Abstract
Cytochrome c is a highly conserved protein, with 20 residues identical in all eukaryotic cytochromes c. Glycine 41 is one of these invariant residues, and is the position of the only reported naturally occurring mutation in cytochrome c (human G41S). The basis, if any, for the conservation of Gly-41 is unknown. The mutation of Gly-41 to Ser enhances the apoptotic activity of cytochrome c without altering its role in mitochondrial electron transport. Here we have studied additional residue 41 variants and determined their effects on cytochrome c functions and conformation. A G41T mutation decreased the ability of cytochrome c to induce caspase activation and decreased the redox potential, whereas a G41A mutation had no impact on caspase induction but redox potential increased. All residue 41 variants decreased the pKa of a structural transition of oxidized cytochrome c to the alkaline conformation, and this correlated with a destabilization of the interaction of Met-80 with the heme iron(III) at physiological pH. In reduced cytochrome c the G41T and G41S mutations had distinct effects on a network of hydrogen bonds involving Met-80, and in G41T the conformational mobility of two Ω loops was altered. These results suggest the impact of residue 41 on the conformation of cytochrome c influences its ability to act in both of its physiological roles, electron transport and caspase activation.
Keywords: Cytochrome c, alkaline transition, apoptosis, redox potential, NMR
Introduction
Cytochrome c (cyt c) is a highly conserved heme protein that has multiple functions within the cell. It is absolutely required as an electron carrier in the intermembrane space of mitochondria, and is an important trigger for caspase activation and subsequent apoptotic cell death when released into the cytosol. Cyts c from many species have been used as model proteins to understand how structure drives function in heme-containing proteins. Five pH-dependent conformations of oxidized cyt c have been characterized by differences in heme ligation and protein folding, and conformational mobility and changes in heme ligation have been correlated with changes in the redox potential of cyt c [1-3]. However the impact of conformational dynamics [4-6] on function is not fully understood, particularly in relation to caspase activation.
During apoptosis cyt c is released from mitochondria into the cytosol where it binds to the apoptotic protease-activating factor 1 (Apaf-1) triggering oligomerization to form the heptameric apoptosome, a platform for caspase activation [7-9]. The interaction between Apaf-1 and cyt c is thought to be predominantly electrostatic, involving surface lysine residues and the solvent-exposed pyrrole ring C of cyt c [10-15]. It has also been reported that during apoptosis cyt c undergoes a conformational change, which may be required for binding to Apaf-1 [16,17].
We have previously reported the identification of a naturally occurring mutation of human cyt c (G41S), in a family with autosomal dominant thrombocytopenia (low platelets) [18]. Glycine 41 is conserved in all eukaryotic cyts c [19] and is located at the beginning of an Ω-loop (residues 40-to-57) [20] which plays an important role in cyt c folding and unfolding [21]. At pH 7 Fe(II)-G41S cyt c (PDB 3NWV) has no significant structural difference when compared to other reduced cyts c [18,22] suggesting that the variant can adopt the native conformation. However the γ-hydroxyl of Ser-41 participates in a hydrogen bond network including propionate 7 of the heme pyrrole ring A that could stabilize both the heme environment and the 40-57 Ω-loop, and thus influence the conformational mobility of cyt c (Figure 1). In vitro the G41S mutation enhances the ability of cyt c to activate caspases [18]. Although the presence of serine at residue 41 alters the electronic structure of the heme cofactor and increases the electron-self-exchange rate of the protein [22], the redox potential is not altered and the respiratory activity of cyt c in the mitochondrial electron transport chain is unaffected [18].
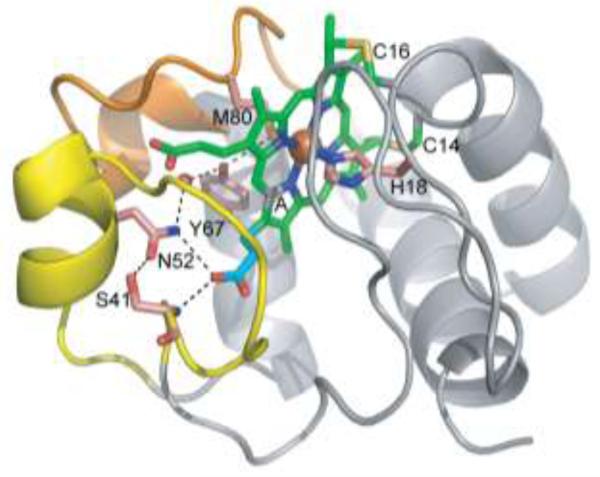
FIG. 1.
Ser-41 introduces an additional H-bond in the network linking Ω loop 40-to-57 (yellow) to propionate 7 in Fe(II) cyt c. An additional H-bond between the γ-hydroxyl of Ser-41 and the δ-NH2 of Asn-52 is introduced to the H-bond network that also includes propionate 7 (cyan) of the heme (green) pyrrole ring A (A), Tyr-67, Met-80 and a water molecule (red sphere). Selected sidechains are labeled and shown as sticks with carbon in salmon, oxygen in red, nitrogen in blue and sulfur in yellow. The 71-to-85 Ω loop is shown in orange. Drawn from PDB entry 3NWV.
To further investigate the role of residue 41 in determining biochemical and biophysical properties of cyt c, additional Gly-41 variants, G41A and G41T, were made. Here we report a biophysical characterization of these variants along with assessment of their apoptotic activities.
Materials and Methods
Expression and purification of cyt c
The G41S, G41A and G41T mutations were introduced into the human cyt c expression vector pBTR (HumanCc) by site-directed mutagenesis as previously described [18]. Cyts c were expressed and purified as previously described [22]. Protein concentrations were measured spectrophotometrically using a Nanodrop UV-visible spectrophotometer. Concentrations were calculated using the molar extinction coefficient 106.1 mM−1cm−1 at 410 nm [23].
