Abstract
Understanding the principles of abiotic and biotic stress responses, tolerance and adaptation remains important in plant physiology research to develop better varieties of crop plants. Better understanding of plant stress response mechanisms and application of knowledge derived from integrated experimental and bioinformatics approaches are gaining importance. Earlier, we showed that compiling a database of stress-responsive transcription factors and their corresponding target binding sites in the form of Hidden Markov models at promoter, untranslated and upstream regions of stress-up-regulated genes from expression analysis can help in elucidating various aspects of the stress response in Arabidopsis. In addition to the extensive content in the first version, STIFDB2 is now updated with 15 stress signals, 31 transcription factors and 5,984 stress-responsive genes from three species (Arabidopsis thaliana, Oryza sativa subsp. japonica and Oryza sativa subsp. indica). We have employed an integrated biocuration and genomic data mining approach to characterize the data set of transcription factors and consensus binding sites from literature mining and stress-responsive genes from the Gene Expression Omnibus. STIFDB2 currently has 38,798 associations of stress signals, stress-responsive genes and transcription factor binding sites predicted using the Stress-responsive Transcription Factor (STIF) algorithm, along with various functional annotation data. As a unique plant stress regulatory genomics data platform, STIFDB2 can be utilized for targeted as well as high-throughput experimental and computational studies to unravel principles of the stress regulome in dicots and gramineae. STIFDB2 is available from the URL: http://caps.ncbs.res.in/stifdb2
Keywords: Abiotic and biotic stress, Biocuration, Biological data mining, Stress response, Transcriptional regulatory cascade
Introduction
Among the world population, around 3.1 billion people from developing countries live in rural areas. For a large subset of this population (∼2.5 billion people), agriculture is the primary source for their livelihood and it also contributes to economic growth as 30% of the gross domestic product (GDP) (FAO 2012). By the middle of the 21st century, the expected world population will be about 10 billion and we will witness serious food shortages (Smith et al. 2010). The increasing pressure on global food productivity due to climate change, combined with the drastic increase in population, results in a demand for crop varieties that are adaptive and resistant to a variety of stresses. Considering these various socio-economic and agro-economic factors, sustainable agricultural production is an urgent issue to meet these challenges (Takeda and Matsuoka 2008, Turner et al. 2009, Newton et al. 2011).
Plants are sessile, and they are often exposed to a wide range of both biotic and abiotic stresses, and they have developed intricate mechanisms to detect precise environmental changes, allowing optimal responses to adverse conditions (Atkinson and Urwin 2012). Biotic stress factors including bacteria, fungi, viruses, nematodes and herbivorous insects, and abiotic stress factors such as drought (Dubouzet et al. 2003, Mahajan and Tuteja 2005, Chaves et al. 2009, Fleury et al. 2010, Harb and Perreira 2011), cold (Warren 1998), heat, (Kang et al. 2011) salinity (Ulm et al. 2002, Mahajan and Tuteja 2005, Wallia et al. 2005), dehydration (Urao et al. 1993, Shinozaki and Yamaguchi-Shinozaki 2000, Tran et al. 2004), UV-B (Kilian et al. 2007), wounding (Cheong et al. 2002) and heavy metals (Jonak et al. 2004) cause 30–60% yield losses every year globally (Mantri et al. 2010). Stress response and tolerance towards these stresses were propagated by complex biological pathways and regulatory events involving mutliple molecular components (Shinozaki and Yamaguchi-Shinozaki 2000, Zhu 2001, Xiong et al. 2002, Abe et al. 2003, Jayasekaran et al. 2006, Liu et al. 2007, Agrarwal and Jha 2010, Baena-Gonzalez 2010, Tran and Mochida 2010, Walley and Dehesh 2010, Sinha et al 2011). The effects of abiotic or biotic stress may occur singularly or in combination and induce cellular damage at multiple stages of plant growth and development, and may induce varying degrees of phenotypic lethality including reduction in growth, wilting, loss of leaves, etc. (Chinnusamy et al. 2004). However, this is achieved by activating distinct signal transduction cascades, which in turn activate stress-responsive genes, ultimately leading to survival by transcriptional reprogramming at multiple stages of plant growth and development (Xiong and Zhu 2001). Understanding the basic biological mechanisms by which plants respond to multiple stresses is a prerequisite for the development of stress tolerance in crop plants.
