Abstract
Human longevity and personality traits are both heritable and are consistently linked at the phenotypic level. We test the hypothesis that candidate genes influencing longevity in lower organisms are associated with variance in the five major dimensions of human personality (measured by the NEO-FFI and IPIP inventories) plus related mood states of anxiety and depression. Seventy single nucleotide polymorphisms (SNPs) in six brain expressed, longevity candidate genes (AFG3L2, FRAP1, MAT1A, MAT2A, SYNJ1 and SYNJ2) were typed in over one thousand 70-year old participants from the Lothian Birth Cohort of 1936 (LBC1936). No SNPs were associated with the personality and psychological distress traits at a Bonferroni corrected level of significance (p < 0.0002), but there was an over-representation of nominally significant (p < 0.05) SNPs in the synaptojanin-2 (SYNJ2) gene associated with agreeableness and symptoms of depression. Eight SNPs which showed nominally significant association across personality measurement instruments were tested in an extremely large replication sample of 17 106 participants. SNP rs350292, in SYNJ2, was significant: the minor allele was associated with an average decrease in NEO agreeableness scale scores of 0.25 points, and 0.67 points in the restricted analysis of elderly cohorts (most aged > 60 years). Because we selected a specific set of longevity genes based on functional genomics findings, further research on other longevity gene candidates is warranted to discover whether they are relevant candidates for personality and psychological distress traits.
Keywords: NEO personality, IPIP personality, anxiety, depressive symptoms, ageing, genetics
Individual differences in personality traits are influenced by genetic variation, and these genetic effects mostly endure across the lifespan [1, 2, 3]. Specific personality traits (e.g., high conscientiousness and low neuroticism) are predictive of longevity (which itself shows familial influence) [4], so it follows that candidate genes for longevity might be associated with various personality dimensions. There is likely no single mechanism to explain the link between personality traits and longevity. It might be that physiological changes in old age result in personality changes (e.g., worsening physical health leads someone to be more depressed) or that certain personality types engage in behaviours conducive to survival. But if either of the causal traits shows genetic variability that influences intermediary behaviours then this should also be detected in the correlated trait. Alternately, there may be a genetic correlation between personality and longevity resulting from genetic pleiotropy, for instance, a gene that influences the stress response could have effects on personality and longevity. In this study, we test the association of six longevity candidate genes—that were identified through a comparative functional genomics study [5]—with measures of personality and related measures of anxiety and depression in an elderly cohort and in replication cohorts.
The five major dimensions of personality include neuroticism (characterised by e.g., emotional sensitivity, and a tendency toward anxiety), extraversion (e.g., outgoingness, sociability), openness to experience (e.g., interest in intellectual pursuits, arts, ideas), conscientiousness (e.g., dutifulness, organisation) and agreeableness (e.g., non-confrontational, easy to get along with) [6]. Each has been studied in relation to longevity (in both normally ageing and exceptionally long-lived samples), with conscientiousness being the most consistently associated personality trait. A meta-analysis of 20 studies confirmed a significant positive correlation (r = .11, 95% confidence interval of .05-.17) between conscientiousness and longevity [7]. With regard to the other traits, a study of 246 offspring (mean age of 75 years) of centenarians showed that they were more extraverted and less neurotic, and women more agreeable, than the normative mean population levels [8]. This agreed with findings from the Baltimore Longitudinal Study of Aging: among deceased participants, those scoring above one standard deviation from the mean on the general activity facet of extraversion, emotional stability, or conscientiousness had lived two to three years longer than those in the reversed tail of the distributions [4]. In Weiss and Costa’s [9] analysis of the NEO-PI-R, agreeableness and conscientiousness (and a trend for neuroticism) were protective for survival over three years in 66 to 102 year-olds. Such personality-longevity associations might even be stronger if gene by environment interaction effects are present.
Animal studies confirm a role of genes in longevity [10, 11], which complements human twin studies of deaths from age-related disease, and family studies of centenarians that show genetic effects explaining between one quarter to half of the variability in longevity [12, 13, 14]. The identification of genes involved in age-related diseases has focussed predominantly on pathways involved in cell cycle control, oxidative stress, insulin, other endocrine signalling, and inflammation [15]. In comparative functional genomics, Smith and colleagues [5] were the first to assess the extent to which longevity genes are conserved between highly divergent eukaryotic species by comparing the yeast Saccharomyces cerevisiae and the nematode Caenorhabditis elegans. They identified 25 ageing genes in C. elegans that were conserved in their yeast relatives and were therefore good candidate genes for ageing in humans. We selected a subset of these candidate genes comprising all those that are brain-expressed and evolutionary-conserved—AFG3L2, FRAP1, MAT1A, MAT2A, SYNJ1 and SYNJ2—which we hypothesised would be relevant for personality and mood traits in an ageing cohort. The first gene found to alter ageing—located in the insulin-like signalling pathway—was discovered in C. elegans and subsequent studies suggest that innate immunity is influenced by this pathway and that it is conserved from yeast to humans [16]. There are known associations between inflammatory biomarkers and personality in humans [17], illustrative of potential mechanisms which might underlie personality-longevity associations, that is, through pleiotropic effects acting on the immune and central nervous systems. The recently discovered set of conserved genes in C. elegans might also uncover biological pathways of relevance to individual differences in human ageing and their associations with personality differences .
We test a specific set of candidate longevity genes—identified by comparative functional genomics—in humans in relation to non-cognitive psychological traits and states. Haplotype-tagging SNPs in each of these genes were genotyped in an elderly Scottish cohort (mean age of 70 years) and tested for their association with measures of personality and psychological distress, which are predictive of longevity. Associations were followed up in independent replication cohorts, including elderly samples from Finland, Italy, USA and Germany.
