Abstract
We report the development of a bidirectional affinity system for the identification of cancer–related natural products and their biological targets. In this study, we apply a resin format to selectively identify aplysqualenol A as a ligand of the dynein light chain, DYNLL1. The unique combination of forward and reverse affinity methods suggests that both small molecule isolation and target identification can be conducted using conventional molecular biological methods.
Keywords: natural products isolation, bidirectional affinity chromatography, aplysqualenol A, dynein light chain, mode of action
Natural products continue to serve as a hotbed for cancer chemotherapy, providing vital leads, probes, and clinical entries.[1] The advance of spectroscopic tools[2] and analytical techniques[3] has led our understanding into the dynamic yet complex structural diversity of marine secondary metabolites. Even given the expanse of this molecular diversity, we have only identified the biomolecular targets of a small subset of the known natural products,[4] and many of those that have been identified arise due to the fact that they share common targets (examples include actin or microtubules).[5] The fact remains that we have only begun to understand the myriad of biological pathways engaged by secondary metabolites.
Despite the fact that a variety of methods exist for evaluating a natural product’s biological targets,[6] the lack in this knowledge has arisen due to a combination of reasons, including: (a) the fact that many natural products were isolated before the onset of key biological techniques; (b) the lack of advanced biological screens during the discovery process; and (c) the gradual shift away from natural products as therapeutic leads.[7] While target identification studies are only one factor in the drug discovery process, the lost of these data has a significant impact on our understanding of the relationship between the structure and function of small molecules, and therefore it plays an essential role.
As isolation efforts are an ongoing process, we focused our program on developing methods that allow one to screen a natural product’s biological targets in conjunction with its isolation and structure elucidation. Recently, we described the use of a bidirectional affinity platform to identify the binding of the dimeric pyrrole–imidazole alkaloid sceptrin to a prokaryotic homolog to actin, MreB.[8] This study demonstrated the ability to identify metabolites and their bacterial protein targets in harmony. Here, we describe a bidirectional approach (outlined in Figure 1) to sequentially identify cancer targets and their corresponding natural product ligands.
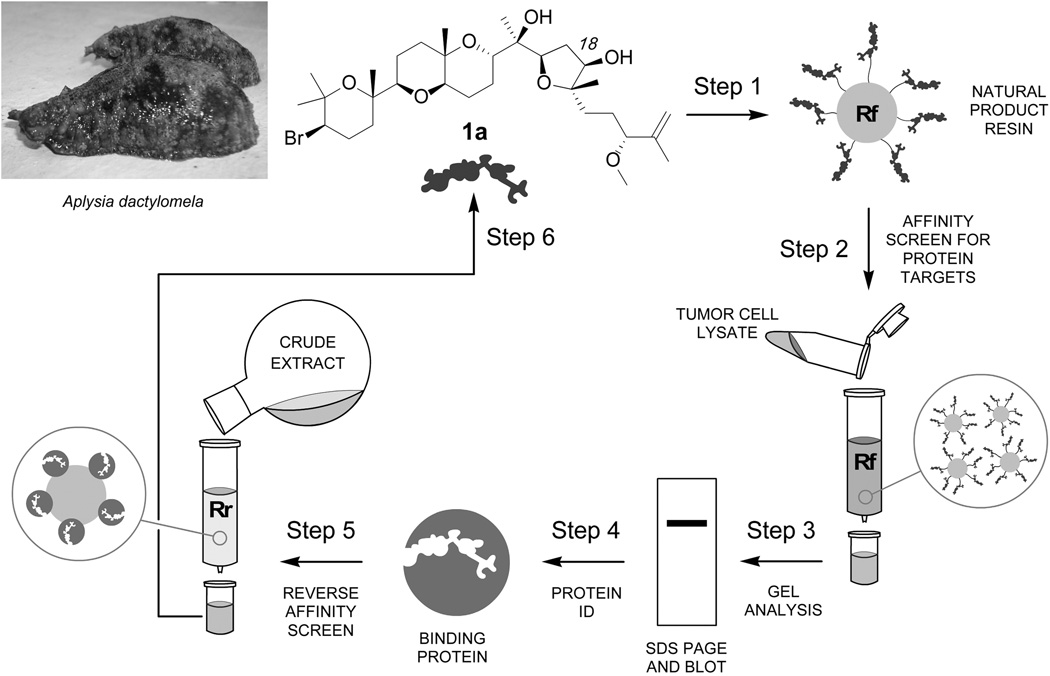
Figure 1.
Protocol for bidirectional affinity–guided studies. (Step 1) Aplysqualenol A (1a) was covalently linked to a resin (Rf). (Step 2) Resin Rf was then used to screen for aplysqualenol A (1a) binding proteins. (Steps 3–4) The bound proteins were examined by SDS PAGE, identified by proteomic analyses, and validated by western blot analysis. (Steps 5–6) The isolated binding proteins were then attached to resin and used in a reverse–affinity sense using resin (Rr) to isolate 1a from its corresponding crude natural product extract.
