophoresed onto SDS-PAGE, transferred to nitrocellulose membrane and developed by fluorography (top panel) to confirm MTA1 methylation status. Western blot for MTA1 serves as loading control (Second panel from top). Lines, different gels from the same experiment.
(F) In vitro methylation using indicated peptide substrates and recombinant G9a. Reactions contained 50 mM glycine (pH 9.6), 2.0 µM [methyl- 3H] AdoMet, 0.50 µM G9a, and 60 µM peptide and were incubated at 37°C for 45 min. Reaction mixtures were precipitated using TCA and quantitated by liquid scintillation. Values are represented as tritium label (CPM)/µg of peptide. Error bars represent SD.
(G) In-vitro methylation assay using Flag-G9a enzyme immunoprecipitates, bulk histones and increasing concentrations of His-MTA1 (1×= 0.5ug). Reaction products were analyzed by fluorography and immunoblotting analysis for anti-H3Me2K9 and histone H3. 1/10th of reaction mixture was electrophoresed on a separate gel to confirm the presence of G9a and MTA1 using anti-Flag and anti-His antibodies.
(H) Recombinant G9a and GST-MTA1 domains were incubated in HMTase buffer containing 3H-labeled AdoMet and 5µg of histones. Reaction was carried out for 1 hr at 37°C and stopped using SDS-loading buffer and analyzed by fluorography. G9a inhibitor, Bix-01294 (1µM).
G9a-mediated methylation of MTA1 is essential for its corepressor activity
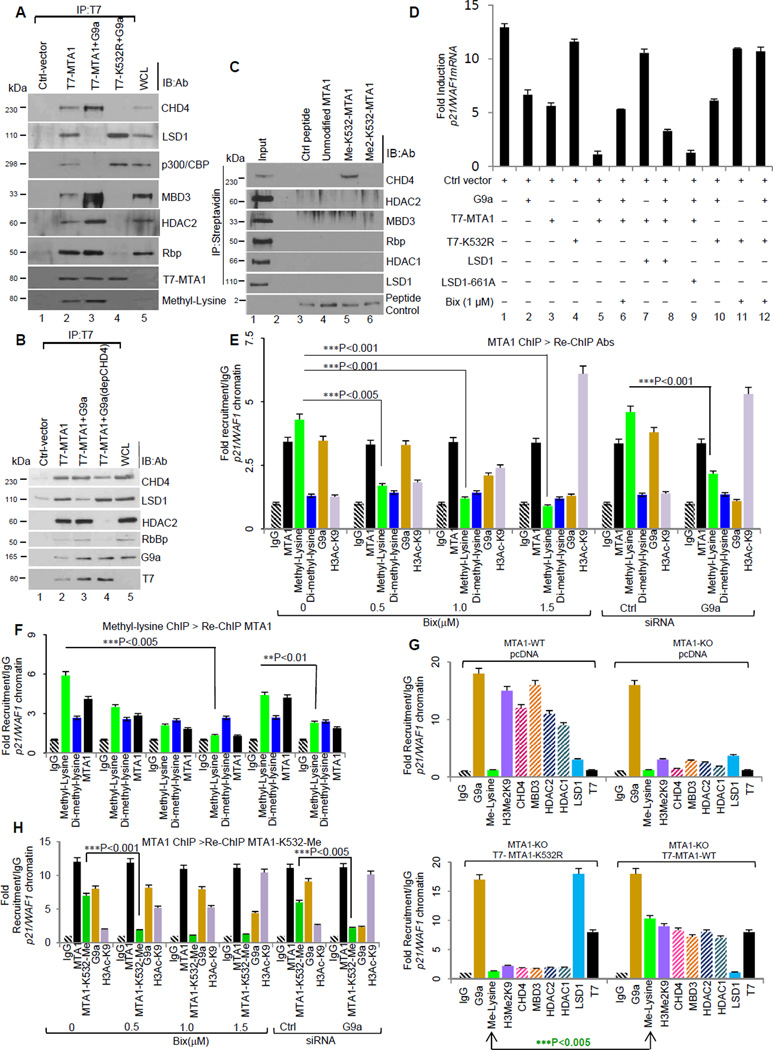
Recombinant G9a directly interacts with the amino acids 388–535 of MTA1 (Figure S2A). Functional interaction between the MTA1 and G9a resulted in a potentiating effect on G9a methyltransferase activity. Deletion analysis confirmed that potentiating effect was due to catalytic ELM2-SANT domain (aa 165–360) of MTA1 (Figures 1G and H). Accordingly, a subset of G9a-repressed genes exemplified by SCN1A, SERAC1 and IMAG2 (Collins et al., 2008) were significantly de-repressed in the absence of MTA1 (Figure S2B), suggesting that G9a-mediated methylation of MTA1 may be essential for the corepressor functions of MTA1 as well as of G9a. G9a methylated MTA1 but not MTA1-K532R interacts with CHD4 protein to form a functional NuRD complex (Figure 2A). Intriguingly, during the course of these studies, we consistently noticed the presence of LSD1 in the demethylated MTA1 complex, suggesting that methylation status dictates interaction with functionally distinct MTA1 interacting proteins (Figure 2A). Consistent with this notion, immunodepletion of CHD4 from HeLa cells nuclear lysates resulted in the dissociation of the NuRD complex components from the methylated MTA1 (Figure 2B), suggesting CHD4 preferentially recognizes methylated MTA1 to direct an orderly association of the NuRD components. Further results from streptavidin-biotin peptide pull down assay using biotin-tagged control, unmodified, or methylated MTA1 peptides (with mono and dimethyl-lysine substitution at K532) and HeLa cells’ nuclear extracts revealed that methylated lysine 532 of MTA1 serve as docking site for CHD4. We did not find other members of the NuRD complex in the peptide pull down assay (Figure 2C).
Figure 2. Methylated MTA1 directs functional association of the NuRD components.
(A) HeLa cells were transfected with T7-MTA1 or methylation-deficient T7-K532R and Flag- G9a. T-7 immunoprecipitates from the nuclear lysates were analyzed by immunoblotting using indicated antibodies. Methylation status of the precipitated MTA1 was determined using anti-methyl lysine antibody (Me-K) in an identical gel. WCL,lysates from transfected cells.
(B) Nuclear lysates from the HeLa cells transfected with T7-MTA1 and Flag-G9a were immunoprecipitated using T7-affinity resin. Immunoprecipitate material was analyzed by immunoblotting using indicated antibodies. WCL, lysates from untransfected HeLa cells. Lane 4, CHD4-immunodepleted nuclear lysates.
(C) Streptavidin-biotin peptide pull down assay was performed using biotin-tagged control, unmodified, or methylated MTA1 peptides (with mono and dimethyl-lysine substitution at K532) and HeLa cell nuclear extracts. Peptide bound proteins were eluted and analyzed on polyacrylamide gels using indicated antibodies. Lines, different gels from the same experiment.
(D) Total RNA was isolated from HeLa cells expressing T7-MTA1 or -MTA1-K532R and 1ug of RNA was reverse transcribed and used in a quantitative PCR reaction using specific primers for endogenous p21/WAF1. Data is presented as fold induction over 18 sRNA (internal control). Error bars represent SD. See also Figure S2C.
(E&F) Chromatin prepared from the HeLa cells were subjected to ChIP analysis at p21/WAF1 promoter, using anti-MTA1 (E) or ChIP analysis using anti-Methyl-lysine antibody (F) followed by re-ChIP using indicated antibodies.. Recruitment profiles were generated in quantitative-PCR assay specific primers encompassing MTA1 recruitment region on p21-WAF1 promoter. Values are represented as fold recruitment/IgG. **p<0.01,***p<0.001, ***p<0.005. Error bars represent SD.
(G) Quantitative ChIP relative to IgG control at p21/WAF1 promoter using indicated antibodies and chromatin from MTA1-WT and KO cells transfected with T7-MTA1 or -methylation deficient K532R. Methylation status of MTA1 at target chromatin is evaluated using anti-Methyl lysine antibody. ***p<0.005. Error bars represent SD.
