SUMMARY
DNA double-strand breaks (DSBs) activate a DNA damage response (DDR) that coordinates checkpoint pathways with DNA repair. ATM and ATR kinases are activated sequentially. Homology-directed repair (HDR) is initiated by resection of DSBs to generate 3′ ssDNA overhangs. How resection and HDR are activated during DDR or the roles of ATM and ATR in HDR are not known. Here, we show that CtIP undergoes ATR-dependent hyperphosphorylation in response to DSBs. ATR phosphorylates an invariant threonine, T818 of Xenopus CtIP (T859 in human). Non-phosphorylatable CtIP (T818A) does not bind to chromatin or initiate resection. Our data support a model in which ATM activity is required for an early step in resection leading to ATR activation, CtIP-T818 phosphorylation, and accumulation of CtIP on chromatin. Chromatin binding by modified CtIP precedes extensive resection and full checkpoint activation.
INTRODUCTION
DNA double-strand breaks (DSBs) can arise during normal cell metabolism as a consequence of interaction with reactive oxygen species, collapse of stalled replication forks, telomere dysfunction, chromosome breakage during anaphase, or following programmed genomic rearrangements during immune and germ cell maturation. Additionally, DSBs are formed after exposure to exogenous insults such as ionizing radiation (IR) or chemotherapeutic agents. Cells have evolved pathways, collectively termed the DNA damage response (DDR), to sense, signal, and repair these lesions. Failure to repair DSBs properly is associated with cancer development, radiation sensitivity, immune deficiencies, and developmental disabilities (Hoeijmakers, 2009).
DSBs are sensed by the Mre11-Rad50-Nbs1 complex (MRN), which binds to DNA ends and activates ATM protein kinase (Lee and Paull, 2007). ATM, ATR and DNA-PK are all members of the PIKK family of kinases that controls the DDR. ATM activation triggers multiple signaling pathways, causing changes in cell-cycle progression (damage checkpoints), gene expression, chromatin structure, and recruitment of repair proteins to sites of DNA damage (Derheimer and Kastan, 2010). DSBs can be repaired by non-homologous end-joining (NHEJ), which requires very minimal or no end-processing. Alternatively, DNA ends are resected to form 3′ single-stranded DNA (ssDNA) overhangs that allow annealing of the ends or strand invasion and homology search (HDR; (Symington and Gautier, 2011)). Repair pathway choice depends on cell-cycle phase, the structure of the damaged DNA ends, and the availability of DNA homologous to the damaged sequence (Shrivastav et al., 2008). HDR and NHEJ compete for DNA ends: binding of the NHEJ factor KU impairs resection, whereas resection prevents KU binding (Langerak et al., 2011; Sun et al., 2012). By generating RPA-coated ssDNA filaments, resection also activates a second protein kinase, ATR, which is recruited to ssDNA-RPA through the ATRIP adaptor protein (Zou and Elledge, 2003). Activation of Chk1 downstream of ATR requires a signaling complex that includes TopBP1, Rad9-Rad1-Hus1, and claspin. Activated Chk1 then spreads the checkpoint signal throughout the nucleus (Nam and Cortez, 2011). Thus, resection promotes a switch from ATM to ATR activation that reflects the conversion of dsDNA to ssDNA (Shiotani and Zou, 2009).
There are at least three distinct resection pathways. MRN-CtIP initiates resection whereas Exo1 exonuclease both initiates and extends resection tracts. In addition, DNA2 nuclease, in association with a RecQ helicase homolog (Sgs1 in yeast, WRN or BLM in vertebrates) and Top3-Rmi1/2, can extend resection tracks. Studies of DSB repair often utilize restriction endonucleases to create DSBs with a free 5′ phosphate and 3′ hydroxyl group. Repair of these DSBs can occur in the absence of CtIP or MRN (Sartori et al., 2007), and is due to the activity of Exo1 exonuclease and the RecQ helicase in cooperation with DNA2-Top3α-Rmi1/2 (Budd and Campbell, 2009; Liao et al., 2008a; Tomimatsu et al.; Zhu et al., 2008). In contrast, resection of DSBs induced by IR, chemotherapeutic agents or meiotic recombination, as well as those containing modified bases, altered chemistry, or covalent protein adducts (Barker et al., 2005; Henner et al., 1983; Keeney and Neale, 2006; Lawley and Phillips, 1996), must be initiated by the endonucleolytic activity provided by MRN in complex with CtIP (Paull, 2010). Thus, cells defective in Mre11 endonuclease activity or CtIP are highly sensitive to topoisomerase poisons and IR, and are unable to repair Spo11-capped meiotic DSBs. (Akamatsu et al., 2008; Hartsuiker et al., 2009b; Langerak et al., 2011; Limbo et al., 2007; Milman et al., 2009; Rothenberg et al., 2009; Sartori et al., 2007; Williams et al., 2008).
CtIP activation requires Cdk2/Cdk1 phosphorylation of a conserved residue, T847 in humans and T806 in Xenopus. This modification restricts CtIP activity to the S, G2 and M phases of the cell cycle (Huertas and Jackson, 2009; Peterson et al., 2011), ensuring that HDR is not initiated before DNA replication provides a homologous template for repair. Many substrates of ATM and ATR have been identified, including proteins that regulate DSB repair such as Mre11, Nbs1 or CtIP, but the functional impact of these modifications on HDR is not known. S. cerevisiae Sae2, the budding yeast ortholog of CtIP, is phosphorylated by ATM (Tel1) as well as ATR (Mec1), principally by the latter, and these modifications are required for Sae2 activity (Baroni et al., 2004). The functional consequences of CtIP phosphorylation by PIKKs are not known.
The DDR can be recapitulated in cell-free extracts derived from Xenopus laevis eggs (Garner and Costanzo, 2009; Srinivasan and Gautier, 2011). For example, DSB resection can been studied in this setting using small DNA templates, (Liao et al., 2012b; Liao et al., 2008a, 2011; Taylor et al., 2010), as well as in the context of chromosomal DSBs (Peterson et al., 2011; You et al., 2009). Low-speed supernatant (LSS), a cytosolic egg extract that contains all soluble proteins and lipids, supports nuclear envelope assembly around added sperm chromatin, which then undergoes semi-conservative DNA replication. High-speed supernatant (HSS), obtained by ultracentrifugation of LSS, is membrane-free, and does not replicate DNA. However, HSS still supports many DNA transactions, such as pre-replication complex formation, ATM and ATR activation, resection, and checkpoint signaling in response to DSBs.
Here, we use HSS and LSS extracts to evaluate the contribution of the three different resection pathways towards resection and the roles of ATM and ATR. We show that EXO1 can initiate resection of DSBs generated by endonucleases in the absence of ATM. In contrast, ATM is essential for CtIP-MRN-dependent resection in EXO1-defective extract. We identify CtIP-T818 (corresponding to human T859) by mass spectrometry as a conserved target of phosphorylation by PIKKs. Characterization of this site using phospho-specific antibodies and mutational analysis reveals that it is phosphorylated by ATR and is required for binding of CtIP to chromatin and subsequent processive resection. ATM activation precedes ATR phosphorylation of CtIP, which allows DNA2-dependent long-range resection. Therefore, our work provides a functional link between the activation of checkpoint kinases and repair of DSBs.
