Abstract
Protein kinases are a large family of approximately 530 highly conserved enzymes that transfer a γ-phosphate group from ATP to a variety of amino acid residues such as tyrosine, serine and threonine which serves as a ubiquitous mechanism for cellular signal transduction. The clinical success of a number of kinase-directed drugs and the frequent observation of disease causing mutations in protein kinases suggest that a large number of kinases may represent therapeutically relevant targets. To-date the majority of clinical and preclinical kinase inhibitors are ATP-competitive, non-covalent inhibitors that achieve selectivity through recognition of unique features of particular protein kinases. Recently there has been renewed interest in the development of irreversible inhibitors that form covalent bonds with cysteine or other nucleophilic residues in the ATP-binding pocket. Irreversible kinase inhibitors have a number of potential advantages including prolonged pharmacodynamics, suitability for rational design, high potency and ability to validate pharmacological specificity through mutation of the reactive cysteine residue. Here we review recent efforts to develop cysteine-targeted irreversible protein kinase inhibitors and discuss their modes of recognizing the ATP-binding pocket and their biological activity profiles. In addition, we provided an informatics assessment of the potential ‘kinase-cysteinome’ and discuss strategies for the efficient development of new covalent inhibitors.
Keywords: Protein Kinases, Irreversible Kinase inhibitors
Introduction
Kinases are one of the largest gene families whose function is to catalyse the transfer of the γ-phosphate from adenosine triphosphate (ATP) to a specific target molecule bearing nucleophilic hydroxyl or imidazo-groups including lipids, carbohydrate and proteins. Protein phosphorylation can induce a myriad of consequences including modulation of enzymatic activity, conformation, stability, localization and association with other proteins or small molecules.(Cohen, 2002) The approximately 530 kinases present in the human genome have been named the ‘kinome’ and include 420 serine/threonine, 94 tyrosine, 25 atypical and 49 ‘pseudo kinases’ that putatively lack the ability to catalyse the phospho-transfer reaction.(Manning et al., 2002a; Manning et al., 2002b) Kinases have been found to function in virtually every biological process and pathway and perhaps not surprisingly deregulation of kinase function through environmental and genetic alterations is a hallmark of many pathological conditions.(Hanahan and Weinberg, 2011) Since the mid-1980s when v-SRC was initially discovered to be a tyrosine kinase and capable of functioning as an ‘oncogene’, basic research and clinical investigation has implicated the deregulation of approximately 180 kinases in diverse pathology associated with metabolism, development, immunology, cancer and infectious disease.(Hunter and Sefton, 1980) The development of kinase inhibitors has attracted an enormous amount of drug discovery attention, primarily in the oncology and inflammatory diseases areas. Currently approximately 40 kinases are actively being pursued as therapeutic targets and 14 kinase inhibitors have received regulatory approval.(Barf and Kaptein, 2012; Kontzias et al., 2012) Interestingly by far the most significant success has been achieved in oncology by targeting mutationally activated ‘oncogenic’ driver kinases including Bcr-Abl, EGFR, c-Kit, PDGFR, ALK and b-RAF in tumours that are ‘addicted’ to the constitutive activation of these kinases.(Haber et al., 2011)
Currently the vast majority of protein kinase inhibitors (PKIs) are reversible ATP competitive inhibitors, which achieve target selectivity by recognizing unique features of particular ATP binding pockets. This class of inhibitor typically occupy the adenine binding region of the ATP binding pocket and extend into surrounding regions not occupied by ATP and inhibitor binding often induces dramatic conformational rearrangements to the pocket. Compounds that bind to the ATP binding site with the kinase assuming an active conformation, or a conformation otherwise conducive to ATP-binding, are called ‘type I’ inhibitors while those that induce a ‘flip’ of the DFG-motif that marks the start of the activation loop are called ‘type II’.(Liu and Gray, 2006) Multiple other examples of ‘induced-fit’ have been observed providing clear evidence for the plasticity of the ATP-binding pocket.(Changeux and Edelstein, 2011) However given the high sequence conservation of the ATP-binding site and the very large number of kinases, achieving a high degree of kinase selectivity has proven extremely challenging. While there are some examples of exquisitely selective kinase ATP-competitive inhibitors such as lapatinib which targets Her2 and SB239063 which targets p38 kinase, most inhibitors have a spectrum of targets that widens as inhibitor concentration increases.(Rusnak et al., 2001; Underwood et al., 2000) There have been tremendous advances in our ability to measure kinase inhibitor selectivity across near-comprehensive panels using biochemical kinase assays, competition binding assays, and chemical proteomic approaches.(Karaman et al., 2008; Liu et al., 2012b; Okerberg et al., 2005) These efforts have enabled both prospective and retrospective matching of compounds with targets and have greatly furthered our understanding of the mechanism of action of a number of inhibitors.
The majority of new kinase inhibitors are being developed in the pharmaceutical sector typically targeting kinases that have been intensively investigated previously in academia. The requirement for significant pre-validation of potential kinase targets prior to the development of inhibitors in the pharmaceutical sector has resulted in a shortage of inhibitors targeting the kinases whose biological function have received less attention. Therefore new approaches are needed for the rapid generation of inhibitors of poorly understood kinases that can be used as pharmacological probes of mechanism. Covalent kinase inhibitors provide an ideal platform for this endeavour as we discuss further below.
Covalent kinase inhibitors have typically been developed by structure-guided incorporation of an electrophilic moiety into an inhibitor that already possesses sub-micromolar binding affinity to the target of interest.(Potashman and Duggan, 2009) The majority of covalent inhibitors have been designed to target the highly nucleophilic thiol group of cysteine residues.(Leproult et al., 2011). While a large number of kinases have cysteine residues in and around the ATP-binding site, there are no cysteine residues that are conserved amongst kinases that serve a key catalytic function to our knowledge. Covalent inhibitors will initially bind non-covalently and then if the trajectory of the reactive moiety is appropriate, covalent bond formation will take place permanently disabling enzymatic activity. Kinase function is only restored following expression of new protein, the kinetics of which can vary dramatically for different kinases. One major advantage of covalent kinase inhibitors is that high selectivity for a given target kinase can be obtained using a combination of both non-covalent and covalent binding. An irreversible inhibitor that has one dominant binding mode will typically only form a covalent bond with a kinase that possesses a cysteine at a particular position in the ATP-binding site. Therefore non-covalent recognition only needs to enable discrimination between kinases that possess an equivalently placed cysteine residue.
Cysteine residues possess an aliphatic thiol (SH) which has unique reactivity amongst the naturally occurring amino acids. For example, the deprotonated thiolate anion is a potent nucleophile that is exploited as the key catalytic residue in phosphatases and cysteine proteases. Cysteines also serve key non- catalytic functions such as stabilizing protein tertiary structure through formation of disulphide cross-links and coordination of enzyme cofactors such as metals (Jacob et al., 2012). Cysteine residues can be targeted by numerous post-translational modifications including S-nitrosylation, S-prenylation, oxidation to sulfenic and sulfonic acids. (Chalker et al., 2009). Most covalent inhibition strategies that have been explored to-date target the highly nucleophilic cysteine thiolate.
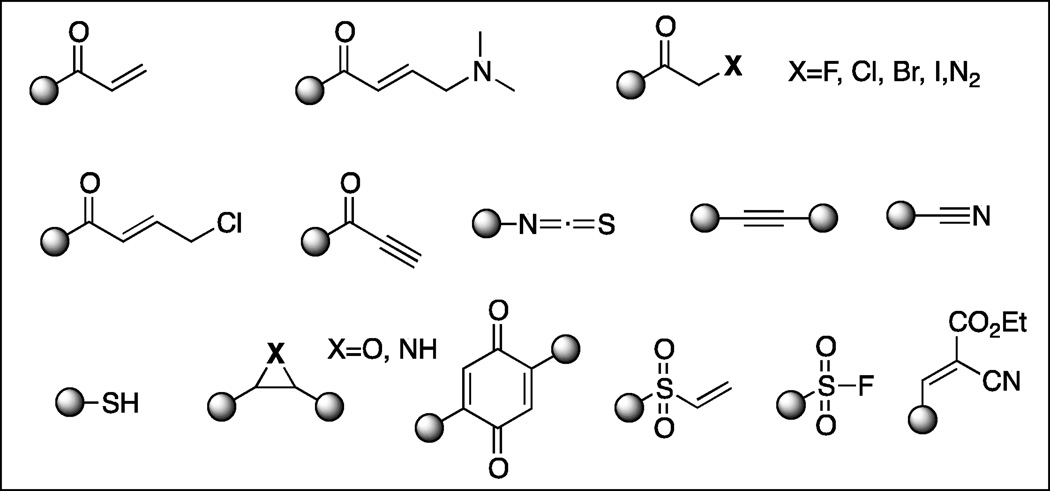
A number of electrophilic “warheads” that can react with nucleophiles such as cysteine, lysine or tyrosine have been explored in the design of irreversible kinase inhibitors. The Michael addition reaction is the most widely utilized reaction to achieve irreversible binding. Functional groups typically introduced to undergo this addition reaction include acrylamides, vinyl sulfonates, quinones, alkynyl amides and propargylic acid derivatives (Figure 1). A second frequently employed chemistry uses nucleophilic displacement or addition to α-halo ketones, thiocyanates, alkynes, nitriles, epoxides, sulfonyl fluoride and sometimes thiol itself. Electrophiles such as cyclic 1,3-diketone have also been developed that specifically react with sulfenic acids. (Leonard et al., 2011) The ability to target differentially modified cysteine residues may afford further means of achieving selectivity.
Fig. 1.
Electrophiles used in irreversible kinase inhibitors.
Historically, there has been considerable reluctance by many organizations to pursue covalent inhibition strategies due to risks of haptenization, the most studied examples being haptens generated by reactive metabolites.(Uetrecht, 2008) Unfortunately, there do not currently exist preclinical or even clinical tests that are reliable predictors of drug safety. Interestingly, a recent retrospective analysis suggested that idiosyncratic toxicities have not been observed for any inhibitors administered at doses of less than 10 mg/Kg.(Nakayama et al., 2009) This suggests that well designed, and highly potent covalent inhibitors might have adequate safety profiles. Irreversible inhibitors are widely used in clinical practice including some of the most important medicines such as the anti-inflammatory drug Aspirin and the broad-class of anti-bacterial beta-lactam antibiotics such as penicillin. Other widely used metabolic activation mechanism based covalent drugs include the proton pump inhibitor omeprazole and anti-platelet drug clopidogrel.(Potashman and Duggan, 2009) Although it may be counter intuitive to create selectivity via a covalent mechanism, the inhibitor electrophilicity can be fine-tuned such that the reaction only occurs in the target binding site. Additionally, covalency can also provide extended pharmacodynamic duration without the need to maintain high levels of drug to achieve continuous target-engagement.(Smith et al., 2009)
Recently there has been a resurgence of interest in irreversible inhibitors and this topic has been excellently reviewed in several publications from a historical perspective(Singh et al., 2011), from a risk-benefit perspective(Barf and Kaptein, 2012; Johnson et al., 2010) and in terms of the current irreversible inhibitors that are in preclinical or clinical development(Garuti et al., 2011; Singh et al., 2010). Leproult et al has also provided a bioinformatic mapping of the potential cysteine containing kinases that could potentially be covalently targeted based upon available X-ray crystal structures.(Leproult et al., 2011) In this review we summarize recent efforts to develop potent and selective irreversible protein kinase inhibitors (PKIs) and describe their modes of recognition of the ATP-binding site and a description of their biological profiles from the perspective of a medicinal chemist. We also provide an analysis of the types of approaches that can be employed to efficiently generate these inhibitors and present a bioinformatics analysis of the potentially targetable cysteines in and around the ATP-binding pocket based on a combination of Pfizer’s in-house and publically available crystal structures. This information is complementary to the previously published articles and we encourage the interested reader to these references for additional information.