(H) Chromatin prepared from the HeLa cells were subjected to ChIP analysis onto p21/WAF1 using MTA1-MeK532 antibody followed by Re-ChIP using indicated antibodies. Quantitative-PCR using specific primers encompassing MTA1 recruitment region on p21-WAF1 promoter were used to generate recruitment profiles. Values are represented as fold recruitment/IgG. ***p<0.001, ***p<0.005. Error bars represent SD.
Functional NuRD complex requires MTA1 methylation
Functional association of methylated MTA1 with the NuRD corepressor complex prompted us to study the impact of methylation on its corepressor activity. Co-expression of MTA1 and Flag- G9a in the HeLa cells resulted in a significant decrease in the levels of the endogenous p21/WAF1 transcripts (Figure 2D). Similar effects were also observed using a synthetic p21/WAF1-LUC promoter reporter plasmid (Li et al., 2010) in a transfection based luciferase assay (Figures 2D and S2C). Results from ChIP and re-ChIP analyses using MTA1 antibody followed by re-ChIP with antibodies against methyl-lysine, dimethyl-lysine, G9a, or H3Ac-K9 or vice-versa confirmed that MTA1 is methylated at the endogenous p21/WAF1 chromatin under physiological conditions (Figures 2E and 2F). Further over-expression of MTA1-K532R mutant but not WT-MTA1 in the MTA1-knockout (KO) MEFs was accompanied by the loss of recruitment of CHD4 and other NuRD subunits onto the p21WAF1 promoter (Figure 2G). These findings suggest that G9a-mediated methylation of MTA1 is important for the assembly of a functional NURD co-repressor complex at the target promoters. Analysis of methylation status of the endogenous MTA1 at its target p21/WAF1 promoter using MTA1 methylation site specific MTA1-MeK532-Ab further corroborated the hypothesis that MTA1 methylation triggers transcriptional corepressive events (Figure 2H).
Earlier studies have highlighted the significance of ELM2-SANT domain in interaction with histones (Wang et al., 2008). Because MTA1 contains a well conserved ELM2-SANT domain (aa 165–360) and the fact that it potentiates G9a methyltransferase activity on core histones (Figure 1H), we predicted that the ELM2-SANT domain in MTA1 may be important for histone H3MeK9 recognition and could play a crucial role in the permissive substrate recognition for G9a methyltransferase activity. As G9a-mediated methylation of MTA1 results in a selective loss of histone H3Me2K9 at its target p21/WAF1 (Figure S2D and S2E), we reasoned that methylated MTA1 may remain bound to the unmodified histone H3 (amino acids 12–21) and thus, expose histone H3 lysine 9 site within N-terminal histone H3 (amino acids 1–11) for G9a methylation. Consistent with this notion, we found an increased level of promoter-bound H3K9 methylation when methylated MTA1 and G9a are co-recruited to its target p21/WAF1 promoter (Figures 2G, S2D and S2E). Interestingly, this effect was significantly reversed by the inclusion of G9a inhibitor Bix-01294 or by the use of MTA1-K532R. As expected, G9a-mediated methylation of MTA1 also results in a selective loss of histone H3Me2K9 but not of H3Me2K4; co-recruitment of MTA1 and G9a to the target chromatin along with an increased histone H3 lysine 9 methylation (Figure 2G). These findings suggested that MTA1 facilitates the formation of repressive chromatin. We observed a significant increase in the level of p21/WAF1 mRNA when methylation deficient mutant MTA1-K532R was re-expressed in MTA1-KO MEF’s, further suggesting the dependency of corepressor activity of MTA1 on its lysine 532 methylation (Figure S2F) .
Lysine specific demethylase-1 (LSD1) demethylates MTA1
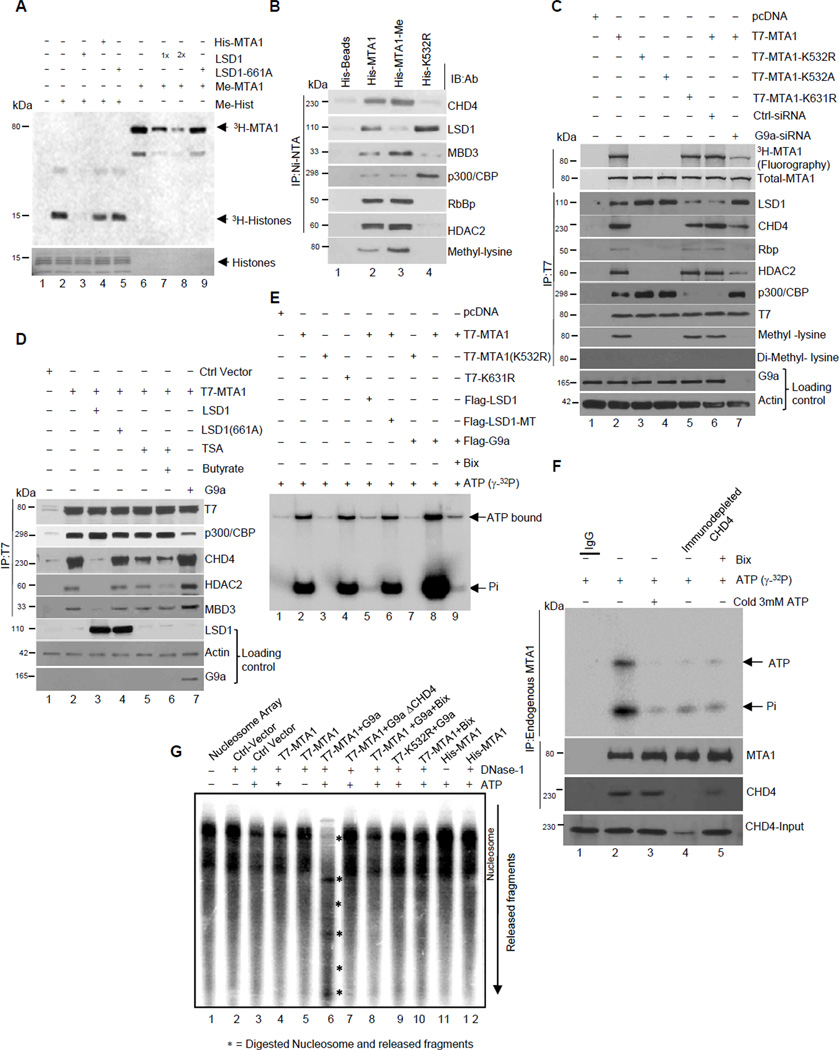
Because we consistently observed that LSD1 associates with demethylated MTA1 (Figure 2A) and the fact that LSD1 demethylates lysine residues in non-histone proteins (Shi et al., 2004), we next showed that G9a-methylated MTA1 could be effectively demethylated by LSD1 (Figure 3A). Overexpression of LSD1 in the HeLa cells resulted in demethylation of the endogenous MTA1 and these effects are compromised with demethylase-deficient LSD1-K661A mutant (Figure S3A). LSD1 interacts with the amino acid 441–535 region of MTA1 (Figure S3B). These findings suggest the possibility that MTA1 methylation status is dynamically modulated by G9a and LSD1. Accordingly, increased methylation of His-MTA1 by purified G9a enhances the ability of MTA1 to interact with the corepressive components of the NuRD complex in the nuclear extracts prepared from the HeLa cells (Figure 3B). In the same vein, modulation of MTA1 methylation status by siRNA-mediate G9a-depletion inhibited MTA1’s interaction with CHD4 or HDAC2 (Figure 3C). Similarly, overexpression of LSD1 and treatment of the HeLa cells with TSA or sodium butyrate results in the destabilization of the MTA1/NuRD corepressor complex but enhanced MTA1’s association with the p300/CBP complex (Figure 3D). These results suggest that stoichiometric levels of the MTA1-containing complexes are subject to alteration and destabilization by factors that inhibit MTA1 methylation.