RESULTS
Initiation of DSB resection in cell-free Xenopus extracts
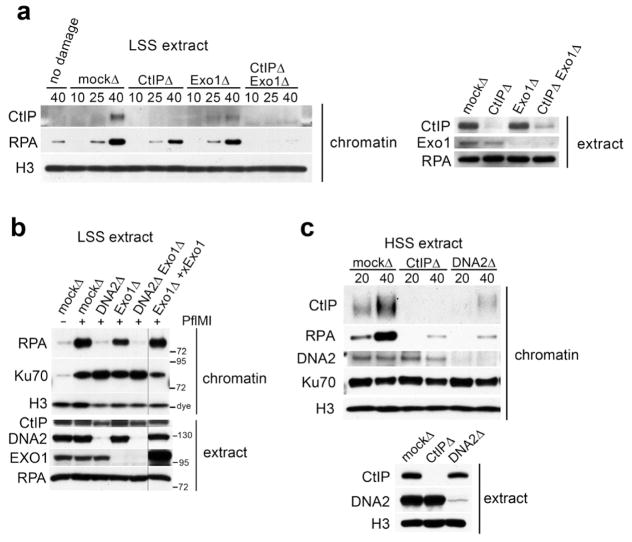
Studies in yeast and our work in Xenopus extracts suggest that at least two pathways can initiate DNA DSB resection in the context of chromatin. In Low Speed Supernatant (LSS), inactivation of the CtIP-MRN pathway delays but does not abrogate resection (Peterson et al., 2011) suggesting that another factor(s) can initiate resection. In contrast, membrane-free cytosol, referred to as High Speed Supernatant (HSS), exclusively initiates resection via the CtIP-MRN pathway (Peterson et al., 2011; You et al., 2009). Our studies also established that the EXO1, DNA2 and CtIP-MRN pathways all participate in processing DSBs on small DNA templates (Liao et al., 2012a; Liao et al., 2008b, 2011). We wished to elucidate the molecular basis for the difference between extracts, and to delineate the contribution of the three known resection pathways, CtIP-MRN, EXO1, and DNA2, towards initiation and long-range chromosomal DNA resection. We quantitatively depleted EXO1 from LSS using specific antibodies (Liao et al., 2011). EXO1 depletion inhibited, but did not abolish, ssDNA formation from PflMI-induced DSBs, as detected by chromatin-binding of RPA (Fig. 1a). As we reported previously (Peterson et al., 2011), CtIP depletion likewise reduced, but did not abrogate, resection (Fig. 1a). Importantly, no resection was detected in the absence of both EXO1 and CtIP. These results indicate that CtIP-MRN and EXO1 independently initiate resection of DSBs (Fig. 1a). We next evaluated the contribution of the DNA2 pathway, which processively extends resection tracts, but, unlike EXO1, cannot initiate resection. Depletion of DNA2 nearly abrogated resection (Fig. 1b), as did immunodepletion of both EXO1 and DNA2 (Fig. 1b). The resection defect in EXO1-depleted extracts was rescued by addition of purified recombinant EXO1 protein. However, addition of purified DNA2 protein did not rescue DNA2 depletion presumably because DNA2 functions in a complex with BLM or WRN, RMI1, and/or TOP3α. We conclude that EXO1 and DNA2 pathways are not redundant for resection, that DNA2 resection depends upon initial DSB processing, and that resection in LSS can be accounted for by the CtIP-MRN, DNA2 and EXO1 pathways.
Figure 1.
Contribution of individual resection pathways to resection of chromosomal DNA DSBs. (a) Chromatin was incubated in extract that was depleted of CtIP, Exo1, both, or mock-depleted and isolated at the indicated time (minutes) after addition of 0.05 U/μL PflMI restriction endonuclease, and immunoblotted with the indicated antibodies (“chromatin”, top panels). Aliquots of the depleted extract were immunoblotted with the indicated antibodies to access immunodepletion (“extract”, lower panel). (b) Chromatin was incubated in mock-depleted LSS or LSS immunodepleted of DNA2, Exo1, or DNA2 and Exo1, and chromatin-isolation was performed as in (a). Recombinant Exo1 was added to Exo1-depleted extract as indicated. (c) HSS extract was mock-depleted, or depleted of either CtIP or DNA2, and chromatin-isolation was performed as in (a).
In contrast to LSS, CtIP depletion abrogates resection in HSS (Peterson et al., 2011; You et al., 2009). This suggests that the EXO1 pathway is inactive in HSS, and that processive recession is promoted by DNA2. As predicted, depletion of DNA2 from HSS eliminated resection (Fig. 1c). This establishes that DNA2 requires CtIP to perform long-range resection in the absence of EXO1.
Hyperphosphorylation of CtIP in response to DNA DSBs
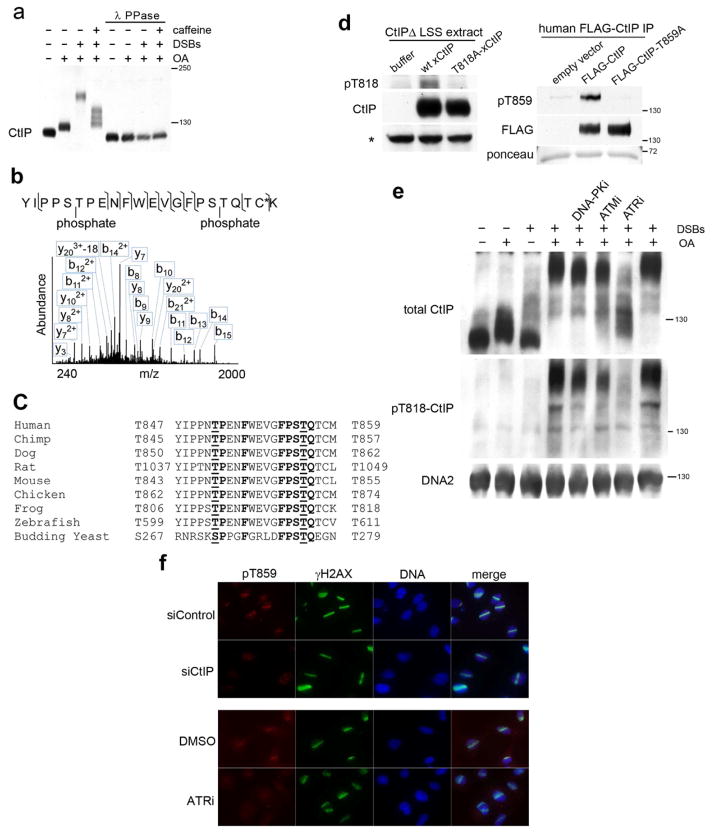
Phosphorylation of CtIP following DNA damage has been reported in yeast and mammalian cells, both in functional studies as well as in phospho-proteomic screens (Baroni et al., 2004; Kim et al., 1999; Li et al., 2000; Matsuoka et al., 2007). We therefore asked whether CtIP was also phosphorylated in a DSB-dependent manner in Xenopus extracts. LSS was incubated with or without linear DNA fragments to trigger DSB signaling. The phosphatase inhibitor okadaic acid (OA) was added to trap labile phosphorylation intermediates (Fig. 2a). In the absence of DSBs, OA treatment revealed a single species of phosphorylated CtIP, whereas DSBs and OA induced a striking electrophoretic mobility shift of soluble CtIP. This mobility shift was reduced with caffeine, an inhibitor of ATM and ATR. Remarkably, all isoforms of CtIP collapsed to a single fast-migrating band following lambda phosphatase treatment, confirming that the electrophoretic mobility change was due exclusively to phosphorylation (Fig. 2a).