Overview of the currently developed irreversible PKIs
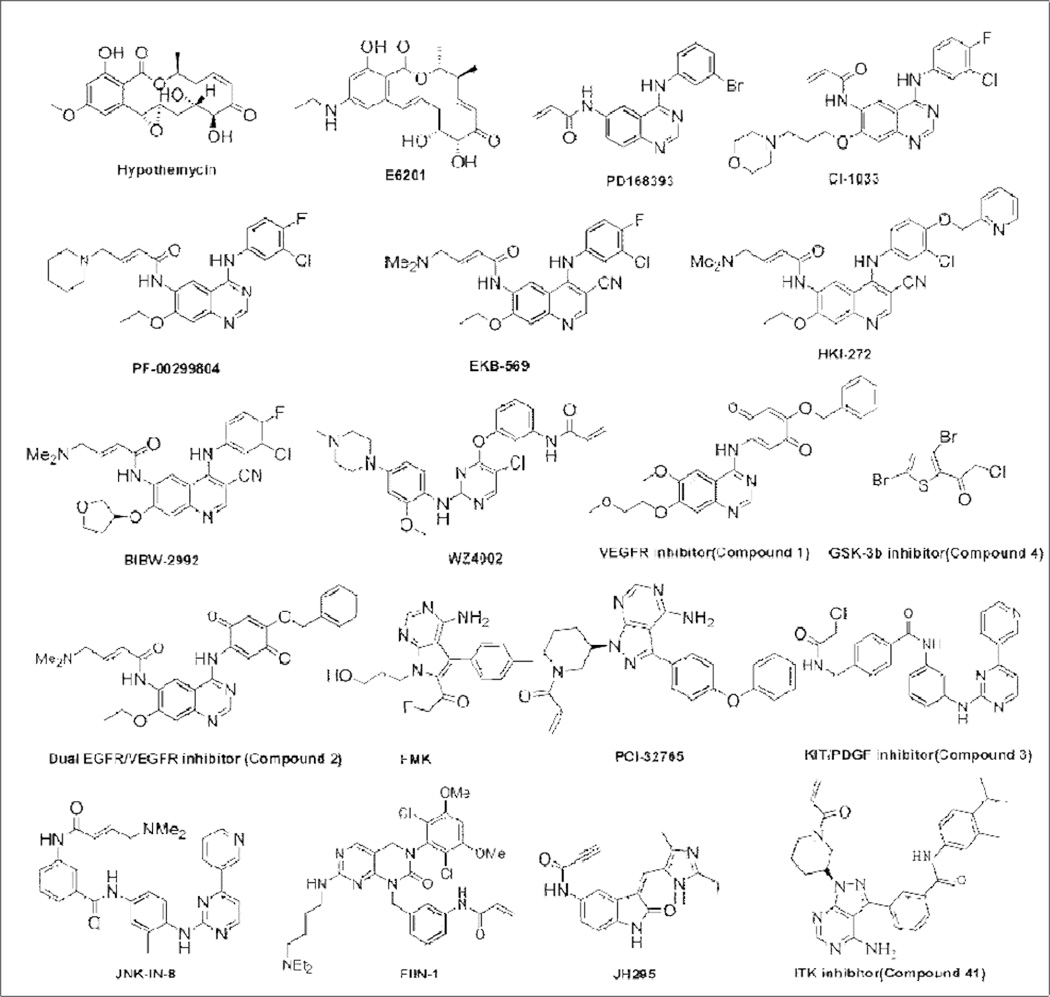
Although most recently reported covalent inhibitors are synthetic, a number of natural products have evolved that covalently modify cysteine residues in kinase ATP-binding sites.(Liu et al., 2012a) One of the most well characterized classes of covalent kinase inhibitors are the resorcylic acid lactones (RALs) with hypothemycin being the most well-known member.(Sonoda et al., 1999) Hypothemycin was originally isolated based on its anti-fungal activity and subsequent investigations demonstrated it to be a covalent protein kinase inhibitor. Covalent bond formation is achieved through reaction of its base cis-enone function with cysteine residues (Figure 2). Santi and co-workers used sequence alignment to identify a conserved cysteine residue immediately preceding the conserved ‘DFG-motif’ that marks the start of the kinase activation loop that is present in a number of kinases inhibited by hypothemycin including MEK1/2, ERK1/2, PDGFRs, FLT3, and VEGFRs.(Schirmer et al., 2006) A co-crystallized structure of ERK2 with hypothemycin (PDB: 2E14) demonstrated a covalent bond between Cys166 of ERK2 and the cis-enone moiety of the inhibitor (Figure. 3A).(Ohori et al., 2007) The phenolic hydroxyl group of hypothemycin forms two hydrogen bonds with Met108 in the kinase hinge segment. Two additional hydrogen bonds are formed between Lys114 in the solvent exposed area and the methoxy group and between Tyr36 located in the p-loop with the hydroxyl group in the marocyclic ring. Several hypothemycin analogues, including FR148083, LL-Z1640-2 and LL-782277, are believed to share the same inhibitory mechanism with TAK1 and MEK kinases.(Winssinger and Barluenga, 2007) Starting from hypothemycin, a focused medicinal chemistry effort to improve its drug like properties resulted in variety of analogues including the structurally similar drug candidate E6201.(Barluenga et al., 2010; Goto et al., 2009; Jogireddy et al., 2009) E6201 inhibits MEK1 biochemically with a low nanomolar IC50 and exhibits strong anti-inflammatory and anti-proliferation activities. E6201 is currently in Phase I clinical trials for advanced solid tumours and in a Phase II trial for psoriasis (Table 1). (Goto et al., 2009; Muramoto et al., 2010)
Fig. 2.
Representative chemical structures of reported irreversible protein kinase inhibitors
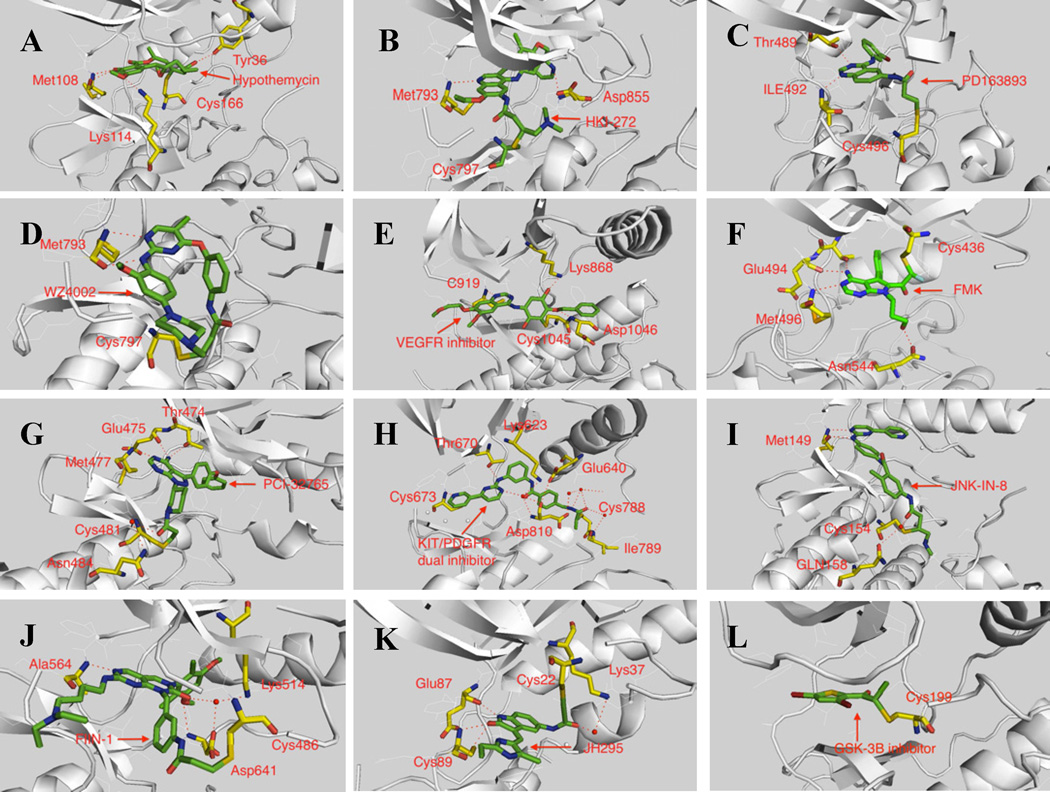
Figure 3.
Representative binding modes of irreversible protein kinase inhibitors A. ERK(PDB: 2E14), B. EGFR(PDB: 2JIV), C. BMX(Modeling based PDB: 3SXS), D.EGFR(PDB: 3IKA), E. VEGFR(modeling based PDB: 1VR2), F. RSK2(Modeling based PDB: 2QR7), G. BTK(Modeling based PDB:3GEN), H. KIT( Modeling based PDB:1T46), I. JNK(PDB:3V6S), J. FGFR(PDB:2FGI), K. NEK2(Modeling based PDB: 2JAV), L.GSK3-β (Modeling based PDB:1I09)
Table 1.