We recently reported the isolation of the bromotriterpene polyethers aplysqualenol A (1a) and B (1b) (Figure 2) from the Caribbean sea slug Aplysia dactylomela, collected in Puerto Rico.[9] Samples of 1a were submitted to the NCI for evaluation in the NCI–60, one–dose primary anticancer assay. It displayed potent in vitro activity against SNB–19 (IC50 value of 0.4 µM), a CNS tumor cell line, and T–47D (IC50 value of 0.4 µM), a breast cancer cell line.[9] Comparable analyses indicated that 1a was also active in HCT–116 colon carcinoma cells (IC50 value for 1a in HCT–116 cells of 20.6 µM via NCI screening and 12.2±0.3 via an in house MTT screen).[10]
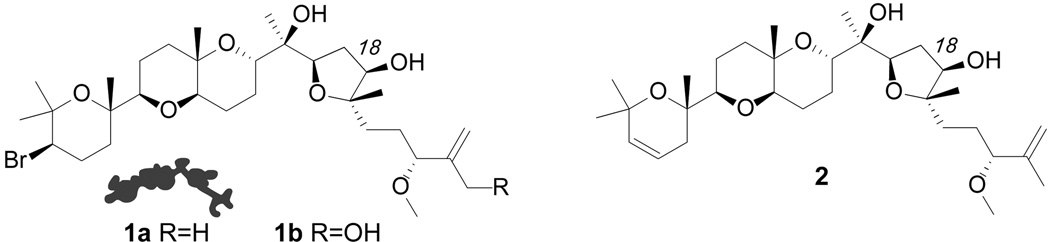
Figure 2.
Structures of aplysqualenol A (1a), aplysqualenol B (1b) and elimination product 2.
We began by developing methods for the isolation of proteins that bound to 1a by preparing resins with covalently linked 1a (resin Rf, Figure 1). While protocols existed for installation of a tag at the hindered secondary hydroxyl group at C18,[9] these methods contain semi–synthetic steps to convert the natural product into a functional derivative.[11] In order to minimize the use of 1a, we examined procedures that allowed the natural product to be attached in one step.
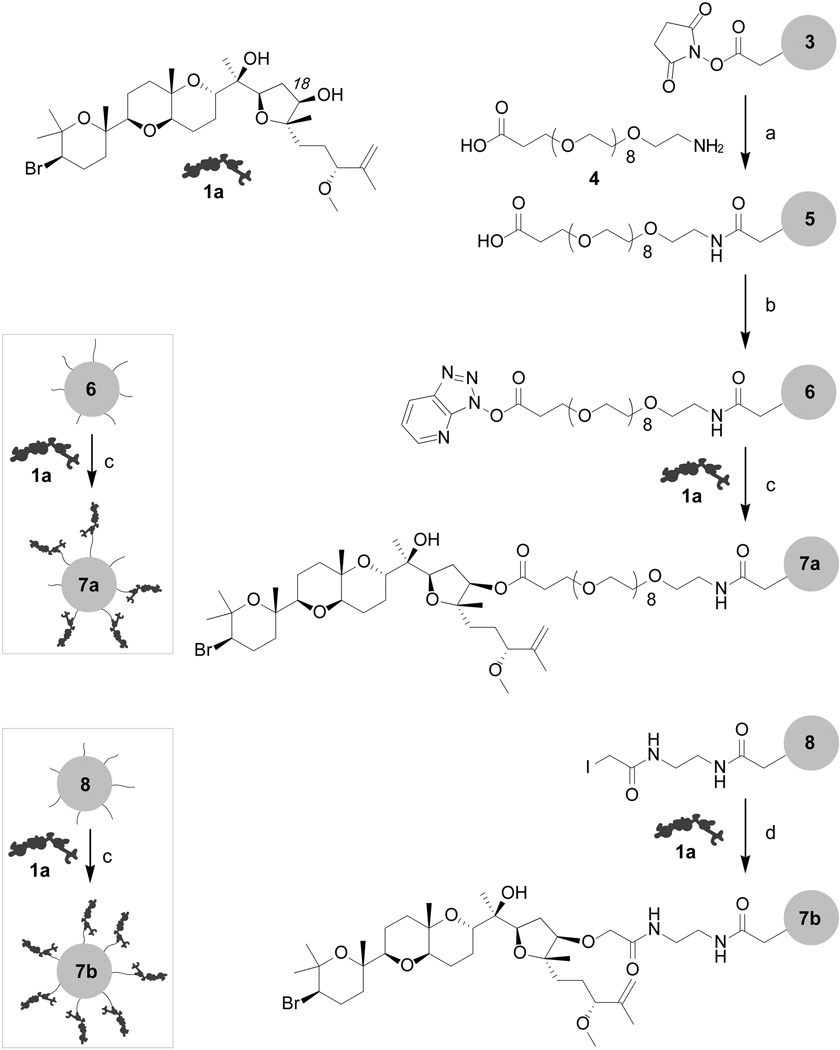
Our studies focused on preparation of two resins 7a and 7b (Figure 3). Preparation of resin 7a began with the installation of spacer 4[12] onto Affigel 10 resin 3 to afford resin 5. We then activated resin 5 by treatment with HATU and coupled 1a onto the activated resin 6 under DMAP catalysis.[13] While the yield was modest (0.5±0.1 mg/mL[14]), we were able to confirm the loading of 1a on resin 7a hydrolytically.[15]
Figure 3.