Materials and Methods
Subjects – LBC1936
All participants were born in 1936 and had taken part in the Scottish Mental Survey of 1947; they were tested on psychological (including mood) and medical traits at about 70 years of age at the Wellcome Trust Clinical Research Facility (Western General Hospital, Edinburgh) and completed some questionnaires (including personality) at home [18]. The sample was Caucasian and lived independently in the Lothian region (Edinburgh city and surrounding area) of Scotland. Further description about participant recruitment can be found elsewhere [18]. DNA samples and psychological distress data were available for 1 078 participants. The available personality data gave an analysis sample of N = 951 for these traits of 470 men and 481 women. The mean age of the sample was 69.5 years (SD = 0.8). The mood states of anxiety and depression were assessed using the Hospital Anxiety Depression Scale (HADS) [19]. The personality traits of the Five Factor model (see Introduction) were measured using the NEO Five-Factor Inventory [20]. The NEO Five-Factor Inventory (NEO-FFI) is a 60-item inventory consisting of 12 items for each of the five factors. Participants also completed the IPIP Big-Five 50-item inventory [21], consisting of 10 items for each of the Big-Five personality factors. The participants were given the personality questionnaires with written instructions at the end of their clinic visit and asked to return them by post.
SNP selection
SNPs with a minor allele frequency greater than .05 were selected using Tagger [22] in Haploview v 4.1 [23] based on the Hapmap CEPH population (Release 22) data. Using pairwise tagging (r2=.80), 70 SNPs tagged haplotypes from the specific gene regions and 5kb either side of the gene, although one of these SNPs was excluded due to high linkage disequilibrium with another tagged SNP. They served as direct proxies to all other untyped SNPs in the six genes because they are highly correlated with one another. Non-synonymous SNPs were also included as haplotype-tagging SNPs. The UCSC genome browser [24] was used to identify non-synonymous SNPs—rs2502601 in exon 27 of SYNJ2 a missense substitution (G=Gly, A=Glu, Glu1468Gly) and rs2254562 in exon 8 of SYNJ1 a missense substitution (G=Arg, A=Lys, Lys295Arg), giving 70 SNPs in total.
Genotyping
Genomic DNA was isolated from whole blood by standard procedure at the WTCRF Genetics Core, Western General Hospital, Edinburgh. Forty-nine SNPs were genotyped using a competitive allele specific PCR system (KASPar) by Kbiosciences, Herts, UK. A further 21 SNPs—all in SYNJ2—were genotyped using the Illumina Human610-Quadv1 Chip, for more detail see Luciano and colleauges [25].
Genotype data
The mean genotyping rate was 99% (range 92%-100%) and minor allele frequencies (MAF) were > 0.04 (see Online Resource Table 1 for marker position and MAFs). Genotype frequencies were similar to the HapMap CEPH population (mean difference in genotype frequencies = .03, minimum .002, maximum .08) , and all SNPs were in Hardy Weinberg equilibrium (HWE) as judged by the HWE exact SNP Tests (all p-values > .001) in Haploview.
Statistical Analysis and Power
Association tests were performed for individual SNPs in PLINK [26] using the regression option (additive model) and including sex and age as covariates. The statistical power to detect a genetic effect size of 1% (for MAF of 0.25) was 90.8% [27]. Because a large number of measures and SNPs were investigated, a Bonferroni correction was applied that resulted in a new significance criterion of .0002. This was based on correcting for five independent factors; that is, the five major personality dimensions (anxiety and depression are aligned with the neuroticism factor) multiplied by the number of independent SNPs. Nyholt’s [28] Single Nucleotide Polymorphism Spectral Decomposition program was used to estimate SNP independence within genes, resulting in a correction for 46 independent SNPs.
Replication Cohorts
Personality Traits
Eleven cohorts were available in which to replicate any associations for NEO personality measures. These cohorts were of European descent and are described in de Moor et al [29]. In brief, they comprised samples of varying age range (15 – 87 years) from Italy (Cilento, SardiNIA), The Netherlands (NTR/NESDA, ERF), United States (BLSA, SAGE), Finland (HBCS), Australia (QIMR, NAG/IRPG), Estonia (EGPUT) and Germany, totalling 17 106 participants with mean age ranging from 19.4 ±3 (Australia: QIMR) to 78.9 ±5.4 (Italy: Cilento) years. Personality scores for the five factors were based on the 60 items of the NEO-FFI (12 items per factor)[6]. SNP data were available from varying sized genome-wide scans which had all been imputed to ~2.4M SNPs using HAPMAP II data. Association analysis of each SNP (under an additive model) had been performed using either PLINK, SNPTEST or MERLIN[29]. The results for selected longevity candidate gene SNPs were meta-analysed using METAL [30]. Two separate meta-analyses of older cohorts (HBCS, BLSA, Cilento, Germany: mean age > 63.4 years; N = 2555) and younger cohorts (SardiNIA, NTR/NESDA, ERF, SAGE, NAG/IRPG, QIMR, EGPUT: mean age < 49.3 years; N = 14 551), then a combined analysis, were performed.
Anxiety and Depression Traits
HADS data were available in an independent cohort of 517 participants from the Lothian region, born in 1921 (LBC1921) and assessed on psychological distress at ~79 years [31]. Further details of the mood inventory data collection can be found in Gow and colleagues [32]. SNP genotyping was performed with the Illumina Human610-Quadv1 Chip with SNPs imputed to ~2.4M based on HAPMAP II data. For further details see Luciano and colleagues [25]. Replication testing, using PLINK, was performed for association findings from the main analysis.
