Abstract
Background
A Pyrosequencing assay has been used in identification of fungal species such as Candida or Aspergillus and diagnosis of pathogenic bacteria such as Helicobacter pylori but there has been no report on successful isolation and identification of Malassezia yeasts using the pyrosequencing method.
Objective
Examine the applicability and plausibility of the pyrosequencing method in identification of the Malassezia species.
Methods
At internal transcribed spacer (ITS) sites 1 and 2, three primers were developed using Pyrosequencing Assay Design Software (Biotage AB). Pyrosequencing was performed on 11 standard strains and 83 genomic DNA samples obtained from 66 healthy controls aged from 1 to 80.
Results
The eleven Malassezia standard species and 83 genomic DNA samples were successfully identified using the pyrosequencing assay.
Conclusion
The pyrosequencing method is a new tool for analysis of Malassezia yeasts, and its precision and rapidity suggests its clinical applicability.
Keywords: Malassezia yeasts, Pyrosequencing assay
INTRODUCTION
Malassezia yeasts are lipophilic fungi that are recovered in 75~98% of healthy adults1,2. The yeasts, since being first introduced in 1889, have been linked to various skin conditions such as pityriasis versicolor, seborrheic dermatitis, and Malassezia folliculitis, and most recently, atopic dermatitis3,4. Its pathogenic ability is drawing attention more than ever as cases of confluent and reticulated papillomatosis5,6 and Malassezia onychomycosis, as well as systemic Malassezia infection in immunocompromised adults and neonates receiving intravenous fluid replacement7,8 have recently been reported9,10.
Conventional studies and identification on Malassezia yeasts have traditionally been based on morphological and biochemical analyses11. However, these methods often have dubious criteria, and environmental factors and genetic mutations are giving rise to new species. Therefore, new molecular biological methods, which would overcome these limitations, are now in demand.
The authors have already reported successful identification of Malassezia yeasts using 26S rDNA polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP)12. Restriction fragment length polymorphism (RFLP) methods enable us to analyse the pattern and size of fragmented amplified ribosomal DNA with the use of two restriction enzymes, Hha1, and BstF1. With these methods, genetic diversity can be examined, and it can be widely used in the rapid diagnosis and epidemiological study of fungal species because it is rapid, precise and cost-effective. In addition, the pyrosequencing method, which has recently been brought into the spotlight, enables us to identify the species with only a 30~40 bp sequence. Even though it has been used in the identification of fungal species such as Candida or Aspergillus13 and diagnosis of pathogenic bacteria such as Helicobacter pylori14, there has been no report on successful isolation and identification of Malassezia yeasts using the pyrosequencing method.
Thus, the authors were prompted to examine the applicability and plausibility of the pyrosequencing method in the identification of the Malassezia species and rapid and precise diagnosis with effective therapy of Malassezia yeasts-related infections, and compare its efficacy to the RFLP method.