Figure 3. MTA1 methylation status directs NuRD dependent chromatin remodeling.
(A) In vitro demethylation assay using G9a methylated His-MTA1, Methylated histones and LSD1. COS1 cells were transfected with Flag-LSD1-WT (1×=2.5µg) and -demethylase deficient LSD-661A. Flag immunoprecipitates were used in demethylase reaction. Reaction products were analyzed by fluorography.
(B) Ni-NTA-bound His-MTA1 or His-MTA1-K532R was used in methyltransferase assay with G9a. Ni-NTA beads were washed several times in the immunoprecipitation buffer, bound nonmethylated His-MTA1; methylated His-MTA1 and His-MTA1-K532R were incubated with 1 mg of the HeLa cells’ nuclear extracts. Bound proteins were eluted in the SDS-loading buffer and analyzed onto SDS-PAGE using indicated antibodies. An aliquot of reaction was electrophoresed on separate gel and analyzed using anti- methyl lysine antibody to determine MTA1 methylation status.
(C) HeLa cells expressing T7-MTA1 or -MTA1-K532R were treated with either control or G9a siRNA or transfected with Flag-G9a and -LSD1. Nuclear lysates were incubated with anti-T7- antibody or control IgG and analyzed by immunoblotting using indicated antibodies. Methylation status of MTA1 under all conditions was evaluated by fluorography (top panel). Western blot for Anti-MTA1 serves as loading control (Second panel from top). Lines, different gels from the same experiment.
(D) HeLa cells expressing various expression vectors were treated with TSA and sodiumbutyrate for 12 hr. Nuclear lysates were immunoprecipitated by T7 affinity resin, resolved onto a SDS-PAGE and immunoblotted with the indicated antibodies. Lines, different gels from the same experiment.
(E) ATP binding of T7-MTA1, -MTA1-K532R, and -MTA1-K631R precipitated from HeLa cells with 1µCi of γ-32ATP. ATP binding and hydrolysis was evaluated using formaldehyde-urea gels. See also Figure 3C.
(F) ATP binding of the endogenous MTA1 immunoprecipitated from the HeLa cells. IgG or MTA1 immunoprecipitates from CHD4-immunodepleted or Bix-treated nuclear lysates were incubated with either 1 µCi of γ-32ATP alone or in combination with 3-fold excess cold non-hydrolysable ATP. ATP binding and hydrolysis was evaluated using formaldehyde-urea gels. Presence of CHD4 in MTA1 immunoprecipitates were analyzed using anti-CHD4 antibody. (also see Figure S3C for immunoblotting using anti-T7, -G9a, and -LSD1 antibodies). Lines, different gels from the same experiment.
(G) In vitro mono-nucleosome mobilization and chromatin remodeling assay using reconstituted nucleosomes and T7 immunoprecipitates containing methylated and de-methylated MTA1 containing complexes from the COS-1 cells transfected with indicated expression plasmids.
MTA1 methylation orchestrates chromatin remodeling
Findings presented in the preceding sections establish that MTA1 methylation is a prerequisite for its binding to CHD4. We next investigated the impact of MTA1 methylation on the ability of the MTA1/CHD4 complex to bind ATP (Tong et al., 1998). We found that methylated MTA1-associated complex efficiently binds ATP while non-methylated and the methylation-deficient MTA1 K532R mutant associated complexes exhibits minimal to nonexistent ATP binding (Figure 3E). Interestingly, MTA1-containing complexes were capable of hydrolyzing ATP while such binding was completely abrogated in complex containing a methylation-deficient MTA1-K532R mutant. Consistent with these findings, overexpression of LSD1 or treatment with Bix-01294 also results in the loss of the ATP binding ability of the MTA1 complex, while G9a overexpression leads to a robust ATP hydrolysis by MTA1-bound complexes but not by MTA1- K532R (Figure 3E and S3C).
To further substantiate our conclusion that endogenous levels of methylated MTA1 drive recruitment of CHD4 ATPase, we first demonstrated ATPase enzymatic activity with MTA1-associated immunoprecipitated protein complex in the HeLa cells. We observed a robust hydrolysis of ATP upon its binding to MTA1-asscociated protein complex (Figure 3F, compare lanes 1 and 2) and such binding was compromised upon the use of non-hydrolysable cold ATP (Figure 3F, compare lanes 2 and 3). To implicate the suspected contribution of CHD4 ATPase in the above assay, we showed that the prior immunodepletion of CHD4 from the nuclear extracts compromises the ability of the incubated precipitated MTA1 to bind and hydrolyze ATP (Figure 3F, compare lanes 2 and 4). This suggested that CHD4 is the core ATPase that binds to methylated MTA1. Further immuprecipitation of MTA1 from the nuclear extracts prepared from Bix-treated HeLa cells also exhibited a clear abolition of ATP binding of MTA1 (Figure 3F, lane 5). These results support the notion that methylation of the endogenous MTA1 enhances its binding to CHD4 (Figure 3F, lower panel), and thus, a functional methylated-MTA1:CHD4 complex may probably utilize energy from ATP hydrolysis to remodel chromatin.
We next investigated whether the methylated- or demethylated-MTA1-containing remodeling complexes utilize energy from ATP hydrolysis to remodel chromatin using rotationally phased mononucleosome cores as substrate (Tong et al., 1998; Xue et al., 1998). We found that mononucleosomes treated with MTA1 immunoprecipitates alone exhibit a distinct pattern of DNase1 digestion that could be effectively disrupted by the use of immunoprecipitates containing methylated MTA1. Interestingly, the pattern of DNAse1 hypersensitivity of mononucleosome cores with the methylated MTA1 containing complex was also lost upon immune-depletion of CHD4 from the nuclear lysates, reinforcing the notion that CHD4 acts as a chromatin remodeling factor for the methylated MTA1-containing complex; while there was no effect of CHD4 immunodepletion on the DNAse1 hypersensitity of the demethylated MTA1-immunoprecipitates (Figure 3G, compare lanes 6 with 7). These findings suggest that MTA1 methylation orchestrates the recruitment of CHD4 as well as ATP-binding driven chromatin remodeling function of the NURD complex.