Preparation of aplysqualenol A resins 7a and 7b. Reagents and conditions: (a) PEG spacer 4, H2O, pH 8.5, 4 °C, 8 h; (b) HATU, 1,4–dioxane, 12 h, 4 °C to rt; (c) 1a, DMAP, 1,4–dioxane, 18 h, 4 °C to rt, followed by capping with 0.1 M glycine methyl ester in PBS pH 8.0; (d) 1a, 1,4–dioxane, 18 h, 4 °C to rt, followed by capping with 0.05 M 2–mercaptoethanol in PBS pH 8.0.
A second resin 7b was prepared by adapting protocols developed by Mitcheson and Walzack.[17] Affigel 10 resin 3 was converted to iodoacetamide resin 8 (Figure 3) by sequential treatment with ethylenediamine and iodoacetic acid N–hydroxysuccinimide ester. Aplysqualenol A (1a) was then coupled to resin 8 by incubation in dry dioxanes followed by capping with 2–mercaptoethanol. This procedure was far more effective providing resins with 1.8±0.3 mg/mL of 1a.[14]
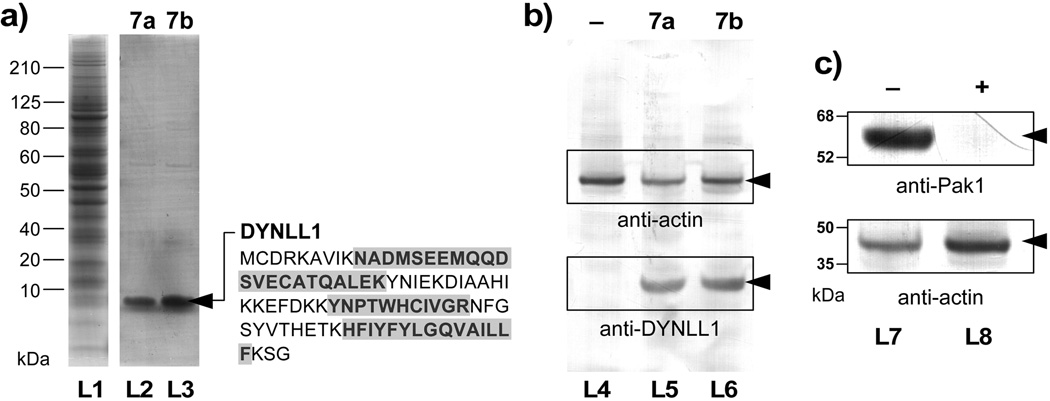
We then applied resins 7a and 7b to screen tumor cell lysates for binding proteins. Using lysates freshly prepared from HCT–116 cells (lane L1, Figure 4a), we identified a band from a <10 kDa protein over multiple repetitions using ether resin 7a or 7b. After optimization, a single band could be obtained in the bound fractions (lanes L2 or L3, Figure 4a). While both resins were functional, resin 7b provided nearly twice the return.
Figure 4.
Forward affinity studies identify dynein light chain type 1 (DYNLL1) as a target of aplysqualenol A (1a). (a) SDS–PAGE gels of HCT–116 cell lysate (lane L1) and the fractions that were pulled down with resins 7a (lane L2) and 7b (lane L3). A band corresponding to a <10 kDa protein was observed in the bound fractions from both resins 7a and 7b. (b) Western blot analysis using an anti–DYNLL1 mAb confirms the presence of DYNLL1 protein in the bound fractions from 7a (lane L5) and 7b (lane L6). Resins that lack 1a (lane L4) as given by a sample of resin 5 that was capped with glycine methyl ester did not contain DYNLL1. Actin was added to each fraction as loading control. (c) Affigel resin bearing 2.4±0.2 mg/mL of DYNLL1 was capable of isolating Pak1 from HCT–116 cell lysate. Western blots depict the isolation of Pak1 in DYNLL1–affinity fraction of pure lysates (–, lane L7). In contrast, the same lysate treated with 20 µM 1a lost the ability to sequester Pak1 (+, lane L7), indicating that 1a blocked the interaction between DYNLL1 and Pak1.[16]
Samples of these bands were excised and submitted for protein identification. Tryspin digestion followed by LC–MS/MS analyses indicated that this protein was the light chain of dynein type 1 (DYNLL1) with coverage by three peptides (blue sequences in Figure 4a). We validated this identification using three methods. First, western blot analysis using antibodies against human DYNLL1 confirmed the presence of DYNLL1 in the bound fractions (lanes L5 and L6, Figure 4b). Second, isothermal calorimetry (ITC) indicated that 1a bound to recombinant human DYNLL1[18] with a Kd value of 0.84±0.31 µM. Finally, we confirmed this binding constant using surface plasmon resonance (SPR) measurements, which returned a comparable Kd of 0.52±0.09 µM (see Supporting Information).