A wide variety of protein-coding genes, non-coding genes including microRNAs (Liu et al. 2008), transcription factors, epigenetic mechanisms and various biological pathways have been attributed to stress response (Singh et al. 2002, Hirayamna and Shinozaki 2007, Shinozaki and Yamaguchi-Shinozaki 2007, Kilian et al. 2012), tolerance (Sreenivasulu et al. 2007, Roy et al. 2011) and adaptation (Mirouze and Paszkowski 2011). Some genes have been reported to respond to both biotic and abiotic stress signals (Mantri et al 2010). Transcription factors are the master regulatory elements that directly bind to their distinct cis-regulatory elements and activate expression of many downstream genes, resulting in various mechanisms including stress tolerance (Agrarwal and Jha 2010). Various transcription factors such as AREB/ABF, MYB, AP2/EREBP, bZIP, MYC,HSF, DREB1/CBF, NAC, HB and WRKY were shown to influence stress response in plants (Singh et al. 2002, Shameer et al. 2009). Traditional methods to characterize stress-responsive transcription factors include nitrocellulose binding assays, DNA footprinting methods, gel-shift analysis, Southwestern blotting of both DNA and protein and high-throughput techniques such as chromatin immunoprecipitation chip (ChIp-chip) or ChIP-sequence, etc. While experimental methods are highly accurate, identification and characterization of the role of a given gene in a given stress response event will often be laborious and time consuming (Bulyk 2003, Zhou et al. 2010, Krasensky and Jonak 2012). To overcome this, computational approaches offer a platform to gather information by integrating various public data sets and sensitive prediction algorithms. Concurrent mining of public databases and analysis using robust algorithms would provide a novel platform to understand the major molecular activities involved in stress response, adaptation and tolerance (Hu et al. 2003, Fernandez-Suarez and Birney 2008, Perez-Rodriguez et al. 2010, Guberman et al. 2011, Kinsella et al. 2011, Sucaet and Deva 2011, Spooner et al. 2012).
Genomic technologies are revolutionizing 21st century plant biology with the advent of high-throughput experimental platforms, optimized assay systems and advanced bioinformatics approaches. DNA microarray is a high-throughput technology extensively used to investigate plant model organisms such as Arabidopsis and rice varieties to detect expression levels of multiple transcripts quantitatively in parallel. Transcriptomic studies were employed for quantitative analysis of thousand of genes expressed at germination, growth and development, fertilization, flowering, and under biotic and abiotic stress conditions, thereby providing a convenient medium to characterize the stress regulome in plants (Oktem et al. 2008). An extensive collection of the expression data is currently available from public microarray databases such as Gene Expression Omnibus (GEO) (Barrett et al. 2001), ArrayExpress (Parkinson et al. 2007), Gene Expression Atlas (Kapushesky et al. 2010), etc. Mining such large-scale expression databases and integrating them with diverse data categories, using data interpretation algorithms, enables robust biological discoveries. The completion of the Arabidopsis thaliana (Arabidopsis Genome Initiative 2000) and Oryza sativa L. genomes (Yu et al. 2002, Zhao et al. 2004, International Rice Genome Sequencing Project 2005) further enhances the application of sensitive sequence search algorithms to predict putative transcription factor binding sites at the whole-genome level and to understand stress regulation via multiple transcription factors in plants.
The stress-responsive mechanism in plants involves complex regulation of multiple genes and transcription factors. In the response to abiotic stresses such as ABA, drought, dehydration, cold, salinity, high light, heat, heavy metals, 10 specific families of transcription factors were known to be involved in A. thaliana and six specific families of transcription factors were known to be involved in O. sativa L. In plant biology, a detailed knowledge of the mechanisms of the transcriptional regulation of genes in response to biotic and abiotic stresses is an important paradigm. We hypothesize that identifying putative transcription factor binding sites for these stress-responsive transcription factors, upstream regions of genes differentially up-regulated in microarray studies due to multiple stress signals, could enable better understanding of genes and regulatory events mediating stress response mechanisms. Earlier, we described the Stress-responsive Transcription factor database, STIFDB (Shameer et al. 2009), a database that catalogued information of stress-responsive genes and transcription factor binding sites for abiotic stress-responsive genes in A. thaliana. Multiple experimental studies that profiled stress-responsive analysis in plants (Kang et al. 2011, Sanghera et al. 2011, Babitha et al. 2012) and computational studies (Mishra et al, 2009, Georgii et al. 2012) on stress-specific gene regulation have utilized the data compiled in STIFDB. In this paper, we report a new version of the database, called ‘STIFDB2’, as a data platform for the investigation of the plant stress regulome. To enable the utility of such a database for a wider plant research community, we have now updated the database with new features such as: the addition of agriculturally important crop species, new stress signals, curated transcription factors and their binding sites, additional stress-responsive genes from microarray experiments and orthologs recorded from other important crop plants. We have also integrated predicted orthologs for other agriculturally important crop species, i.e. maize, sorghum and soybean.
Results
The data compiled using biocuration and genomic data mining, stress-responsive transcription factor prediction and data integration were compiled as a web-based database called STIFDB2.