Cell culture, and preparation of cell cytosolic extracts
Cell-free cytosolic extracts were prepared from human leukemic monocyte lymphoma cells (U937) as previously described [18]. Briefly, cells were grown in RPMI 1640 media with 10% fetal bovine serum in 5% CO2 at 37°C. Cells were collected and ice-cold cell extraction buffer (Hepes-KOH (pH 7.5), 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol (DTT), 250 μM PMSF and Complete protease inhibitor cocktail (Roche)) was added to give 8 × 107 cells/mL. Cells were allowed to swell on ice for 60 minutes and lysed by passing through a 28-gauge needle. Lysates were centrifuged for 10 minutes at 10,000 g, followed by ultracentrifugation at 186,007 g for 60 minutes. Lysates were stored in aliquots at −80°C until required.
Caspase assays
U937 cytosolic extracts (100 μg protein) were incubated at 37°C in the presence of 10, 50 or 100 nM cyt c and 1 mM dATP in a final volume of 60 μL caspase assay buffer (100 mM Hepes-KOH (pH 7.25), 10% w/v sucrose, 0.1% w/v 3-[(3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate (CHAPS) and 5 mM DTT). Caspase 3-like activity was measured fluorimetrically with an excitation wavelength of 390 nm and an emission wavelength of 460 nm by the addition of Ac-DEVD-AMC (50 μM, AMC is 7-amino-4-methylcoumarin) at 37°C in 96-well plates using a POLARstar OPTIMA Microplate Reader (BMG LABTECH). To derive a measure of apoptosome activation for each cyt c variant, the maximum slope of a plot of rate of AMC production versus time was calculated (in effect, the second derivative of AMC concentration vs. time). The slope was determined by least squares fitting of a line to at least 30 data points and the standard deviation error of the slope was calculated according to Taylor [24].
Circular dichroism spectroscopy and differential scanning calorimetry
Purified ferricyt c was dialyzed into 50 mM citric acid buffer pH 3.8 (thermal denaturation studies) or 50 mM sodium phosphate pH 7.5 (CD spectra) and diluted to 30 μM. CD data were obtained using an Olis DSM 10 spectrophotometer equipped with a thermostat (Olis, Bogart, GA) with a 1 mm path length quartz cuvette. Spectra were recorded at wavelengths of 260-200 nm with a 1.0 nm step size and a slit bandwidth of 1.5 nm at 25°C. Signal averaging time was 3 s/point and ellipticities reported as mean residue ellipticity (θ) in degree cm2/dmol. Thermal denaturation profiles were acquired by monitoring the ellipticity at 222 nm as a function of temperature between 4 and 96°C in 2°C increments. The midpoint of thermal denaturation (Tm) was determined using a two-state model as described previously [25].
Differential scanning calorimetry (DSC) experiments were performed using a VP-DSC. Calorimic cells (0.5 mL) were kept under an excess pressure of 30 psi and at a scan rate of 60°C h−1. Data from the scanning calorimeter were analyzed using the Orion software supplied by MicroCal. The buffer baseline was subtracted, and the data were converted to molar excess heat capacity by using the protein concentration and the cell volume [26].
Redox potential of cyt c
The redox potentials of the cyt c variants were determined in 50 mM citrate buffer at pH 6.5 by measuring the change in absorbance at 550 and 570 nm while altering the redox state of the system by addition of increasing amounts of ascorbate [27]. Data were analyzed as described [27].
Monitoring Met-80 ligation by A695
Cyt c was dialyzed into 0.1 M sodium phosphate (pH 7.5). Absorbance at 695 nm was measured using 250 μM ferricyt c in a 1 cm pathlength cuvette with a Varian Cary UV-Vis spectrophotometer; pH titrations were carried out by 1 μL additions of 3 M NaOH at 25°C in 50 mM sodium phosphate. The pH values were measured directly in cuvettes using a Mettler Toledo InLab Micro Pro Electrode connected to an Orion pH meter. Analysis of the data was done using the curve fitting program SigmaPlot (v11). Changes in the absorbance of the 4 cyts c were analyzed using an expression for a two state transition where AH695 is the absorbance at 695 nm of the alkaline form and AL695 is that of the native form (equation 1) [28].
| (1) |
NMR Spectroscopy
All NMR experiments were performed at 25 °C on a Varian Inova 500-MHz spectrometer equipped with a triple-resonance probe. Samples for 1-D 1H NMR contained 1 mM cyt c in either 45 mM sodium phosphate (10 % D2O, pH 7.0) or 45 mM sodium acetate (10 % D2O, pH 5.2). 1H-1H total correlation spectroscopy (TOCSY) and 1H-1H nuclear Overhauser enhancement spectroscopy (NOESY) experiments on reduced proteins contained 3 mM cyt c in 100 mM sodium phosphate buffer, pH 7.0. Oxidized protein samples contained 15 mM K3[Fe(CN)6] (Sigma), and reduced samples were deoxygenated and contained 15 mM Na2S2O4 (Fluka).
1-D 1H NMR spectra used a relaxation delay of 1 s, a water pre-saturation delay of 1 s, an acquisition time of 1 s (total recycle time = 3 s). A sweep width of 40 kHz was used for oxidized proteins, and 10 kHz for reduced proteins. TOCSY experiments used a recycle time of 1.2 s and a spin-lock time of 45 ms while NOESY experiments used a recycle time of 1.1 s and a mixing time of 100 ms. 1-D NMR spectra were processed in the ACD/Labs 12.0 software package with 10 Hz exponential line broadening. 2-D NMR spectra were processed in NMRPipe with polynomial solvent correction of the time-domain data. All spectra were referenced to the residual H2O peak at 4.77 ppm (pH 7.0) or 4.74 ppm (pH 5.2). Assignments were made using standard methods, assisted by assignments available in the literature [29,30].