Summary of clinically developed irreversible kinase inhibitors
| Drug name | Primary Target |
Activity IC50(nM) |
Warhead | Active Site |
Clinical stage |
Developer |
|---|---|---|---|---|---|---|
| HKI-272 | EGFR/Her2 | 59 /92 | Acrylamide | C797 | Phase III | Pfizer, licensed to Puma |
| BIBW-2992 | EGFR/Her2 | 14 | Acrylamide | C797 | Phase III | Boehringer Ingelheim |
| PF-00299804 | Pan-EGFR | 15 | Acrylamide | C797 | Phase III | Pfizer |
| CO1686 | EGFR/T790M | 0.5 | Acrylamide | C797 | Phase II | Avila/Clovis |
| E6201 | MEK1 | 5.2 | Enone | C297 | Phase II | EiSai |
| PCI-32765 | BTK | 0.46 | Acrylamide | C481 | Phase III | Pharmacyclics |
| AVL-292 | BTK | 0.5 | Acrylamide | C481 | Phase I | Avila/Celgene |
The development of synthetic irreversible PKIs was initiated at Parke-Davis and Wyeth (now Pfizer) in the early 1990’s with the goal of targeting EGFR for the treatment of cancers with covalent inhibitors.(Singh et al., 1997; Wissner and Mansour, 2008) PD168393 was one of the first reported synthetic irreversible PKIs with a reported IC50 of 2 nM against EGFR and an IC50 of 114 nM against Her2.(Fry et al., 1998) PD168393 is a 4-amino-quinazoline with an acrylamide electrophile installed at the 6-position designed to target Cys797 located a few residues C-terminal to the kinase hinge binding region of EGFR. PD168393 inhibits the proliferation of EGFR and Her2 dependent cell lines A431 and SKBR3 with 95 nM and 15 nM EC50s, respectively, while not inhibiting the proliferation of the non-EGFR and Her dependent cell line SW620 at concentrations of up to 4 µM.(Tsou et al., 2005) Further development of this scaffold resulted in a number of compounds that advanced to clinical trials including CI-1033(clinical development terminated) and PF-00299804 (phase III trial).(Engelman et al., 2007; Smaill et al., 2000) Replacement of one of the quinazoline nitrogens with a nitrile resulted in another set of compounds that advanced to clinical trials: EKB569 (phase II trial completed), HKI-272 (phase III trial) and BIBW-2992 (phase III trial).(Li et al., 2008; Yoshimura et al., 2006) All of these compounds target Cys797, a residue conserved amongst EGFR, Her2 and Her4 which can be visualized in the HKI-272-EGFR co-structure (PDB: 2JIV). The co-structure suggests that a key hydrogen bond exists between the quinoline nitrogen and Met793 in the kinase hinge segment and between Asp855 in the highly conserved DFG motif and the pyridine side chain of the inhibitor. A covalent bond is evident between the N,N-dimethyl-butenoic amide and Cys797 (Figure 2B). HKI-272 exhibits IC50s of 59 nM and 92 nM for inhibition of Her2 and EGFR kinase activity, respectively. HKI-272 inhibits the proliferation of A431 and SKBR3 cells with EC50s of 86 nM and 118 nM, respectively, while demonstrating selectivity over the non EGFR-addicted cell line SW620(730 nM). Kinome wide selectivity profiling using KinomeScan™ methodology demonstrated HKI-272 to be a very selective inhibitor: in addition to EGFR and Her2, it only exhibited potent binding to MAP4K3/5 and MST3/4 (Kd <10 nM).(Davis et al., 2011) These additional targets do not appear to have been validated in cellular assays to-date.
Kinase sequence alignment indicates that there are 8 non-EGFR family kinases that also possess a cysteine at the same position as Cys797 of EGFR. These 8 kinases include all five members of the Tec-family of kinases (BMX, BTK, ITK, TEC, TXK), one Src-family member (BLK), MKKa7 and JAK3. Not surprisingly, some compounds cross-react with kinases in this group. PD168393 for example exhibits 1.1 µM IC50 against BMX and inhibits BMX dependent cell growth in the Ba/F3 cell system with an EC50 of 0.3 µM.(Hur et al., 2008) As discussed further below, more recent efforts have been expended to develop covalent inhibitors of the Tec-family kinase BTK (Figure 3C).
Despite lung cancer patients that express mutant EGFR (exon 19 deletion or L858R) displaying dramatic responses to first generation ATP-competitive inhibitors initially, all patients relapse after 12–18 months.(Rosell et al., 2009) In approximately 50% of cases, resistance is the result of mutation of the so-called gatekeeper residue from a threonine to a methionine (T790M). This mutation is believed to induce resistance by decreasing the Km for ATP thereby increasing the concentration of inhibitor needed to efficiently inhibit signaling.(Yun et al., 2007) While the first generation covalent inhibitors including CI-1033 and HKI-272 can inhibit activated alleles of EGFR harboring T790M they do so at a concentration 10–100 times higher than required to inhibit the non gatekeeper mutants. To overcome this limitation a second-generation of covalent inhibitor from the aminopyrimidine class, exemplified by WZ-4002, was discovered that potently inhibits activated alleles of EGFR harboring T790M.(Zhou et al., 2009) This inhibitor also targets Cys797 but approaches the thiol group from a different trajectory relative to the first generation quinazoline acrylamide inhibitors. Interestingly, an interaction between the chlorine on the pyrimidine of the inhibitors with the methionine gatekeeper, an interaction that has been termed a ‘halogen bond,’ results in the inhibitors possessing selectivity for T790M versus wild-type EGFR (Figure 3D).(Hernandes et al., 2010) WZ4002 overall possesses quite good kinase selectivity but does show activity on some Tec-family kinases as expected based on the cysteine position.
The same 4-aminoquinazoline scaffold present in the first generation EGFR inhibitors was used by scientists at Wyeth to develop a covalent inhibitor of the vascular endothelial growth factor receptor (VEGFR) (Compound 1) using a structure-based design approach (Figure 2).(Wissner et al., 2005) Compound 1 uses an electrophilic 1,4-benzoquinone to target Cys1045 located immediately before the ‘DFG’ motif in VEGFR. Molecular modeling suggests the quinazoline N1 makes the expected hydrogen bond to the hinge region with additional hydrogen-bonds formed between the inhibitor and Asp1046 and Lys868 (Figure 3E). This irreversible inhibitor exhibited a 50 nM IC50 for VEGFR. Further elaboration of this compound allowed the construction of a dual covalent inhibitor of VEGFR and EGFR (Compound 2).(Wissner et al., 2007) An acrylamide was installed to target Cys797 in EGFR and the quinone was maintained to target Cys1045 in VEGFR (Figure 2). The resulting compound inhibits the kinase activity of EGFR and VEGFR with IC50s of 18 nM and 102 nM respectively.
Another successful example of structure-guided covalent inhibitor design is the development of FMK, a fluoromethyl ketone derivatized inhibitor of RSK1, 2, 4 (Figure 2).(Cohen et al., 2005) FMK achieves selectivity for the RSKs by combining two selectivity filters: the presence of a small gatekeeper amino acid (residue T493) in Rsk and covalent bond formation with Cys463 located in the P-loop which resides on the ‘roof’ of the ATP-binding site. There are only 11 kinases (RSKs, MSKs, PLKs, NEK2 and MEKK1) that possess a cysteine at the same position as Cys463 in Rsk and all of these kinases have a larger gatekeeper amino acid. FMK inhibits Rsk kinase activity with an IC50 of 15 nM. Molecular modeling indicates that the pyrollopyrimidine core forms two hydrogen bonds in the hinge binding area with Met496 and Glu494 (Figure 3F). In addition, a hydrogen bond is predicted to exist between the exocyclic amino group and gatekeeper residue Thr494. The hydroxyl group in the butenol side chain forms a hydrogen bond with Asn544 which helps to fix the binding conformation. A biotin-labeled FMK probe was capable of efficiently purifying only RSK1 and RSK2 from HEK293 cell at a concentration of 1 µM. The discovery and characterization of FMK provides a compelling example of the strength of structure-guided design of highly selective covalent kinase inhibitors.
Another group of kinases that have been successfully targeted by covalent inhibitors are the TEC family kinase and include TEC, ITK, TXK, BMX and BTK. These kinases possess a cysteine in an identical location to Cys797 of EGFR. Researchers at Pharmacyclics developed a pyrazolopyrimidine compound, PCI-32765, that is a highly potent inhibitor of BTK.(Honigberg et al., 2010) Molecular modeling predicts that the aminopyrazolopyrimidine forms three hydrogen bonds in the hinge area with Met477, Glu475 and Thr474 (Figure 3G). The bisphenol ether is directed to the inner hydrophobic pocket, analogous to the binding mode of FMK. A water-bridged tri-hydrogen bond between Cys481, Asn484 and PCI-32761 also helps to fix the binding conformation. The acrylamide electrophile forms a covalent bond with Cys481. PCI-32765 inhibits BTK activity with an IC50 of 0.5 nM and is also inhibitory towards several other protein kinases including BLK, BMX, EGFR, Her2, ITK, JAK3 and TEC, that contains an analogous cysteine but does not show activity against most of other protein kinases. PCI-32765 is currently in phase II clinical trials for various B-cell related lymphomas such as DLBCL, MCL and INHL. A compound (AVL-292, structure not disclosed) developed by Avila therapeutics (now Celgene) is believed to share the covalent mechanism as PCI-32765 and is currently in Phase I clinical trials for NHL and CLL.
Researchers from Pfizer recently reported the development of the first covalent inhibitors of ITK derived from a privileged ATP-site directed pyrazollopyrimidine(Zapf et al., 2012) Starting from an acrylamide containing screening ‘hit’, structure-based design was used to guide the development of a potent inhibitor that was active at nanomolar concentration in whole blood and that effectively silenced T-cell receptor signaling for 24 hours.
A dual c-KIT/PDGFR inhibitor (Compound 3) (Figure 2) was rationally designed based on the systematic analysis of available X-ray structures of protein c-KIT, PDGFR, and Abl kinases.(Leproult et al., 2011) Starting from the well-known type II kinase inhibitor imatinib, the methyl piperazine ring in the tail was replaced with a chloroacetamide which resulted in a compound possessing an IC50 of 788 nM for c-KIT and an IC50 of 1 µM for PDGFRα, but that lost potency against Abl (IC50 > 20 µM). Both c-KIT and PDGFRα possess a cysteine residue in the imatinib tail binding area. Molecular docking to PDGFR demonstrated that this new inhibitor adopted the same type II binding mode as observed for imatinib bound to ABL.(Cowan-Jacob et al., 2007) One hydrogen bond was formed in the hinge binding area between the pyridine and Cys673 (Figure 3H). Several additional amino acids including Thr670, Lys623, Glu640, Asp810 and Ile789 also formed hydrogen bonds with the inhibitor. The docking model places Cys788 within 3Å distance of the chloroacetamide; mass spectrometry has verified that Cys788 can indeed form a covalent bond with the newly designed inhibitor. Kinome wide selectivity profiling showed that this compound only potently inhibited seven others including JNK1-3, DDR1, BRAF(V600E) and CSF1R. To our knowledge this is the first reported irreversible PKI that adopts a type II binding conformation.