MTA1 reads modified histone marks
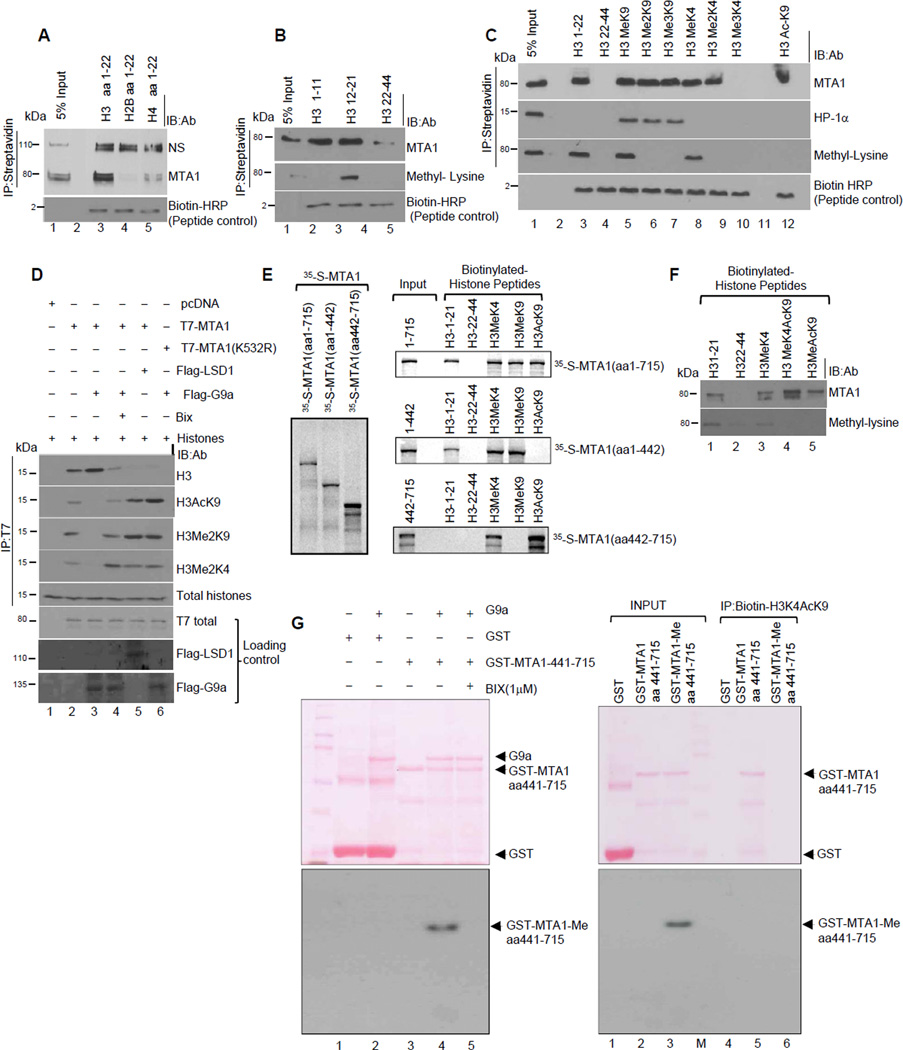
High chromatin affinity of MTA1 prompted us to further define the inner working of MTA1 at the nucleosomal level. We noticed that MTA1 binds to both poly- and mono-nucleosomal fractions prepared from the HeLa cell nuclei (Figure S4). Since MTA1 does not directly bind to DNA, we speculated that MTA1 interaction with the nucleosomal template may be mediated through core histones. Results from biotinylated peptide-affinity assays (Wysocka et al., 2006) revealed that MTA1 preferentially binds to the histone H3 but not with the histone H2B or histone H4 (Figure 4A). We discovered a substantial association of the methylated MTA1 with the histone H3 amino acid 12–21 but not with the histone H3 amino acid 1–11 or amino acid 22–44 (Figure 4B), and that MTA1 effectively interacts with both H3 mono-methyl K9 and monomethyl K4 peptides (Figure 4C). However, we unexpectedly discovered a robust MTA1 binding to the histone H3 Acetyl-K9 peptide (Figure 4C, lane 12).
Figure 4. MTA1 is a reader of modified histones.
(A-C) Peptide pull down assays were carried out using the HeLa cells’ nuclear extracts and biotinylated histone H3, H2B and H4 peptides (aa1–22) (A)or biotinylated Histone H3 (aa1–11, aa12–21, aa22–44) peptides (B) or N-terminal histone H3 peptides with specific methyl and acetyl modifications (C). HP1-alpha pulldown, was positive control for H3K9Me3 binding. The material was immunoprecipitated using Streptavidin where indicated. See also Figure S4.
(D) Histone binding assay using T7-WT MTA1 or T7-MTA1-K532R from the COS1 cells and core histones from the HeLa cells. Bound histones were electrophoresed onto a 15% PAGE and analyzed using indicated antibodies. Immunoblotting analyses using anti-flag (G9a and LSD1) and anti-T7 is shown here. The material was immunoprecipitated using an anti-T7 antibody where indicated.
(E) In vitro translated 35S labeled MTA1 domains were used in a peptide pull-down assay using various histone H3 biotinylated peptides.
(F) Biotinylated peptide pull down assay was performed using control unmodified and biotinylated Histone H3Acetyl lysine-9 and Histone H3 Methyl lysine-4-Acetyl-lysine-9 peptide and HeLa cell nuclear extracts.
(G) In vitro methylation assay with G9a enzyme and GST and GST-MTA1-441–715. Methylation of GST-MTA1-441–715 was confirmed by fluorography (left panel). Inclusion of Bix (1µM) completely abolished methylation. Equal amount of GST, unmethylated and G9a methylated MTA1-441–715 was used in a streptavidin-biotin peptide pull-down assay. 50ng of biotinylated histone H3K4AcK9 was incubated with equal amount of GST, unmethylated and G9a-methylated GST-MTA1-445–715 in histone binding buffer (20mM Hepes, ph 8.0, 200mM KCL, 0.5% TritonX-100, 0.2mM EDTA) for 3 hr at room temperature. Beads were washed with high stringent wash buffer (histone binding buffer supplemented with 400 mM KCL) and bound proteins were eluted using glycine buffer (pH:3.0). Eluted proteins were analyzed onto a 10% SDS-PAGE and transferred to nitrocellulose membrane and stained with Ponceau. Blots were scanned and further treated with fluorgraphy reagent to detect methylated GST-fusion proteins.
Because methylation of the histone H3K4 and/or acetylation at H3K9 have been shown to disrupt NuRD-nucleosomal binding (Nishioka et al., 2002; Zegerman et al., 2002), our findings are apparently not consistent with the corepressive nature of the NuRD complex as both acetylation of histone H3-K9 and/or methylation of K4 did not disrupt the binding of the MTA1/NuRD complex to nucleosomes (Figures 4C). These results suggest that MTA1 may be a novel reader of modified histone marks and that its interaction with acetylated histones might have a yet undiscovered role in transcriptional control. To validate the impact of MTA1 methylation upon histone binding, we observed that methylated MTA1 did not bind to dimethylated histone H3K9 while demethylated MTA1 effectively binds to acetylated histone H3K9 (Figure 4D). We found that the ELM2-SANT domain of MTA1 recognizes methylatedhistone- H3K9 and -H3K4 marks while the C-terminal MTA1 domain amino acids 442–715 participates in the recognition of acetyl-K9 of histone H3 (Figure 4E). Thus MTA1 interacts with modified histones using two separate effector domains.
Having tested the ability of MTA1 to bind to histone H3MeK4 and Acetyl-lysine 9 we next evaluated if MTA1 can bind combinatorial H3MeK4-AcK9 modification. We observed a significant binding of demethylated MTA1 to bivalent histone H3K4-AcK9 (Figure 4F) as was the case with the methylation-deficient MTA1-K532R mutant (Figure 4D). To further strengthen this observation, we used an equal amount of GST, unmethylated, and G9a-methylated MTA1 amino acids 441–715 fragment in a streptavidin-biotin peptide pull-down assay with a biotinylated bivalent H3K4-AcK9 peptide. We found that MTA1 methylation compromises its binding to bivalent H3K4AcK9 peptide. (Figure 4G right panel; compare lanes 3 and 6) Accordingly, unmethylated MTA1 showed a very high affinity to bivalent histone H3K4AcK9 (Figure 4G right panel, compare lanes 2 and 5). Collectively these findings suggest that MTA1- mediated specific histone recognition may be largely guided by post-translational methylation of MTA1 on lysine 532 which triggers a multivalent engagement of chromatin modifications.