MATERIALS AND METHODS
1) Standard strains
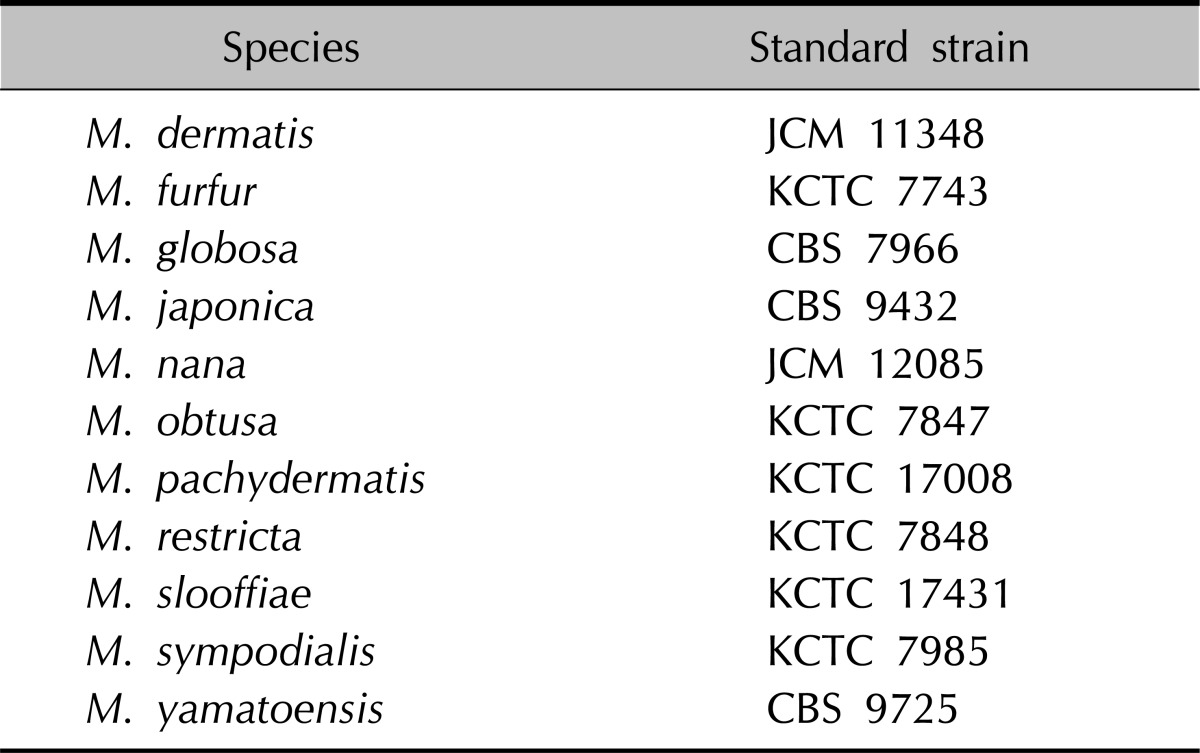
The 11 standard strains of the Malassezia species reported to date have been used in this study (Table 1). Six strains, i.e, M. furfur (KCTC 7743), M. obtusa (KCTC 7847), M. pachydermatis (KCTC 17008), M. restricta (KCTC 7848), M. slooffiae (KCTC 17431), M. sympodialis (KCTC 7985) have been obtained from the Department of Life Resources, Korea Research Institute of Biosciences and Biotechnology (KRIBB) while M. globosa (CBS 7966) and M. yamatoensis (CBS 9725) have been purchased from Centraalbureau voor Schimmelcultures (CBS) and M. dermatis (JCM 11348), M. nana (JCM 12085), and M. japonica (CBS 9432) from the Japanese Collection of Microorganisms (JCM).
Table 1.
Malassezia species used as standard strain
2) Specimen sampling from healthy subjects
Specimens were gathered from 6 different body locations (scalp, forehead, cheeks, anterior chest, inner aspect of upper arm, and upper inner portion of the thigh) of healthy adults. They were rubbed thoroughly with a sterile cotton swab measuring 5 cm in diameter with detergent (8.7 mM NaH2PO4, 12.3 mM Na2HPO4, 0.1% Triton X-100 [pH 7.9]) for 10 seconds and then streaked on LNA (Leeming & Notman). They were cultured for 2 weeks at 34℃.
Methods
1) DNA extraction
The glass beads method was used for isolation of genomic DNA from Malassezia yeasts. After sampling of species, 400 µl of lysis buffer (100 mM Tris-HCl pH 8.0, 1.0% SDS, 2.0% Triton X-100, 10 mM EDTA, 100 mM NaCl) was added and thoroughly mixed for 10 seconds before adding 400µl P/C/I (phenol:chloroform:isoamyl alcohol= 25:24:1, v/v) and 400 mg of glass beads approximately 0.5 mm in diameter. The resultant was stirred at room temperature for 10 minutes before being centrifuged at 15,000 rpm for 10 minutes. After the supernatant was removed, an identical amount of C/I (chloroform:isoamyl alcohol=24:1, v/v) was added and stirred at room temperature for 10 minutes before being centrifuged at 15,000 rpm for 10 minutes. The supernatant was removed, and an equal amount of isopropanol was added and DNA was concentrated at -80℃ for 1 hour. The mixture was then centrifuged at 13,000 rpm for 20 minutes so that DNA would precipitate, before washing with 70% ethanol and centrifuging again at 13,000 rpm for 5 minutes. The supernatant was removed and the DNA was dried at 37℃ for 20 minutes, and it was dissolved in distilled water and preserved at -20℃. The concentration of the extracted DNA was measured with a NanoDrop spectrophotometer ND-1000 (NanoDrop Technologies, Wilmington, DE, USA), and 50 ng was taken to be used on polymerase chain reaction (PCR).
2) Primer for pyrosequencing
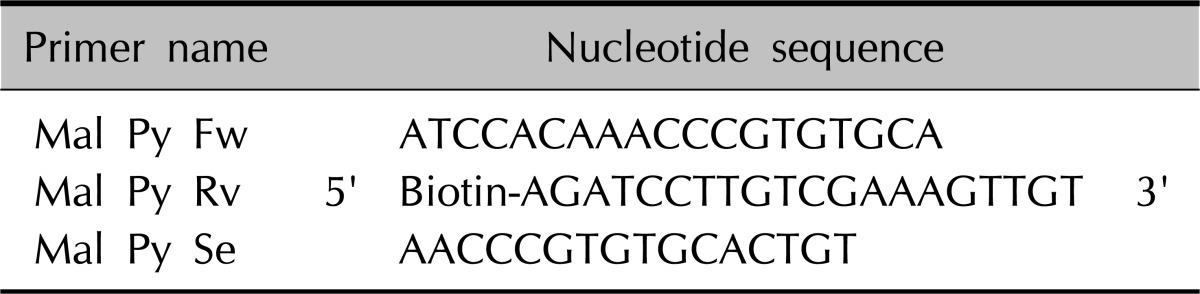
Based on information gathered from Blast of NCBI (http://www.ncbi.nlm.nih.gov) inter-specific homology of 18S, 5.8S, 28S ribosomal DNA of 11 standard strains has been compared at Cluster W. At internal transcribed spacer (ITS) sites 1 and 2, which exhibit disparity due to low interspecific homology, three primers were developed using pyrosequencing Assay Design Software (Biotage AB, Uppsala, Sweden). The primer designed for pyrosequencing was produced by the synthesis of a forward primer, sequence primer and reverse primer which contains biotin (Cosmogenetech, Seoul, Korea) at 5' end (Table 2).