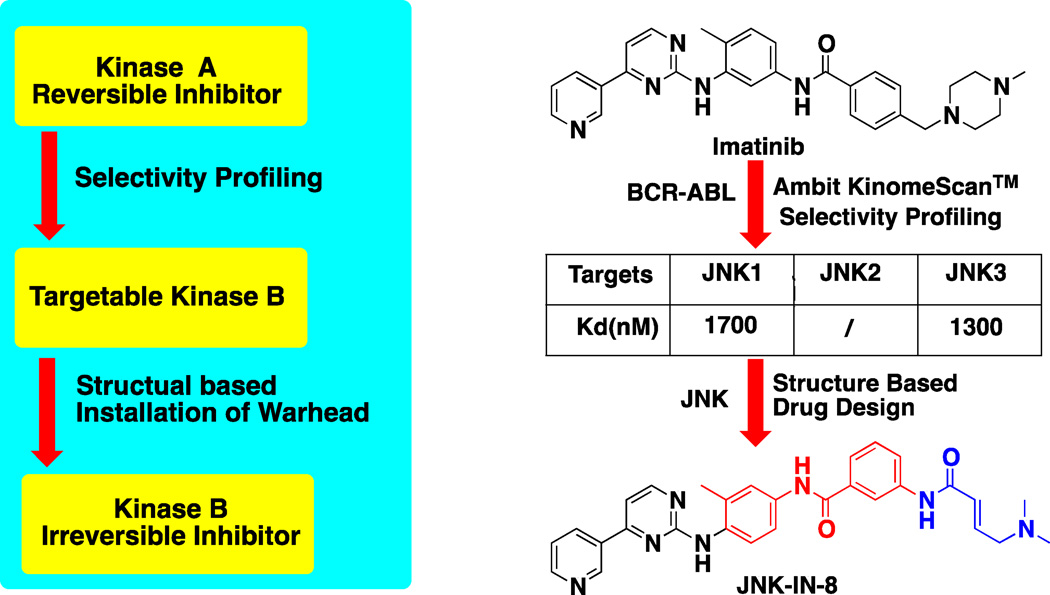
The imatinib scaffold has also been elaborated to create the selective pan-JNK inhibitor, JNK-IN-8.(Zhang et al., 2012) JNK-IN-8 was discovered to inhibit JNK kinase by broad-base kinase selectivity profiling of a library of acrylamide kinase inhibitors based on the structure of imatinib using the KinomeScan™ approach. This discovery was perhaps not entirely unexpected as imatinib itself possesses a Kd of 1.7 µM and 1.3 µM for JNK1 and JNK3, respectively. JNK-IN-8 possess distinct regio-chemistry of the 1,4-dianiline and 1,3-aminobenzoic acid substructures relative to imatinib and uses an N,N-dimethyl butenoic actemide warhead to covalently target Cys154. JNK-IN-8 potently inhibits JNK1, 2 and 3 enzymatic activity with IC50s of 4.7 nM, 18.7 nM and 1.0 nM respectively and inhibits a c-Jun, a direct phosphorylation substrate of JNK, with an EC50 of 486 nM in Hela cell. Broad-based profiling using the chemical proteomics approach termed ‘Kinativ™’(Patricelli et al., 2007) demonstrated exclusive inhibition of JNK amongst the approximately 150 kinases detected using the approach. In contrast to imatinib and the covalent c-KIT/PDGFR inhibitor described above, JNK-IN-8 adopts an L-shaped type I binding conformation to access Cys 154 located towards the lip of the ATP-binding site (Figure 3I, PDB:3V6S). This finding demonstrates how a kinase inhibitor class can adopt two completely distinct binding conformations when binding to different kinases.
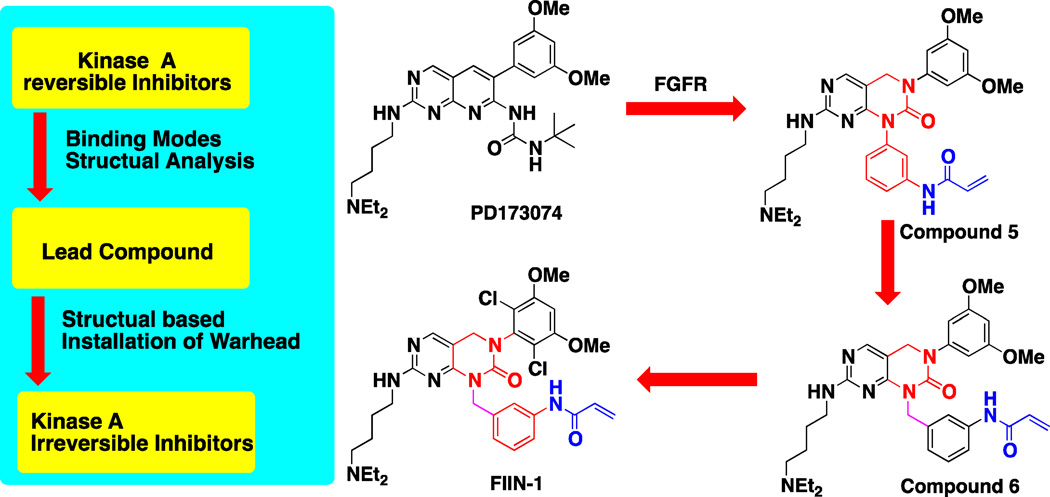
A pan-FGFR1 ,2 ,3 and 4 covalent inhibitor FIIN-1 was developed starting from the well-established non-covalent inhibitor PD173074 using a structure-guided approach.(Bansal et al., 2003; Zhou et al., 2010) FIIN-1 is a potent FGFR family covalent inhibitor which inhibits kinase activity with IC50s of 9.2 nM, 6.2 nM, 11.9 nM and 189 nM for FGFR1, 2, 3 and 4 respectively. Kinome wide selectivity profiling demonstrated that FIIN-1 is quite selective and may also bind to FLT1/4 and VEGFR but no other protein kinases. Molecular modeling suggests that a pair of hydrogen bonds is present between the amino-pyrimidine core structure and Ala564 located in the kinase hinge region (Figure 3J). Several other hydrogen bonds are predicted including a water-mediated tri-hydrogen bond involving Lys514, Asp641 and the carbonyl in the pyrimidine core structure and a hydrogen bond between Asp641 and the methoxy oxygen in the 2,6-dichloro-3,5-dimethoxy aniline side chain. The covalent binding takes place between Cys486 in the P-loop and the acrylamide warhead.
A structure guided approach, was also used to develop an irreversible NEK2 PKI named JH295.(Henise and Taunton, 2011) Starting from a relatively non-selective oxindole–derived inhibitor SU11652, a propargyl acid warhead was installed to target Cys22 located in the P-loop. JH295 is a potent NEK2 inhibitor with a biochemical IC50 of 770 nM. Molecular modeling suggests that three hydrogen bonds are formed between the kinase ‘hinge’ residues Glu87 and Cys89 and the oxindole core (Figure 3K). A water-bridged hydrogen bond involving Lys37 and propargyl amide carbonyl helps to fix the binding conformation putting the electrophilic acetylene in close proximity to Cys22 in the glycine-rich loop (P-loop).
A thienylhalomethylketone based selective irreversible GSK3-β inhibitor was discovered using high throughput screening.(Perez et al., 2009) After optimization of the lead compound, a new GSK3β inhibitor with an IC50 of 0.5 µM and exhibiting selectivity relative to other protein kinases up to a concentration of 10 µM was developed. Molecular modeling suggests that Cys199 in the DFG motif may react with the chloroacetate to form a covalent bond.(Figure 3L).
Several natural products such as celastrol(Lee et al., 2006), 17-acetoxyjolkinolide B (Yan et al., 2008), PGA1 (Rossi et al., 2000), Parthenolide (Kwok et al., 2001), and Manumycin A (Bernier et al., 2006) have been reported to irreversibly bind IKKβ kinases. Irreversible binding has been indicated using mutagenesis or biochemical experiments (Liu et al., 2012a), however crystallographic information is not yet available.
Recently, the Taunton laboratory has explored making “reversible” covalent inhibitors of RSK2. (Serafimova et al., 2012) Here a 2-cyanoacrylate, which allows reversible addition of a cysteine thiol to an activated α,β-unsaturated ester, was introduced onto FMK(Cohen et al., 2005), a scaffold which binds to the ATP-binding site of RSK2. Competition assays and crystallographic analyses were consistent with covalent bond formation. Further investigation is required to determine whether ‘reversible’ covalence will help mitigate pharmacology that derives from covalent modification of unintended targets.
Chemoinformatics analysis of the cysteinome
Many protein kinases possess cysteine residues in and around the ATP-binding site. However, to-date only six distinct cysteine sites have been unequivocally demonstrated to be targeted by a covalent inhibitor: the cysteine at the lip of the ATP binding site targeted by the covalent EGFR, BMX and BTK inhibitors; the cysteine in the P-loop region targeted by FGFR; the cysteine in the roof region of the pocket targeting by NEK2 and RSK inhibitors; the cysteine immediately preceding the DFG motif targeted by VEGFR, ERK2 and GSK3β inhibitors; the cysteine in the solvent area targeted by JNKs inhibitors; and the cysteine located in the catalytic loop targeted by inhibitors of c-KIT and PDGFR. In total, potent and selective inhibitors exist for fewer than 20 kinases with no examples for two classes of kinases. (Table 2 and Figure 4B).
Table 2.
Kinases targeted by covalent inhibitors organized by kinase family
| AGC | CAMK | CKI | CMGC | STE | TK | TKL | Other |
|---|---|---|---|---|---|---|---|
| RSK | None reported | None reported | GSK-3β, JNK, ERK2 | MEK1 | EGFR, BTK, VEGFR, FGFR, BMX, KIT, ITK | TAK1 | IKKb, NEK2 |
Figure 4.