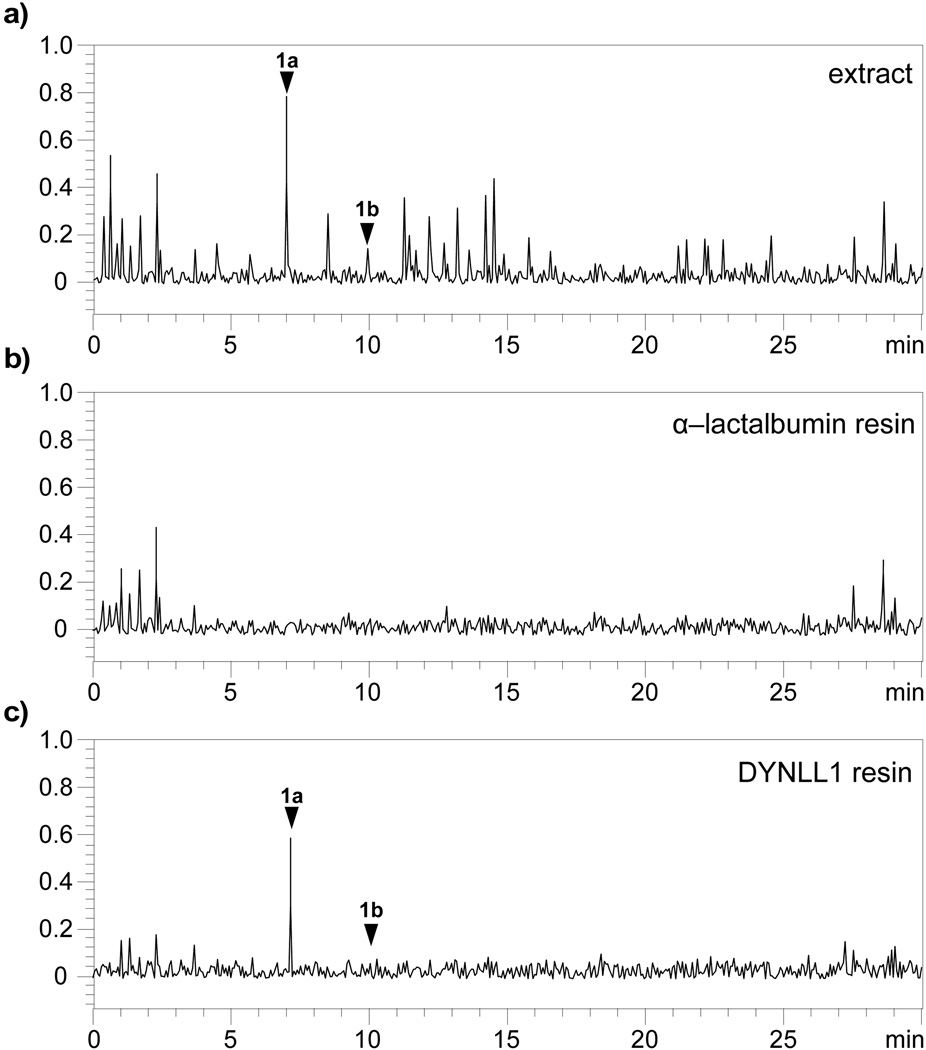
We further validated the binding event by reversing the affinity method. Samples of recombinant DYNLL1 protein[18] were loaded on Affigel 10 resin at 2.4±0.2 mg/mL[19] and screened for their ability to isolate aplysqualenol A (1a) from extracts of A. dactylomela (Figure 5a). By using 100 µL of this resin, we were able to observe the sequestration of 1a from 10 mg of crude extract (Figure 5c). Comparable extraction using α–lactalbumin,[20] a negative control, failed to return 1a (Figure 5b).
Figure 5.
Reverse affinity confirms the interaction between 1a and DYNLL1. (a) Metabolic profile obtained from the crude A. dactylomela extract using LC/MS analysis. The y–coordinate denotes the net signal detected by the mass spectrometer. Samples of this extract were then incubated with resins containing either (b) 3.2±0.1 mg/mL[19] of α–lactalbumin[20] on Affigel 10 resin as a negative control or (c) 2.4±0.2 mg/mL[19] of DYNLL1 on Affigel 10 resin. Aplysqualenol A (1a) and aplysqualenol B (1b) are noted by arrows and were confirmed by the presence of [M-HBr]+ of 536 and 552 m/z for 1a and 1b, respectively.