LSD1 demethylation of MTA1, a prerequisite for its coactivator function
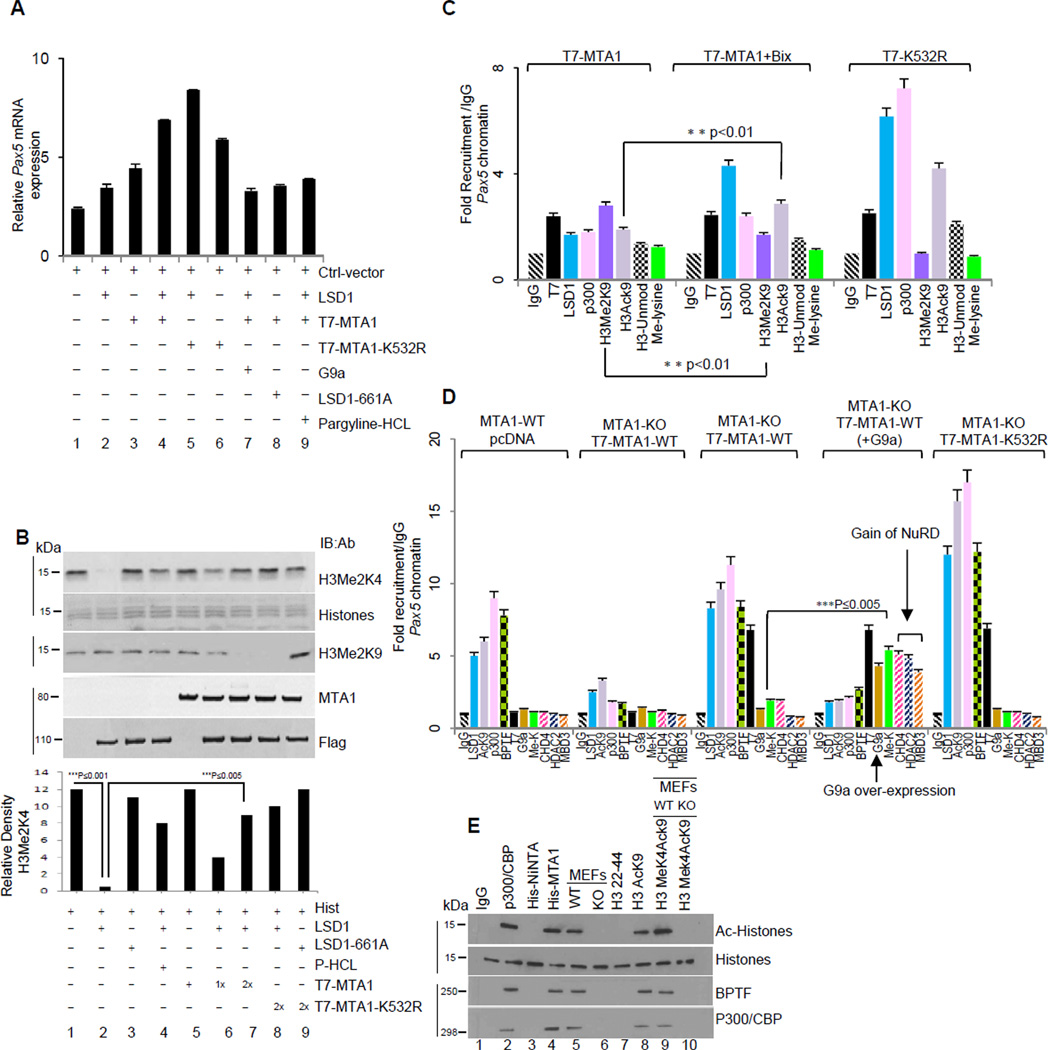
LSD1 could act both as a coactivator or corepressor depending on its ability to demethylate histone H3 at lysine 4 or lysine 9 (Metzger et al., 2005). Demethylation of lysine 4 leads to gene repression while demethylation of lysine 9 correlates with gene activation events (Lan et al., 2007). To understand the impact of MTA1 demethylation on its coactivator versus corepressor function, we used the Pax5-luc and p21/WAF1-luc as prototypes of stimulated or repressed target genes, respectively (Balasenthil et al., 2007; Li et al., 2010). We found that MTA1 coexpression with Flag-LSD1 results in stimulation of Pax5 transcription in a G9a sensitive manner (Figure 5A and S5A), suggesting that while MTA1 methylation is an important determinant of its co-repressor function. In addition, LSD1-mediated demethylation of MTA1 abolishes its corepressive activity upon its target p21WAF1 (Figures 2D and S2C), implying that LSD1-mediated demethylation of MTA1, is a prerequisite for its coactivator function.
Figure 5. MTA1 demethylation recruits co-activator functions.
(A) Total RNA from HeLa cells transiently expressing T7-MTA1 or -MTA1-K532R were transfected was reverse transcribed and used in a quantitative PCR reaction using specific primers for endogenous Pax5. Data is represented as fold induction over 18 sRNA (internal control). Where indicated small molecular inhibitor of G9a, Bix-01294 and pargyline-hydrochloride a specific small molecule inhibitor of LSD1 were used at a final concentrations of 1µM and 1mM, respectively. Error bars represent SD. See also Figure S5A
(B) Flag-LSD1 or –LSD1-K661A from the COS1 cells were used in the histone demethylase assay. Where indicated recombinant His-MTA1 was included in the assay. Histones from demethylase reaction were analyzed independently using anti-H3Me2K4 and anti-H3-Me2K9 antibodies. Lines, different gels from the same experiment.
(C) HeLa cells were transfected with the indicated expression vectors were used for ChIP assay to evaluate changes at the Pax5 promoter, using anti-T7 antibody followed by Re-ChIP using indicated antibodies. Recruitment profiles were generated using quantitative-PCR and specific primers encompassing MTA1 recruitment region on Pax5 promoter. Values are represented as fold recruitment/IgG. **p<0.01.Error bars represent SD. See also Figure S5B.
(D) Quantitative ChIP relative to IgG control at the Pax5 promoter using indicated antibodies and chromatin from MTA1-WT and MTA1-KO cells transfected with T7-MTA1 or -MTA1- K532R. (also see Supplementary Figure 4C). ***p<0.005. Error bars represent SD. See also Figure S5C.
(E) Histone acetyltransferase assay (HAT) using immunoprecipitates from Ni-NTA or streptavidin-biotin pull down from the HeLa cells nuclear extracts were incubated with His-MTA1 protein or biotinylated peptides. Lines, different gels from the same experiment.
We next evaluated the underlying mechanism by which LSD1 converts MTA1’s corepressive activity to coactivator activity on its target chromatin. Since MTA1 demethylation alone and/or destabilization of the repressor complex might not be sufficient for the stimulation of target gene transcription, we suspected that derepression of target genes can be facilitated by events that promote rapid demethylation of lysine 9 or methylation at lysine 4 of histone H3. LSD1 can demethylate mono-methyl and dimethyl Histone H3lysine4 to repress target genes (Metzger et al., 2005). LSD1 also has been shown to demethylate histone H3 lysine 9 under certain scenario (Lan et al., 2007; Nair et al., 2010). We determined the influence of MTA1 on the demethylase activity of LSD1 upon nucleosomes. To our surprise, we found that MTA1 effectively repressed LSD1-mediated demethylation of histone H3Me2K4 while promoting demethylation of H3Me2K9 (Figure 5B, compare lanes 1–2 with lanes 6–8). These findings suggested that demethylated MTA1 binds to di-methyl-H3K4 leading to non-availability of this residue for LSD1-mediated demethylation. Our in vivo histone binding studies suggested that demethylated MTA1 binds H3Me2K9 with high affinity (Figure 4D). It is possible that different histone binding affinity of demethylated MTA1 might result in substrate preference and demethylation of histone H3Me2K9 by LSD1. Demethylated histone H3K9 is now a good substrate for p300/CBP mediated acetylation and thus generating an active chromatin state.
Using the Pax5 promoter as a prototype for coactivator function of MTA1 (Balasenthil et al., 2007), we found that MTA1 demethylation results in a robust recruitment of LSD1 and demethylation of histone H3K9. This, in-turn, leads to, in-parallel, histone H3K9 acetylation as well as recruitment of p300/CBP at the Pax5 promoter chromatin (Figure 5C and S5B). Quantitative-ChIP analysis of the Pax5 promoter chromatin from the wild type (WT) and MTA1-KO MEFs revealed co-recruitment of LSD1 and BPTF3 (a member of the NURF activator complex) onto the Pax5 promoter as well as increased acetylation of the associated histone H3K9 (Figures 5D and S5C). Overexpression of G9a with T7-MTA1 in MTA1-KO MEFs resulted in MTA1 methylation at the Pax5 chromatin and destabilization of coactivator complex (Figure 5D). Interestingly, levels of H3AcK9 and H3Me2K4 were readily enhanced at the Pax5 promoter when we used chromatin from cells expressing the methylation-deficient MTA1-K532R mutant. These observations establish that demethylated MTA1 promotes gene transcription by recruiting factors that can acetylate histone H3K9 and by preventing rapid demethylation of H3K4 by LSD1. Accordingly we noticed an increase in the levels of Pax5 mRNA upon re-expression of K532R in the MTA1-KO MEFs (Figure S5D). These findings suggest that methylation and demethylation of MTA1 coordinates the recruitment of critical effectors at the target chromatin to influence the transcriptional outcome.