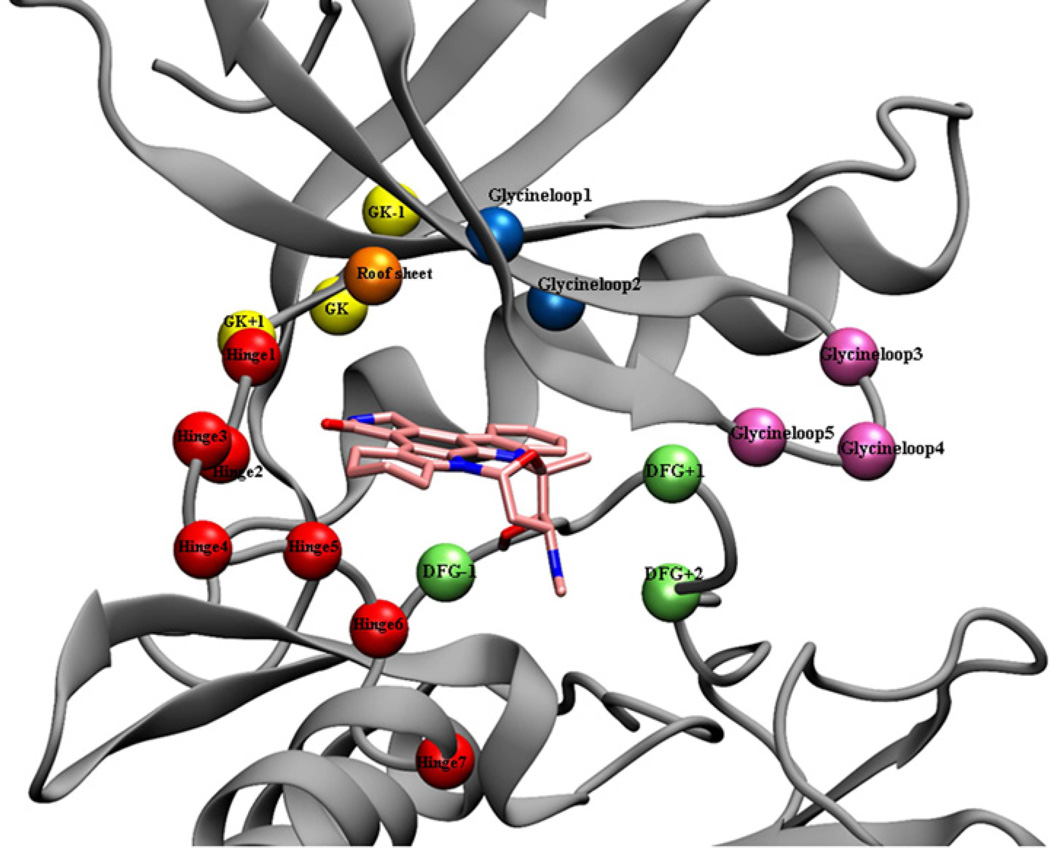
Representative positions of accessible cysteines in the active site, 1YVJ – kinase domain used for depiction (cyan = staurosporine). The various colored circles indicate relative positions of Cys residues depicted on top of 1YVJ. Red = hinge region, Yellow = gatekeeper and neighboring residues, Blue = glycine loop closer to the ATP-site, Mauve = flexible region of glycine loop, Green = activation loop (DFG-area), Orange = roof region
This analysis suggests that currently available irreversible inhibitors only target a very small fraction of the kinases that possess potentially reactive cysteine residues. Previously we reported that a comprehensive analysis of the literature revealed accessible cysteines in the ATP binding pocket of more than 200 protein kinases sites.(Zhang et al., 2009) Recently, Leproult et. al. mined available X-ray crystal structures in the public domain and informatics analyses on different conformations of the protein kinases revealed there are: 27 kinases retaining an active conformation and bearing 211 cysteine residues that can be accessible in theory; 10 protein kinases retaining the DFG-out conformation and bearing 127 cysteines; and 6 protein kinases retaining the c-helix-out conformation and bearing 66 cysteines.(Leproult et al., 2011) These studies have provided a good starting point for developing cysteine targeted irreversible kinase inhibitors. Nonetheless, due to the database limitations, a full spectrum of the targetable cysteine has yet to be fully mapped.
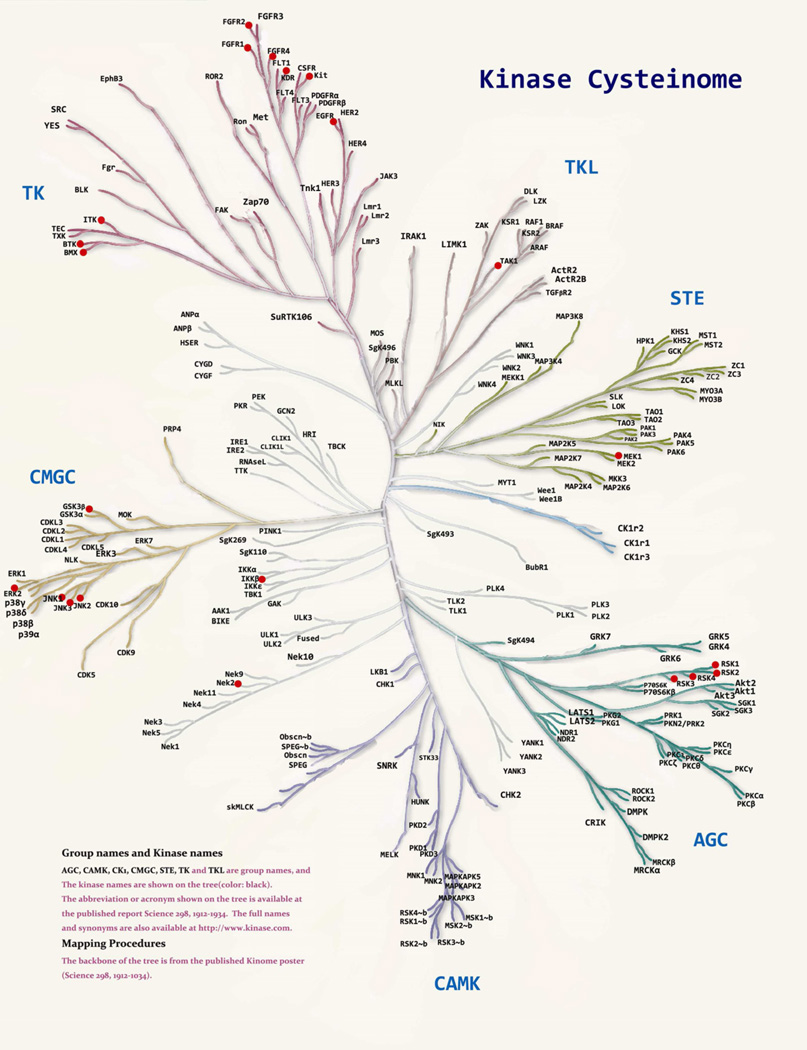
In order to gain a complete picture of the accessible cysteines in the kinome and generate a “kinase cysteinome” to facilitate the systematic exploration for irreversible inhibitors, we performed an informatics study based on the kinome’s primary sequence alignment with PFAAT, a Java-based multiple sequence alignment editor and viewer designed for protein family analysis. (Caffrey et al., 2007) Based on the Pfizer in-house X-ray structure database and RCSB public database, 442 protein kinases were superimposed using CLUSTAW (Thompson et al., 1994) and their ATP binding sites were evaluated (atypical and inactive kinases were excluded). Using this program, 18 spatial cysteine positions were identified (Figure 4) in 200 unique kinases corresponding to 252 positions in total.(Table 3) This estimatation is only approximate as some cysteine thiols maybe inaccessible due to their trajectory or post-translational modification (nitrosylation, disulphide etc). In addition, it may also be there are kinases that possess cysteines that are distant from the ATP-site in sequence space but that are in-fact proximal to the ATP-site due to the overall conformation of the kinase.
Table 3.
Detailed classification of cysteinome
| Position | Subposition | Kinases |
|---|---|---|
| Gate keeper region | GK | MOK |
| GK+1 | SgK494 | |
| GK-1 | MAP2K4 MKK3 MAP2K6 RNAseL KHS1 KHS2 GCK HPK1 MEK2 MEK1 MAP2K5 | |
| DFG region | DFG+1 | MAP3K8 MOS MAP3K4 PINK1 |
| DFG+2 | PKCz PKCi AKT1 AKT2 AKT3 PKCg SGK1F SGK2 ROCK1 ROCK2 WART1 NDR1 NDR2 WART2 PAK4 PAK5 PAK6 PAK2 PAK3 PAK1 PRK1 PRK2 PKN2 PKCt PKCl PKCe PKCd PKCb PKCa DMPK MRCK1 MRCKb DMPK2 P70S6K SGK3 MOK SgK496 P70S6Kb IRE2 IRE1 MELK PINK1 | |
| DFG-1 | PBK TGFbR2 CDKL3 CDKL2 PRP4 MNK2 MNK1 ERK7 CDKL5  ERK1 NLK NIK MAPKAPK5 PKD2 PKD3 PKD1 ERK1 NLK NIK MAPKAPK5 PKD2 PKD3 PKD1  GSK3a Fused MAP2K4 MKK3 MAP2K6 MEK2 GSK3a Fused MAP2K4 MKK3 MAP2K6 MEK2  MAP2K5 ZAK RSK1_Domain2 RSK3_Domain2 RSK4_Domain2 RSK2_Domain2 AAK1 BIKE FLT1 FLT4 KDR PDGFRb PDGFRa CDKL1 CDKL4 GAK SPEG Obscn MAP2K7 Kit FLT3 MAP2K5 ZAK RSK1_Domain2 RSK3_Domain2 RSK4_Domain2 RSK2_Domain2 AAK1 BIKE FLT1 FLT4 KDR PDGFRb PDGFRa CDKL1 CDKL4 GAK SPEG Obscn MAP2K7 Kit FLT3 

|
|
| Glycine rich loop region | Glycineloop | WNK4 WNK1 WNK2 WNK3 HER3 |
| Glycineloop1 | ZAK | |
| Glycineloop2 | SgK496 MEKK1 PLK2 PLK3 PLK1 RSK1_Domain2 RSK3_Domain2 RSK4_Domain2 RSK2_Domain2 NEK2 MSK2_Domain2 MSK1_Domain2 | |
| Glycineloop3 | SgK493 | |
| Glycineloop5 | ||
| Hinge binding region | Hinge1 | FGFR4 TTK MAPKAPK2 MAPKAPK3 P70S6Kb |
| Hinge2 | IKKa 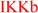 LKB1 NEK4 Obscn_Domain2 ROR2 Wee1 Wee1B AAK1 BIKE SLK Lmr3 Lmr1 Lmr2 NEK9 BRAF RAF1 ARAF HRI FLT1 FLT4 LKB1 NEK4 Obscn_Domain2 ROR2 Wee1 Wee1B AAK1 BIKE SLK Lmr3 Lmr1 Lmr2 NEK9 BRAF RAF1 ARAF HRI FLT1 FLT4 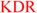 PDGFRb Kit PDGFRa CSFR FLT3 IRE2 IRE1 CLIK1L CLIK1 MYT1 CDKL1 CDKL4 PKR GCN2 PEK GAK SPEG_Domain2 LZK DLK ZC4 ZC1 ZC2 ZC3 MYO3A MYO3B CHK1 TAO2 TAO1 TAO3 SPEG NEK11 MAP3K4 STK33 Obscn CDK5 CDK9 CDK10 TLK1 TLK2 PLK4 TBK1 IKKi ULK3 MELK PKG1 PKG2 FAK NEK5 NEK3 NEK1 ULK1 ULK2 MST1 MST2 LOK HUNK RNAseL KHS1 KHS2 GCK HPK1 PLK2 PLK3 PLK1 PDGFRb Kit PDGFRa CSFR FLT3 IRE2 IRE1 CLIK1L CLIK1 MYT1 CDKL1 CDKL4 PKR GCN2 PEK GAK SPEG_Domain2 LZK DLK ZC4 ZC1 ZC2 ZC3 MYO3A MYO3B CHK1 TAO2 TAO1 TAO3 SPEG NEK11 MAP3K4 STK33 Obscn CDK5 CDK9 CDK10 TLK1 TLK2 PLK4 TBK1 IKKi ULK3 MELK PKG1 PKG2 FAK NEK5 NEK3 NEK1 ULK1 ULK2 MST1 MST2 LOK HUNK RNAseL KHS1 KHS2 GCK HPK1 PLK2 PLK3 PLK1 
|
|
| Hinge3 | Ron FGR SgK494 Kit CSFR FLT3 | |
| Hinge4 | SgK110 BubR1 LKB1 TBK1 | |
| Hinge5 | PINK1 EphB3 | |
| Hinge6 | MAP2K7 TEC TXK  BLK HER2 BLK HER2  HER4 JAK3 HER4 JAK3 |
|
| Hinge7 | ||
| Roof region | Roof sheet | HER3 |
Red colour indicates that highlighted kinases have been irreversibly targeted by small molecule inhibitors.