Figure 2.
CtIP is hyperphosphorylated by ATR in response to DSBs, including conserved residue T818. (a) LSS was incubated with okadaic acid (OA), BstEII-digest of λ DNA (DSBs), and 5 mM caffeine, as indicated. Samples were split, and half were treated with λ phosphatase. Protein size markers are indicated at right (kD). (b) FLAG-xCtIP was incubated in LSS treated with OA with or without DSBs, recovered by affinity purification, and subjected to tandem MS. Shown is a spectrum of a triply protonated diphospho-peptide spanning residues 801–822. The fragmentation pattern localizes a phosphate group to S805/T806, and a second to S817/T818. Asterisk (*): alkylated cysteine. (c) The site of the TP and TQ motif for each model organism is listed to the left and right of the alignment, respectively. (d) Left panels: wt- or T818A-xCtIP was incubated in CtIP-depleted LSS in presence of OA and DSBs, as in (a). * non-specific cross-reactive band used as loading control. Right panels: human wt- or T859A-CtIP was expressed in 293 cells, isolated with anti-FLAG beads, and incubated in LSS in the presence of OA and DSBs. Western blots were probed, as indicated (e) Experiment performed as in (a), but with 50 μM DNA-PKi, 100 μM ATMi, or 50 μM ATRi, as indicated. (f) U2OS cells treated with control or CtIP siRNA 2 days prior to irradiation and treated with DMSO or 10μM ATRi 1 hour prior to localized “stripe” irradiation with high-power UV laser. Cells were fixed and stained with anti-pT818-CtIP and anti-γH2AX antibodies. See also Figure S1.
CtIP is phosphorylated at T818 by ATR in response to DSBs
We then identified the critical damage-dependent phosphorylation sites in CtIP. Recombinant FLAG-tagged CtIP was incubated in LSS with OA in the presence or absence of DSBs, and then affinity-purified and subjected to tandem mass spectrometry analysis. The complete list of phosphorylation sites induced by DSBs is shown in Fig. S1a. Notably, two DSB-induced phosphorylation sites, S200 and T818, conformed to the canonical SQ/TQ PIKK consensus sequence (Fig. 2b). Although S200 is not conserved phylogenetically, T818 lies within the evolutionarily-conserved C-terminus of CtIP. Moreover, this site exists within a TQ sequence among CtIP orthologs ranging from budding yeast Sae2 (T279) to human CtIP (T859) (Fig. 2c). Therefore, we produced and affinity-purified a phospho-specific antibody against pT818, and tested its specificity for phosphorylated T818 in Xenopus and the equivalent T859 in human. Recombinant WT-CtIP or CtIP-T818A (Xenopus) and WT-CtIP or CtIP-T859A (human) proteins were incubated in extracts with OA in the presence of DSBs and then probed for total CtIP and pT818-CtIP (αpT818, Fig. 2d). The antibody recognized phosphorylated Xenopus and human CtIP but not the non-phosphorylatable T818A and T859A variants, demonstrating the specificity of the antibody. Importantly, treatment with λ phosphatase eliminated the pT818 signal (Fig. S1b). To identify the protein kinase(s) responsible for CtIP-T818 phosphorylation in response to DSBs, we used small molecule inhibitors specific for ATM (KU55933; (Hickson et al., 2004)), ATR (ETP-46464; (Toledo et al., 2011)) or DNA-PK (NU7441; (Leahy et al., 2004)) and monitored CtIP electrophoretic mobility shift in LSS supplemented with DSB-containing DNA (Fig. 2e). Inhibition of DNA-PK or ATM only slightly increased the mobility of phosphorylated CtIP. In contrast, inhibition of ATR strongly reduced CtIP phosphorylation. Notably, ATR inhibition also abrogated detection of pCtIP using the anti-pT818 antibody, indicating that ATR phosphorylates CtIP on T818 (Fig. 2e). T818 phosphorylation of soluble CtIP is transient, and is undetectable in the absence of phosphatase inhibitor. Since ATR activates Chk1, we also treated extracts with the Chk1 inhibitor (SB-218078), which did not affect DSB-dependent phosphorylation of T818. Our data are therefore consistent with ATR phosphorylating T818 of CtIP (Fig. S1c).
ATR is activated by RPA-coated ssDNA (Costanzo et al., 2003; Zou and Elledge, 2003). This intermediate is generated by resection of DSBs, as well as by replication fork stalling and subsequent uncoupling of the replicative helicase. To test whether RPA-coated ssDNA in the absence of DSBs was sufficient to cause chromatin-recruitment of CtIP, we treated replicating chromatin with aphidicolin to trigger fork stalling and ATR activation (Byun et al., 2005). As a control, we used HaeIII restriction enzyme to create DSBs, which are resected to form RPA-ssDNA (Fig. S1d). As anticipated, CtIP was recruited to chromatin containing DSBs but not to RPA-ssDNA filaments that form in the absence of DSBs.
We then tested whether DNA damage induced phosphorylation of CtIP in human cells. Since the CtIP domain containing T818 is highly conserved (Fig. 2c) and the pT818 antibody reacts with phosphorylated T859 in human CtIP (Fig. 2d), we induced DSBs by high-power UV laser in human cells in the absence of OA, and probed with anti-pT818 antibody (Peterson et al., 2011). The pT859-CtIP signal colocalized with the DSB marker γH2AX. P859-CtIP signal was absent in cells depleted of CtIP via siRNA, establishing the specificity of the phospho-specific antibody (Fig. 2f). Furthermore, treatment with ATRi prior to irradiation significantly decreased the pT859-CtIP signal compared to treatment with DMSO, despite the presence of DSBs (Fig. 2f). This establishes that regulation of CtIP by ATR phosphorylation of T818/T859 is conserved among vertebrates.
Phosphorylation of CtIP by ATR is required for resection
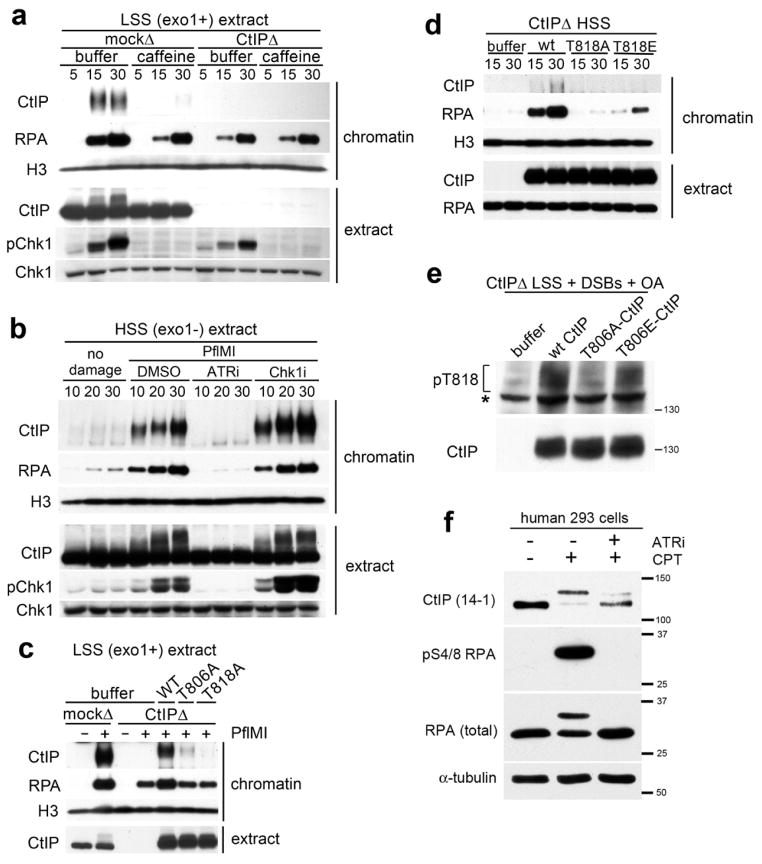
To determine the functional consequences of DSB-dependent CtIP phosphorylation, we evaluated the effect of caffeine on CtIP-dependent resection in LSS. Caffeine almost entirely abolished CtIP chromatin-binding and partially inhibited resection, similar to depletion of CtIP (Fig. 3a). In contrast, resection in CtIP-depleted LSS was not inhibited by caffeine, indicating that EXO1 does not require ATM or ATR activity (Fig. 3a).