Results and Discussion
Ser and Thr substitutions of Gly-41 in human cyt c have opposite effects on caspase activation
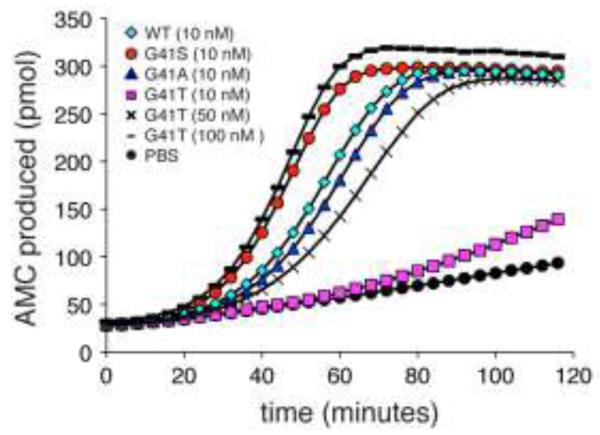
The naturally occurring human cyt c variant, G41S, has an enhanced ability to trigger caspase activation (Figure 2, Table 1, [18]). To further understand the molecular basis of this enhanced activity, additional residue 41 variants of cyt c were studied. The presence of Ser introduces a polar side chain at residue 41. To determine whether the introduced hydroxyl group was an important factor in the enhanced caspase activation, a Thr was introduced at residue 41. In striking contrast to G41S cyt c, G41T cyt c had 15-fold lower caspase activation compared to WT cyt c. The activation of caspases by G41T cyt c was restored by increasing the G41T cyt c concentration 5-10 – fold compared to WT (Figure 2, Table 1). Substitution of Gly-41 by Ala had no impact on caspase activation (Figure 2, Table 1), suggesting that the β-carbon of Ser-41 is not the source of the change in activity.
FIG. 2.
Changes at residue 41 in cyt c modulate caspase activation. Cleavage of the caspase 3 substrate Ac-DEVD-AMC was monitored at 37°C in U937 cytosol with the addition of 10 nM WT, G41S, G41A or G41T, 50 nM G41T, 100 nM G41T cyts c or PBS at pH 7.25. All reactions contained 1 mM dATP and 5 mM DTT. The error bars have been omitted to improve the clarity of the figure. The full figure with error bars is in the Supplementary Material (Figure S1).
Table 1.
Rates of apoptosome activation and biophysical properties of cyt c variants.
| Apoptosome activation | CD | DSC | Redox potential |
Met-80 ε- C1H3 |
Alkaline transition |
||
|---|---|---|---|---|---|---|---|
| pmol.min−1.min−1 | Normalized to WT |
Tm (°C) | Tm (°C) | mV ± SD | ppm | pKa | |
| PBS | 0.003 ± 0.001 | 0.015 | |||||
| WT | 0.200 ± 0.003 | 1 | 71 ± 3 | 66.7 ± 0.2 | 226 ± 3 | −23.9 | 9.3 ± 0.4 |
| G41S | 0.317 ± 0.004 | 1.58 | 69 ± 1 | 66.6 ± 0.8 | 227 ± 4 | −23.2 | 7.8 ± 0.3 |
| G41A | 0.181 ± 0.004 | 0.9 | 69 ± 1 | 67.9 ± 0.5 | 244 ± 3 | −23.5 | 8.1 ± 0.5 |
| G41T | 0.014 ± 0.001 | 0.07 | 68 ± 4 | 67.6 ± 0.5 | 195 ± 1 | −19.9 | 6.7 ± 0.2 |
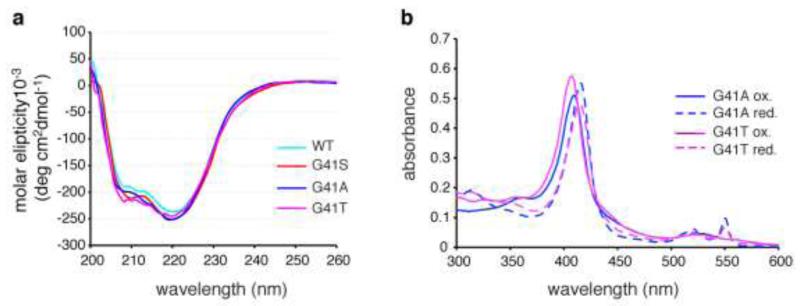
The decreased caspase activation seen with G41T cyt c was not due to a significant change in protein secondary structure. The far-UV CD spectra of oxidized G41T cyt c and the other variants were consistent with a high α-helical content and did not indicate any significant differences in secondary structure (Figure 3a). The narrow lines and excellent chemical shift dispersion in the 1-D NMR spectra of reduced WT, G41S and G41T cyts c (Figure S2) indicate that all are folded with well-defined tertiary structures. The 2-D NMR spectrum of reduced G41T is very similar to WT with the exception of some NOESY peaks (see below), further indicating that the protein is folded with a tertiary structure similar to WT. The 1-D NMR spectra of the oxidized proteins are discussed below. Using conditions under which thermal denaturation of ferricyt c is reversible (pH 3.8), there were no significant differences in Tm between the variants as measured by CD and DSC (Table 1), with values being similar to those previously reported for human ferricyt c under the same conditions [15]. In addition G41T and G41A cyts c had 300-600 nm absorption spectra resembling WT (Figure 3b).
FIG. 3.
Changes at residue 41 do not alter the gross structural features of cyt c. a. Far-ultraviolet circular dichroism spectra of recombinant human ferricyt c variants. The experiments were carried out at 25°C in 50 mM sodium phosphate, pH 7.5. b. UV-Vis absorption spectra of oxidized and reduced G41A and G41T cyts c.
Mutation of Gly41 alters the redox potential of human cyt c
As previously reported there was no significant difference in redox potentials of WT and G41S cyts c (Table 1, [18]). In contrast substitution of Gly-41 by Thr decreased the redox potential of cyt c by 31 mV whereas substitution with Ala increased the redox potential by 18 mV (Table 1). The redox potential of cyt c is determined by the relative stability of the oxidized and reduced states of the heme [31]. Changes in the redox potential of cyt c may indicate changes to the heme environment [2,4,32,33], and alterations in the conformation and H-bond network of Gly-41 have been reported in yeast ferrocyt c variants (N52A and I75M) with reduced redox potentials [34]. To investigate the heme environment in the variants, NMR and UV-Vis absorption spectroscopy were employed.