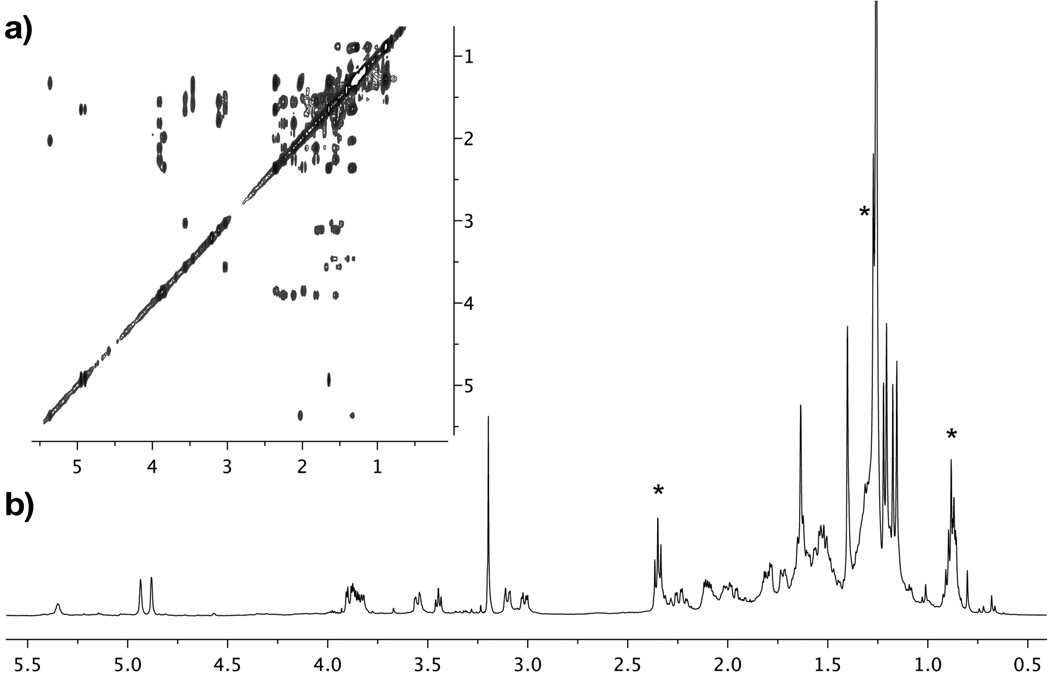
Application of this reverse affinity process to 25 mg of A. dactylomela extract returned sufficient 1a to be evaluated by capillary NMR analysis (Figure 6).[21] Standardization with pure 1a indicated that we obtained 8.4±0.6 μg of 1a, corresponding to a yield of 85 µg/mL resin, and that at least 0.35 mg/g of 1a was in the A. dactylomela extract.[22]
Figure 6.
NMR analysis confirms the isolation of aplysqualenol A (1a) from the DYNLL1 reverse affinity column. (a) gCOSY and (b) 1H–NMR obtained with a 1.7 mm Bruker BioSpin 600 MHz NMR equipped with a cryoprobe using material eluted from the reverse affinity column. These NMR spectroscopic data were comparable to those published for aplysqualenol A (1a) in CDCl3.[9] Lipid impurities are noted by an *.
With this evidence in hand, we turned to screen if the binding of 1a had any effects on proteins known to interact with DYNLL1. In particular, DYNLL1 was recently described to participate in the nuclear transport of Pak1, a serine/threonine kinase implicated in regulation of cell motility, cell survival and in the malignant transformation of mammary epithelia cells.[16, 23] Using western blot analysis (Figure 4c), we determined that treatment of HCT–116 cell lysates with 1a blocked the binding between DYNLL1 and Pak1, as indicated by the loss in the sequestration of Pak1 in lysates treated with 1a (lane L7 versus L8, Figure 4c).[24]
In summary, a bidirectional affinity approach was used to identify the binding of marine polyether aplysqualenol A (1a) to protein DYNLL1. DYNLL1 is a small cytosolic protein that belongs to a large enzymatic complex, dynein, with molecular mass of ~1.5 MDa.[25] In the cytoplasm, dynein plays a major role in motion regulating not only cell movement along microtubules but also guiding the positioning of organelles within the cell.[26] While the function of the heavy and intermediate chains are somewhat established,[27] that of the light chains such as DYNLL1 is less understood.[28] The identification of ligands to DYNLL1, such as 1a, provides a starting point for development of molecular probes.[29]
While samples of 1a or 1b can be obtained naturally they are scarce.[30] Development of a synthetic entry to 1a could provide a secondary source of material as well as expand the scope of probe development. Such probes offer immediate potential to examine the complex interactions of DYNLL1 but also could be used to establish the features of DYNLL1 that differentiate its isoforms.[31] Last, discovery of 1a as a ligand to DYNLL1 suggests new potential for the development of small molecule regulators of the dynein complex,[32] an endeavor which could have applications to cancer,[33] viral infections,[34] and neurodegenerative diseases.[35]
In conclusion, we demonstrated the ability to sequester proteins from small molecules and vice versa. The integration of both approaches into a single tactical (forward and reverse affinity) protocol provided further support for this target identification and supported its specificity. By combining these studies with mass spectrometric and NMR methods, we were able to rapidly identify the binding event, and suggest the further integration of these methods as primary tools for MOA research. While this approach is intrinsically linked to evaluate individual natural products serially, the adaption of multiplexed approaches such as bead libraries may allow one to rapidly integrate each metabolite within a metabolome with a corresponding binding protein, or set of proteins, in a given proteome.
Supplementary Material
Acknowledgments
We gratefully acknowledge the financial support of the National Institutes of Health (1SC1GM086271 and 1SC1GM086271-01A1). We wish to thank Ignacio Rodríguez–Crespo (Universidad Complutense de Madrid) for providing a plasmid containing the DYNLL1 gene, Dr. J. Beld and M. D. Burkart (UC San Diego) for assistance in the production of the recombinant DYNLL1, and Lauge Farnes and Chambers Hughes (UC San Diego) for assistance with the capillary NMR analyses.