Figure 3.
ATR phosphorylation of T818 is required for CtIP chromatin-binding. (a) Chromatin was isolated (at the indicated time after addition of PflMI) from mock- or CtIP-depleted LSS that was incubated with either 5 mM caffeine or buffer (b) Chromatin was incubated in HSS in presence of 50 μM ATRi, 50 μM Chk1i (SB-218078), or DMSO, and isolated at the indicated time (min) after addition of PflMI or buffer (no damage). Samples were processed as in (a). (c) Mock- or CtIP-depleted LSS was incubated with 80 nM recombinant WT-, T806A-, T818A-CtIP protein, or buffer, as indicated. Chromatin was isolated 30 minutes after addition of PflMI (+) or buffer (−). (d) CtIP-depleted HSS was incubated with 40 nM recombinant WT-, T818A-, T818E-CtIP, or buffer, and chromatin was isolated as in (a). (e) Experiment performed as in Fig 2(d) with wt-, T806A-, or T806E-xCtIP incubated in CtIP-depleted extract. *non-specific cross-reacting band. (f) 293 cells were incubated with 10 μM ATRi prior to treatment with DMSO or CPT. Cell lysates were probed with the indicated antibodies. See also Figure S2.
To determine if ATR was the PIKK responsible for activating CtIP-dependent resection in extracts, we used small molecule inhibitors of ATR or its effector kinase Chk1 and monitored RPA recruitment to DSBs (Fig. 3b). To circumvent resection by EXO1, these experiments were performed in HSS. Notably, we found that ATR inactivation abolished both recruitment of CtIP to damaged chromatin and resection. Interestingly, inhibition of Chk1, which is downstream to ATR, had no effect on CtIP phosphorylation, chromatin recruitment, or DSB resection (Fig. 3b). To confirm the specificity of the small molecule ATR inhibitor, we immunodepleted ATR from HSS (Costanzo et al., 2003). As with chemical inhibition, immunodepletion of ATR abrogated CtIP-dependent resection (Fig. S2a).
To test the functional significance of phosphorylation on T818, we compared recombinant xCtIP proteins with or without substitutions at T818, including non-phosphorylatable T818A and phosphomimic T818E. The recombinant proteins were purified from baculovirus-infected insect cells and added to CtIP-depleted LSS. As expected, the purified recombinant proteins (Fig. S2b) did not react with the pT818 antibody (Fig. S2c). Unlike WT CtIP, the CtIP-T818A mutant neither bound to chromatin nor stimulated resection (Fig. 3c). Similarly, CtIP-T818A did not support resection in CtIP-depleted HSS. In contrast, CtIP-T818E partially restored resection in depleted HSS (Fig. 3d), indicating that T818 phosphorylation by ATR is required for CtIP binding to and resection of damaged chromatin. CtIP-T818E did not restore resection in CtIP-depleted extract in the presence of ATRi, suggesting that either additional ATR-dependent phosphorylation events also regulate CtIP activity or that CtIP-T818E is a weak phosphomimic of pT818 (data not shown).
Phosphorylation of CtIP by CDK2 at the onset of S-phase is essential for CtIP-dependent resection (Huertas and Jackson, 2009). In Xenopus, this phosphorylation takes place at T806 and is required for CtIP association to chromatin (Peterson et al., 2011). Since T818 and T806 are in close proximity within the highly conserved domain of CtIP (Fig. 2c), we asked whether CDK phosphorylation is a pre-requisite for ATR phosphorylation. We showed previously that CtIP-T806A is defective in chromatin-binding, whereas CtIP-T806E is functional (Peterson et al., 2011). Therefore, we generated and purified CtIP-T806A and CtIP-T806E and compared phosphorylation of T818 in these mutants to WT-CtIP upon incubation in extracts containing DSBs (Fig. 3e). Notably, WT CtIP and CtIP-T806E became phosphorylated at T818, whereas phosphorylation of CtIP-T806A was greatly reduced, indicating that phosphorylation of T806 by CDK facilitates T818 phosphorylation by ATR.
Lastly, we tested the impact of ATR inhibition on resection in human cells. U2OS cells were treated with camptothecin (CPT), which generates DSBs during S-phase, in the presence or absence of ATRi (Fig. 3f). CPT treatment triggered phosphorylation of CtIP, as evidenced by its reduced electrophoretic mobility, and initiation of DNA resection, as indicated by Ser4/Ser8 phosphorylation of RPA2 (Fig. 3f), (Bunting et al., 2010; Sartori et al., 2007). Moreover, CPT-induced DNA resection was abolished by siRNA-mediated CtIP depletion, indicating that resection is dependent on CtIP (data not shown). Notably, ATRi treatment abrogated both CtIP phosphorylation and CPT-induced DNA resection (Fig. 3f). Therefore, we conclude that phosphorylation of CtIP by ATR is required for resection in human cells.
A distinct role for ATM in resection initiation
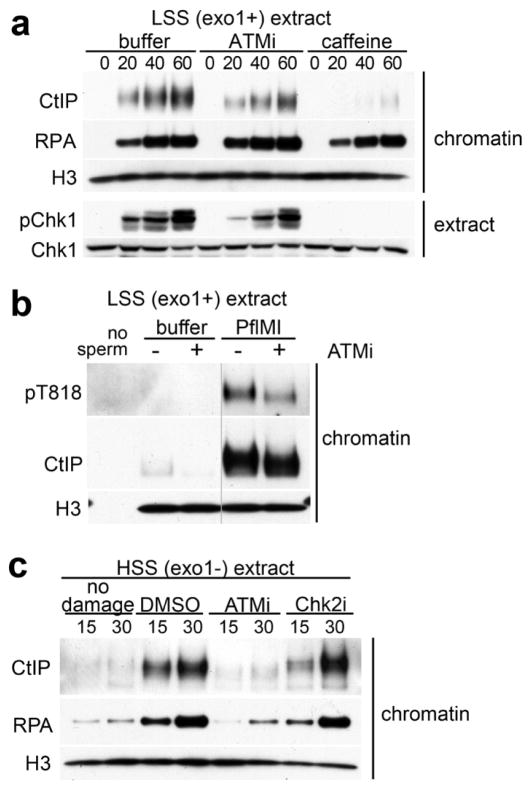
Previous studies in HSS extracts implicated ATM in chromosomal DSB resection (You et al., 2009). We therefore probed the role of ATM in CtIP-dependent resection in HSS as well as in LSS. First, we tested the impact of ATM inhibition on CtIP binding to chromatin in LSS. Inhibition of ATM decreased, but did not abolish, CtIP binding. In contrast, inhibition of both ATM and ATR with caffeine entirely abrogated CtIP binding (Fig. 4a). Inhibition of ATR alone with ATRi in LSS gave the same result (Fig. S3), which is consistent with a requirement for ATR for stable CtIP association to chromatin in HSS (Fig. 3b). Furthermore, ATM inhibition reduced, but did not abolish, phosphorylation of T818, supporting the notion that T818 is modified by ATR (Fig. 2e, Fig. 4b).