Table 2.
Primers for pyrosequencing by pyrosequencing assay design software (Biotage AB, Uppsala, Sweden)
3) PCR for pyrosequencing
For identification of 11 standard strains using pyrosequencing, 50 ng of genomic DNA was taken and PCR was performed at ITS sites 1 and 2. PCR was carried out using 0.5 µM of primer, 0.25 mM deoxynucleoside triphosphate (dNTPs), 10X PCR buffer, and 0.6 U of Taq polymerase (Enzynomics, Daejeon, Korea), based on the genomic DNA as template. The conditions of the reaction were as follows; 30 seconds of denaturation at 94℃ following initial denaturation of 14 minutes at 94℃, 30 seconds of annealing at 54.5℃, and one minute of extension at 72℃. The whole process was repeated for 35 cycles, and the final extension was done at 72℃ for 7 minutes. Amplified bands of template DNA was examined on 3% TAE buffer agarose gel and was sequenced.
4) Pyrosequencing
3µ lof Streptavidin bead (Streptavidin Sepharose High Performance Amersham), 37 µl of 2X Binding buffer solution (10 mM Tris-HCl pH 7.6, 1 mM EDTA, 2 M NaCl, 0.1% Tween 20) and 20µl of distilled water were mixed and 40 µl was placed on each of the 96 well PCR plates. 20 µl of biotinylated PCR product was added and then was vortex at 1,400 rpm for 20 minutes. 100 pM of sequencing primer 0.15 µl was mixed with 40 µl of annealing buffer (200 mM Tris-acetate, 50 mM Mg-acetate), and was placed on PSQ 96 Plate Low (Bitage, Uppsala, Sweden). At the Vacuum Prep Workstation, the bead that had been placed on the 96 Well PCR plate was captured using vacuum, and was washed with 100% ethanol, Denaturation solution (0.2 M NaOH), Washing buffer (10 mM Tris -acetate pH 7.6) and distilled water, and the resultant was placed on the well of PSQ 96 Plate Low (Bitage). The mixture was heated at 87℃ for two minutes and was cooled slowly. Pyrosequencing was performed using a SNP reagent kit (enzyme and substrate mixture, dATP-S, dCTP, dGTP, dTTP).
RESULTS
Pyrosequencing of standard Malassezia strains
The eleven standard Malassezia strains were identified using pyrosequencing assay. Using the biotin-labeled primer set (Table 2) which is directed at the Internal transcribed spacer (ITS) 1~2 region at the genomic DNA, PCR was performed, and the expected fragment about 100 ~200 bp in size was obtained (Fig. 1).
Fig. 1.
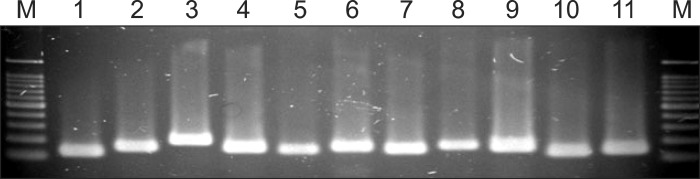
Polymerase chain reaction (PCR) amplification with primer of internal transcribed spacer (ITS) 1~2 from 11 Malassezia standard strains. Lanes: M, molecular marker; 1. M. dermatis (JCM 11348); 2. M. furfur (KCTC 7743); 3. M. globosa (CBS 7966); 4. M. japonica (CBS 9432); 5. M. nana (JCM 12085); 6. M. obtusa (KCTC 7847); 7. M. pachydermatis (KCTC 17008); 8. M. restricta (KCTC 7848); 9. M. slooffiae (KCTC 17431); 10. M. sympodialis (KCTC 7985); 11. M. yamatoensis (CBS 9725).
The result of pyrosequencing was analyzed and compared for homology with genetic target sequence using a SQA Identification Program (Biotage AB). M. dermatis, M. furfur, M. globosa, M. restricta, M. slooffiae and M. yamatoensis showed a 100% homology, followed by M. sympodialis (96%), M. nana (81%), M. pachydermatis (81%), M. japonica (80%), and M. obtusa (80%). Thus, the fact that isolation and identification of Malassezia standard strains is possible has been confirmed by the result that was obtained.