Iron coordination environment in human cyt c variants
In WT human cyt c the heme is covalently bound through thioether linkages with Cys-14 and Cys-17. At physiological pH the Fe is hexacoordinated by four pyrrole nitrogens of the heme, the ε-nitrogen atom of His-18 and the sulfur atom of Met-80, maintaining the Fe in a low-spin state. Compared to WT, the 1H resonance of the upfield Met-80 ε-CH3 group shifted downfield in all oxidized variants, with the largest shift being 4 ppm for G41T cyt c (Table 1). These data suggest either a perturbation to the Fe(III)Met-80 coordination, or another perturbation to the heme electronic structure.
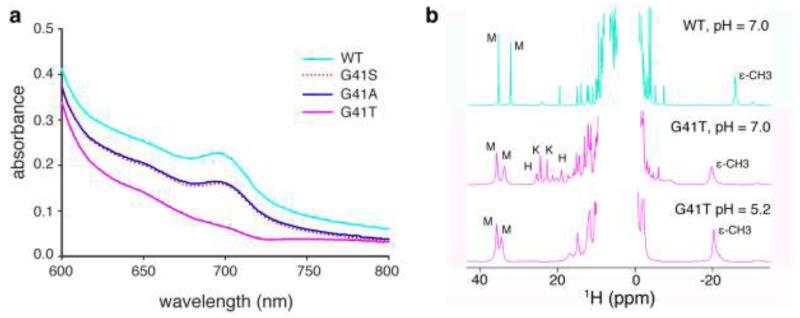
The absorption spectrum of ferricyt c is characterized by a weak absorbance at 695 nm, which is diagnostic for Fe(III)-Met-80 coordination [35]. When assessed at pH 7.5, the 695-nm absorption band was significantly diminished in the UV/Vis spectrum of G41T cyt c compared to WT, G41S and G41T cyts c (Figure 4a). This implies a perturbation to Fe(III)-Met-80 coordination in G41T cyt c, which was confirmed in the 1H NMR spectra (Figure 4b). The 1H NMR spectrum of WT ferricyt c exhibits two intense downfield resonances between 30 and 40 ppm, and one intense upfield resonance at −23.9 ppm, typical of a eukaryotic cyt c with His/Met axial ligation of Fe(III). While these resonances are retained in the NMR spectrum of G41T ferricyt c at pH 7.0, additional peaks are observed between 18 and 26 ppm. These peaks have similar chemical shifts and line widths to those of non-native conformations of horse Fe(III) cyt c under mildly denaturing conditions. By analogy to previous work, likely assignments for the narrow resonances at 24.3 and 22.6 ppm are heme methyls for a non-native form with His/Lys axial ligation, and the broader resonances at 25.2 and 18.9 ppm are heme methyls for a form with His/His ligation (Table S1) [36]. When G41T ferricyt c is exchanged into sodium acetate buffer (pH 5.2), the resonances characteristic of His/Lys and His/His axial ligation are no longer present, whereas the peaks indicating His/Met axial ligation remain (Figure 4b). The increased widths of the hyperfine-shifted resonances for the His/Met form of G41T relative to WT may be a result of heterogeneity and/or mobility in the heme environment. This behaviour is observed at pH 7 and pH 5.2 for the resonances suggesting that it is not just a function of pH but a property of G41T with His/Met axial ligation. The exact position of the 1H resonances diagnostic for His/Met ligation changes from pH 7.0 to 5.2, but this is most likely due to pH-dependent perturbations of the active site environment. These data suggest that when oxidized, the G41T variant exists as an equilibrium of multiple coordination states at pH 7.0 and one coordination state (His/Met) at pH 5.2. The existence of these multiple coordination states may contribute to the decrease observed in the redox potential of G41T cyt c as a result of increased heme solvent exposure and/or the participation of misligated states in the redox equilibrium.
FIG. 4.
Introducing a threonine at residue 41 disrupts Fe(III)-Met-80 coordination. a. Fe(III)-Met-80 coordination was measured by monitoring absorbance from 600-800 nm with 250 μM ferricyt c in 50 mM sodium phosphate pH 7.5. b. Nuclear magnetic resonance spectroscopy at pH 7 (sodium phosphate) and pH 5.2 (sodium acetate). Downfield heme methyl resonances are labeled M (His/Met), K (His/Lys) and H (His/His) as assigned in [36]. Upfield Met-80 ε-CH3 group is labeled ε-CH3.
Substitution of Gly-41 lowers the pKa of the alkaline transition for human cyt c
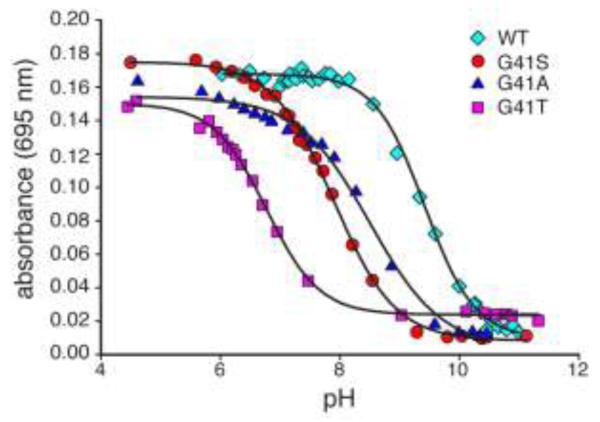
Loss of Fe(III)-Met-80 coordination is characteristic of oxidized cyt c having undergone the alkaline transition [37], which occurs with a pKa of 9.3-9.9 for WT human cyt c [38-40]. The alkaline transition is characterized by replacement of Met-80 axial Fe(III) ligation with Lys-72, Lys-73 or Lys-79, maintaining the iron in a low-spin state [3,4,41-45]. We determined the pKa of the alkaline transition for the 4 variants by monitoring A695 as the pH was increased (Figure 5). Cumulative addition of NaOH caused a progressive decrease in absorbance at 695 nm, reflecting the displacement of the Met-80 ligand. The pKa for this transition was significantly lower for G41T compared to the other variants, although G41S and G41A also displayed lower pKa values than WT (Table 1). Interestingly, there is a clear correlation between changes in the pKa of the alkaline transition and the 1H chemical shift of the Met-80 ε-CH3 group (Table 1). Since the negative NMR hyperfine shift of this resonance arises from spin polarization across the Fe(III)-Met-80 bond [46], the NMR data are consistent with decreasing Fe(III)-Met-80 bond strength from WT>G41A>G41S>G41T.