MTA1 interaction with H3K4-AcK9 drives its coactivator activity
Since MTA1 demethylation correlates with an enhanced recruitment of the histone H3K9 acetylation as well as of nucleosome-remodeling factor BPTF onto the target promoter, we next investigated whether recognition of histone H3K9 acetylation, an epigenetic marker of an active chromatin, by demethylated MTA1 triggers an assembly of a distinct nuclear proteome for its coactivator function. Rapid exchange of coactivator and corepressor complexes is largely driven by effector proteins that modulate the epigenetic landscape.
To understand the biological consequence of the MTA1 binding to histone H3K4-AcK9, we next determined if MTA1 bound to active histone tail can recruit acetyltransferase activity. We found that demethylated MTA1-bound complexes exhibit a significant associated HAT activity (Figure 5E). A detailed analysis of proteins found in MTA1-interacting complexes revealed the presence of distinctive components of coactivator proteins such as p300/CBP and BPTF (Figure 5E). In brief, these findings reveal a previously uncharacterized dynamic role of MTA1 in recognition of histone H3AcK9. Furthermore, MTA1 recognition of the bivalent H3K4-AcK9 marks drives histone H3K9 acetylation and co-activator functions.
MTA1 associates with NuRD or NURF complexes in cyclical- and signaling-dependent manner
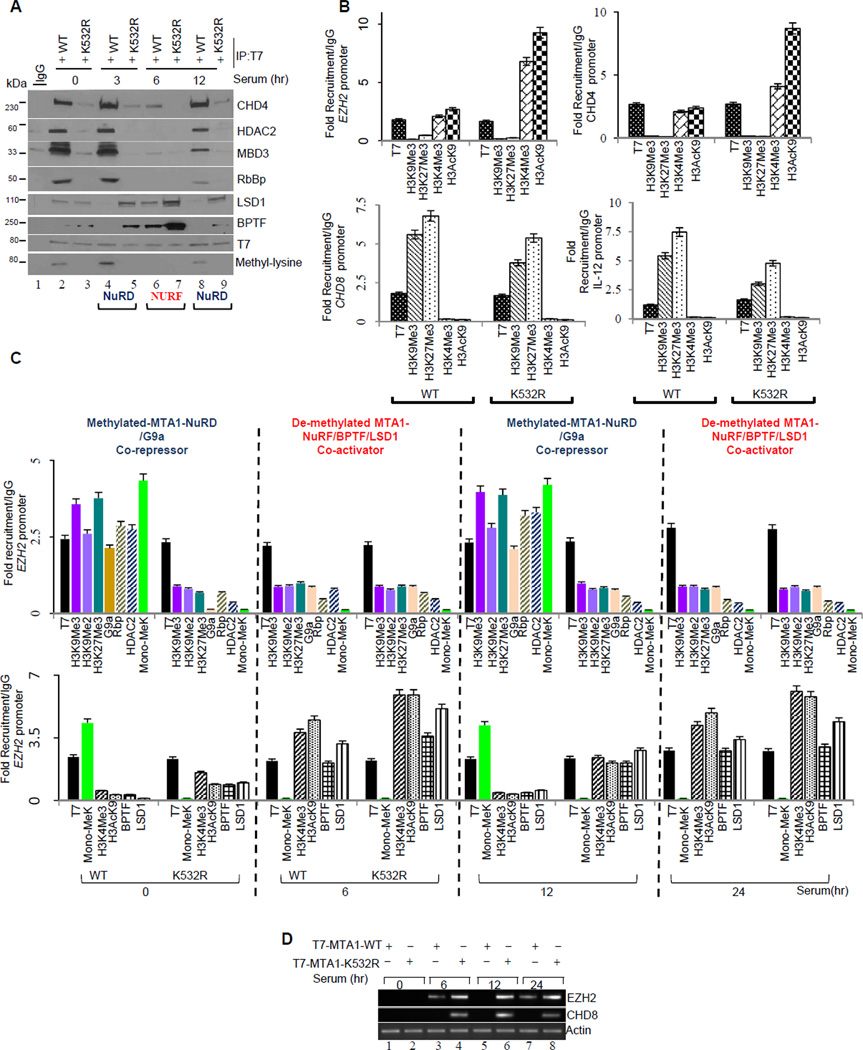
Findings presented above support an interesting notion that methylation-demethylation events direct MTA1 association with coactivator versus corepressor complexes. We next investigated the possibility of methylation-dependent cyclical association of MTA1 with NuRD and NuRF (nucleosome-remodeling factor) complexes in a signaling-dependent fashion. To this end, we first examined the association of MTA1 with the NuRD or NURF complexes in serum-stimulated MCF-7 cells at different time points. We used serum stimulation because serum has been shown to induce dynamic cyclical association of chromatin associated proteins at promoters of cellular genes. Indeed, we discovered a cyclical interaction of MTA1 with the NuRD and NuRF complexes in serum-stimulated cells over the entire time course (Figure 6A). Interestingly, MTA1-methylation appears to be a prerequisite for functional association with NURD components at all-time points tested here, while methylation-deficient MTA1 constitutively associates with the coactivator NuRF complex (Figure 6A).
Figure 6. MTA1 associates with the components of the NuRD and NURF complexes in a cyclical and signaling dependent manner.
(A) MCF-7 cells expressing T7-MTA1 or –MTA1-K532 constructs were serum starved for 48 hours followed by serum stimulation for indicated time points, and lysates were immunoprecipitated with T7-affinity-resin. Bound immunoprecipitates (retained proteins) were analyzed separately on a polyacrylamide gel followed Western blot analysis using indicated antibodies. Methylated MTA1 binds co-repressor complex while demethylated MTA1 recruits activator complex. Lower panel from a different gel from the same experiment.
(B) Chromatin was prepared from the MCF-7 transfected with the T7-MTA1, MTA1-K532R. Quantitative ChIP with T7 antibody followed by re-ChIP with indicated antibodies is represented relative to IgG control at EZH2, CHD4, IL-12 and CHD8 promoters. Error bars represent SD.
(C) Chromatin was prepared from MCF-7 transfected with MTA1-WT, MTA1-K532R and serum treated at indicated time points. Quantitative ChIP with T7 antibody followed by Re-ChIP with indicated antibodies is represented relative to IgG control at EZH2 promoter. Values are represented as fold recruitment/IgG. Green colored bars in graph represent status of methylated MTA1 at indicated time points . Error bars represent SD. See also Figure S6A
(D) Total RNA from MCF-7 cells transfected with the indicated expression vectors were used reverse transcribed and subjected to semi-quantitative PCR using specific primers to detect EZH2 and CHD8 mRNA levels.
To understand the dynamic of MTA1 associated components of the NuRD or NURF complexes at the target chromatin, we evaluated the levels of active or repressed chromatin marks and the core components of NuRD or NuRF complexes at the target gene promoters such as CHD4, CHD8, EZH2 and 1L-12. Chromatin remodeler proteins chromodomain protein-4 (CHD4), chromodomain protein-8 (CHD8) and polycomb group protein Enhancer of Zeste homlog-2 (EZH2) and interleukin-12 have been identified as MTA1 targets from a distinct, ongoing ChIP-seq genome-wide study in our laboratory (SN and RK, unpublished observations). MTA1 acts as a co-activator for EZH2 and CHD4, while it represses CHD8 and IL-12. We first evaluated the chromatin landscape upon recruitment of the wild-type or methylation deficient mutant K532R on the target promoters. We first performed ChIP using an ant-T7 mAb to detect either T7-MTA1 or T7-K532R recruitment before undertaking the second round of ChIP with the indicated antibodies (Figure 6B). Results from ChIP studies validated that methylation of MTA1 triggers a more repressive environment at target gene promoters while de-methylation signals a more active environment (Figure 6B).