Pyrosequencing of clinically isolated samples
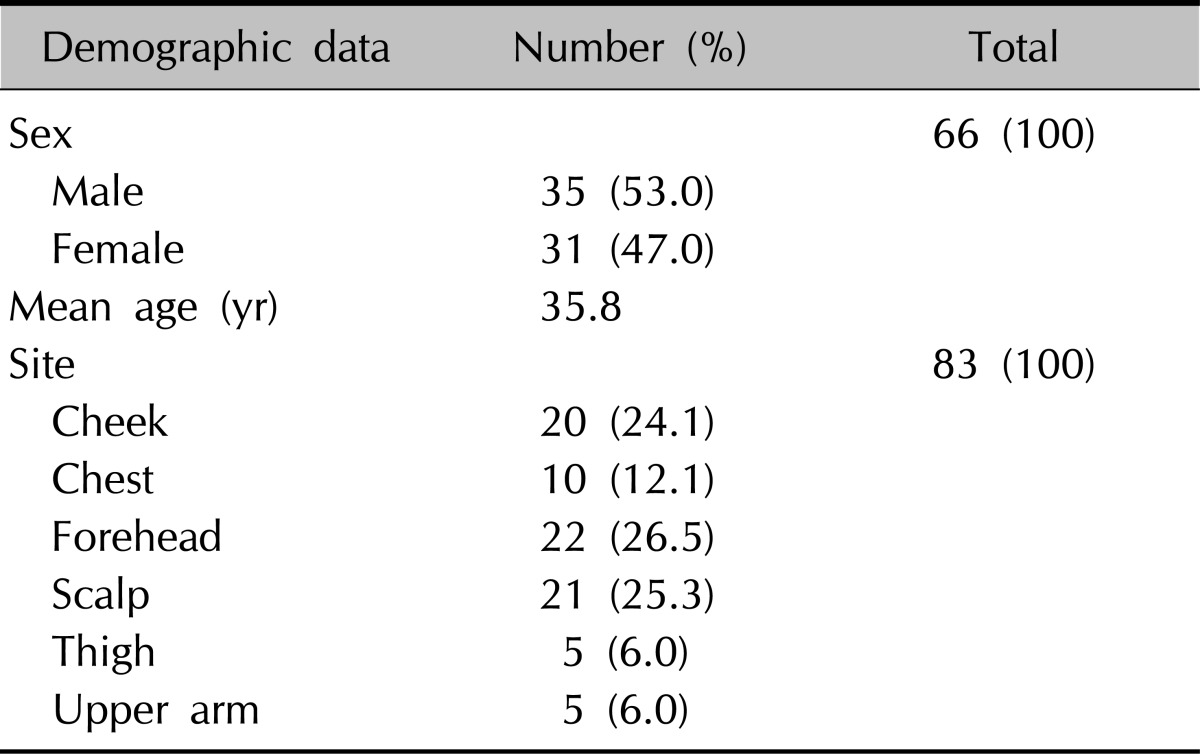
Eighty-three genomic DNA samples obtained from 66 healthy people aged from 1 to 80 were used in this study (35 males and 31 females). The average age of the subjects was 35.8 years. The samples were collected at the scalp in 21 cases, forehead in 22, cheeks in 20, anterior chest in 10, inner aspect of upper arm in 5, and upper inner aspect of thighs in 5 (Table 3).
Table 3.
Characteristics and distribution of patients enrolled in this study

RFLP: restriction fragment length polymorphism.
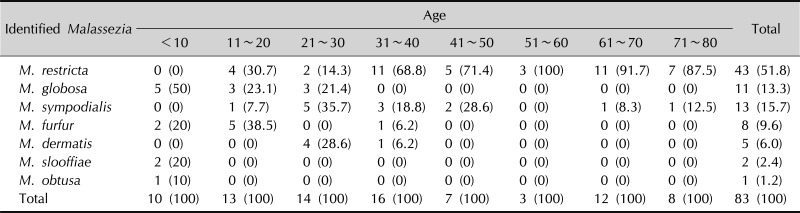
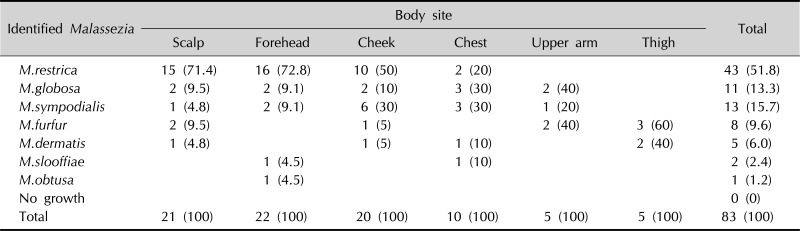
M. restricta was most predominant with 43 cases (51.8%), followed by M. sympodialis in 13 (15.7%), M. globosa in 11 (13.3%), M. furfur in 8 (9.6%), M. dermatis in 5 (6.0%), M. slooffiae in 2 (2.4%), and M. obtusa in 1 (1.2%). By age, M. restricta was most frequently found in the age group of 31~80 years, while M. globosa, M. furfur, and M. sympodialis were the most predominant species in the groups of less than 10 years, 11~20 years, and 21~30 years, respectively (Appendix 1). In the scalp, forehead, and cheek, M. restricta was the predominant species, whereas M. globsa and M. sympodialis were found in the chest in highest frequency, and likewise, M. globsa and M. furfur in the inner aspect of upper arm, and M. furfur in the upper inner portion of the thigh, respectively (Appendix 2).
