Recent studies have implicated epigenetic mechanisms including histone modifications and DNA methylation in various human diseases.1,2 Histone H3 lysine 4 trimethylation (H3K4me3) and H3 lysine 27 trimethylation (H3K27me3) are hallmarks of gene activation and repression, respectively.3 These two histone methylations are now known to regulate gene transcriptions in the brain, including those during human brain development.3,4 In addition, a candidate molecular approach identified an H3K4me3 alteration in the glutamate decarboxylase 1 (GAD1) gene in postmortem brains from patients with schizophrenia (SZ).5 Nonetheless, as far as we are aware, genome-wide unbiased studies for multiple histone modifications are not yet available for SZ.
To enable the detection of histone modification changes relevant to this disease, we exploited olfactory cells obtained from SZ patients and controls. These primary cells were established from dissociated olfactory epithelial tissues collected by nasal biopsy,6 and stained with βIII tubulin (a representative marker for immature neurons), to near homogeneity (Supplementary Figure 1). The olfactory cells are complementary to postmortem brains that have provided insight into the pathophysiology of SZ as these cells may reflect molecular states associated with neurodevelopment and also be used for functional assays. Using the olfactory cells, we explored SZ-associated genome-wide variations in H3K4me3 and H3K27me3 by using ChIP-seq (chromatin immunoprecipitation coupled with high throughput sequencing). We also conducted gene expression microarray analysis with the same cell samples. We then examined which specific genes and gene groups are affected in expression by these epigenetic modifications.
Four SZ patients were compared with four controls matched in age, gender, education and smoking habits (Supplementary Table 1). Chromatin immunoprecipitation was performed with olfactory cell lysates by using antibodies against H3K4me3 and H3K27me3, and the precipitated DNA samples were sequenced with an Illumina/Solexa Genome Analyzer II platform as previously described.7 Sequence alignment and signal quantification were performed using Illumina's ELAND algorithm and CisGenome, a novel integrated software for statistical analysis of ChIP-seq data.8,9
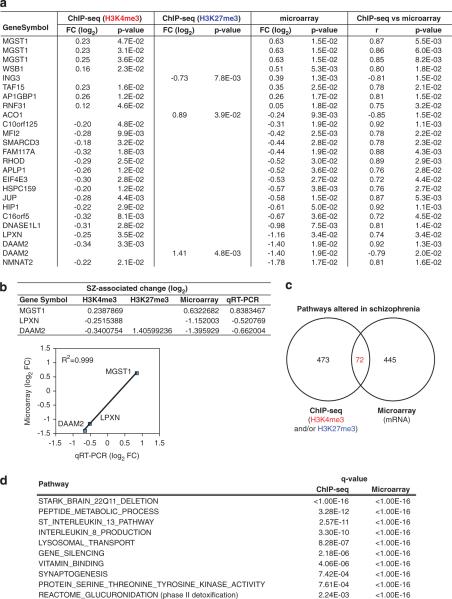
We first compared the levels of H3K4me3 and H3K27me3 signals at the promoter regions (the first 1000 base pairs (bp) upstream and downstream of the transcription start sites (±1000 bp)) between SZ patients and controls. The promoter regions where H3K4me3 or H3K27me3 signals were significantly changed in SZ were then compared with the genes whose expression was significantly altered in SZ. This analysis identified 22 genes whose expression is likely to be epigenetically modified according to the following criteria: (i) both gene expression and histone modification (H3K4me3 and/or H3K27me3) levels are significantly different (P<0.05) between SZ and controls, and (ii) the correlation between gene expression and histone modification (H3K4me3 and/or H3K27me3) levels is significant (P<0.05) (Figure 1a, Supplementary Table 2 and Supplementary Figure 2). SZ-associated increase (or decrease) in gene expression is correlated with increase (or decrease) in H3K4me3 levels or decrease (or increase) in H3K27me3 levels. Although most SZ-associated gene expression changes are associated with changes either in H3K4me3 or H3K27me3 level, DAAM2 gene expression is unique in that it seems to be affected by reciprocal changes in both H3K4me3 and H3K27me3 levels (decreased H3K4me3 levels and increased H3K4me3 levels) at the promoter region. Quantitative reverse transcription PCR (qRT-PCR) analysis was performed for the genes whose mRNA expression levels were most increased/decreased in the microarray analysis. All the three genes selected for the analysis (microsomal glutathione S-transfer-ase 1 (MGST1), disheveled associated activator of morphogenesis 2 (DAAM2) and leupaxin (LPXN)) showed an excellent correlation between qRT-PCR and microarray data (Figure 1b).
Figure 1.
Effects of SZ-associated epigenetic alterations on gene expression. (a) Genes whose expression levels are significantly affected by altered H3K4me3 and/or H3K27me3 levels associated with SZ. See Supplementary Table 2 for the full analysis results. Data from subject 1 were not included in the reference distribution for the H3K4me3 data because it led to artifacts in the resulting distributions. None of the 22 genes survived multiple-testing corrections. (b) qRT-PCR validation of microarray data for MGST1, DAAM2 and LPXN mRNA. qRT-PCR data were normalized by β-actin mRNA expression. (c) Venn diagram of functional groups significantly altered in SZ in pathway analysis: 72 pathways showed significant changes both in ChIP-seq and microarray studies. False discovery rate, q<0.01. See Supplementary Tables 3 and 4 for full analysis data. (d) Representative pathways that are potentially related to schizophrenia among the 72 pathways from c. See Supplementary Table 5 for full analysis data. FC, fold change.