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Brunilda Vera, Department of Chemistry University of Puerto Rico P.O. Box 23346, UPR Station San Juan, Puerto Rico 00931–3346
Abimael D. Rodríguez, Email: abimael.rodriguez1@upr.edu, Department of Chemistry University of Puerto Rico P.O. Box 23346, UPR Station San Juan, Puerto Rico 00931–3346.
James J. La Clair, Xenobe Research Institute P.O. Box 3052, San Diego, CA 92163-3052 i@xenobe.org, http://www.xenobe.org
References
- 1.Excellent reviews include: Blunt JW, Copp BR, Munro MH, Northcote PT, Prinsep MR. Nat. Prod. Rep. 2010;27:165–237. doi: 10.1039/b906091j. Olano C, Méndez C, Salas JA. Mar. Drugs. 2009;7:210–248. doi: 10.3390/md7020210. Newman DJ, Cragg GM, Holbeck S, Sausville EA. Curr. Cancer Drug Targets. 2002;2:279–308. doi: 10.2174/1568009023333791. Molinski TF. Clin. Adv. Hematol. Oncol. 2009;7:383–385. Nagle DG, Zhou YD, Mora FD, Mohammed KA, Kim YP. Curr. Med. Chem. 2004;11:1725–1756. doi: 10.2174/0929867043364991.
- 2. Examples of spectroscopic tools, include: Dalisay DS, Morinaka BI, Skepper CK, Molinski TF. J. Am. Chem. Soc. 2009;131:7552–7553. doi: 10.1021/ja9024929. Molinski TF. Nat. Prod. Rep. 2010;27:321–329. doi: 10.1039/b920545b. Esquenazi E, Yang YL, Watrous J, Gerwick WH, Dorrestein PC. Nat. Prod. Rep. 2009;26:1521–1534. doi: 10.1039/b915674g.
- 3. Examples of analytical techniques, include: Tetala KK, van Beek TA. J. Sep. Sci. 2010;33:422–438. doi: 10.1002/jssc.200900635. Pettit RK. Appl. Microbiol. Biotechnol. 2009;81:19–25. doi: 10.1007/s00253-009-1916-9. Lu Y, Hu R, Pan Y. Anal. Chem. 2010;82:3081–3085. doi: 10.1021/ac100121j.
- 4.La Clair JJ. Nat. Prod. Rep. 2010;27:969–995. doi: 10.1039/b909989c. [DOI] [PubMed] [Google Scholar]
- 5.Saito SY. Prog. Mol. Subcell. Biol. 2009;46:187–219. doi: 10.1007/978-3-540-87895-7_7. [DOI] [PubMed] [Google Scholar]
- 6.Selected examples of new methods for evaluating natural product MOA, include: Lomenick B, Hao R, Jonai N, Chin RM, Aghajan M, Warburton S, Wang J, Wu RP, Gómez F, Loo JA, Wohlschlegel JA, Vondriska TM, Pelletier J, Herschman HR, Clardy J, Clarke CF, Huang J. Proc. Natl. Acad. Sci. U S A. 2009;106:21984–21989. doi: 10.1073/pnas.0910040106. Pukkila–Worley R, Holson E, Wagner F, Mylonakis E. Curr. Med. Chem. 2009;16:1588–1595. doi: 10.2174/092986709788186237. Ho CH, Magtanong L, Barker SL, Gresham D, Nishimura S, Natarajan P, Koh JL, Porter J, Gray CA, Andersen RJ, Giaever G, Nislow C, Andrews B, Botstein D, Graham TR, Yoshida M, Boone C. Nat. Biotechnol. 2009;27:369–377. doi: 10.1038/nbt.1534. Mandrekar N, Thakur NL. Biotechnol. Lett. 2009;31:171–179. doi: 10.1007/s10529-008-9868-1. Crawford AD, Esguerra CV, de Witte PA. Planta Med. 2008;74:624–632. doi: 10.1055/s-2008-1034374. Schmitz K, Haggarty SJ, McPherson OM, Clardy J, Koehler AN. J. Am. Chem. Soc. 2007;129:11346–11347. doi: 10.1021/ja073965s. Hughes CC, Yang YL, Liu WT, Dorrestein PC, La Clair JJ, Fenical W. J. Am. Chem. Soc. 2009;131:12094–12096. doi: 10.1021/ja903149u. Hughes CC, MacMillan JB, Gaudêncio SP, Fenical W, La Clair JJ. Angew. Chem. Int. Ed. Engl. 2009;48:728–732. doi: 10.1002/anie.200804107.
- 7.Li JW, Vederas JC. Science. 2009;325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 8.Rodríguez AD, Lear MJ, La Clair JJ. J. Am. Chem. Soc. 2008;130:7256–7258. doi: 10.1021/ja7114019. [DOI] [PubMed] [Google Scholar]
- 9.Vera B, Rodríguez AD, Avilés E, Ishikawa Y. Eur. J. Org. Chem. 2009:5327–5336. doi: 10.1002/ejoc.200900775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The MTT assay was conducted using protocols recently described, see: Kang M, Jones BD, Mandel AL, Hammons JC, DiPasquale AG, Rheingold AL, La Clair JJ, Burkart MD. J. Org. Chem. 2009;74:9054–9061. doi: 10.1021/jo901826d.