The 196 kinases identified bearing approachable cysteines are well distributed amongst the kinase subfamilies (Table 3, Figure 5). There are 27 kinases in the TK family, 8 kinases in the TKL family, 34 kinases in the STE family, 16 kinases in the CMGC family, 23 kinases in the CAMK family, 33 kinases in the AGC family and 48 kinases in the CK1 and other families that possess an ATP-site cysteine. While this primary sequence analysis is complementary to Leproult’s X-ray based analysis, experimental validation will be required to unequivocally establish which kinases can be targeted by an irreversible PKI.
Fig. 5.
Distribution of the cysteinome in the kinome tree. Red dots represent kinases that have been proven to be targeted irreversibly by small molecule inhibitors.
Strategies to develop irreversible kinase inhibitors
There are two major approaches for developing novel covalent inhibitors. The first is to use existing non-covalent inhibitors in conjunction with structure-guided design. The second is to create libraries of potentially covalent kinase inhibitors in conjunction with broad-based kinase profiling to identify new covalent inhibitors. While we discuss both methods separately, in practise both are complementary and are typically used in an interwoven fashion.
1. Structure-guided design of new covalent inhibitors
The first ‘human-inspired’ covalent inhibitors were constructed by modifying a known non-covalent inhibitor with a reactive electrophile at a position predicted to be in proximity to a reactive cysteine residue. Examples include the first-generation covalent EGFR inhibitor derived from PD168393, the FGFR inhibitor FIIN-1, the VEGFR inhibitor (Compound 1), the NEK2 inhibitor JH295, the RSK inhibitor FMK and the KIT/PDGFR inhibitor (Compound 3). With the massive expansion in the number of kinase-ligand complex structures available in the public domain, there is a wealth of starting points for this approach. The major challenge with this approach is finding templates that bind non-covalently in the micromolar range and that exhibit selectivity amongst all the kinases with an equivalently positioned cysteine. Successful covalent bond formation can then allow selectivity to be achieved relative to all kinases that do not possess an appropriately positioned cysteine residue. A further challenge is to obtain a scaffold that presents a suitable ‘platform’ for installation of an electrophilic moiety at the correct trajectory relative to the nucleophilic cysteine. The less flexibility there is between the ATP-site recognition element and the electrophile, the higher the chances that only a single cysteine residue will be targeted. In this scenario the ideal compound is one whose initial binding mode to the kinase positions the electrophile in a geometry that will allow for rapid bond formation.
The development of FIIN-1 provides a good example of this structure-guided design approach (Zhou et al., 2010). After a survey of the known ATP-competitive FGFR inhibitors including Chir258, Su5402, SU6668, NP603 and PD173074, PD173074 was chosen based upon its potency, selectivity and availability of co-crystal structure with FGFR. Inspection of the structure reveals that Cys486 in the p-loop (this residue needed to be modelled in because it was mutated in the X-ray structure) of FGFR1 is an approachable nucleophile which is located approximately 10 Å away from the pyridine nitrogen of PD173074. A modeling study suggested that attaching a phenyl group bearing a meta-acrylamide to the 1-nitrogen of the pyrimido[4,5]pyridimine might be a reasonable approach. Compound 5 exhibited good selectivity when screened against 402 kinases using the KinomeScan™ approach but its cellular activity against FGFR-dependent cell lines was moderate at 1.5 µM. The loss of 300-fold in cellular activity relative to PD173074 suggested that no covalent bond formation was taking place. Further elaboration involved elongation of the phenyl warhead by one more carbon to afford compound 6, which improved the EC50 to 400 nM. Further elaboration of the structure by incorporation of the previously used 2,6-dichloro-3,5-dimethoxyphenyl group to the core scaffold afforded FIIN-1 which exhibited an EC50 of 14 nM for inhibiting FGFR-dependent cell growth. Although FIIN-1 is a covalent inhibitor, its cellular EC50 is very similar to a corresponding non-covalent analog (FRIN), which possess a propyl amide in place of the acrylamide (Figure 6). This suggests that the scaffold has sufficiently potent non-covalent binding to achieve the degree of target engagement necessary to initiate apoptosis in the tel-FGFR2 transformed Ba/F3 cells used in the study. The major disadvantage of this structure-guided approach is that it often fails, which unfortunately goes unreported in the literature.
Fig. 6.
Schematic representation of reversible PKI scaffold base approach to develop irreversible inhibitors
2. Selectivity profiling data orientated design of new irreversible inhibitors
A large number of ‘new’ kinase inhibitors are developed based upon serendipitous observations of cross-reactivity observed for established kinase inhibitors. Examination of large scale kinase profiling data sets has revealed that each compound class has a particular constellation of kinases that it can efficiently target. These constellations can range in size from very small such as lapatinib related compounds, which primarily only target the EGFR-family, to exceedingly large such as staurosporine-related compounds. By combining information regarding the constellation of targets that can be addressed non-covalently with the subset of these targets that may possess an appropriately positioned cysteine, one can generate new potential covalent inhibitors. The development of JNK-IN-8 provides an instructive example (Zhang et al., 2012). Examination of broad-based profiling of imatinib reveals that the compounds can bind to Abl, c-Kit, PDGFR, DDR1 and DDR2 with relatively high affinity and to JNK1,2,3 and Raf with moderate affinity. Both c-Kit and PDGFR possess a cysteine that looks like it could be accessed by replacing the piperazine ring of imatinib which was realized by the development of compound 3 (Leproult et al., 2011). However broad-based profiling of these types of inhibitors revealed that they are in-fact also inhibitors of JNK1,2,3. Examination of the JNK X-ray structures revealed that Cys116 in JNK1/2 and Cys154 in JNK3 could be targeted if imatinib used a binding conformation crystallographically observed for Syk (PDB: 1XBB). Further structure-activity guided optimization of the linker-arm between the pyridylpyrimidine ATP-pharmacophore of the compound and the acrylamide resulted in the identification of JNK-IN-8 (Figure 7).
Figure 7.
Schematic representation of flowchart for profiling data based approach
Our endeavors to identify an irreversible JNK inhibitor started from the rational design of a type II irreversible inhibitor of KIT and PDGFR. Kinome-wide selectivity profiling serendipitously demonstrated that JNKs are one of the most potently inhibited enzymes by this class of molecules. A key consideration with this broad-based screening approach is the choice of kinase profiling technology. Currently there are several different technologies available: competitive binding based KinomeScan™ developed by DiscoverX (Previously Ambit Biosciences) which can provide more than 450 kinases and related mutants assay;(Fabian et al., 2005; Karaman et al., 2008) Activity based FRET facilitated SelectScreen™ technology developed by LifeScience (Previously Invitrogen) which can test more than 220 different kinases and mutants(Lebakken et al., 2009); proteomic technology based Kinativ™ developed by ActivX which can provide data on up to up to 400 different kinases in a single cell type depending on the kinase expression(Patricelli et al., 2007); and the traditional radiometric activity-based assays provided from multiple commercial and non commercial sources (Table 3). The Kinativ™ approach is a chemical proteomics-based technology that uses a biotin-tagged acyl phosphate ATP/ADP probe that can acylate the conserved catalytic lysines and other lysines located in proximity of the ATP-binding cleft. (Patricelli et al., 2011) By performing competition assays in cells or lysates with the inhibitor of interest it is possible to monitor which kinases are protected from labeling. Because this is one of the only kinase profiling technologies that can be done following treatment of live cells where the inhibitor is exposed to kinases under more ‘native’ conditions, it is ideally suited for the profiling of covalent inhibitors.
Conclusion
Potent and selective kinase inhibitors are highly desirable reagents for use as probes to pharmacologically interrogate kinase biology or for use as potential therapeutics. Despite the great utility of irreversible kinase inhibitors to fulfill both of these goals, there currently exist only a small number of well characterized covalent inhibitors. However, recently there has been a resurgence of interest in covalent inhibitors driven by a number of factors. First is the realization that suitably designed covalent inhibitors that are potent and selective are exhibiting clinical efficacy and safety. Second is the clear potential to use structure-based design and broad-based selectivity profiling to rapidly generate and characterize lead compounds against a large number of additional kinase targets(Serafimova et al., 2012).
Informatic and structural analysis of the cysteinome has identified approximately 200 distinct kinases with cysteines in close proximity to the ATP-binding cleft. Furthermore, it is likely that additional kinases will possess cysteines that are spatially proximal to the ATP-cleft despite being distant in the primary sequence. Despite this abundance of targets, approximately only 20 kinases have been reported to be targeted by an irreversible inhibitor. There is thus tremendous opportunity for development of new inhibitors. From a research perspective the inhibitors will be valuable tools to help understand the biological function of the roughly one-third of the kinome whose function is poorly understood or for which inhibitors with useful levels of selectivity do not exist. Cysteine-directed covalent inhibitors have some key advantages relative to non-covalent inhibitors when they are to be used as pharmacological tools for investigating the biological function of kinases. Most irreversible inhibitors will obligately require covalent bond formation with a particular cysteine residue. Therefore it is possible to create a mutant form of the kinases that is resistant to the effects of the inhibitor by mutating the reactive cysteine to a serine or an alanine. This inhibitor-resistant mutant form of the kinase can then be re-introduced to the biological system of interest by transient or stable expression and the degree to which it can ‘rescue’ the biological effects elicited by the covalent inhibitor can be used to establish the functional selectivity of the compound. This is an extremely powerful control experiment because the ubiquity of ATP-binding sites in biological systems always provides for the potential of unanticipated off-target effects for kinase inhibitors. In addition, it is possible to make non-covalent versions of the inhibitors that can be used to establish the requirement for covalent bond formation to achieve potent inhibition. From a therapeutic perspective, estimates suggest 180 kinases may represent attractive targets for the development of new therapeutics. However, to-date there exist only 11 kinases that are targeted by FDA approved kinase inhibitors which suggests that there exists considerable opportunity for new covalent inhibitor drugs. The advantage of covalent inhibitors from a therapeutic standpoint is the potential to achieve durable target suppression without the necessity of maintaining high continuous drug exposure.
There are a number of key research directions that would extend the potential of covalent inhibitors both as pharmacological ‘tools’ and as potential therapeutics. The greatest challenge is to devise new strategies to improve inhibitor selectivity. The most obvious way to achieve this is to improve inhibitor potency through optimization of reversible binding (Ki) and efficiency of covalent bond formation (kinact). Unlike non-covalent inhibitors, which can be optimized by monitoring Ki or IC50 values, such measurements are not accurate means for establishing potency for covalent inhibitors as they depend upon the incubation time between the protein and inhibitor. Therefore, considerable caution should be exercised when comparing the IC50s of different irreversible inhibitors, and a more appropriate method to assess inhibition is the ‘specificity constant’ kinact/Ki– the larger this value, the more efficient the molecule is at inhibiting the protein, as it takes into account both the initial equilibrium of the non-covalent complex and the rate of inactivation through the formation of a covalent bond.(Schirmer et al., 2006) Unfortunately, there is a paucity of such information in the scientific literature for irreversible inhibitors.