Results
The anxiety and depression scores from the HADS were negatively skewed and were square root transformed to improve their distribution. All personality measures showed distributions close to normality. Descriptive statistics for the NEO-FFI, IPIP and HADS measures appear in Table 1. Consistent with prior evidence, sex differences (women scoring higher) were observed for HADS anxiety (p < .001), NEO and IPIP neuroticism (p < .01) and agreeableness (p < .0001). NEO openness to experience (p < .0001) and extraversion (p < .05) scores were also higher in women but their corresponding IPIP factors showed no sex differences. Age effects were observed for NEO and IPIP conscientiousness (p < .05), such that lower scores tended to be associated with older age. However, the age range in the sample is very small.
Table I.
Sample size, range, mean and standard deviation of personality and psychological distress scales in the LBC1936.
| N | All | Men | Women | |
|---|---|---|---|---|
|
| ||||
| Range; Mean (SD) |
Range; Mean (SD) |
Range; Mean (SD) |
||
| IPIP | ||||
| Emotional Stability | 950 | 1 – 40; 24.6 (7.7) | 1 – 40; 25.5 (7.6) | 1 – 40; 23.7 (7.6) |
| Extraversion | 954 | 0 – 40; 21.3 (7.1) | 0 – 40; 21.0 (7.3) | 0 – 40; 21.6 (6.8) |
| Intellect | 948 | 5 – 40; 23.8 (5.7) | 5 – 40; 23.8 (5.8) | 5 – 40; 23.9 (5.6) |
| Agreeableness | 952 | 5 – 40; 31.1 (5.4) | 5 – 40; 29.0 (5.4) | 14 – 40; 23.1 (4.6) |
| Conscientiousness | 952 | 9 – 40; 28.2 (6.0) | 10 – 40; 28.0 (5.9) | 9 – 40; 28.5 (6.1) |
| NEO-FFI | ||||
| Neuroticism | 954 | 0 – 47; 17.1 (7.6) | 0 – 41; 15.7 (7.6) | 1 – 47; 18.4 (7.4) |
| Extraversion | 943 | 6 – 43; 27.0 (5.9) | 9 – 42; 26.6 (6.1) | 6 – 43; 27.4 (5.8) |
| Openness | 947 | 9 – 43; 26.0 (5.8) | 9 – 42; 25.2 (5.7) | 12 – 43; 26.9 (5.8) |
| Agreeableness | 954 | 17 – 47; 33.5 (5.3) | 17 – 47; 31.8 (5.2) | 22 – 47; 35.0 (4.8) |
| Conscientiousness | 947 | 11 – 48; 34.7 (6.0) | 11 – 48; 34.4 (6.0) | 14 – 48; 34.9 (5.9) |
|
| ||||
|
Psychological
Distress |
||||
| HADS Anxiety | 1089 | 0 – 17; 4.9 (3.2) | 0 – 16; 4.2 (2.9) | 0 – 17; 5.6 (3.3) |
| HADS Depression | 1086 | 0 – 16; 2.8 (2.2) | 0 – 16; 2.9 (2.3) | 0 – 11; 2.7 (2.1) |
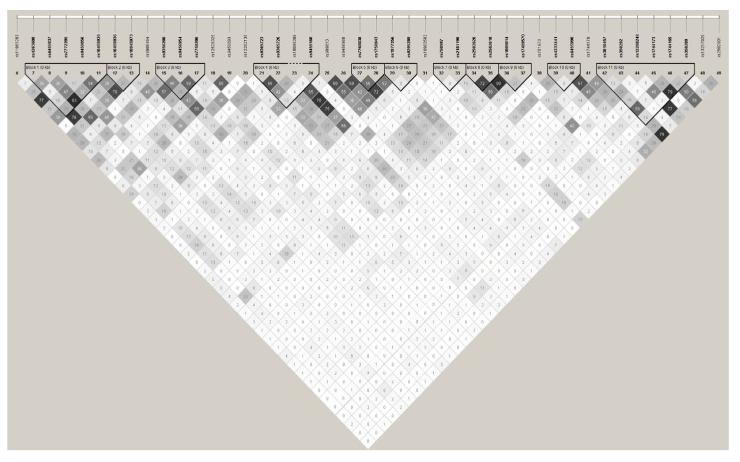
The results of the association tests for personality and psychological distress measures in the LBC1936 are shown in the Online Resource Table 1, these will be described. No SNPs surpassed the Bonferroni corrected significance level for any of the traits. However, for agreeableness, there were a larger number of nominally significant findings (p < .05) than for the other measures: 10 for the IPIP and six for the NEO. For IPIP agreeableness all of the nominally significant SNPs were located in the SYNJ2 gene (in independent SNPs), more than would be expected by chance (with .05 probability) assuming 44 tests. For NEO agreeableness, an additional SNP (rs2254562) was observed in the SYNJ1 gene. The most significant SNP for agreeableness, located in SYNJ2, was rs12202135 (p = .0003 for IPIP; p = .01 for NEO); MAF of this SNP was 0.13. Ten SNPs were nominally significant for depression, with all of these located in SYNJ2 (more than expected by chance); the most significant SNP was rs10046389 (p = .004) with a MAF of 0.40. Another of these SNPs (rs1750043) was also associated with anxiety (p = .01; MAF: 0.48). High conscientiousness and low neuroticism have been especially linked with longevity, but the association results were predominantly null for conscientiousness (three SNPs nominally associated with NEO and one non-overlapping SNP with IPIP). Eight SNPs were associated with IPIP emotional stability, two of which (rs10455936, MAF, 0.22; rs9459093, MAF, 0.43) located in SYNJ2 overlapped with significant SNPs for NEO neuroticism. Because a large number of SNPs (i.e., 44) was typed in SYNJ2 the linkage disequilibrium between them is shown in Figure 1; note that 24 independent SNPs were derived by SNP Spectral Decomposition.