FIG. 5.
Changes at residue 41 alter the pKa of the alkaline transition. The 695 nm absorbance of 250 μM ferricyt c in sodium phosphate (50 mM) at 25°C was monitored as the pH was increased. Curves represent fit to a two-state transition.
It has previously been shown that mutation of Tyr-48 to a phosphotyrosine mimetic (Y48E) or nitration of Tyr-74 decreases the alkaline transition pKa of cyt c (to 7.0 and 7.2 respectively) [38,47]. In both cases there was also a loss of cyt c-induced caspase activation [38,48]. In the alkaline conformation the heme iron(III) is ligated to a lysine ε-amino group in place of the sulfur of Met-80. Mutations of the surface lysine residues implicated in this coordination (Lys-72, -73, -79) also impair the interaction of cyt c with Apaf-1 and inhibit caspase activation in vitro and in vivo [11,14,15]. Thus the sequestration of a lysine residue in coordination with the heme iron has previously been suggested to be responsible for the significant inhibition of caspase activation by Y48E and nitrated cyt c. However the assessment of caspase activation is routinely performed in the presence of 1-5 mM DTT. Under these conditions cyt c is rapidly reduced [49], and reduced cyt c does not undergo a pH-dependent transition to the alkaline conformation. It therefore remains unclear why G41T, or indeed the Y48E or nitrated cyt c, exhibit decreased caspase activation in vitro. The modifications that cause the alkaline transition to occur at physiological pH may also independently reduce apoptotic activity. Whether the G41T mutation, phosphorylation or nitration of cyt c in vivo will cause adoption of the alkaline conformation is an open question. Following mitochondrial outer membrane permeabilization, the cytosolic cyt c pool has been reported to be fully reduced [50] suggesting there is little relevance to caspase activation in vivo for the alkaline transition per se. However others have suggested that cyt c must be oxidized in order to maximize caspase activation [51]. If this is correct then the ability of nitrated or phosphorylated cyt c to adopt the alkaline conformation at a physiological pH could directly provide protection against apoptosis, as has been suggested [48].
The G41T and G41S mutations exert distinct effects on the conformation of reduced cyt c
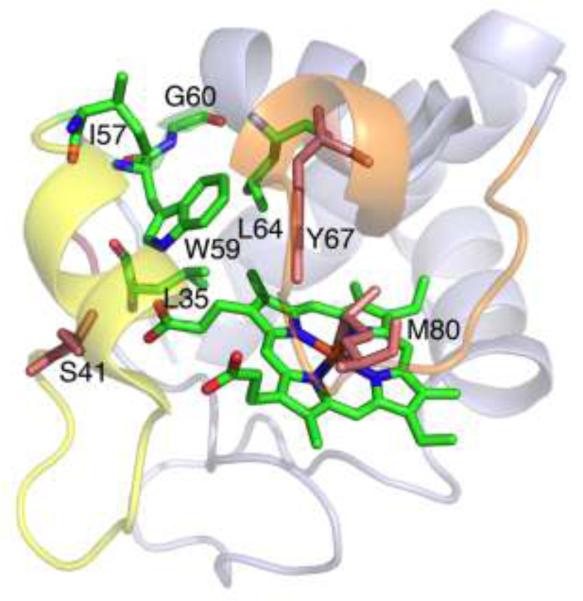
Reduced cyt c is the relevant oxidation state in the apoptosis assay. In order to investigate the impact of residue 41 mutation on reduced cyt c we analysed TOCSY and NOESY spectra of WT, G41T and G41S ferrocyts c. Overall the spectra were very similar, consistent with the lack of widespread structural change or stability indicated from the CD and DSC analyses of the oxidized variants. However, localized changes in NOEs were identified that lend insight into the disparate effects of the G41T and G41S mutations. Although assignment of Thr-41 itself could not be made, suggesting high solvent exposure and/or mobility, effects of the mutation on other nearby residues were evident. In particular, the G41T mutation was found to affect NOEs involving residues at and near the C-terminus of the Ω-loop that spans residues 40-to-57 and in the core of the protein near the heme. The residues analysed are highlighted in Figure 6. In WT cyt c, NOESY cross-peaks are observed between Ile-57 δ••H3 and Trp-59 ring protons (ε3-H, ζ3-H, η2-H). Trp-59 ε3-H also shows NOEs to Gly-60 HN and Leu-64 HN. The Trp-59 ε-NH shows a NOE to heme propionate 7 and to the Leu-35 δ-CH3 (Figure 6). In G41S, these NOEs among Ile-57, Trp-59, Gly-60, Leu-64, Leu-35, and heme propionate-7 are unaltered. In contrast significant disruptions to these interactions are observed for G41T. In G41T the only NOE observed between residues 57 and 59 is a very weak connectivity between Ile-57 δ••H3 and Trp-59 ring ε3-H, in contrast with the three strong NOEs observed between residues 57 and 59 in WT and in G41S cyt c. Also, the NOE between Trp59 ε3-H and Gly-60 HN is lost in G41T, and the NOE from Trp-59 ε3-H to Leu-64 HN is significantly weakened. In addition, the NOE between Leu-35 and Trp-59 is lost.