Figure 4.
ATM activity is required for CtIP-dependent resection initiation. (a) Chromatin was incubated in mock- or CtIP-depleted LSS with buffer, 100 μM ATMi or 5 mM, isolated and probed with the indicated antibodies (b) 100 μM ATMi (+) or DMSO (−) was added to LSS extract treated with PflMI or buffer, and chromatin was isolated after 40 minutes. (c) Chromatin was incubated in HSS in presence of 100 μM ATMi, 100 μM Chk2i (PV1019), or DMSO, isolated and probed with the indicated antibodies. See also Figure S3.
The decrease in pT818 upon exposure to ATMi was commensurate with reduction in chromatin-bound CtIP (Fig. 4a). We suggest that this reflects the delayed kinetics of ATR activation when ATM is inhibited (compare the rates of CtIP chromatin-binding to Chk1 activation in Fig 4a). In contrast to LSS, inhibition of ATM in HSS abrogated CtIP chromatin binding and resection.
This is a direct effect of ATM, since inhibition of the ATM effector, Chk2, had no impact on CtIP binding or resection (Fig. 4c). We conclude that ATM is required for initiating resection in HSS but not in LSS. As mentioned above, a critical difference between these extracts is their EXO1 status. EXO1 is active in LSS and can initiate resection in the absence of active ATM (Fig. 3a, CtIP compare +/− caffeine), generating ssDNA-RPA and activating ATR. We hypothesize that in EXO1-deficient HSS, ATM activity is required for initial processing of DSBs.
DISCUSSION
We have investigated the roles of ATM and ATR in DSB resection of chromosomal DNA using two types of cell-free Xenopus extracts, one lacking (HSS) and one containing (LSS) active EXO1. We show that CtIP is phosphorylated at T818 (T859 in humans) by ATR, and that ATR phosphorylation at T818 and other sites promotes stable association of CtIP to damaged chromosomes, allowing CtIP-dependent resection. This finding is significant, as it provides the first direct functional link between a PIKK and DSB repair by homology-directed repair in vertebrates. Moreover, our observation has several implications for the current model of DSB repair, as will be discussed below. Our findings are summarized in Fig. 5.
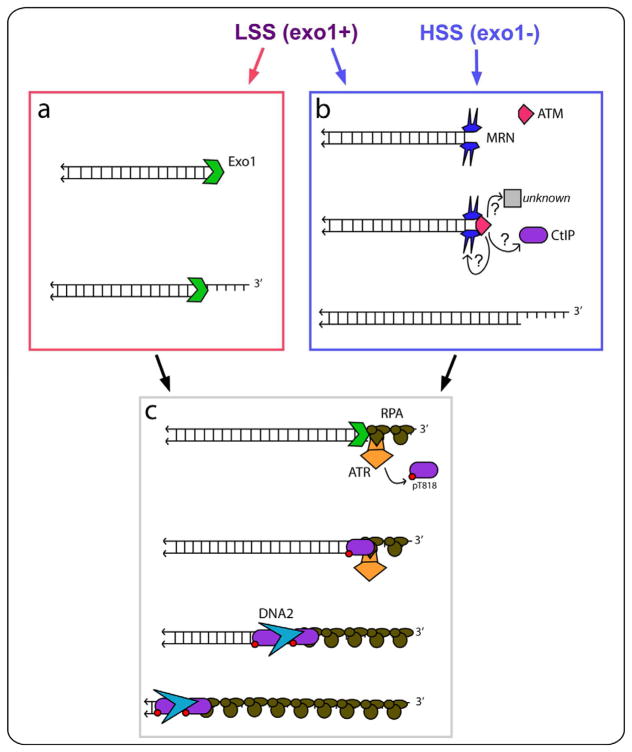
Figure 5.
Model for resection of neat DSBs in LSS and HSS. In LSS, resection can be initiated by Exo1, in an ATM-independent manner (a), or by CtIP-MRN in an ATM-dependent manner (b). In HSS, Exo1 is not active and resection is initiated by CtIP-MRN in an ATM-dependent manner (b). Initiation by either pathway allows clipping of DSBs to provide sufficient RPA-ssDNA to locally activate ATR (c). ATR then phosphorylates CtIP on T818, which is essential for stable CtIP chromatin-binding, and downstream extension of resection tracts by a DNA2-dependent mechanism.
ATM and DSB resection
Crude extract (LSS) supports resection in the absence of CtIP or active ATM and ATR (Fig. 1a, 3a, 4a), whereas co-depletion of EXO1 and CtIP from LSS abolishes all resection (Fig. 1a). Furthermore, depletion of DNA2 nearly abrogates resection. Together, these results suggest that EXO1 can initiate restriction-enzyme generated DSBs independently of CtIP, ATM or ATR but cannot support long-range resection in the absence of DNA2 (Fig. 5a). This is consistent with the fact that EXO1 overexpression can rescue the IR-defect of MRX mutants in yeast (Lewis et al., 2002; Moreau et al., 2001). Additionally, EXO1 and CtIP may cooperate to promote efficient resection, since these proteins can interact (Eid et al., 2010). In contrast, membrane-free cytosol (HSS), derived from high-speed centrifugation of crude extract (LSS), supports resection only when CtIP is present and ATM and ATR are active (Fig. 1 and Fig. 4c), supporting the notion that HSS is EXO1-defective (Fig. 5b).
The requirement for ATM in the absence of EXO1 (Fig. 4c; (You et al., 2009), suggests that initiation of resection requires ATM phosphorylation of a component of MRN, CtIP, or of another factor (Fig. 5b). In support of this notion, phospho-proteomic screens in humans reveals phosphorylation of CtIP, Mre11, Rad50 and Nbs1 in response to DNA damage (Kim et al., 1999; Matsuoka et al., 2007; Mu et al., 2007). However, the functional consequences of these phosphorylation events have not been determined. Consistent with a role for ATM in initiating resection, ATM-deficient mammalian cells have a defect in RPA foci formation and Chk1 activation in response to IR (Cuadrado et al., 2006; Jazayeri et al., 2006; Myers and Cortez, 2006; Shiotani and Zou, 2009).