Figure 1.
The linkage disequilibrium structure (including r2) for the region genotyped in SYNJ2 in the LBC1936.
Across all traits, standardised coefficients for nominally significant associations ranged between .06 and .11. A reduced set of SNPs which were nominally associated with both NEO and IPIP scales or with both psychological distress and emotional stability (and therefore deemed more reliable associations) in the LBC1936 are shown in Table 2. Replication of these SNPs was sought in independent cohorts. Table 3 shows the replication results for the personality traits. One of these SNPs, rs350292 (located in intron 18 of SYNJ2; MAF: 0.11), was significantly associated with agreeableness in the older cohorts (p = .0059), and at a reduced level in the combined older and younger cohorts meta-analysis. Within the eleven individual cohorts tested, nominal significance was observed in three of these (ERF, p = .049; HBCS, p = .038; BSA, p = .048), with two being ageing cohorts where the mean ages were 63.4 ±3 years (HBCS) and 68.5 ±17 years (BSA). The allele effects in these cohorts were in the same direction as the original association in LBC1936: the unstandardised regression coefficient was −.67 in the older cohort replication meta-analysis versus −.93 in the LBC1936 cohort (the A allele relating to lower agreeableness scores). For psychological distress measures there were no significant associations in the LBC1921 cohort, p > 0.18.
Table II.
Effect size (standardised regression coefficient) and significance of haplotype-tagging longevity gene SNPs that are nominally significant for at least two correlated traits (i.e., the same NEO and IPIP scale or psychological distress and neuroticism measures) in the LBC1936. P-values < 0.05 appear in bold and p < .005 are denoted with an asterisk.
| Anxiety | Depression | NEO N | IPIP ES | NEO E | IPIP E | NEO O | IPIP I | NEO A | IPIP A | NEO C | IPIP C | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||||||||||
| GENE | Bet | Bet | Bet | Bet | Bet | Bet | Bet | Bet | Bet | Bet | Bet | Bet | ||||||||||||
| SNP | a | p | a | p | a | p | a | p | a | p | a | p | a | p | a | p | a | p | a | p | a | p | a | p |
| MAT2A | ||||||||||||||||||||||||
| rs1446669 | − .01 |
.8 5 |
.00 | 1 | − .01 |
.8 0 |
.02 | .65 | − .03 |
.4 3 |
− .03 |
.3 2 |
−
.07 |
.0
4 |
−
.06 |
.0
5 |
−.01 | .8 7 |
− .05 |
.10 | − .03 |
.3 4 |
− .02 |
.5 2 |
| SYNJ2 | ||||||||||||||||||||||||
| rs4263608 a |
.05 | .1 2 |
.07 | .03 | .07 | .0 4 |
−. 06 |
.09 | − .06 |
.1 0 |
− .05 |
.1 3 |
.00 | .9 2 |
− .03 |
.3 1 |
− .01 |
.7 9 |
− .02 |
.51 | − .05 |
.1 3 |
− .05 |
.1 6 |
| rs6455937 b |
.07 |
.0
3 |
.02 | .49 | .04 | .2 5 |
− .07 |
.04 | − .01 |
.8 3 |
−
.07 |
.0 5 |
− .03 |
.4 3 |
− .04 |
.2 2 |
.04 | .2 5 |
.04 | .20 | .00 | .9 9 |
− .02 |
.6 5 |
| rs1045593 5b |
.07 | .0 2 |
.00 | .91 | .05 | .1 2 |
− .08 |
.02 | .01 | .8 9 |
− .07 |
.0 3 |
− .03 |
.3 2 |
− .04 |
.2 0 |
.02 | .5 3 |
.06 | .07 | .01 | .7 6 |
.00 | .9 5 |
| rs1045593 6a |
.06 | .0 7 |
.05 | .09 | .07 | .0 3 |
− .07 |
.04 | − .05 |
.1 2 |
− .07 |
.0 3 |
− .01 |
.7 5 |
− .05 |
.1 6 |
− .01 |
.7 2 |
− .01 |
.79 | − .02 |
.5 1 |
− .02 |
.4 7 |
| rs1094597 3 |
− .08 |
.0 1 |
−.01 | .75 | − .06 |
.0 8 |
.07 | .03 | − .02 |
.5 0 |
.06 | .0 6 |
.03 | .4 2 |
.03 | .3 0 |
− .03 |
.3 3 |
− .03 |
.34 | − .01 |
.7 7 |
.00 | .9 4 |
| rs1220213 5 |
.02 | .5 3 |
−.07 | .03 | − .02 |
.6 4 |
.02 | .54 | .02 | .6 6 |
.01 | .7 4 |
.01 | .6 7 |
− .03 |
.3 9 |
.09 | .0 1 |
.11 | .00 * |
.05 | .1 5 |
.04 | .2 9 |
| rs9459093 | .05 | .0 8 |
.04 | .18 | .07 | .0 3 |
−
.10 |
.00 * |
− .02 |
.6 1 |
− .04 |
.2 1 |
− .03 |
.2 9 |
− .06 |
.0 8 |
.02 | .6 4 |
.01 | .80 | − .04 |
.2 7 |
.03 | .3 9 |
| rs9459160 | .04 | .1 7 |
−.03 | .35 | .02 | .4 6 |
.01 | .70 | .05 | .1 7 |
.04 | .1 8 |
.03 | .3 5 |
− .04 |
.2 0 |
.07 | .0 3 |
.08 | .01 | .02 | .5 2 |
− .02 |
.6 5 |
| rs1750043 | .08 | .0 1 |
.06 | .04 | .03 | .3 8 |
− .04 |
.23 | − .03 |
.3 6 |
− .06 |
.1 1 |
.01 | .8 0 |
− .04 |
.3 0 |
− .05 |
.1 0 |
− .07 |
.02 | − .04 |
.2 9 |
− .05 |
.1 4 |
| rs9295289 | .05 | .1 2 |
.06 | .05 | .04 | .2 1 |
− .07 |
.04 | − .01 |
.8 5 |
− .05 |
.1 3 |
.00 | .9 3 |
.01 | .8 3 |
− .05 |
.1 6 |
− .03 |
.37 | − .01 |
.7 4 |
.00 | .9 7 |
| rs350292 | − .03 |
.3 5 |
.03 | .37 | − .01 |
.7 2 |
− .01 |
.76 | .01 | .8 5 |
.03 | .3 3 |
.00 | .9 1 |
.00 | .9 0 |
− .08 |
.0 1 |
− .07 |
.03 | − .02 |
.5 9 |
− .03 |
.4 3 |
| rs1744173 c |
.03 | .3 6 |