FIG. 6.
Interaction of W59, the 40-to-57 Ω-loop (yellow) and the 71-to-85 Ω-loop (orange) in human Fe(II) G41S cyt c (PDB: 3NWV). The amino acid residues analyzed in NMR spectra, and the heme, are shown in green, residues 41, 67, and 80 in salmon, oxygen in red and nitrogen in blue. View is rotated ~100° around a horizontal axis, relative to figure 1.
The changes in NOEs reveal that mutation of residue 41 has effects on the C-terminal end of the 40-to-57 Ω-loop and on core residues nearby (Trp-59 in particular). The most significant change observed in G41T is the loss of most NOEs involving Trp-59, suggesting a change in structure or an increase in mobility. The chemical shifts of the residues analyzed here show little difference among the variants (Table S2), suggesting that an increase in mobility rather than a change in structure accounts for the loss of NOEs. In particular, proximity of Ile-57 to Trp-59 is revealed in all variants by the unusual upfield chemical shifts of Ile-57 induced by the Trp-59 ring current. We thus conclude that the G41T, but not the G41S mutation, increases Trp-59 mobility. Trp-59 and Tyr-67 lie between the 40-to-57 and the 71-to-85 Ω-loops; the latter rearranges in the alkaline form [4]. Importantly, Tyr-67 forms a hydrogen bond to Met-80 and is involved in a hydrogen-bonding network in the interior of the protein that also includes Trp-59. A useful comparison is the Y67F mutant of yeast iso-1-cyt c, which has a lowered pKa for the alkaline transition. The X-ray crystal structure of the reduced Y67F mutant reveals increased mobility of Trp-59 and small changes in backbone structure for Gly41 and Trp-59 [52]. Here, we find that the G41T mutation results in increased mobility for Trp-59, disruption of the H-bonding network in the heme pocket including the Tyr-67-Met-80 H-bond, and weakening of the Met-Fe bond. Another mutation involving the 40-to-57 Ω-loop, N52G, has been reported to destabilize the loop and drop the pKa for the alkaline transition to 7.4 [53]. Finally, another study in which mutations at residue 67 were introduced revealed that these mutations weakened the Met-80-Fe bond [54].
It is surprising that the G41T mutation exerts significantly different effects on protein conformational mobility than does G41S, given the similarity of the substituted residues. We suggest that the increased size and hydrophobicity of the Thr side chain, although slight, is enough to disrupt the interaction between the neighbouring Thr-40 with Ile-57; the two residues that form a hydrogen bond to close the Ω-loop. This disruption could come from a steric clash of adjacent beta-branched residues (Thr-40, Thr-41), propagating structural change to residues 57 and 59. The G41S mutation also yields a change in internal hydrogen-bonding, but these changes are seen for the internal water molecule, heme propionate 7, and Arg-38, whereas Trp-59 is not perturbed [22]. The distinct effects of the G41S and G41T mutations on cyt c conformational mobility may explain why the mutations have opposite effects on caspase activation.
The role of the 40-57 Ω-loop in cyt c functions is intriguing and not fully understood. Previous studies have indicated that destabilization of this loop induces cyt c to more easily undergo the alkaline transition [53,55]. It has also been hypothesised that cyt c must undergo a conformational change to bind to Apaf-1 [16,17], with the region encompassing residue 44 implicated in this change [16]. Finally this region, and residue 41 specifically, has been implicated in a putative interaction with Bcl-xL to form an antiapopotic complex [56]. The diverse effects of different mutations at residue 41 that we observe support an important role of this residue and the surrounding region in cyt c functions, and are consistent with the complete conservation of this residue across all eukaryotic cyts c. Addition of a β-carbon at this position (G41A) has little effect on caspase activation, but increases the redox potential and causes a decrease of ~1 pH unit in the pKa of the alkaline transition. Further addition of a γ-hydroxyl group (G41S) enhances caspase activation, further decreases the alkaline transition pKa, but the redox potential is indistinguishable from WT. The effect of an additional methyl group (G41T) is striking, dramatically reducing caspase activation to ~7% that of WT or the G41A variant, decreasing the redox potential by 31 mV, and decreasing the pKa of the alkaline transition to 6.7. Analysis of the NMR spectra reveals that the G41T and G41S mutations have distinct effects on the 40-to-57 Ω-loop, with the G41S mutant resembling WT more closely, and the G41T mutant showing destabilization of the loop and increased mobility of Trp-59. Taken together these results suggest that the conformational mobility of the 40-to-57 Ω-loop, and its interaction with the rest of the protein, is a major factor in determining the properties and activities of cyt c.
Supplementary Material
Acknowledgements
We gratefully acknowledge the Macromolecular Interactions Facility (The University of North Carolina at Chapel Hill) and the Centre for Protein Research (University of Otago) for resources. We thank Prof Gary Pielak (Dept of Chemistry, The University of North Carolina at Chapel Hill) and Dr Ashutosh Tripathy (Macromolecular Interactions Facility, The University of North Carolina at Chapel Hill) for assistance with the DSC, Moira Hibbs (University of Otago) for technical assistance with protein expression and purification, and Rob Weeks (University of Otago) for assistance with cloning.
This work was supported by the Health Research Council (NZ), Marsden Fund (NZ), University of Otago Research Grant and HS and JC Anderson Charitable Trusts (NZ) (E.C.L), Elman Poole Travelling Scholarship (T.M.J.) and the NIH (GM63170 to K.L.B and F32 GM089016 to M.D.L).