- 11.a) Alexander MD, Burkart MD, Leonard MS, Portonovo P, Liang B, Ding X, Joullié MM, Gulledge BM, Aggen JB, Chamberlin AR, Sandler J, Fenical W, Cui J, Gharpure SJ, Polosukhin A, Zhang HR, Evans PA, Richardson AD, Harper MK, Ireland CM, Vong BG, Brady TP, Theodorakis EA, La Clair JJ. Chembiochem. 2006;7:409–416. doi: 10.1002/cbic.200500466. [DOI] [PubMed] [Google Scholar]; b) Peddibhotla S, Dang Y, Liu JO, Romo D. J. Am. Chem. Soc. 2007;129:12222–12231. doi: 10.1021/ja0733686. [DOI] [PubMed] [Google Scholar]; c) Leslie BJ, Hergenrother PJ. Chem. Soc. Rev. 2008;37:1347–1360. doi: 10.1039/b702942j. [DOI] [PubMed] [Google Scholar]
- 12.Iwaoka E, Mori T, Shimizu T, Hosoya K, Tanaka A. Bioorg. Med. Chem. Lett. 2009;19:1469–1472. doi: 10.1016/j.bmcl.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 13.The addition of 4–dimethylaminopyridine (DMAP) serves to accelerate the coupling of alcohols to activated esters, see: Neises B, Steglich W. Angew. Chem. Int. Ed. Engl. 1977;17:522–524. The use of anhydrous conditions permitted the coupling of 1a to the resin via its C18 hydroxyl group. In our isolation efforts, we demonstrated that this hydroxyl group was modestly hindered, as we were able to acylate 1a under mild conditions (Ac2O, pyridine, DMAP, rt).
- 14.Concentrations of 1a on resins were determined by monitoring the loss of 1a via LC/MS analysis. Aliquots of the reaction mixture were sampled before and after coupling and compared to stock solutions of 1a. Samples of unreacted 1a were recycled.
- 15.A 50 µL aliquot of resin 7a was treated with 0.05 N NaOH at rt for 4 h followed by neutralization to pH 7. Mass spectrometric analysis on this material identified a compound with a parent ion m/z of 537. HR–EI–MS analysis returned an m/z of 536.3691 that matched the elimination product 2 (calcd. 536.3713 for C31H52O7). A comparable loss of HBr was also observed in the HR–ESI–MS on 1a as noted in reference 9
- 16.a) Lightcap CM, Kari G, Arias–Romero LE, Chernoff J, Rodeck U, Williams JC. PLoS One. 2009;4:e6025. doi: 10.1371/journal.pone.0006025. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lightcap CM, Sun S, Lear JD, Rodeck U, Polenova T, Williams JC. J. Biol. Chem. 2008;283:27314–27324. doi: 10.1074/jbc.M800758200. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Song C, Wen W, Rayala SK, Chen M, Ma J, Zhang M, Kumar R. J. Biol. Chem. 2008;283:4004–4013. doi: 10.1074/jbc.M704512200. [DOI] [PubMed] [Google Scholar]
- 17.Field CM, Oegema K, Zheng Y, Mitchison TJ, Walczak CE. Methods Enzymol. 1998;298:525–541. doi: 10.1016/s0076-6879(98)98043-0. [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez–Crespo I, Yélamos B, Roncal F, Albar JP, Ortiz de Montellano PR, Gavilanes F. FEBS Lett. 2001;503:135–141. doi: 10.1016/s0014-5793(01)02718-1. [DOI] [PubMed] [Google Scholar]
- 19.Protein concentrations were determined using the Bradford assay, see: Noble JE, Bailey MJ. Methods Enzymol. 2009;463:73–95. doi: 10.1016/S0076-6879(09)63008-1.
- 20.α–Lactalbumin was used as a standard due to its low molecular weight and use as an affinity matrix. For example, see: Trayer IP, Barker R, Hill RL. Method. Enzymol. 1974;34:359–360. doi: 10.1016/s0076-6879(74)34038-4.
- 21.a) Molinski TF. Curr. Opin. Drug Discov. Devel. 2009;12:197–206. [PubMed] [Google Scholar]; b) Dossey AT, Walse SS, Rocca JR, Edison AS. ACS Chem. Biol. 2006;19:511–514. doi: 10.1021/cb600318u. [DOI] [PubMed] [Google Scholar]
- 22.Dalisay DS, Molinski TF. J. Nat. Prod. 2009;72:739–744. doi: 10.1021/np900009b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dummler B, Ohshiro K, Kumar R, Field J. Cancer Metastasis Rev. 2009;28:51–63. doi: 10.1007/s10555-008-9168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.While there have yet to be efforts to target this protein, studies have been conducted to develop inhibitors of the Pak1 enzymes, see: Yi C, Maksimoska J, Marmorstein R, Kissil JL. Biochem. Pharmacol. 2010;80:683–689. doi: 10.1016/j.bcp.2010.03.012.