There is ample evidence in yeast and vertebrates that CtIP-MRN promotes limited processing of DSBs. Budding yeast cells lacking EXO1 and Sgs1 undergo slow, Sae2-MRX-dependent degradation of DSBs, indicating that Sae2-MRX can clip oligonucleotides from the 5′ strand (Mimitou and Symington, 2008; Shim et al., 2010; Zhu et al., 2008). Moreover, Sae2 is required for processing Spo11-capped DSBs during meiosis (McKee and Kleckner, 1997; Prinz et al., 1997), which yields a short oligonucleotide covalently linked to Spo11 at the 5′ terminus (Hartsuiker et al., 2009a; Neale et al., 2005). This suggests that Sae2-MRX initiates resection by endonuclease attack on the 5′ strand. A similar mechanism has been proposed for S. pombe Ctp1 and vertebrate CtIP in removal of protein adducts from 5′ DNA ends (Hartsuiker et al., 2009b; Milman et al., 2009; Nakamura et al., 2010; Rothenberg et al., 2009). Furthermore, Mre11-bound oligonucleotides have been detected in Xenopus extracts following incubation with DSB-containing DNA. These complexes are sufficient to activate ATM (Jazayeri et al., 2008). Finally, in S. pombe, the endonuclease activity of Mre11, but not its exonuclease activity, is important for resistance to IR, which requires processing of modified DNA ends (Williams et al., 2008).
We cannot detect chromatin-bound CtIP in the absence of ATR activity, (Fig. 3b, Fig. S3), suggesting that the initial ATM-dependent step does not require stable chromatin association of CtIP. However, EXO1-defective HSS depleted of DNA2 supports modest recruitment of CtIP to chromatin (Fig. 1c), suggesting that slow processing of DSBs by CtIP-MRN generates sufficient ssDNA to allow binding of RPA, ATR, and subsequent stabilization of CtIP. However, we cannot rule out the possibility that this effect is due to incomplete depletion of DNA2.
ATR and DSB resection
We propose that CtIP-MRN-dependent initial processing generates sufficient ssDNA to allow binding of RPA and ATRIP-ATR, which requires at least 60 nt of ssDNA (Zou and Elledge, 2003). Limited activation of ATR is then responsible for CtIP phosphorylation at T818, which in turns promotes stable chromatin association of CtIP and robust resection. We note that T818 is conserved within a TQ sequence from yeast Sae2 to human CtIP. Damage-induced T859 phosphorylation is observed in Xenopus extracts and human cells (Fig. 2d, 2f), suggesting that phosphorylation of CtIP by ATR is an evolutionarily-conserved mechanism linking DDR to activation of DSB repair by HDR. Indeed, inhibition of CtIP phosphorylation with ATRi abrogates resection in human cells (Fig. 3f), Mutation of all 9 putative ATM/ATR phosphorylation sites, including the invariant threonine (S. cerevisiae T279, Xenopus T818), inactivates S. cerevisiae Sae2 whereas mutation of any single site, including the T818 equivalent (T279), has no functional consequence (Baroni et al., 2004). Phosphorylation of human CtIP at two ATM/ATR sites, S664 and S745, has been reported (Li et al., 2000; Matsuoka et al., 2007), but whether modification of these sites affects CtIP activity is not known (Li et al., 2000; Wu-Baer and Baer, 2001). Phosphorylation of human CtIP-T859 (corresponding to Xenopus T818) has not been previously described.
Given its importance in DDR, redundant mechanisms of CtIP recruitment to damaged chromatin may exist. You et al. have described a “DR motif” within the central region of the protein that promotes CtIP recruitment to sites of DNA damage, but does not contain T818/T859 (You et al., 2009). Thus, both elements of CtIP (T818/T859 phosphorylation and the DR motif) may contribute to the recruitment of CtIP to damage sites. We note that the DR motif is not fully conserved in Xenopus and is absent entirely in yeast. In contrast, the T818/T859 phosphorylation site is conserved as a TQ sequence from yeast to human (Fig. 2c). The greater conservation of the T818/T859 phosphorylation site may reflect its key role as a reversible modification that allows CtIP recruitment in response to ATR signaling. Moreover, structural data show that CtIP interaction with Nbs1 – through a N-terminal motif of CtIP – is critical for CtIP recruitment (Lloyd et al., 2009; Williams et al., 2009).
ATM inhibition does not prevent CtIP chromatin-binding when resection is initiated by EXO1; i.e., when generation of the ssDNA-RPA substrate to activate ATR occurs independently of ATM activity (Fig. 4a, 4b). Thus, if ATM phosphorylates CtIP, this phosphorylation is not required for ATR-dependent association of CtIP with chromatin.
Implications for ATR activation
Our model suggests that can ATR promote its own activation by phosphorylating and activating CtIP, which in turn generates additional ssDNA-RPA (Fig. 3b, 5c), (Cimprich and Cortez, 2008). Starting this feedback loop would require some initial ATR activation. We favor the idea that this initial step is triggered by limited end-processing of DSBs dependent on ATM, MRN and CtIP (Fig 5b), and/or Exo1 (Fig. 5a).
A positive feedback loop for ATR activation is consistent with the fact that ATR kinase activity is important for the localization of RPA and ATR to IR-induced foci (Barr et al., 2003). Furthermore, ssDNA-RPA and ATRIP are sufficient for ATR autophosphorylation, which leads to full ATR activation by providing a docking site for TopBP1 recruitment prior to checkpoint activation (Liu et al., 2011). We propose that Chk1 activation only occurs after CtIP stably binds to chromatin and initiates processive resection.
CtIP or EXO1 can initiate resection but do not support processive resection. Conversely, DNA2-dependent resection requires initiation by either Exo1 or CtIP-MRN (Fig. 1a, Fig. 5c). CtIP depletion abolishes resection in EXO1-defective HSS, despite the presence of active DNA2 (Fig. 1c). In the absence of DNA2, resection is nearly abolished in EXO1-proficient LSS. By initiating resection, CtIP-MRN could generate the preferred substrate for DNA2 recruitment. Indeed, yeast Dna2 requires Sgs1 and RPA to function in vitro (Cejka et al., 2010), implying that DNA2 starts resection on an RPA-ssDNA intermediate (Fig 5c). However, it is also possible that DNA2 recruitment requires chromatin-bound CtIP. In support of this notion, incubation of linear DNA containing 3′ overhangs with purified Xenopus DNA2 and RPA does not promote resection (Yan et al., 2011), suggesting that RPA-bound ssDNA alone cannot promote DNA2-dependent resection.
Mutations of either ATR or CtIP in humans causes Seckel Syndrome
ATR is an essential gene, but inherited hypomorphic mutations in ATR have been identified. These mutations cause Seckel syndrome in humans, which is characterized by dwarfism, microcephaly and mental retardation (O’Driscoll et al., 2003). ATR-mutant Seckel cells display defects in the G2/M damage checkpoint, reduced phosphorylation of ATR targets, and replication stress-induced genome instability characterized by nuclear fragmentation (Alderton et al., 2004). Seckel syndrome is genetically heterogenous, with five loci identified to date. Recently, SCKL2 was identified as a recessive mutation in CtIP (Qvist et al., 2011). The mutation alters splice site selection and produces low-level expression of a truncated form of CtIP in which the last 134 amino acids are replaced by 20 amino acids of novel sequence (truncation after H762). Notably, the deleted region includes both the ATR and CDK phosphorylation sites. Therefore, the same human disease is caused either by hypomorphic mutation of ATR, or expression of a truncated CtIP lacking the two phosphorylation sites required for ATR activation. This suggests that regulation of CtIP activity at this site by ATR and/or CDK has profound cellular as well as physiological consequences.