Comparison of results obtained by 26S rDNA PCR-RFLP and the Pyrosequencing method
The results from 26S rDNA PCR-RFLP and the pyrosequencing method were compared. In the RFLP method, M. restricta was found in 43.5%, M. globosa in 26.6%, M. sympodialis in 16.7%, M. furfur in 4.8% and M. obtusa in 0% while in the pyrosequencing method M. restricta was isolated in 51.8%, M. globosa in 13.3%, M. sympodialis in 15.7%, M. furfur in 9.6% and M. obtusa in 1.2% (Table 4). Among the 83 samples, the results obtained by the two methods were in agreement with each other in 68 cases. The other 15 samples which showed different results were performed with sequencing analysis for confirmative identification. The result was found to be the same as the one obtained by the pyrosequencing method (Table 5).
Table 4.
The results from PCR-RFLP and pyrosequencing method
PCR-RFLP: polymerase chain reaction-restriction length fragment polymorphism.
Table 5.
Differences between identified Malassezia species from PCR-RFLP and pyrosequencing method
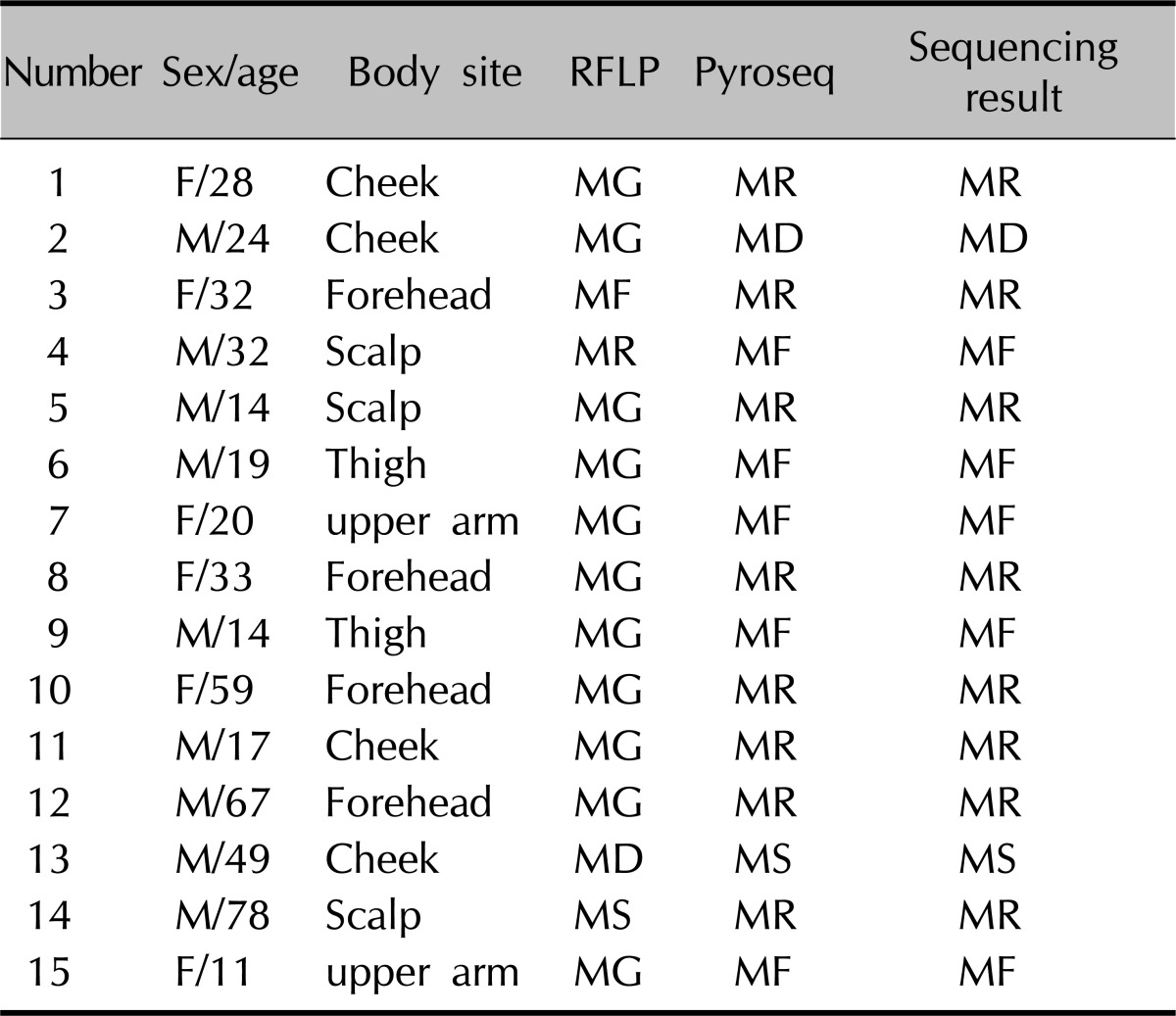
PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism, RFLP: restriction fragment length polymorphism, F: female, M: male.