−.03 | .42 | .08 | .0 2 |
− .03 |
.41 | − .09 |
.0 1 |
− .09 |
.0 1 |
.01 | .6 8 |
− .06 |
.0 9 |
− .03 |
.31 | − .05 |
.11 | − .07 |
.0 4 |
− .03 |
.4 6 |
| rs350289c | .04 | .1 8 |
.00 | .89 | .09 | .0 1 |
− .05 |
.16 | − .09 |
.0 1 |
− .09 |
.0 1 |
.02 | .5 2 |
− .04 |
.2 6 |
− .04 |
.1 7 |
− .07 |
.03 | − .07 |
.0 3 |
− .02 |
.5 0 |
N = Neuroticism; ES = Emotional Stability (reverse pole of N); E = Extraversion; I = Intellect; O = Openness; A = Agreeableness; C = Conscientiousness
= Linkage Disequilibrium (LD) r2 = .76;
= LD r2 = .83;
= LD r2 = .79
Table III.
Replication p-values and unstandardised betas in the NEO consortium of SNPs in MAT2A and SYNJ2 that were identified in the LBC1936 as nominally associated with measures of both NEO and IPIP personality. P-values < 0.05 are highlighted in bold.
| Measure | GENE | Older Cohorts | Younger Cohorts | Combined | |||
|---|---|---|---|---|---|---|---|
| SNP | Beta (SE) | p | Beta (SE) | p | Beta (SE) | p | |
| MAT2A | |||||||
| Openness | rs1446669 | .00 (.19) | .98 | .03 (.11) | .80 | .05 (.08) | .56 |
| SYNJ2 | |||||||
| Neuroticism | rs10455936 | −.08 (.27) | .76 | −0.09 (.15) | .54 | −.13 (.12) | .26 |
| Agreeableness | rs12202135 | −.02 (.23) | .92 | .07 (.14) | .64 | .02 (.11) | .83 |
| Neuroticism | rs9459093 | .01 (.21) | .96 | −.02 (.13) | .86 | .01 (.10) | .91 |
| Agreeableness | rs9459160 | −.26 (.28) | .36 | .15 (.14) | .31 | .10 (.11) | .37 |
| Agreeableness | rs350292 | −.67 (.25) | .01 § | −.17 (.14) | .22 | −.25 (.11) * | .02 |
| Extraversion | rs1744173 | .25 (.23) | .27 | −.08 (.15) | .60 | .00 (.11) | .96 |
| Extraversion | rs350289 | .11 (.25) | .66 | −.13 (.16) | .42 | −.07 (.12) | .58 |
p-value = 0.0059
8 of 11 cohorts showed beta effect in the same direction; note that the unstandardised beta estimate in LBC1936 was −.93 with a standard error of .36.
Discussion
A priori evidence of an association between personality and longevity [4] led us to test the association between a specific set of longevity candidate genes—established via a functional genomics study—and personality, anxiety and depression measures. Of the six genes tested, SYNJ2 showed a heightened number of SNPs that were nominally significant at an uncorrected level for measures of agreeableness and depression, and to a lesser extent, neuroticism.
Due to their increased association with longevity, conscientiousness and neuroticism were especially hypothesised to associate with longevity candidate genes, but neither of these traits showed strong evidence of association with any of the genes. Most notable were the SNPs in SYNJ2 that showed p-values lower than .05 for NEO neuroticism (five SNPs) and IPIP emotional stability (six SNPs). SNPs in this same gene were also associated with agreeableness and depression scores, although they too did not exceed the corrected probability threshold. There is biological evidence to suggest SYNJ2 may have a plausible role in disordered mood. Decreased expression of SYNJ2 has been shown in the temporal cortex of major depressive disorder patients [33] and in a rodent model of depression overlap between cingulate cortex gene expression, stress behaviour and anti-depressant response identified SYNJ2 as a candidate gene for therapeutic targets [34]. While this is aligned with our findings of association with depression and neuroticism, our strongest support for SYNJ2 was with agreeableness, where rs350292 showed association in our replication sample of more than 17 000 participants and the direction of the effect was consistent between the original and replication sample. Of note, the meta-analysis of older cohorts—but not the younger cohorts—showed a significant effect of this SNP with agreeableness. Agreeableness, particularly its straightforwardness facet, is predictive of survival in Americans older than 66 years [9]. Six other SNPs in SYNJ2 were in high linkage disequilibrium with this SNP but, like rs350292, they were all intronic SNPs, with no documented previous associations. Ours is the first association study to investigate SYNJ2 and personality traits. Synaptojanins are a family of phosphoinositide phosphatases; and like synaptojanin 1, mammalian synaptojanin 2 is involved in dynamin and clathrin-mediated synaptic vesicle recycling but, distinct from SYNJ1 it is expressed more widely (both are concentrated in nerve terminals) and is linked to membrane trafficking and signal transduction pathways [35, 36].