EXPERIMENTAL PROCEDURES
Extract & Sperm Chromatin Preparation, assay to monitor the response to chromosomal DSBs
Preparation of extracts (LSS and HSS) and resection assays were performed as described (Peterson et al., 2011). See also Supplemental information.
Generation and purification of phospho-spcific pT818-CtIP antibody
Rabbits were immunized with CtIP-phosT818 peptide (CWEVGFPSpTQTCKDRG) and serum was purified by two-step affinity chromatography using SulfoLink Immobilization kit (Thermo cat#44999). One column was conjugated to the phosT818 peptide, and another was conjugated to the non-phosT818 peptide (CWEVGFPSTQTCKDRG), according to manufacturer’s instructions. 0.5 mL of crude serum was diluted to 4 mL with 0.1M sodium phosphate buffer, 150 mM NaCl, pH 7.2. Diluted serum was incubated with phosT818 column, and flow-thru was discarded. Antibody was eluted sequentially by acid (0.1M glycine, pH 2.5) then base (0.1M triethylamine, pH 11.5) into 1M tris pH 7.5. This eluate was incubated with the non-phosT818 column and the flow-thru was saved as the purified anti-phosT818-CtIP antibody. The purified antibody is a 1:20 dilution relative to the serum, and it is used at 1:200 final dilution for western blotting and immunofluorescence.
Immunodepletions and antibodies
Immunodepletions were performed as described (Peterson et al., 2011). See also Supplemental information.
Site-directed mutagenesis of xCtIP
xCtIP mutants were generated by two-step PCR, as described previously (Peterson et al., 2011). See Supplemental information for details about the oligonucleotides.
Recombinant protein purification
See (Peterson et al., 2011). See also Supplemental information.
Immunofluorescence of UV laser-irradiated human U2OS cells
Immunofluorescence and laser irradiation were performed as described (Peterson et al., 2011). See also Supplemental information.
Isolation of recombinant xCtIP from control and DSB-containing extract. and Tandem MS/MS analysis of xCtIP
Purification of recombinant from extract and mass spectrometry analysis were performed as described in (Peterson et al., 2011).
ATRi inhibition of damage-induced CtIP phosphorylation in human cells
293 human cells were seeded at 0.3 × 106 cells per 100 mm plate. Two days later, the cells were treated with or without 10uM ATRi for one hour, and then incubated for an additional hour with or without 1uM of camptothecin (CPT) prior to harvest. The cells lysed in NETN150 buffer (20mM Tris-Cl, 150 mM NaCl, 1 mM EDTA, 0.5% NP40) supplemented with complete Mini EDTA-Free protease inhibitor cocktail (Roche), 0.5 mM PMSF, 1 mM DTT and 50 mM NaF. 50 μg of each protein lysate were subjected to SDS-PAGE and Western analysis.
Phosphorylation of recombinant proteins
CtIP-depleted LSS extract was incubated with 10 nM recombinant xCtIP and treated with 4 μM okadaic acid and 33.3 μg/mL BstEII-digest of lambda DNA, or buffer, for 30 minutes at 21°C. Phos phorylation of human proteins: Anti-FLAG beads were added to 50 μL of lysate from U20S cells that were treated with siRNA against CtIP and transfected with vector alone, WT-CtIP, or T859A-CtIP (as described above). The washed FLAG-beads were incubated in Xenopus LSS extract treated with okadaic acid and BstEII-digest of lambda DNA, as described above. The beads were washed three times, boiled in Laemmli buffer, and processed for Western blotting with anti-pT818 and anti-FLAG antibodies.
Supplementary Material
HIGHLIGHTS.
ATR phosphorylates CtIP at a conserved site (T818 in Xenopus, T859 in human)
This is required for stable CtIP chromatin-binding and CtIP-dependent resection
ATM activity is required prior to ATR for resection and leads to ATR activation
This demonstrates a direct link between checkpoint activation and DSB repair
Acknowledgments
We thank Dr. Fernandez-Capetillo for the generous gift of ATRi and Dr. J. Sible for anti-Chk1 antibodies. We thank L. Phillips for help with antibodies screening. We thank Dr. M. Jasin for her support during the peer-review process. This work was supported by grants RO1CA092245 and RO1GM077495 to J. Gautier and RR00862 and RR022220 to B.T. Chait.
Footnotes
Authors contributions: SEP performed all experiments, except MS of purified CtIP, which was designed by YYL and BC, and performed by YYL, and ATRi inhibition of resection in U2OS cells, which was performed by FWB. SEP, MEG, and JG designed all other experiments. HY, RB and FWB generated critical reagents. SEP, MEG, RB, HY and JG discussed results. SEP, MEG and JG wrote the manuscript, which was read by all authors.
Contains Supplemental Figures S1–S3, Supplemental Figure Legends, Experimental Procedures, and Supplemental References.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akamatsu Y, Murayama Y, Yamada T, Nakazaki T, Tsutsui Y, Ohta K, Iwasaki H. Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex. Molecular and cellular biology. 2008;28:3639–3651. doi: 10.1128/MCB.01828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderton GK, Joenje H, Varon R, Borglum AD, Jeggo PA, O’Driscoll M. Seckel syndrome exhibits cellular features demonstrating defects in the ATR-signalling pathway. Human molecular genetics. 2004;13:3127–3138. doi: 10.1093/hmg/ddh335. [DOI] [PubMed] [Google Scholar]
- Barker S, Weinfeld M, Zheng J, Li L, Murray D. Identification of mammalian proteins cross-linked to DNA by ionizing radiation. The Journal of biological chemistry. 2005;280:33826–33838. doi: 10.1074/jbc.M502477200. [DOI] [PubMed] [Google Scholar]
- Baroni E, Viscardi V, Cartagena-Lirola H, Lucchini G, Longhese MP. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Molecular and cellular biology. 2004;24:4151–4165. doi: 10.1128/MCB.24.10.4151-4165.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr SM, Leung CG, Chang EE, Cimprich KA. ATR kinase activity regulates the intranuclear translocation of ATR and RPA following ionizing radiation. Curr Biol. 2003;13:1047–1051. doi: 10.1016/s0960-9822(03)00376-2. [DOI] [PubMed] [Google Scholar]
- Budd ME, Campbell JL. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS ONE. 2009;4:e4267. doi: 10.1371/journal.pone.0004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nature reviews Molecular cell biology. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J. An ATR-and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell. 2003;11:203–213. doi: 10.1016/s1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- Cuadrado M, Martinez-Pastor B, Murga M, Toledo LI, Gutierrez-Martinez P, Lopez E, Fernandez-Capetillo O. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J Exp Med. 2006;203:297–303. doi: 10.1084/jem.20051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derheimer FA, Kastan MB. Multiple roles of ATM in monitoring and maintaining DNA integrity. FEBS letters. 2010;584:3675–3681. doi: 10.1016/j.febslet.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid W, Steger M, El-Shemerly M, Ferretti LP, Pena-Diaz J, Konig C, Valtorta E, Sartori AA, Ferrari S. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO reports. 2010;11:962–968. doi: 10.1038/embor.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner E, Costanzo V. Studying the DNA damage response using in vitro model systems. DNA Repair (Amst) 2009;8:1025–1037. doi: 10.1016/j.dnarep.2009.04.015. [DOI] [PubMed] [Google Scholar]