Abbreviations used
- Ac-DEVD-AMC
acetyl-Asp-Glu-Val-Asp-7-amino-4-methylcoumarin Apaf-1, apoptosis protease activating factor 1
- CHAPS
3-[(3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate
- cyt c
cytochrome c
- DSC
differential scanning calorimetry
- DTT
dithiothreitol
- NOE
nuclear Overhauser enhancement
- NOESY
NOE spectroscopy
- NMR
nuclear magnetic resonance
- Tm
midpoint of thermal denaturation
- TOCSY
total correlation spectroscopy
- WT
wild-type
References
- 1.Duncan MG, Williams MD, Bowler BE. Protein Sci. 2009;18:1155–1164. doi: 10.1002/pro.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michel LV, Ye T, Bowman SE, Levin BD, Hahn MA, Russell BS, Elliott SJ, Bren KL. Biochemistry. 2007;46:11753–11760. doi: 10.1021/bi701177j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dopner S, Hildebrandt P, Rosell FI, Mauk AG. J Am Chem Soc. 1998;120:11246–11255. [Google Scholar]
- 4.Assfalg M, Bertini I, Dolfi A, Turano P, Mauk AG, Rosell FI, Gray HB. J Am Chem Soc. 2003;125:2913–2922. doi: 10.1021/ja027180s. [DOI] [PubMed] [Google Scholar]
- 5.Baxter SM, Fetrow JS. Biochemistry. 1999;38:4493–4503. doi: 10.1021/bi982742z. [DOI] [PubMed] [Google Scholar]
- 6.Sakamoto K, Kamiya M, Uchida T, Kawano K, Ishimori K. Biochem Biophys Res Commun. 2010;398:231–236. doi: 10.1016/j.bbrc.2010.06.065. [DOI] [PubMed] [Google Scholar]
- 7.Ow YP, Green DR, Hao Z, Mak TW. Nat Rev Mol Cell Biol. 2008;9:532–542. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- 8.Riedl SJ, Salvesen GS. Nat Rev Mol Cell Biol. 2007;8:405–413. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- 9.Jiang X, Wang X. Annu Rev Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- 10.Abdullaev Z, Bodrova ME, Chernyak BV, Dolgikh DA, Kluck RM, Pereverzev MO, Arseniev AS, Efremov RG, Kirpichnikov MP, Mokhova EN, Newmeyer DD, Roder H, Skulachev VP. Biochem J. 2002;362:749–754. doi: 10.1042/0264-6021:3620749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao Z, Duncan GS, Chang C-C, Elia A, Fang M, Wakeham A, Okada H, Calzascia T, Jang Y, You-Ten A, Yeh W-C, Ohashi P, Wang X, Mak TW. Cell. 2005;121:579–591. doi: 10.1016/j.cell.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Kluck RM, Ellerby LM, Ellerby HM, Naiem S, Yaffe MP, Margoliash E, Bredesen D, Mauk AG, Sherman F, Newmeyer DD. J Biol Chem. 2000;275:16127–16133. doi: 10.1074/jbc.275.21.16127. [DOI] [PubMed] [Google Scholar]
- 13.Sharonov GV, Feofanov AV, Bocharova OV, Astapova MV, Dedukhova VI, Chernyak BV, Dolgikh DA, Arseniev AS, Skulachev VP, Kirpichnikov MP. Apoptosis. 2005;10:797–808. doi: 10.1007/s10495-005-0366-9. [DOI] [PubMed] [Google Scholar]
- 14.Yu T, Wang X, Purring-Koch C, Wei Y, McLendon GL. J Biol Chem. 2001;276:13034–13038. doi: 10.1074/jbc.M009773200. [DOI] [PubMed] [Google Scholar]
- 15.Olteanu A, Patel CN, Dedmon MM, Kennedy S, Linhoff MW, Minder CM, Potts PR, Deshmukh M, Pielak GJ. Biochem Biophys Res Commun. 2003;312:733–740. doi: 10.1016/j.bbrc.2003.10.182. [DOI] [PubMed] [Google Scholar]
- 16.Jemmerson R, Liu J, Hausauer D, Lam K-P, Mondino A, Nelson RD. Biochemistry. 1999;38:3599–3609. doi: 10.1021/bi9809268. [DOI] [PubMed] [Google Scholar]
- 17.Patriarca A, Eliseo T, Sinibaldi F, Piro MC, Melis R, Paci M, Cicero DO, Polticelli F, Santucci R, Fiorucci L. Biochemistry. 2009;48:3279–3287. doi: 10.1021/bi801837e. [DOI] [PubMed] [Google Scholar]
- 18.Morison IM, Cramer Borde EM, Cheesman EJ, Cheong PL, Holyoake AJ, Fichelson S, Weeks RJ, Lo A, Davies SM, Wilbanks SM, Fagerlund RD, Ludgate MW, da Silva Tatley FM, Coker MS, Bockett NA, Hughes G, Pippig DA, Smith MP, Capron C, Ledgerwood EC. Nat Genet. 2008;40:387–389. doi: 10.1038/ng.103. [DOI] [PubMed] [Google Scholar]
- 19.Banci L, Bertini I, Rosato G, Varani G. J Biol Inorg Chem. 1999;4:824–837. doi: 10.1007/s007750050356. [DOI] [PubMed] [Google Scholar]
- 20.Leszczynski JF, Rose GD. Science. 1986;234:849–855. doi: 10.1126/science.3775366. [DOI] [PubMed] [Google Scholar]
- 21.Krishna MM, Lin Y, Rumbley JN, Englander SW. J Mol Biol. 2003;331:29–36. doi: 10.1016/s0022-2836(03)00697-1. [DOI] [PubMed] [Google Scholar]
- 22.Liptak MD, Fagerlund RD, Ledgerwood EC, Wilbanks SM, Bren KL. J Am Chem Soc. 2011;133:1153–1155. doi: 10.1021/ja106328k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Gelder B, Slater EC. Biochim Biophys Acta. 1962;58:593–595. doi: 10.1016/0006-3002(62)90073-2. [DOI] [PubMed] [Google Scholar]
- 24.Taylor JR. An introduction to error analysis: the study of uncertainties in physical measurements. University Science Books; 1982. [Google Scholar]