To identify molecular networks that are particularly affected by altered histone methylations in SZ patients, we performed pathway analysis on both the ChIP-seq and gene expression microarray data. For ChIP-seq, we computed ΔK4 and ΔK27 for each gene, defined as difference in the mean log2 ChIP-seq signals between SZ and normal controls for H3K4me3 and H3K27me3, respectively. As H3K4me3 and H3K27me3 are major indicators for gene activation and repression, respectively, we summarized their overall histone modification changes at each gene using d=ΔK4–ΔK27. A positive d should correlate with an upregulation of gene expression associated with SZ, and a negative d should correlate with a downregulation. d=0 implies no difference between SZ and controls. We then asked which gene groups (pathways) are significantly associated with SZ by applying the generalized Wald test (see Supplementary Methods). Out of 6767 gene groups in total, 545 gene groups showed SZ-associated changes in the levels of histone methylation signals (H3K4me3 and/or H3K27me3) and 517 gene groups had SZ-associated gene expression changes in microarray (false discovery rate cutoff point at q<0.01), respectively (Figure 1c and Supplementary Tables 3 and 4 for full analysis data). As an overlap between 545 and 517 gene groups, we identified 72 gene groups whose expression is affected by SZ-associated H3K4me3 and/or H3K27me3 changes (Figure 1d and Supplementary Table 5 for full analysis data).
Although the individual gene analysis and pathway analysis were performed independently, genes in the category of `phase II detoxification' were significantly altered in the SZ group compared with normal controls in both analyses. These genes encode enzymes that transfer cofactor groups such as glutathione or uridine 5'-diphospho-glucuronosyltransferase (UDP-glucuronosyltransferase) to xenobiotic substrates and inactivate/detoxify them. Microsomal glutathione S-transferase 1 (MGST1), identified in the individual gene analysis, is involved in the conjugation of glutathione S-transferase to these substrates. Gene groups of `Reactome_glucuronidation' consist of the UDP-glucuronosyltransferases that catalyze a wide range of endogenous and xenobiotic compounds by the transfer of a glucuronate group of uridine diphosphoglucuronate. It has been increasingly recognized that oxidative stress is involved in SZ pathophysiology.10 As phase II detoxification is involved in cellular protection against oxidative stress, epigenetic alterations in the expression of these enzymes may underlie at least in part a pathophysiological mechanism for SZ.
This discovery study comprises several advantages and also limitations. Although we enriched olfactory cells with neuronal traits to near homogeneity, these cells may not fully represent brain neurons. Many confounding factors, such as medications and diet, may be washed out by many changes of media in the course of cell culture. Furthermore, treatment of olfactory cells with clozapine, a representative neuroleptic, did not affect the expression of MGST1 gene, one of our major findings in this study (Supplementary Figure 3). However, we cannot completely exclude persistent hallmarks that might be imprinted before the biopsy. Generation of induced pluripotent stem cell (iPS)-derived neurons or induced neurons (iN) from `drug-naïve' SZ patients may be an alternative approach that might overcome these limitations. Nonetheless, these iPS/iN cells artificially generated by introduction of exogenous factors also include substantial confounds, such as reprogramming-related epigenetic alterations and heterogeneity of cells in culture. Although our samples are not completely matched by race, additional analysis shows that racial differences are not significantly associated with any gene groups (pathways) (Supplementary Tables 6 and 7). Finally, we also fully acknowledge that the sample size in this study is small. Pathway analysis on the basis of unbiased profiling of multiple histone modifications in homogeneous cells with neuronal trait, however, enables us to obtain a scientifically meaningful notion.
Supplementary Material
Footnotes
CONFLICT OF INTEREST The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
REFERENCES
- 1.Feinberg AP. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 2.Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS. Nat Rev Endocrinol. 2009;5:401–408. doi: 10.1038/nrendo.2009.102. [DOI] [PubMed] [Google Scholar]
- 3.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung I, Shulha HP, Jiang Y, Matevossian A, Wang J, Weng Z, et al. Proc Natl Acad Sci USA. 2010;107:8824–8829. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang HS, Matevossian A, Whittle C, Kim SY, Schumacher A, Baker SP, et al. J Neurosci. 2007;27:11254–11262. doi: 10.1523/JNEUROSCI.3272-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tajinda K, Ishizuka K, Colantuoni C, Morita M, Winicki J, Le C, et al. Mol Psychiatry. 2010;15:231–232. doi: 10.1038/mp.2009.73. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Z, Yuan Q, Mash DC, Goldman D. Proc Natl Acad Sci USA. 2011;108:6626–6631. doi: 10.1073/pnas.1018514108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji H. Methods Mol Biol. 2010;674:143–159. doi: 10.1007/978-1-60761-854-6_9. [DOI] [PubMed] [Google Scholar]
- 10.Do KQ, Cabungcal JH, Frank A, Steullet P, Cuenod M. Curr Opin Neurobiol. 2009;19:220–230. doi: 10.1016/j.conb.2009.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.