DISCUSSION
Since the fact that Malassezia yeasts are isolated in 75~98% of healthy adults and are associated with various skin disorders, there has been a number of studies on the quantitative characteristics and differential distribution of the yeasts in physiologic and pathological settings, in an effort to establish the link between the yeasts and the skin disorders. A number of studies of the past on the mycological aspects of Malassezia yeasts had been based on biochemical and morphological aspects, such as the size of the colonies, surface condition, color, and shape. This is time-consuming, is subject to individual interpretations, has dubious criteria for judgment, and is not sufficient to provide objective evidence for isolation and identification of new species. Furthermore, it has its limitations in that there may even exist genetically different species among the ones that share the identical morphological and biochemical characteristics.
With the advance of molecular biological analysis and identification techniques, new identification tools focusing on genetic structure and characteristics, and various softwares are being developed15-23. Among these, sequencing analysis (Sanger's method)24 involves synthesizing targeted DNA with a template and analyzing sequences about 500 bp in size. With this method, identification of species, polymorphism, and taxonomical analysis are possible, but it is time-consuming and complex, and thus not suitable for clinical applications. Nested PCR, which has recently been attempted for identification and isolation of Malassezia species, is advantageous in that results are obtainable in a short period of time, but when compared to 26S rDNA PCR-RFLP, the distinction between species is difficult when the difference in a single band is between 1 and 10 bp25. In contrast, the RFLP method is relatively more accurate than nested PCR because it analyzes multiple band patterns produced by restriction enzymes26,27. In this study, the results produced by 26S rDNA PCR-RFLP and the pyrosequencing method were in concordance with each other in 68 cases out of 83 samples. For accurate identification of the other 15 samples, we performed the sequencing analysis and the results turned out to be identical with the ones of the pyrosequencing method, but not those of 26S rDNA PCR-RFLP. Thus, the authors concluded that the pyrosequencing method is more accurate than the traditional 26S rDNA PCR-RFLP. Furthermore, the fact that only 4 hours are required from extraction of DNA to final results adds to its efficiency in clinical application. Its drawbacks may be the high cost of the pre-treatment process of pyrosequencing. However, since the entire kit is purchased for use, comparing the exact costs (per mg or µg) for each method is not very feasible, although in general, the cost is roughly 2 to 3 times higher for pyrosequencing (Table 6).
Table 6.
Comparison of pyrosequencing and RFLP in terms of the time required for identification, cost, and accuracy
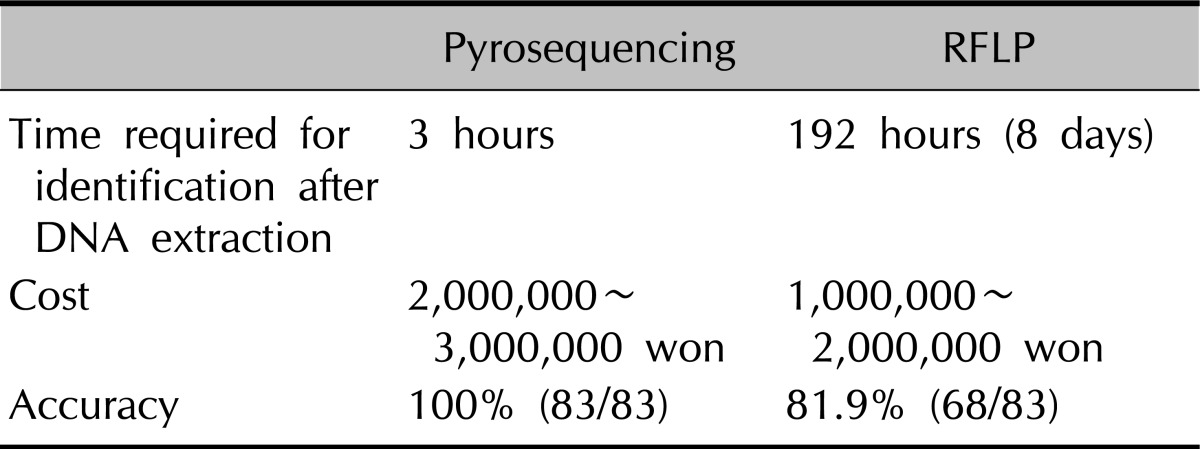
RFLP: restriction fragment length polymorphism.