The only gene that has been tested previously for association with traits relevant to our study was the SYNJ1 gene. It is a phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2) involved in clathrin-coated pit dynamics, which are required for efficient synaptic vesicle endocytosis and re-availability at nerve terminals [37]. It was identified in a linkage region for bipolar disorder, but the association analysis results of this gene with depression have been inconsistent. Stopkova and colleagues [38] did not find differences in allele frequencies between controls and bipolar patients for a common mutation, and previous rare variants in this gene were not detected in their sample. However, one of the homozygote groups was overrepresented among bipolar patients in one of their samples. In our study, no SNPs in SYNJ1 were associated with depression or associated measures of anxiety and neuroticism, suggesting that the effects of this gene on depression may be limited to rare mutations if they are important at all.
Our SNPs were chosen using information from HapMap, but one study which re-sequenced longevity candidates (including FRAP1) in healthy old adults showed that only 19% of variants in their sequencing set were observed in HapMap [39]. It is possible, then, that we are missing important uncommon variants that do have effects on the longevity-related traits of personality and psychological distress. While it is possible that other genes or other factors may influences the relationship between personality and longevity, we can fairly confidently rule out common variants in the AFG3L2, FRAP1, MAT1A, MAT2A, and SYNJ1 genes. Follow-up of further variants in SYNJ2, and especially in older cohorts, is needed before we can dismiss it as a candidate gene for personality and psychological distress in old-age. Genome-wide association studies of both of longevity and personality [40, 29] will serve as important repositories of gene associations that can be interrogated systematically for longevity candidate genes, such as SYNJ2. Furthermore, strategies focussed on rare variants in longevity candidate genes could prove a fruitful approach.
Supplementary Material
Acknowledgements
We thank the LBC1936 and LBC1921 cohort members and study Secretary Paula Davies. We thank Alan Gow, Michelle Taylor, Janie Corley, Caroline Brett and Caroline Cameron for data collection and data entry. We thank the nurses and other staff at the Wellcome Trust Clinical Research Facility where the data were collected. We thank the staff at Lothian Health Board, and the staff at the SCRE Centre, University of Glasgow. The research was supported by a programme grant (to IJD and JMS) from Research Into Aging. Phenotype collection in the Lothian Birth Cohort 1936 was supported by Research Into Ageing. Phenotype collection in the Lothian Birth Cohort 1921 was supported by the BBSRC, The Royal Society, and The Chief Scientist Office of the Scottish Government. The work was undertaken by The University of Edinburgh Centre for Cognitive Aging and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative (G0700704/84698). Funding (to IJD and JMS) from the BBSRC, EPSRC, ESRC and MRC is gratefully acknowledged. ML is a Royal Society of Edinburgh/Lloyds TSB Foundation for Scotland Personal Research Fellow. LML is the beneficiary of a post-doctoral grant from the AXA Research Fund. We would like to acknowledge the contributions of research participants and staff on all the replication samples (SardiNIA, NTR/NESDA, ERF, SAGE, HBCS, NAG/IRPG, QIMR, BLSA, EGPUT, Germany), specific details of which (including grant funding) can be found in de Moor et al (in press).
Contributor Information
Michelle Luciano, Centre for Cognitive Aging and Cognitive Epidemiology, The University of Edinburgh, Scotland, UK; Department of Psychology, The University of Edinburgh, Scotland, UK.
Lorna M. Lopez, Centre for Cognitive Aging and Cognitive Epidemiology, The University of Edinburgh, Scotland, UK; Department of Psychology, The University of Edinburgh, Scotland, UK
Marleen H.M. de Moor, Department of Biological Psychology, VU University Amsterdam, 1081 BT, Amsterdam, The Netherlands
Sarah E. Harris, Centre for Cognitive Aging and Cognitive Epidemiology, The University of Edinburgh, Scotland, UK; Medical Genetics Section, The University of Edinburgh, Scotland, UK
Gail Davies, Centre for Cognitive Aging and Cognitive Epidemiology, The University of Edinburgh, Scotland, UK; Department of Psychology, The University of Edinburgh, Scotland, UK.
Teresa Nutile, Institute of Genetics and Biophysics A. Buzzati-Traverso – CNR, Naples, Italy.
Robert F. Krueger, Department of Psychology, University of Minnesota, Minneapolis, USA
Tõnu Esko, Institute of Molecular and Cell Biology & Estonian Genome Center, University of Tartu and Estonian Biocenter, Estonia.
David Schlessinger, National Institute on Aging, NIH, DHHS, Baltimore, USA.
Tanaka Toshiko, National Institute on Aging, NIH, DHHS, Baltimore, USA.
Jaime L. Derringer, Department of Psychology, University of Minnesota, Minneapolis, USA
Anu Realo, Department of Psychology, University of Tartu, Estonia.