For example, these values can be used to delineate structure-activity (and selectivity) relationships across a range of covalent inhibitors. Prospectively then, optimisation of both binding affinity and the resulting rate of reaction with the target residue in the kinase, will ultimately lead to the development of highly efficient, and selective, inhibitors. When the covalent inhibitors are used in cellular assays or in vivo, further considerations become important. First, the cellular environment possesses a large number of potentially reactive cysteine residues and modification of the desired target always occurs competitively with these additional off-targets. A second consideration is that a covalent inhibitor only inactivates the enzyme-target until protein synthesis generates new protein. Proteins are synthesized at a range of rates and therefore it is important to determine the time it takes to recover protein function after exposure to the covalent inhibitor (Johnson et al., 2010; Singh et al., 2011).
A second technological break-though would be to develop new classes of ‘electrophilic warheads’ that are specifically ‘tuned’ to a thiol based on its pKa. For example, it maybe possible to develop ‘mechanism-based’ covalent inhibitors where the electrophile is only unleashed upon binding the kinase target. This could be achieved by designing an electrophile whose reactivity is enhanced due to a protein-induced conformational change to the inhibitor. Another significant challenge is to devise methods to better annotate the subset of the proteome that is reactive to a given covalent inhibitor class (i.e. the ‘protein reactome’) and to establish targets associated with toxicology in various organs. We anticipate that a large number of new covalent kinase inhibitors will be developed in the near future which will provide the means to obtain new therapeutic insights.
Table 4.
Commercially available kinase inhibitor selectivity profiling services
| Companies | Website | Technology | Kinase detected |
|---|---|---|---|
| ActivX Biosciences | www.activx.com | Kinativ™ | 400 |
| Bellbrook Labs | www.bellbrooklabs.com | Transcreener® | / |
| Carna Biosciences | www.carnabio.com | QuickScout™ | 305 |
| DiscoverRx | www.discoverx.com | KinomeScan™ | 451 |
| Invitrogen | www.invitrogen.com | SelectScreen™ | 224 |
| MDS Pharma Services | www.mdsps.com | IMAP™ | 155 |
| Millipore | www.millipore.com | KinEASY FP | / |
| MRC Protein Phosphorylation Unit | www.kinase-screen.mrc.ac.uk | (33P-ATP) filter-binding assay | 137 |
| ProQinase | www.proqinase.com | ADP Glo™ | 239 |
| Reaction Biology Corporation | www.reactionbiology.com | HotSpotSM | 440 |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Bansal R, Magge S, Winkler S. Specific inhibitor of FGF receptor signaling: FGF-2-mediated effects on proliferation, differentiation, and MAPK activation are inhibited by PD173074 in oligodendrocyte-lineage cells. J. Neurosci. Res. 2003;74:486–493. doi: 10.1002/jnr.10773. [DOI] [PubMed] [Google Scholar]
- Barf T, Kaptein A. Irreversible Protein Kinase Inhibitors: Balancing the Benefits and Risks. J. Med. Chem. 2012 doi: 10.1021/jm3003203. [DOI] [PubMed] [Google Scholar]
- Barluenga S, Jogireddy R, Koripelly GK, Winssinger N. In vivo efficacy of natural product-inspired irreversible kinase inhibitors. Chembiochem. 2010;11:1692–1699. doi: 10.1002/cbic.201000205. [DOI] [PubMed] [Google Scholar]
- Bernier M, Kwon YK, Pandey SK, Zhu TN, Zhao RJ, Maciuk A, He HJ, Decabo R, Kole S. Binding of manumycin A inhibits IkappaB kinase beta activity. J. Biol. Chem. 2006;281:2551–2561. doi: 10.1074/jbc.M511878200. [DOI] [PubMed] [Google Scholar]
- Caffrey DR, Dana PH, Mathur V, Ocano M, Hong EJ, Wang YE, Somaroo S, Caffrey BE, Potluri S, Huang ES. PFAAT version 2.0: a tool for editing, annotating, and analyzing multiple sequence alignments. BMC. Bioinformatics. 2007;8:381. doi: 10.1186/1471-2105-8-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker JM, Bernardes GJ, Lin YA, Davis BG. Chemical modification of proteins at cysteine: opportunities in chemistry and biology. Chem. Asian. J. 2009;4:630–640. doi: 10.1002/asia.200800427. [DOI] [PubMed] [Google Scholar]
- Changeux JP, Edelstein S. Conformational selection or induced fit? 50 years of debate resolved. F1000 Biol. Rep. 2011;3:19. doi: 10.3410/B3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MS, Zhang C, Shokat KM, Taunton J. Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science. 2005;308:1318–1321. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. The origins of protein phosphorylation. Nat. Cell Biol. 2002;4:E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- Cowan-Jacob SW, Fendrich G, Floersheimer A, Furet P, Liebetanz J, Rummel G, Rheinberger P, Centeleghe M, Fabbro D, Manley PW. Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia. Acta. Crystallogr D Biol. Crystallogr. 2007;63:80–93. doi: 10.1107/S0907444906047287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Gale CM, Lifshits E, Gonzales AJ, Shimamura T, Zhao F, Vincent PW, Naumov GN, Bradner JE, et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res. 2007;67:11924–11932. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- Fry DW, Bridges AJ, Denny WA, Doherty A, Greis KD, Hicks JL, Hook KE, Keller PR, Leopold WR, Loo JA, et al. Specific, irreversible inactivation of the epidermal growth factor receptor and erbB2, by a new class of tyrosine kinase inhibitor. Proc. Natl. Acad. Sci. USA. 1998;95:12022–12027. doi: 10.1073/pnas.95.20.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garuti L, Roberti M, Bottegoni G. Irreversible protein kinase inhibitors. Curr. Med. Chem. 2011;18:2981–2994. doi: 10.2174/092986711796391705. [DOI] [PubMed] [Google Scholar]
- Goto M, Chow J, Muramoto K, Chiba K, Yamamoto S, Fujita M, Obaishi H, Tai K, Mizui Y, Tanaka I, et al. E6201 [(3S,4R,5Z,8S,9S,11E)-14-(ethylamino)-8, 9,16-trihydroxy-3,4-dimethyl-3,4,9,19-tetrahydro-1H-2-benzoxacyclotetradecine-1,7 (8H)-dione], a novel kinase inhibitor of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase (MEK)-1 and MEK kinase-1: in vitro characterization of its anti-inflammatory and antihyperproliferative activities. J. Pharmacol. Exp. Ther. 2009;331:485–495. doi: 10.1124/jpet.109.156554. [DOI] [PubMed] [Google Scholar]
- Haber DA, Gray NS, Baselga J. The evolving war on cancer. Cell. 2011;145:19–24. doi: 10.1016/j.cell.2011.03.026. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Henise JC, Taunton J. Irreversible Nek2 kinase inhibitors with cellular activity. J. Med. Chem. 2011;54:4133–4146. doi: 10.1021/jm200222m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandes MZ, Cavalcanti SM, Moreira DR, de Azevedo Junior WF, Leite AC. Halogen atoms in the modern medicinal chemistry: hints for the drug design. Curr. Drug. Targets. 2010;11:303–314. doi: 10.2174/138945010790711996. [DOI] [PubMed] [Google Scholar]
- Honigberg LA, Smith AM, Sirisawad M, Verner E, Loury D, Chang B, Li S, Pan Z, Thamm DH, Miller RA, et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc. Natl. Acad. Sci. USA. 2010;107:13075–13080. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T, Sefton BM. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc. Natl. Acad. Sci. USA. 1980;77:1311–1315. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur W, Velentza A, Kim S, Flatauer L, Jiang X, Valente D, Mason DE, Suzuki M, Larson B, Zhang J, et al. Clinical stage EGFR inhibitors irreversibly alkylate Bmx kinase. Bioorg. Med Chem. Lett. 2008;18:5916–5919. doi: 10.1016/j.bmcl.2008.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob C, Battaglia E, Burkholz T, Peng D, Bagrel D, Montenarh M. Control of oxidative posttranslational cysteine modifications: from intricate chemistry to widespread biological and medical applications. Chem. Res. Toxicol. 2012;25:588–604. doi: 10.1021/tx200342b. [DOI] [PubMed] [Google Scholar]
- Jogireddy R, Dakas PY, Valot G, Barluenga S, Winssinger N. Synthesis of a resorcylic acid lactone (RAL) library using fluorous-mixture synthesis and profile of its selectivity against a panel of kinases. Chemistry. 2009;15:11498–11506. doi: 10.1002/chem.200901375. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Weerapana E, Cravatt BF. Strategies for discovering and derisking covalent, irreversible enzyme inhibitors. Future Med. Chem. 2010;2:949–964. doi: 10.4155/fmc.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- Kontzias A, Laurence A, Gadina M, O'Shea JJ. Kinase inhibitors in the treatment of immune-mediated disease. F1000 Med. Rep. 2012;4:5. doi: 10.3410/M4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok BH, Koh B, Ndubuisi MI, Elofsson M, Crews CM. The anti-inflammatory natural product parthenolide from the medicinal herb Feverfew directly binds to and inhibits IkappaB kinase. Chem. Biol. 2001;8:759–766. doi: 10.1016/s1074-5521(01)00049-7. [DOI] [PubMed] [Google Scholar]
- Lebakken CS, Riddle SM, Singh U, Frazee WJ, Eliason HC, Gao Y, Reichling LJ, Marks BD, Vogel KW. Development and applications of a broad-coverage, TR-FRET-based kinase binding assay platform. J. Biomol. Screen. 2009;14:924–935. doi: 10.1177/1087057109339207. [DOI] [PubMed] [Google Scholar]
- Lee JH, Koo TH, Yoon H, Jung HS, Jin HZ, Lee K, Hong YS, Lee JJ. Inhibition of NF-kappa B activation through targeting I kappa B kinase by celastrol, a quinone methide triterpenoid. Biochem. Pharmacol. 2006;72:1311–1321. doi: 10.1016/j.bcp.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Leonard SE, Garcia FJ, Goodsell DS, Carroll KS. Redox-based probes for protein tyrosine phosphatases. Angew. Chem. Int. Ed. Engl. 2011;50:4423–4427. doi: 10.1002/anie.201007871. [DOI] [PubMed] [Google Scholar]