The pyrosequencing method is a new tool for analysis of Malassezia yeasts, and its precision and rapidity suggests its clinical applicability. However, further studies focusing on the identification of fungal species and epidemiology of fungal diseases is necessary.
ACKNOWLEDGMENT
This work was subsidized by a research grant from the Hanmi pharmaceutical company in 2008.
Appendix 1
Identified Malassezia species from the age groups
Values are presented as number (%).
Appendix 2
Identified Malassezia species from the body sites
Values are presented as number (%).
References
- 1.Janik MP, Heffernan MP. Yeast infections: Candidiasis, pityriasis (tinea) versicolor. In: Wolff K, Goldsmith LA, Katz SI, Gilchrest BA, Paller AS, Leffell DJ, editors. Fitzpatrick's dermatology in general medicine. 7th ed. New York: McGraw-Hill; 2008. pp. 1822–1830. [Google Scholar]
- 2.Ahn KJ. Taxonomy of the genus malassezia. Korean J Med Mycol. 1998;3:81–88. [Google Scholar]
- 3.Ljubojević S, Skerlev M, Lipozencić J, Basta-Juzbasić A. The role of Malassezia furfur in dermatology. Clin Dermatol. 2002;20:179–182. doi: 10.1016/s0738-081x(01)00240-1. [DOI] [PubMed] [Google Scholar]
- 4.Kanda N, Tani K, Enomoto U, Nakai K, Watanabe S. The skin fungus-induced Th1- and Th2-related cytokine, chemokine and prostaglandin E2 production in peripheral blood mononuclear cells from patients with atopic dermatitis and psoriasis vulgaris. Clin Exp Allergy. 2002;32:1243–1250. doi: 10.1046/j.1365-2745.2002.01459.x. [DOI] [PubMed] [Google Scholar]
- 5.Ginarte M, Fabeiro JM, Toribio J. Confluent and reticulated papillomatosis (Gougerot-Carteaud) successfully treated with tacalcitol. J Dermatolog Treat. 2002;13:27–30. doi: 10.1080/09546630252775216. [DOI] [PubMed] [Google Scholar]
- 6.Yesudian P, Kamalam S, Razack A. Confluent and reticulated papillomatosis (Gougerot-Carteaud). An abnormal host reaction to Malassezzia furfur. Acta Derm Venereol. 1973;53:381–384. [PubMed] [Google Scholar]
- 7.Chowdhary A, Randhawa HS, Sharma S, Brandt ME, Kumar S. Malassezia furfur in a case of onychomycosis: colonizer or etiologic agent? Med Mycol. 2005;43:87–90. doi: 10.1080/13693780400006070. [DOI] [PubMed] [Google Scholar]
- 8.Gupta AK, Batra R, Bluhm R, Boekhout T, Dawson TL., Jr Skin diseases associated with Malassezia species. J Am Acad Dermatol. 2004;51:785–798. doi: 10.1016/j.jaad.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 9.Devlin RK. Invasive fungal infections caused by Candida and Malassezia species in the neonatal intensive care unit. Adv Neonatal Care. 2006;6:68–77. doi: 10.1016/j.adnc.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Curvale-Fauchet N, Botterel F, Legrand P, Guillot J, Bretagne S. Frequency of intravascular catheter colonization by Malassezia spp. in adult patients. Mycoses. 2004;47:491–494. doi: 10.1111/j.1439-0507.2004.01047.x. [DOI] [PubMed] [Google Scholar]
- 11.Senczek D, Siesenop U, Böhm KH. Characterization of Malassezia species by means of phenotypic characteristics and detection of electrophoretic karyotypes by pulsed-field gel electrophoresis (PFGE) Mycoses. 1999;42:409–414. doi: 10.1046/j.1439-0507.1999.00478.x. [DOI] [PubMed] [Google Scholar]
- 12.Lee YW, Lim SH, Ahn KJ. The application of 26S rDNA PCR-RFLP in the identification and classification of Malassezia yeast. Korean J Med Mycol. 2006;11:141–153. [Google Scholar]
- 13.Gharizadeh B, Norberg E, Löffler J, Jalal S, Tollemar J, Einsele H, et al. Identification of medically important fungi by the Pyrosequencing technology. Mycoses. 2004;47:29–33. doi: 10.1046/j.1439-0507.2003.00949.x. [DOI] [PubMed] [Google Scholar]
- 14.Nilsson I, Shabo I, Svanvik J, Monstein HJ. Multiple displacement amplification of isolated DNA from human gallstones: molecular identification of Helicobacter DNA by means of 16S rDNA-based pyrosequencing analysis. Helicobacter. 2005;10:592–600. doi: 10.1111/j.1523-5378.2005.00361.x. [DOI] [PubMed] [Google Scholar]