Narelle K. Hansell, Queensland Institute of Medical Research, Brisbane, Australia
Michele L. Pergadia, Department of Psychiatry, Washington University School of Medicine, St. Louis, USA
Anu-Katriina Pesonen, Insitute of Behavioral Sciences, University of Helsinki, Helsinki, Finland.
Serena Sanna, Istituto di Neurogenetica e Neurofarmacologia, Consiglio Nazionale delle Ricerche, Monserrato, Cagliari, Italy.
Antonio Terracciano, National Institute on Aging, NIH, DHHS, Baltimore, USA.
Pamela A.F. Madden, Department of Psychiatry, Washington University School of Medicine, St. Louis, USA
Brenda Penninx, VU University Medical Center, Amsterdam, The Netherlands.
Philip Spinhoven, Leiden University, Leiden, The Netherlands.
Catherine Hartman, University Medical Center Groningen, Groningen, The Netherlands.
Ben A. Oostra, Department of Clinical Genetics, Erasmus University Medical Center, Rotterdam, The Netherlands
A. Cecile J.W. Janssens, Department of Epidemiology, Erasmus University Medical Center, Rotterdam, The Netherlands
Johan G Eriksson, Department of General Practice and Primary Health Care, University of Helsinki, Helsinki, Finland.
John M. Starr, Centre for Cognitive Aging and Cognitive Epidemiology, The University of Edinburgh, Scotland, UK; Department of Geriatric Medicine, The University of Edinburgh, Royal Victoria Hospital, UK
Alessandra Cannas, Istituto di Neurogenetica e Neurofarmacologia, Consiglio Nazionale delle Ricerche, Monserrato, Cagliari, Italy.
Luigi Ferrucci, National Institute on Aging, NIH, DHHS, Baltimore, USA.
Andres Metspalu, Institute of Molecular and Cell Biology & Estonian Genome Center, University of Tartu and Estonian Biocenter, Estonia.
Margeret J. Wright, Queensland Institute of Medical Research, Brisbane, Australia
Andrew C. Heath, Department of Psychiatry, Washington University School of Medicine, St. Louis, USA
Cornelia M. van Duijn, Department of Epidemiology, Erasmus University Medical Center, Rotterdam, The Netherlands
Laura J. Bierut, Department of Psychiatry, Washington University School of Medicine, St. Louis, USA
Katri Raikkonen, Insitute of Behavioral Sciences, University of Helsinki, Helsinki, Finland.
Nicholas G. Martin, Queensland Institute of Medical Research, Brisbane, Australia
Marina Ciullo, Institute of Genetics and Biophysics A. Buzzati-Traverso – CNR, Naples, Italy.
Dan Rujescu, Department of Psychiatry, University of Munich (LMU), Germany.
Dorret I. Boomsma, Department of Biological Psychology, VU University Amsterdam, 1081 BT, Amsterdam, The Netherlands
Ian J. Deary, Centre for Cognitive Aging and Cognitive Epidemiology, The University of Edinburgh, Scotland, UK; Department of Psychology, The University of Edinburgh, Scotland, UK
References
- 1.Bratko D, Butkovic A. Stability of genetic and environmental effects from adolescence to young adulthood: results of Croatian longitudinal twin study of personality. Twin Res Hum Genet. 2007;10(1):151–7. doi: 10.1375/twin.10.1.151. [DOI] [PubMed] [Google Scholar]
- 2.Viken RJ, Rose RJ, Kaprio J, et al. A developmental genetic analysis of adult personality: extraversion and neuroticism from 18 to 59 years of age. J Pers Soc Psychol. 1994;66(4):722–30. doi: 10.1037//0022-3514.66.4.722. [DOI] [PubMed] [Google Scholar]
- 3.Blonigen DM, Carlson MD, Hicks BM, et al. Stability and change in personality traits from late adolescence to early adulthood: a longitudinal twin study. J Pers. 2008;76(2):229–66. doi: 10.1111/j.1467-6494.2007.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terracciano A, Lockenhoff CE, Zonderman AB, et al. Personality predictors of longevity: activity, emotional stability, and conscientiousness. Psychosom Med. 2008;70(6):621–7. doi: 10.1097/PSY.0b013e31817b9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith ED, Tsuchiya M, Fox LA, et al. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 2008;18(4):564–70. doi: 10.1101/gr.074724.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costa PTJ, McCrae RR. Revised NEO Personality Inventory and NEO Five-Factor Inventory: Professional Manual. Psychological Assessment Resources, Inc.; Lutz, FL: 1992. [Google Scholar]
- 7.Kern ML, Friedman HS. Do conscientious individuals live longer? A quantitative review. Health Psychol. 2008;27(5):505–12. doi: 10.1037/0278-6133.27.5.505. [DOI] [PubMed] [Google Scholar]
- 8.Givens JL, Frederick M, Silverman L, et al. Personality traits of centenarians’ offspring. J Am Geriatr Soc. 2009;57(4):683–5. doi: 10.1111/j.1532-5415.2009.02189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss A, Costa PT., Jr. Domain and facet personality predictors of all-cause mortality among Medicare patients aged 65 to 100. Psychosom Med. 2005;67(5):724–33. doi: 10.1097/01.psy.0000181272.58103.18. [DOI] [PubMed] [Google Scholar]
- 10.Brown-Borg HM. Hormonal regulation of longevity in mammals. Ageing Res Rev. 2007;6(1):28–45. doi: 10.1016/j.arr.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuningas M, Mooijaart SP, van Heemst D, et al. Genes encoding longevity: from model organisms to humans. Aging Cell. 2008;7(2):270–80. doi: 10.1111/j.1474-9726.2008.00366.x. [DOI] [PubMed] [Google Scholar]
- 12.Melzer D, Hurst AJ, Frayling T. Genetic variation and human aging: progress and prospects. J Gerontol A Biol Sci Med Sci. 2007;62(3):301–7. doi: 10.1093/gerona/62.3.301. [DOI] [PubMed] [Google Scholar]