Next we analyzed the nature of chromatin modifications at two endogenous target gene promoters EZH2 and CHD8, as prototypes for activated and repressed, target genes by MTA1. Chromatin from serum stimulated MCF-7 was prepared at indicated time points and used in ChIP and Re-ChIP analysis (Figure 6C). We observed a signaling- and methylation- dependent cyclical association of MTA1 with the coactivator or corepressor complexes at EZH2 (Figure 6C) and CHD8 (Figure S6A) target genes and thus, confirming our biochemical data. Methylated-MTA1 associates with the repressed chromatin (H3K9Me2/Me3) and NuRD components while demethylated-MTA1 preferentially interacts with the active chromatin (H3K4Me3-H3AcK9) and BPTF, a member of NURF-trithorax activator complex (Figures 6C and S6A). Functional relevance of MTA1 methylation in triggering an optimal transcriptional response from endogenous targets was revealed by evaluating the expression of prototypic EZH2 and CHD8 mRNAs under identical experimental conditions by semi-quantitative PCR in the MCF-7 cells (Figure 6D). These results suggest that methylation proficient MTA1-WT and methylation deficient MTA1-K532R proteins alter transcriptional status at the endogenous promoters by associating with different chromatin remodeling-histone modifying enzymes.
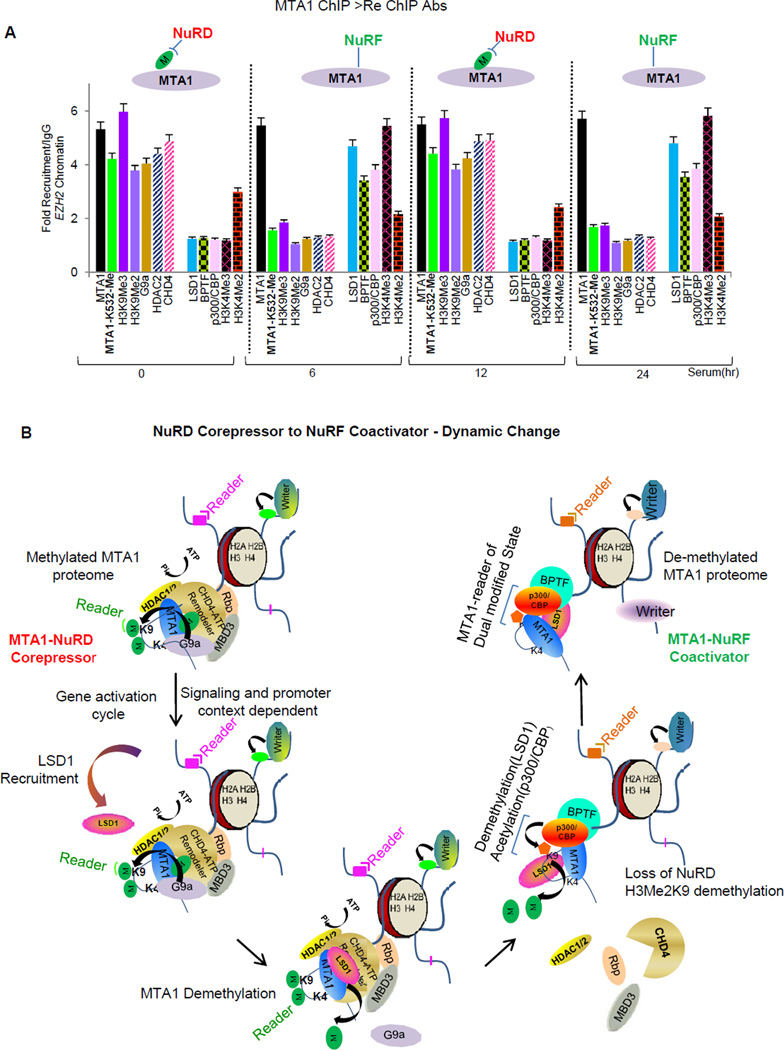
We next addressed if fluctuation in the methylation status of the endogenous MTA1 triggers a similar chromatin signaling as was the case with ectopically expressed proteins. We evaluated changes in MTA1 methylation in serum stimulated cells in a time course-study. Western blot analyses were performed using either methyl-lysine antibody (Figure S6B) or MTA1-K532-Me antibody (Figure S6C). We reproducibly observed that the levels of MTA1 methylation fluctuate in a signal and time dependent manner in vivo (Figure S6B and S6C). Consistent with these results, analysis of MTA methylation status at the endogenous EZH2 promoter in serum stimulated MCF-7 cells, using site specific MTA1-K532-Me antibody, revealed that while recruitment of MTA1 was similar at all-time points post serum stimulation; the combinatorial code of histone modifications and nature of chromatin modifying proteins correlated well with the changes in MTA1 methylation pattern (Figure 7A and S7). As expected methylated MTA1 was associated with the co-repressor NuRD components while demethylated MTA1 with the the coactivator complex components (Figure 7A and S7). Thus dynamic fluctuation of MTA1 methylation status correlates well with the changes in chromatin landscape at the endogenous target promoters.
Figure 7. MTA1 engages with the NuRD and NURF complexes in a cyclic manner.
(A) Cells were stimulated with serum for indicated time points. Recruitment profiles of MTA1 and repressor/activator marks at EZH2 promoter was generated by ChIP using a MTA1-antibody followed by Re-ChIP using MTA1 methylation site specific (MTA1-MeK532) antibody. Values are represented as fold recruitment/IgG. Error bars represent SD. See also Figure S7.
(B) Model of MTA1 dual-coregulator action (Also graphical abstract). At repressive chromatin target, G9a methylated MTA1 is associated with histone H3 aa11–21. Methylation of MTA1 serves as a docking site for CHD4. Binding of CHD4 directs a combinatorial association of NuRD chromatin remodeling complex. MTA1 potentiates G9a methyltransferase activity on HDAC2 deacetylated nucleosomes to create a repressive H3K9Me2 environment. During gene activation cycle, LSD1 is recruited to the NuRD repressor complex and demethylates MTA1. Loss of MTA1-methylation destabilizes the NuRD repressor complex. Increased affinity of demethylated MTA1 for H3K9Me2 triggers rapid demethylation of H3-K9Me2 mark by LSD1 and acetylation by p300/CBP. MTA1 functions as an epigenetic reader of H3K4AcK9 and engages in a co-activator function. Methylated and demethylated states of MTA1 engage distinct histone modifications and factors to facilitate gene repression (NuRD) or activation (NuRF) events.
Discussion
The mammalian NuRD complex is enriched with proteins that act synergistically to silence genes during resting and stimulated conditions. This unique macromolecular complex is composed of six core-subunit proteins and has retained subunit composition during the course of evolution while other ATP-dependent CRCs like SWI/SNF and INO80 families have shuffled subunits to acquire functional specialization over time (Perissi et al., 2010; Lai et al., 2011) Evolution has favored specialization of the core subunits of NuRD complex. The need for combinatorial association of regulatory complexes has been evolutionarily favored as such a strategy provides proximity to couple reactions and presents a unique flexibility for diversification of functions. However, the mechanism that triggers an ordered association of NURD subunits is not well understood. For the first time we found that MTA1 methylation plays a crucial role in formation of NURD repressor complex. Further, loss of MTA1 methylation coincides with gene-activation cycle and promotes recruitment of co-activator complex. This cyclic association of MTA1 with NURD-NURF complex presumably determines a balanced gene transcription in response to cellular signals, and explains dual-coregulatory role of MTA1 (Figure 7B). During gene repression event, chromatin bound MTA1 is methylated by G9a on lysine 532 which increases MTA1’s affinity for histone H3-(aa11–21), making lysine 9 of histone H3 available for methylation by G9a. Methylation of H3K9 by G9a creates a repressive H3K9Me2 chromatin landscape. Methylated MTA1 now amplifies this repressive signal recruiting chromodomain protein CHD4 and formation of a stable NuRD complex involving other members like HDAC1/2, leading to gene repression.