- 25.Cohen DS, Pielak GJ. Protein Sci. 1994;3:1253–1260. doi: 10.1002/pro.5560030811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liggins JR, Sherman F, Mathews AJ, Nall BT. Biochemistry. 1994;33:9209–9219. doi: 10.1021/bi00197a024. [DOI] [PubMed] [Google Scholar]
- 27.Cammack R. Redox states and potentials. In: Brown GC, Cooper CE, editors. Bioenergetics, A Practical Approach. IRL Press; Oxford: 1995. pp. 85–105. [Google Scholar]
- 28.Davis LA, Schejter A, Hess GP. J Biol Chem. 1974;249:2624–2632. [PubMed] [Google Scholar]
- 29.Wand AJ, Di Stefano DL, Feng YQ, Roder H, Englander SW. Biochemistry. 1989;28:186–194. doi: 10.1021/bi00427a026. [DOI] [PubMed] [Google Scholar]
- 30.Jeng WY, Chen CY, Chang HC, Chuang WJ. J Bioenerg Biomembr. 2002;34:423–431. doi: 10.1023/a:1022561924392. [DOI] [PubMed] [Google Scholar]
- 31.Lett CM, Guillemette JG. Biochem J. 2002;362:281–287. doi: 10.1042/0264-6021:3620281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barker PD, Mauk AG. J Am Chem Soc. 1992;114:3619–3624. [Google Scholar]
- 33.Tezcan FA, Winkler JR, Gray HB. J Am Chem Soc. 1998;120:13383–13388. [Google Scholar]
- 34.Rafferty SP, Guillemette JG, Berghuis AM, Smith M, Brayer GD, Mauk AG. Biochemistry. 1996;35:10784–10792. doi: 10.1021/bi960430v. [DOI] [PubMed] [Google Scholar]
- 35.Theorell H, Akesson A. J Am Chem Soc. 1941;63:1804–1820. [Google Scholar]
- 36.Russell BS, Melenkivitz R, Bren KL. Proc Natl Acad Sci U S A. 2000;97:8312–8317. doi: 10.1073/pnas.150239397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silkstone GG, Cooper CE, Svistunenko D, Wilson MT. J Am Chem Soc. 2005;127:92–99. doi: 10.1021/ja045719b. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Heredia JM, Diaz-Quintana A, Salzano M, Orzaez M, Perez-Paya E, Teixeira M, De la Rosa MA, Diaz-Moreno I. J Biol Inorg Chem. 2011;16:1155–1168. doi: 10.1007/s00775-011-0804-9. [DOI] [PubMed] [Google Scholar]
- 39.Osheroff N, Borden D, Koppenol WH, Margoliash E. J Biol Chem. 1980;255:1689–1697. [PubMed] [Google Scholar]
- 40.Ying T, Zhong F, Xie J, Feng Y, Wang ZH, Huang ZX, Tan X. J Bioenerg Biomembr. 2009;41:251–257. doi: 10.1007/s10863-009-9223-9. [DOI] [PubMed] [Google Scholar]
- 41.Rosell FI, Ferrer JC, Mauk G. J Am Chem Soc. 1998;120:11234–11245. [Google Scholar]
- 42.Ferrer JC, Guillemette TG, Bogumil R, Inglis SC, Smith M, Mauk AG. J Am Chem Soc. 1993;115:7507–7508. [Google Scholar]
- 43.Pollock WB, Rosell FI, Twitchett MB, Dumont ME, Mauk AG. Biochemistry. 1998;37:6124–6131. doi: 10.1021/bi972188d. [DOI] [PubMed] [Google Scholar]
- 44.Berghuis AM, Brayer GD. J Mol Biol. 1992;223:959–976. doi: 10.1016/0022-2836(92)90255-i. [DOI] [PubMed] [Google Scholar]
- 45.Hong XL, Dixon DW. FEBS Lett. 1989;246:105–108. doi: 10.1016/0014-5793(89)80262-5. [DOI] [PubMed] [Google Scholar]
- 46.Liptak MD, Wen X, Bren KL. J Am Chem Soc. 2010;132:9753–9763. doi: 10.1021/ja102098p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abriata LA, Cassina A, Tortora V, Marin M, Souza JM, Castro L, Vila AJ, Radi R. J Biol Chem. 2009;284:17–26. doi: 10.1074/jbc.M807203200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Heredia JM, Diaz-Moreno I, Nieto PM, Orzaez M, Kocanis S, Teixeira M, Perez-Paya E, Diaz-Quintana A, De la Rosa MA. Biochim Biophys Acta. 2010;1797:981–993. doi: 10.1016/j.bbabio.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 49.Hampton MB, Zhivotovsky B, Slater AFG, Burgess DH, Orrenius S. Biochem J. 1998;329:95–99. doi: 10.1042/bj3290095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ripple MO, Abajian M, Springett R. Apoptosis. 2010;15:563–573. doi: 10.1007/s10495-010-0455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borutaite V, Brown GC. J Biol Chem. 2007;282:31124–31130. doi: 10.1074/jbc.M700322200. [DOI] [PubMed] [Google Scholar]
- 52.Berghuis AM, Guillemette JG, Smith M, Brayer GD. J Mol Biol. 1994;235:1326–1341. doi: 10.1006/jmbi.1994.1086. [DOI] [PubMed] [Google Scholar]
- 53.Baddam S, Bowler BE. Biochemistry. 2006;45:4611–4619. doi: 10.1021/bi0524971. [DOI] [PubMed] [Google Scholar]
- 54.Ying T, Wang ZH, Lin YW, Xie J, Tan X, Huang ZX. Chem Commun (Camb) 2009:4512–4514. doi: 10.1039/b904347k. [DOI] [PubMed] [Google Scholar]
- 55.Hoang L, Maity H, Krishna MM, Lin Y, Englander SW. J Mol Biol. 2003;331:37–43. doi: 10.1016/s0022-2836(03)00698-3. [DOI] [PubMed] [Google Scholar]
- 56.Bertini I, Chevance S, Del Conte R, Lalli D, Turano P. PLoS ONE. 2011;6:e18329. doi: 10.1371/journal.pone.0018329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.