- Hartsuiker E, Mizuno K, Molnar M, Kohli J, Ohta K, Carr AM. Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Molecular and cellular biology. 2009a;29:1671–1681. doi: 10.1128/MCB.01182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsuiker E, Neale MJ, Carr AM. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell. 2009b;33:117–123. doi: 10.1016/j.molcel.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henner WD, Rodriguez LO, Hecht SM, Haseltine WA. gamma Ray induced deoxyribonucleic acid strand breaks. 3′ Glycolate termini. The Journal of biological chemistry. 1983;258:711–713. [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. DNA damage, aging, and cancer. The New England journal of medicine. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. The Journal of biological chemistry. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J. 2008 doi: 10.1038/emboj.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Keeney S, Neale MJ. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochemical Society transactions. 2006;34:523–525. doi: 10.1042/BST0340523. [DOI] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. The Journal of biological chemistry. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- Langerak P, Mejia-Ramirez E, Limbo O, Russell P. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 2011;7:e1002271. doi: 10.1371/journal.pgen.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley PD, Phillips DH. DNA adducts from chemotherapeutic agents. Mutation research. 1996;355:13–40. doi: 10.1016/0027-5107(96)00020-6. [DOI] [PubMed] [Google Scholar]
- Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, Smith GC. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg Med Chem Lett. 2004;14:6083–6087. doi: 10.1016/j.bmcl.2004.09.060. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- Lewis LK, Karthikeyan G, Westmoreland JW, Resnick MA. Differential suppression of DNA repair deficiencies of Yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase) Genetics. 2002;160:49–62. doi: 10.1093/genetics/160.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ting NS, Zheng L, Chen PL, Ziv Y, Shiloh Y, Lee EY, Lee WH. Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature. 2000;406:210–215. doi: 10.1038/35018134. [DOI] [PubMed] [Google Scholar]
- Liao S, Guay C, Toczylowski T, Yan H. Analysis of MRE11’s function in the 5′-->3′ processing of DNA double-strand breaks. Nucleic Acids Res. 2012a;40:4496–4506. doi: 10.1093/nar/gks044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Guay C, Toczylowski T, Yan H. Analysis of MRE11’s function in the 5′->3′ processing of DNA double-strand breaks. Nucleic Acids Res. 2012b doi: 10.1093/nar/gks044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Toczylowski T, Yan H. Identification of the Xenopus DNA2 protein as a major nuclease for the 5′->3′ strand-specific processing of DNA ends. Nucleic Acids Res. 2008a;36:6091–6100. doi: 10.1093/nar/gkn616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Toczylowski T, Yan H. Identification of the Xenopus DNA2 protein as a major nuclease for the 5′->3′ strand-specific processing of DNA ends. Nucleic Acids Res. 2008b doi: 10.1093/nar/gkn616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Toczylowski T, Yan H. Mechanistic analysis of Xenopus EXO1’s function in 5′-strand resection at DNA double-strand breaks. Nucleic Acids Res. 2011;39:5967–5977. doi: 10.1093/nar/gkr216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Shiotani B, Lahiri M, Marechal A, Tse A, Leung CC, Glover JN, Yang XH, Zou L. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell. 2011;43:192–202. doi: 10.1016/j.molcel.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, Carr AM, Jackson SP, Smerdon SJ. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- McKee AH, Kleckner N. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics. 1997;146:797–816. doi: 10.1093/genetics/146.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman N, Higuchi E, Smith GR. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Molecular and cellular biology. 2009;29:5998–6005. doi: 10.1128/MCB.01127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Morgan EA, Symington LS. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001;159:1423–1433. doi: 10.1093/genetics/159.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu JJ, Wang Y, Luo H, Leng M, Zhang J, Yang T, Besusso D, Jung SY, Qin J. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. The Journal of biological chemistry. 2007;282:17330–17334. doi: 10.1074/jbc.C700079200. [DOI] [PubMed] [Google Scholar]
- Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Kogame T, Oshiumi H, Shinohara A, Sumitomo Y, Agama K, Pommier Y, Tsutsui KM, Tsutsui K, Hartsuiker E, et al. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 2010;6:e1000828. doi: 10.1371/journal.pgen.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam EA, Cortez D. ATR signalling: more than meeting at the fork. The Biochemical journal. 2011;436:527–536. doi: 10.1042/BJ20102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nature genetics. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- Paull TT. Making the best of the loose ends: Mre11/Rad50 complexes and Sae2 promote DNA double-strand break resection. DNA Repair (Amst) 2010;9:1283–1291. doi: 10.1016/j.dnarep.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SE, Li Y, Chait BT, Gottesman ME, Baer R, Gautier J. Cdk1 uncouples CtIP-dependent resection and Rad51 filament formation during M-phase double-strand break repair. The Journal of cell biology. 2011;194:705–720. doi: 10.1083/jcb.201103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S, Amon A, Klein F. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics. 1997;146:781–795. doi: 10.1093/genetics/146.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qvist P, Huertas P, Jimeno S, Nyegaard M, Hassan MJ, Jackson SP, Borglum AD. CtIP Mutations Cause Seckel and Jawad Syndromes. PLoS Genet. 2011;7:e1002310. doi: 10.1371/journal.pgen.1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg M, Kohli J, Ludin K. Ctp1 and the MRN-complex are required for endonucleolytic Rec12 removal with release of a single class of oligonucleotides in fission yeast. PLoS Genet. 2009;5:e1000722. doi: 10.1371/journal.pgen.1000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, Lee SE. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 2010 doi: 10.1038/emboj.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiotani B, Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell research. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- Srinivasan SV, Gautier J. Study of cell cycle checkpoints using Xenopus cell-free extracts. Methods Mol Biol. 2011;782:119–158. doi: 10.1007/978-1-61779-273-1_10. [DOI] [PubMed] [Google Scholar]
- Sun J, Lee KJ, Davis AJ, Chen DJ. Human Ku70/80 protein blocks exonuclease 1-mediated DNA resection in the presence of human Mre11 or Mre11/Rad50 protein complex. The Journal of biological chemistry. 2012;287:4936–4945. doi: 10.1074/jbc.M111.306167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Taylor EM, Cecillon SM, Bonis A, Chapman JR, Povirk LF, Lindsay HD. The Mre11/Rad50/Nbs1 complex functions in resection-based DNA end joining in Xenopus laevis. Nucleic Acids Res. 2010;38:441–454. doi: 10.1093/nar/gkp905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18:721–727. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomimatsu N, Mukherjee B, Deland K, Kurimasa A, Bolderson E, Khanna KK, Burma S. Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair (Amst) doi: 10.1016/j.dnarep.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, Tainer JA. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Baer F, Baer R. Effect of DNA damage on a BRCA1 complex. Nature. 2001;414:36. doi: 10.1038/35102118. [DOI] [PubMed] [Google Scholar]
- Yan H, Toczylowski T, McCane J, Chen C, Liao S. Replication protein A promotes 5′-->3′ end processing during homology-dependent DNA double-strand break repair. J Cell Biol. 2011;192:251–261. doi: 10.1083/jcb.201005110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Shi LZ, Zhu Q, Wu P, Zhang YW, Basilio A, Tonnu N, Verma IM, Berns MW, Hunter T. CtIP links DNA double-strand break sensing to resection. Mol Cell. 2009;36:954–969. doi: 10.1016/j.molcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.