During the gene activation cycle, methylated MTA1 is rapidly demethylated by LSD1 which eliminates the methyl-moiety on K532, the docking interface for CHD4. CHD4 and G9a are displaced from the MTA1-bound complex which results in the destabilization of the NuRD complex. Importantly, demethylated MTA1 has high affinity for H3Me2K9 and LSD1 bound MTA1 demethylates H3Me2K9. Since we observed an increased demethylation of H3Me2K9 at LSD1-MTA1 target promoters, we presumed that MTA1 promotes specific substrate recognition and H3K9Me2 demethylation by LSD1. Demethylated MTA1 recognizes the bivalent histone H3K4-AcK9 mark and recruits histone p300/CBP acetyltransferase activity and the BPTF- a member of the NURF chromatin remodeling complex. Recruitment of the p300/CBP onto the target promoters results in a rapid acetylation of demethylated histone H3K9 to stimulate gene activation.
In summary, for the first time findings presented here introduce a set of novel characteristics of an evolutionary conserved coregulator as a chromatin/histone amplifier. MTA1 chromatin amplifier represents a switch between gene activation and repression events involving sequential recruitment of components with distinct functions. One important characteristic of histone amplifier proteins like MTA1 would be that they interact with various histone modifying enzymes that can inscribe specific histone marks. We believe that evolution has favored repressor complexes like NuRDs to retain proteins like MTA1 that display a unique opportunity for diversification of NURD-dependent processes for optimal biological outcome which at-time, demands gene activation. Another important characteristic of histone amplifier MTA1 is the multivalent engagement of chromatin modifications and unique ability to facilitate cross-talk between chromatin remodeling complexes and histone modifying enzymes. In conclusion, studies presented here offer a new perspective to the molecular governance of a dual coregulator wherein sequential methylation and demethylation resets the physiological switch between active and repressed states of MTA1 through chromatin signaling.
Experimental Procedures
Details of cell lines and transfection, inhibitors, small Interfering RNAs, plasmids and site directed mutagenesis, salt solubilization assay, histone methyl transferase assay, histone acetyltransferase assay, RT-PCR analysis, peptide pull down assays, luciferase assays, coimmunoprecipitation assays and Western blot analyses, preparation of mononucleosomes and nucleosome remodeling assay, ATP binding assay, and statistical analysis are provided in the Supplementary Section.
Supplementary Material
Highlights.
MTA1 methylation dictates the chromatin remodeling activity of the NuRD complex.
MTA1 interprets the “nucleosome code” by engaging multivalent chromatin modifications.
Activator modification mark H3K4-AcK9 does not disrupt MTA1-nucleosomal binding.
Methylated and de-methylated MTA1 nucleate complexes with opposite functions.
MTA1’s methylation status plays an instructive role in the transcriptional outcome
Acknowledgements
We thank all members of the Kumar laboratory for their comments. The proteomic studies presented here were supported by the McCormick Genomic and Proteomic Center. This study was supported by National Institutes of Health Grants CA98823 to R.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests: The authors declare that they have no competing interests.
Author Contributions: RK directed all aspects of this project; SSN and RK designed the experiments; SSN carried out the bulk of experiments; SSN, DQL and RK analyzed all experimental data; and SSN and RK wrote the paper.
References
- Balasenthil S, Gururaj AE, Talukder AH, Bagheri-Yarmand R, Arrington T, Haas BJ, Braisted JC, Kim I, Lee NH, Kumar R. Identification of Pax5 as a target of MTA1 in B-cell lymphomas. Cancer Res. 2007;67:7132–7138. doi: 10.1158/0008-5472.CAN-07-0750. [DOI] [PubMed] [Google Scholar]
- Bochar DA, Savard J, Wang W, Lafleur DW, Moore P, Côté J, Shiekhattar R. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc Natl Acad Sci U S A. 2000;97:1038–1043. doi: 10.1073/pnas.97.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RE, Northrop JP, Horton JR, Lee DY, Zhang X, Stallcup MR, Cheng X. The ankyrin repeats of G9a and GLP histone methyltransferases are mono-and dimethyllysine binding modules. Nat Struct Mol Biol. 2008;15:245–250. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanta KS, Li DQ, Eswaran J, Kumar R. Gene profiling of MTA1 identifies novel gene targets and functions. PLoS One. 2011;6(2):e17135. doi: 10.1371/journal.pone.0017135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Liang G, Sharma S, Lin JC, Choi SH, Han H, Yoo CB, Egger G, Yang AS, Jones PA. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol. 2009;29:5366–5376. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AY, Wade PA. Cancer biology and NuRD : a multifaceted chromatin remodeling complex. Nat Rev Cancer. 2011;7 doi: 10.1038/nrc3091. 588-7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- Li DQ, Pakala SB, Reddy SD, Ohshiro K, Peng SH, Lian Y, Fu SW, Kumar R. Revelation of p53-independent function of MTA1 in DNA damage response via modulation of the p21/WAF1-proliferating cell nuclear antigen pathway. J. Biol. Chem. 2010;285:10044–10052. doi: 10.1074/jbc.M109.079095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DQ, Pakala SB, Nair SS, Eswaran J, Kumar R. Metastasis-associated protein1/nucleosome remodeling and histone deacetylase complex in cancer. Cancer Res. 2012;72:387–394. doi: 10.1158/0008-5472.CAN-11-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavathi B, Kumar R. Metastasis tumor antigens, emerging family of multifaceted master coregulators. J Biol Chem. 2007;19:1529–1533. doi: 10.1074/jbc.R600029200. [DOI] [PubMed] [Google Scholar]
- Mazumdar A, Wang RA, Mishra SK, Adam L, Bagheri-Yarmand R, Mandal M, Vadlamudi RK, Kumar R. Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat Cell Biol. 2001;3:30–37. doi: 10.1038/35050532. [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Nair SS, Nair BC, Cortez V, Chakravarty D, Metzger E, Schüle R, Brann DW, Tekmal RR, Vadlamudi RK. PELP1 is a reader of histone H3 methylation that facilitates oestrogen receptor-alpha target gene activation by regulating lysine demethylase 1 specificity. EMBO Rep. 2010;11:438–444. doi: 10.1038/embor.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, Peng J, Cheng X, Vertino PM. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Alzayady K, Stewart R, Ye P, Yang S, Li W, Shi Y. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol Cell Biol. 2010;30:1997–2005. doi: 10.1128/MCB.01116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- Utley RT, Owen-Hughes TA, Juan LJ, Côté J, Adams CC, Workman JL. In vitro analysis of transcription factor binding to nucleosomes and nucleosome disruption/displacement. Methods Enzymol. 1996;274:276–291. doi: 10.1016/s0076-6879(96)74024-7. [DOI] [PubMed] [Google Scholar]
- Wang L, Charroux B, Kerridge S, Tsai CC. Atrophin recruits HDAC1/2 and G9a to modify histone H3K9 and to determine cell fates. EMBO Rep. 2008;9:555–562. doi: 10.1038/embor.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J. Identifying novel proteins recognizing histone modifications using peptide pull-down assay. Methods. 2006;40:339–343. doi: 10.1016/j.ymeth.2006.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Côté J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;6:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Zegerman P, Canas B, Pappin D, Kouzarides T. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complexJBiol. Chem. 2002;277:11621–11624. doi: 10.1074/jbc.C200045200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.