- 15.Gupta AK, Boekhout T, Theelen B, Summerbell R, Batra R. Identification and typing of Malassezia species by amplified fragment length polymorphism and sequence analyses of the internal transcribed spacer and largesubunit regions of ribosomal DNA. J Clin Microbiol. 2004;42:4253–4260. doi: 10.1128/JCM.42.9.4253-4260.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theelen B, Silvestri M, Guého E, van Belkum A, Boekhout T. Identification and typing of Malassezia yeasts using amplified fragment length polymorphism (AFLP), random amplified polymorphic DNA (RAPD) and denaturing gradient gel electrophoresis (DGGE) FEMS Yeast Res. 2001;1:79–86. doi: 10.1111/j.1567-1364.2001.tb00018.x. [DOI] [PubMed] [Google Scholar]
- 17.Gandra RF, Simão RC, Matsumoto FE, da Silva BC, Ruiz LS, da Silva EG, et al. Genotyping by RAPD-PCR analyses of Malassezia furfur strains from pityriasis versicolor and seborrhoeic dermatitis patients. Mycopathologia. 2006;162:273–280. doi: 10.1007/s11046-006-0063-4. [DOI] [PubMed] [Google Scholar]
- 18.Gaitanis G, Velegraki A, Alexopoulos EC, Chasapi V, Tsigonia A, Katsambas A. Distribution of Malassezia species in pityriasis versicolor and seborrhoeic dermatitis in Greece. Typing of the major pityriasis versicolor isolate M. globosa. Br J Dermatol. 2006;154:854–859. doi: 10.1111/j.1365-2133.2005.07114.x. [DOI] [PubMed] [Google Scholar]
- 19.Gemmer CM, DeAngelis YM, Theelen B, Boekhout T, Dawson TL., Jr Fast, noninvasive method for molecular detection and differentiation of Malassezia yeast species on human skin and application of the method to dandruff microbiology. J Clin Microbiol. 2002;40:3350–3357. doi: 10.1128/JCM.40.9.3350-3357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guillot J, Deville M, Berthelemy M, Provost F, Guého E. A single PCR-restriction endonuclease analysis for rapid identification of Malassezia species. Lett Appl Microbiol. 2000;31:400–403. doi: 10.1046/j.1472-765x.2000.00839.x. [DOI] [PubMed] [Google Scholar]
- 21.Makimura K, Tamura Y, Kudo M, Uchida K, Saito H, Yamaguchi H. Species identification and strain typing of Malassezia species stock strains and clinical isolates based on the DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J Med Microbiol. 2000;49:29–35. doi: 10.1099/0022-1317-49-1-29. [DOI] [PubMed] [Google Scholar]
- 22.Gaitanis G, Velegraki A, Frangoulis E, Mitroussia A, Tsigonia A, Tzimogianni A, et al. Identification of Malassezia species from patient skin scales by PCR-RFLP. Clin Microbiol Infect. 2002;8:162–173. doi: 10.1046/j.1469-0691.2002.00383.x. [DOI] [PubMed] [Google Scholar]
- 23.Mirhendi H, Makimura K, Zomorodian K, Yamada T, Sugita T, Yamaguchi H. A simple PCR-RFLP method for identification and differentiation of 11 Malassezia species. J Microbiol Methods. 2005;61:281–284. doi: 10.1016/j.mimet.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 24.Sugita T, Kodama M, Saito M, Ito T, Kato Y, Tsuboi R, et al. Sequence diversity of the intergenic spacer region of the rRNA gene of Malassezia globosa colonizing the skin of patients with atopic dermatitis and healthy individuals. J Clin Microbiol. 2003;41:3022–3027. doi: 10.1128/JCM.41.7.3022-3027.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim SW, Shin MG, Lim JY, Yun SJ, Kim SJ, Lee SC, et al. Nested PCR for detection of Malassezia species from patient skin scales and clinical strains. Korean J Dermatol. 2008;46:446–452. [Google Scholar]
- 26.Jang SJ, Lim SH, Ko JH, Oh BH, Kim SM, Song YC, et al. The investigation on the distribution of malassezia yeasts on the normal Korean skin by 26S rDNA PCR-RFLP. Ann Dermatol. 2009;21:18–26. doi: 10.5021/ad.2009.21.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh BH, Song YC, Lee YW, Choe YB, Ahn KJ. Comparison of nested PCR and RFLP for identification and classification of malassezia yeasts from healthy human skin. Ann Dermatol. 2009;21:352–357. doi: 10.5021/ad.2009.21.4.352. [DOI] [PMC free article] [PubMed] [Google Scholar]