- 25.a) Carter AP, Cho C, Jin L, Vale RD. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Akhmanova A, Hammer JA., 3rd Curr. Opin. Cell Biol. 2010;22:479–487. doi: 10.1016/j.ceb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Saxton WM. Methods Cell Biol. 1994;44:279–288. doi: 10.1016/s0091-679x(08)60919-x. [DOI] [PubMed] [Google Scholar]
- 26.a) Kardon JR, Vale RD. Nat. Rev. Mol. Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Thyberg J, Moskalewski S. Exp. Cell Res. 1999;246:263–279. doi: 10.1006/excr.1998.4326. [DOI] [PubMed] [Google Scholar]; c) Gonatas NK, Stieber A, Gonatas JO. J. Neurol. Sci. 2006;246:21–30. doi: 10.1016/j.jns.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 27.a) Asai DJ, Wilkes DE. J. Eukaryot. Microbiol. 2004;51:23–29. doi: 10.1111/j.1550-7408.2004.tb00157.x. [DOI] [PubMed] [Google Scholar]; b) Vallee RB, Höök P. J. Struct. Biol. 2006;156:175–181. doi: 10.1016/j.jsb.2006.02.012. [DOI] [PubMed] [Google Scholar]; c) Sakato M, King SM. J. Struct. Biol. 2004;146:58–71. doi: 10.1016/j.jsb.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 28.a) Mohan PM, Hosur RV. J. Biosci. 2009;43:465–479. doi: 10.1007/s12038-009-0052-0. [DOI] [PubMed] [Google Scholar]; b) Harrison A, King SM. Essays Biochem. 2000;35:75–87. doi: 10.1042/bse0350075. [DOI] [PubMed] [Google Scholar]
- 29.Carlson EE. ACS Chem. Biol. 2010;16:639–653. doi: 10.1021/cb100105c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.To date, we have isolated ~25 mg from two A. dactylomela specimens grazing upon the red alga Laurencia obtusa at 0.6 m depth near Bahia Salinas, Cabo Rojo, Puerto Rico. We expended nearly all of this material conducting our isolation efforts (see reference 9) and the work in this manuscript. We recently obtained additional specimens, however, the collection and extraction of A. dactylomela does not represent a sustainable source of 1a. Moreover, A. dactylomela may not be the biosynthetic producer of 1a. Note also that the digestive glands of Aplysia species are known to concentrate and transform natural products, see: Stallard MO, Faulkner DJ. Comp. Biochem. Physiol. B. Comp. Biochem. 1974;49:37–41. doi: 10.1016/0305-0491(74)90219-3. Kamiya H, Sakai R, Jimbo M. Prog. Mol. Subcell. Biol. 2006;43:215–239. doi: 10.1007/978-3-540-30880-5_10.
- 31.a) Radnai L, Rapali P, Hódi Z, Süveges D, Molnár T, Kiss B, Bécsi B, Erdödi F, Buday L, Kardos J, Kovács M, Nyitray L. J. Biol. Chem. 2010;285:38649–38657. doi: 10.1074/jbc.M110.165894. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lu J, Sun Q, Chen X, Wang H, Hu Y, Gu J. Biochem. Biophys. Res. Commun. 2005;331:153–158. doi: 10.1016/j.bbrc.2005.03.128. [DOI] [PubMed] [Google Scholar]; c) Susalka SJ, Nikulina K, Salata MW, Vaughan PS, King SM, Vaughan KT, Pfister KK. J. Biol. Chem. 2002;277:32939–32946. doi: 10.1074/jbc.M205510200. [DOI] [PubMed] [Google Scholar]
- 32.Purealin is an excellent example of a natural product that targets dynein. Recent studies have shown that purealin, as well as its synthetic analogs, inhibit the heavy chain of cytoplasmic dynein, see: Zhu G, Yang F, Balachandran R, Höök P, Vallee RB, Curran DP, Day BW. J. Med. Chem. 2006;49:2063–2076. doi: 10.1021/jm051030l.
- 33.Alberti C. Eur. Rev. Med. Pharmacol. Sci. 2009;13:13–21. [PubMed] [Google Scholar]
- 34.a) Schneider MA, Spoden GA, Florin L, Lambert C. Cell Microbiol. 2011;13:32–46. doi: 10.1111/j.1462-5822.2010.01515.x. [DOI] [PubMed] [Google Scholar]; b) Hsieh MJ, White PJ, Pouton CW. Curr. Opin. Biotechnol. 2010;21:633–639. doi: 10.1016/j.copbio.2010.06.009. [DOI] [PubMed] [Google Scholar]; c) Greber UF, Way M. Cell. 2006;124:741–754. doi: 10.1016/j.cell.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 35.a) Banks GT, Fisher EM. Genome Biol. 2008;9:214. doi: 10.1186/gb-2008-9-3-214. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) El–Kadi AM, Soura V, Hafezparast M. J. Neurosci. Res. 2007;85:2557–2566. doi: 10.1002/jnr.21188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.