- Leproult E, Barluenga S, Moras D, Wurtz JM, Winssinger N. Cysteine mapping in conformationally distinct kinase nucleotide binding sites: application to the design of selective covalent inhibitors. J. Med. Chem. 2011;54:1347–1355. doi: 10.1021/jm101396q. [DOI] [PubMed] [Google Scholar]
- Li D, Ambrogio L, Shimamura T, Kubo S, Takahashi M, Chirieac LR, Padera RF, Shapiro GI, Baum A, Himmelsbach F, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27:4702–4711. doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hu Y, Waller DL, Wang J, Liu Q. Natural products as kinase inhibitors. Nat. Prod. Rep. 2012a;29:392–403. doi: 10.1039/c2np00097k. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kirubakaran S, Hur W, Niepel M, Westover K, Thoreen CC, Wang J, Ni J, Patricelli MP, Vogel K, et al. Kinome-wide selectivity profiling of ATP-competitive mammalian target of rapamycin (mTOR) inhibitors and characterization of their binding kinetics. J. Biol. Chem. 2012b;287:9742–9752. doi: 10.1074/jbc.M111.304485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 2002a;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002b;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Muramoto K, Goto M, Inoue Y, Ishii N, Chiba K, Kuboi Y, Omae T, Wang YJ, Gusovsky F, Shirota H. E6201, a novel kinase inhibitor of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase-1 and mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase-1: in vivo effects on cutaneous inflammatory responses by topical administration. J. Pharmacol. Exp. Ther. 2010;335:23–31. doi: 10.1124/jpet.110.168583. [DOI] [PubMed] [Google Scholar]
- Nakayama S, Atsumi R, Takakusa H, Kobayashi Y, Kurihara A, Nagai Y, Nakai D, Okazaki O. A zone classification system for risk assessment of idiosyncratic drug toxicity using daily dose and covalent binding. Drug Metab Dispos. 2009;37:1970–1977. doi: 10.1124/dmd.109.027797. [DOI] [PubMed] [Google Scholar]
- Ohori M, Kinoshita T, Yoshimura S, Warizaya M, Nakajima H, Miyake H. Role of a cysteine residue in the active site of ERK and the MAPKK family. Biochem. Biophys. Res. Commun. 2007;353:633–637. doi: 10.1016/j.bbrc.2006.12.083. [DOI] [PubMed] [Google Scholar]
- Okerberg ES, Wu J, Zhang B, Samii B, Blackford K, Winn DT, Shreder KR, Burbaum JJ, Patricelli MP. High-resolution functional proteomics by active-site peptide profiling. Proc. Natl. Acad. Sci. USA. 2005;102:4996–5001. doi: 10.1073/pnas.0501205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli MP, Nomanbhoy TK, Wu J, Brown H, Zhou D, Zhang J, Jagannathan S, Aban A, Okerberg E, Herring C, et al. In situ kinase profiling reveals functionally relevant properties of native kinases. Chem. Biol. 2011;18:699–710. doi: 10.1016/j.chembiol.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, Aban A, Chun D, Tanner S, Kozarich JW. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry. 2007;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- Perez DI, Conde S, Perez C, Gil C, Simon D, Wandosell F, Moreno FJ, Gelpi JL, Luque FJ, Martinez A. Thienylhalomethylketones: Irreversible glycogen synthase kinase 3 inhibitors as useful pharmacological tools. Bioorg. Med. Chem. 2009;17:6914–6925. doi: 10.1016/j.bmc.2009.08.042. [DOI] [PubMed] [Google Scholar]
- Potashman MH, Duggan ME. Covalent modifiers: an orthogonal approach to drug design. J. Med. Chem. 2009;52:1231–1246. doi: 10.1021/jm8008597. [DOI] [PubMed] [Google Scholar]
- Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N. Engl. J. Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- Rossi A, Kapahi P, Natoli G, Takahashi T, Chen Y, Karin M, Santoro MG. Anti-inflammatory cyclopentenone prostaglandins are direct inhibitors of IkappaB kinase. Nature. 2000;403:103–108. doi: 10.1038/47520. [DOI] [PubMed] [Google Scholar]
- Rusnak DW, Lackey K, Affleck K, Wood ER, Alligood KJ, Rhodes N, Keith BR, Murray DM, Knight WB, Mullin RJ, et al. The effects of the novel, reversible epidermal growth factor receptor/ErbB-2 tyrosine kinase inhibitor, GW2016, on the growth of human normal and tumor-derived cell lines in vitro and in vivo. Mol. Cancer Ther. 2001;1:85–94. [PubMed] [Google Scholar]
- Schirmer A, Kennedy J, Murli S, Reid R, Santi DV. Targeted covalent inactivation of protein kinases by resorcylic acid lactone polyketides. Proc. Natl. Acad. Sci. USA. 2006;103:4234–4239. doi: 10.1073/pnas.0600445103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafimova IM, Pufall MA, Krishnan S, Duda K, Cohen MS, Maglathlin RL, McFarland JM, Miller RM, Frodin M, Taunton J. Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles. Nat. Chem. Biol. 2012;8:471–476. doi: 10.1038/nchembio.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Dobrusin EM, Fry DW, Haske T, Whitty A, McNamara DJ. Structure-based design of a potent, selective, and irreversible inhibitor of the catalytic domain of the erbB receptor subfamily of protein tyrosine kinases. J. Med. Chem. 1997;40:1130–1135. doi: 10.1021/jm960380s. [DOI] [PubMed] [Google Scholar]
- Singh J, Petter RC, Baillie TA, Whitty A. The resurgence of covalent drugs. Nat. Rev. Drug Discov. 2011;10:307–317. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- Singh J, Petter RC, Kluge AF. Targeted covalent drugs of the kinase family. Curr. Opin. Chem. Biol. 2010;14:475–480. doi: 10.1016/j.cbpa.2010.06.168. [DOI] [PubMed] [Google Scholar]
- Smaill JB, Rewcastle GW, Loo JA, Greis KD, Chan OH, Reyner EL, Lipka E, Showalter HD, Vincent PW, Elliott WL, et al. Tyrosine kinase inhibitors. 17. Irreversible inhibitors of the epidermal growth factor receptor 4-(phenylamino)quinazoline- and 4-(phenylamino)pyrido[3,2-d]pyrimidine-6-acrylamides bearing additional solubilizing functions. J. Med. Chem. 2000;43:1380–1397. doi: 10.1021/jm990482t. [DOI] [PubMed] [Google Scholar]
- Smith AJ, Zhang X, Leach AG, Houk KN. Beyond picomolar affinities: quantitative aspects of noncovalent and covalent binding of drugs to proteins. J. Med. Chem. 2009;52:225–233. doi: 10.1021/jm800498e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda H, Omi K, Hojo K, Nishida K, Omura S, Sugita K. Suppression of oncogenic transformation by hypothemycin associated with accelerated cyclin D1 degradation through ubiquitin-proteasome pathway. Life Sci. 1999;65:381–394. doi: 10.1016/s0024-3205(99)00259-3. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou HR, Overbeek-Klumpers EG, Hallett WA, Reich MF, Floyd MB, Johnson BD, Michalak RS, Nilakantan R, Discafani C, Golas J, et al. Optimization of 6,7-disubstituted-4-(arylamino)quinoline-3-carbonitriles as orally active, irreversible inhibitors of human epidermal growth factor receptor-2 kinase activity. J. Med. Chem. 2005;48:1107–1131. doi: 10.1021/jm040159c. [DOI] [PubMed] [Google Scholar]
- Uetrecht J. Idiosyncratic drug reactions: past, present, and future. Chem. Res. Toxicol. 2008;21:84–92. doi: 10.1021/tx700186p. [DOI] [PubMed] [Google Scholar]
- Underwood DC, Osborn RR, Kotzer CJ, Adams JL, Lee JC, Webb EF, Carpenter DC, Bochnowicz S, Thomas HC, Hay DW, et al. SB 239063, a potent p38 MAP kinase inhibitor, reduces inflammatory cytokine production, airways eosinophil infiltration, and persistence. J. Pharmacol. Exp. Ther. 2000;293:281–288. [PubMed] [Google Scholar]
- Winssinger N, Barluenga S. Chemistry and biology of resorcylic acid lactones. Chem. Commun.(Camb) 2007:22–36. doi: 10.1039/b610344h. [DOI] [PubMed] [Google Scholar]
- Wissner A, Floyd MB, Johnson BD, Fraser H, Ingalls C, Nittoli T, Dushin RG, Discafani C, Nilakantan R, Marini J, et al. 2-(Quinazolin-4-ylamino)-[1,4]benzoquinones as covalent-binding, irreversible inhibitors of the kinase domain of vascular endothelial growth factor receptor-2. J. Med. Chem. 2005;48:7560–7581. doi: 10.1021/jm050559f. [DOI] [PubMed] [Google Scholar]
- Wissner A, Fraser HL, Ingalls CL, Dushin RG, Floyd MB, Cheung K, Nittoli T, Ravi MR, Tan X, Loganzo F. Dual irreversible kinase inhibitors: quinazoline-based inhibitors incorporating two independent reactive centers with each targeting different cysteine residues in the kinase domains of EGFR and VEGFR-2. Bioorg. Med. Chem. 2007;15:3635–3648. doi: 10.1016/j.bmc.2007.03.055. [DOI] [PubMed] [Google Scholar]
- Wissner A, Mansour TS. The development of HKI-272 and related compounds for the treatment of cancer. Arch. Pharm (Weinheim) 2008;341:465–477. doi: 10.1002/ardp.200800009. [DOI] [PubMed] [Google Scholar]
- Yan SS, Li Y, Wang Y, Shen SS, Gu Y, Wang HB, Qin GW, Yu Q. 17-Acetoxyjolkinolide B irreversibly inhibits IkappaB kinase and induces apoptosis of tumor cells. Mol. Cancer Ther. 2008;7:1523–1532. doi: 10.1158/1535-7163.MCT-08-0263. [DOI] [PubMed] [Google Scholar]
- Yoshimura N, Kudoh S, Kimura T, Mitsuoka S, Matsuura K, Hirata K, Matsui K, Negoro S, Nakagawa K, Fukuoka M. EKB-569, a new irreversible epidermal growth factor receptor tyrosine kinase inhibitor, with clinical activity in patients with non-small cell lung cancer with acquired resistance to gefitinib. Lung Cancer. 2006;51:363–368. doi: 10.1016/j.lungcan.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapf CW, Gerstenberger BS, Xing L, Limburg DC, Anderson DR, Caspers N, Han S, Aulabaugh A, Kurumbail R, Shakya S, et al. Covalent Inhibitors of Interleukin-2 Inducible T Cell Kinase (Itk) with Nanomolar Potency in a Whole-Blood Assay. J. Med. Chem. 2012 doi: 10.1021/jm301190s. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Inesta-Vaquera F, Niepel M, Zhang J, Ficarro SB, Machleidt T, Xie T, Marto JA, Kim N, Sim T, et al. Discovery of potent and selective covalent inhibitors of JNK. Chem. Biol. 2012;19:140–154. doi: 10.1016/j.chembiol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, Cortot AB, Chirieac L, Iacob RE, Padera R, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Hur W, McDermott U, Dutt A, Xian W, Ficarro SB, Zhang J, Sharma SV, Brugge J, Meyerson M, et al. A structure-guided approach to creating covalent FGFR inhibitors. Chem. Biol. 2010;17:285–295. doi: 10.1016/j.chembiol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]