- 13.Christensen K, Johnson TE, Vaupel JW. The quest for genetic determinants of human longevity: challenges and insights. Nat Rev Genet. 2006;7(6):436–48. doi: 10.1038/nrg1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herskind AM, McGue M, Holm NV, et al. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870-1900. Hum Genet. 1996;97(3):319–23. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- 15.Cluett C, Melzer D. Human genetic variations: Beacons on the pathways to successful ageing. Mech Ageing Dev. 2009 doi: 10.1016/j.mad.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Amrit FR, May RC. Younger for longer: insulin signalling, immunity and ageing. Curr Aging Sci. 2010;3(3):166–76. doi: 10.2174/1874609811003030166. [DOI] [PubMed] [Google Scholar]
- 17.Sutin AR, Terracciano A, Deiana B, et al. High neuroticism and low conscientiousness are associated with interleukin-6. Psychological Medicine. 2010;40(9):1485–93. doi: 10.1017/S0033291709992029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deary IJ, Gow AJ, Taylor MD, et al. The Lothian Birth Cohort 1936: a study to examine influences on cognitive ageing from age 11 to age 70 and beyond. BMC Geriatrics. 2007;7:28. doi: 10.1186/1471-2318-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zigmond AS, Snaith RP. The hospital anxiety and depression scale. Acta Psychiatr Scand. 1983;67(6):361–70. doi: 10.1111/j.1600-0447.1983.tb09716.x. [DOI] [PubMed] [Google Scholar]
- 20.Costa PTJ, McCrae RR. NEO PI-R Professional Manual. Psychological Assessment Resources; Odessa: 1992. [Google Scholar]
- 21.Goldberg L. A broad-bandwidth, public-domain, personality inventory measuring the lower-level facets of several Five-Factor models. In: Mervielde I, Deary IJ, de Fruyt F, et al., editors. Personality Psychology in Europe. Vol 7. Tilburg University Press; Tilburg: 1999. pp. 7–28. [Google Scholar]
- 22.de Bakker PI, Yelensky R, Pe’er I, et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37(11):1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Karolchik D, Kuhn RM, Baertsch R, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36(Database issue):D773–9. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luciano M, Hansell NK, Lahti J, et al. Whole genome association scan for genetic polymorphisms influencing information processing speed. Biol Psychol. 86(3):193–202. doi: 10.1016/j.biopsycho.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–50. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 28.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74(4):765–9. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Moor MH, Costa PT, Terracciano A, et al. Meta-analysis of genome-wide association studies for personality. Mol Psychiatry. doi: 10.1038/mp.2010.128. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deary IJ, Whiteman MC, Starr JM, et al. The impact of childhood intelligence on later life: following up the Scottish mental surveys of 1932 and 1947. J Pers Soc Psychol. 2004;86(1):130–47. doi: 10.1037/0022-3514.86.1.130. [DOI] [PubMed] [Google Scholar]
- 32.Gow AJ, Whiteman MC, Pattie A, et al. Goldberg’s ‘IPIP’ Big-Five factor markers: internal consistency and concurrent validation in Scotland. Personality and Individual Differences. 2005;39(2):317–29. [Google Scholar]
- 33.Aston C, Jiang L, Sokolov BP. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Mol Psychiatry. 2005;10(3):309–22. doi: 10.1038/sj.mp.4001565. [DOI] [PubMed] [Google Scholar]
- 34.Surget A, Wang Y, Leman S, et al. Corticolimbic transcriptome changes are state-dependent and region-specific in a rodent model of depression and of antidepressant reversal. Neuropsychopharmacology. 2009;34(6):1363–80. doi: 10.1038/npp.2008.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nemoto Y, Arribas M, Haffner C, et al. Synaptojanin 2, a novel synaptojanin isoform with a distinct targeting domain and expression pattern. J Biol Chem. 1997;272(49):30817–21. doi: 10.1074/jbc.272.49.30817. [DOI] [PubMed] [Google Scholar]
- 36.Nemoto Y, Wenk MR, Watanabe M, et al. Identification and characterization of a synaptojanin 2 splice isoform predominantly expressed in nerve terminals. J Biol Chem. 2001;276(44):41133–42. doi: 10.1074/jbc.M106404200. [DOI] [PubMed] [Google Scholar]
- 37.Perera RM, Zoncu R, Lucast L, et al. Two synaptojanin 1 isoforms are recruited to clathrin-coated pits at different stages. Proc Natl Acad Sci U S A. 2006;103(51):19332–7. doi: 10.1073/pnas.0609795104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stopkova P, Vevera J, Paclt I, et al. Analysis of SYNJ1, a candidate gene for 21q22 linked bipolar disorder: a replication study. Psychiatry Res. 2004;127(1-2):157–61. doi: 10.1016/j.psychres.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Halaschek-Wiener J, Amirabbasi-Beik M, Monfared N, et al. Genetic variation in healthy oldest-old. PLoS One. 2009;4(8):e6641. doi: 10.1371/journal.pone.0006641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newman AB, Walter S, Lunetta KL, et al. A meta-analysis of four genome-wide association studies of survival to age 90 years or older: the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. J Gerontol A Biol Sci Med Sci. 65(5):478–87. doi: 10.1093/gerona/glq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.