STIFDB2: a database for analysis of stress regulome in plants
STIFDB2 provides information on stress-responsive genes from a dicot (A. thaliana) and two gramineae species (O. sativa subsp. japonica and O. sativa subsp. indica). A total of 31 transcription factors (Table 1) were identified using the biocuration approach (see Table 1). A library of 15 stress signals that affect plants (ABA, aluminum, bacterial blight, cold, cold–drought–salt, dehydration, drought, heat, high light, iron, NaCl, osmotic stress, oxidative stress, UV-B and wounding) was also compiled (Table 2; details about transcription factors and consensus binding site data for A. thaliana were provided in Shameer et al. 2009). Transcription factor family/subfamily, cis-element, consensus binding site data and corresponding references (PubMed identifiers) used to perform transcription factor binding site prediction using stress-responsive genes in rice species are provided. Using the genomic data mining approach, a data set of 5,984 unique genes were identified and annotated with stress signals. STIFDB2 has 38,798 associations of stress signal, stress-responsive gene, transcription factor binding site, orientation of binding site and z-scores predicted using the STIF (Stress-responsive Transcription Factor) algorithm. Tables 3–5 provide a stress signal- and transcription factor-based summary of stress-responsive genes in STIFDB2. A summary of orthologs retrieved from sorghum, maize and soybean using stress-responsive genes in STIFDB2 is provided in Table 6. The percentage of multiple stress-responsive transcription factors responsive to various stress signals in STIFDB2 is provided in Fig. 1. Chromosomal distributions of genes in STIFDB2 for three species are provided in Fig. 2.
Table 1.
Summary of species, stress-responsive genes, stress-responsive transcription factors and stress signals in STIFDB2
| Species | Stress-responsive genes in STIFDB2 | Stress-responsive transcription factors | Stress signals |
|---|---|---|---|
| Arabidopsis thaliana | 3,150 | ABRE_ABI3_VP1 | ABA, aluminum, cold, cold–drought–salt, dehydration, drought, high light, iron, NaCl, osmotic stress, oxidative stress, UV-B and wounding |
| AuxRE_ARF | |||
| C_ABRE_bZIP | |||
| DREB_AP2_EREBP | |||
| GCC_box_AP2_EREBP | |||
| G_ABRE_bZIP | |||
| G_box1_bZIP | |||
| G_box2_bZIP | |||
| G_box_bHLH | |||
| HBE_HB | |||
| HSE1_HSF | |||
| Myb_box1_MYB | |||
| Myb_box2_MYB | |||
| Myb_box3_MYB | |||
| Myb_box4_MYB | |||
| Myb_box5_MYB | |||
| N_box_bHLH | |||
| Nac_box_NAC | |||
| W_box_WRKY | |||
| Oryza sativa subsp. japonica | 1,118 | ABRE_ABI3_VP1 | ABA, bacterial blight, cold, drought, heat, iron and NaCl |
| ABRE_bZIP | |||
| DREB_AP2_EREBP | |||
| G_box1_bZIP | |||
| Nac_box_NAC | |||
| OsIRO2_bHLH | |||
| PRE2_WRKY | |||
| PRE4_WRKY | |||
| Oryza sativa subsp. indica | 1,716 | DREB_AP2_EREBP | Cold, drought and NaCl |
| ABRE_bZIP | |||
| G_box1_bZIP | |||
| Nac_box_NAC |
Transcription factors are named using the name of the cis-element followed by the transcription family name or subfamily name.
Table 2.
Transcription factor family/subfamily, cis-element, consensus binding site data and corresponding references (PubMed identifiers) used to perform transcription factor binding site prediction using stress-responsive genes curated from the literature (PubMed identifiers of studies are provided) in Oryza sativa subsp. japonica and Oryza sativa subsp. indica
| Transcription factor family name | Stress signal | Reference (Stress signal) | Name of the cis-element | Cis-element | Reference (Cis-element) |
|---|---|---|---|---|---|
| Oryza sativa subsp. japonica | |||||
| ABI3/VP1 | Iron toxicity | 19737364 | ABRE | CATGC | 19737364 |
| AP2/EREBP | Cold, drought, NaCl | 12609047 | CRT/DRE | RCCGAC | 12609047 |
| 15834008 | (ACCGAC | ||||
| 18470484 | GCCGAC) | ||||
| bHLH | Iron toxicity | 16887895 | OsIRO2 | CACGTGG | 16887895 |
| bZIP | ABA, NaCl, drought, heat | 18236009 | G-box1 | CCACGTGTC | 18236009 |
| (C/T)ACGTGGC | 11828032 | ||||
| 11828032 | ABRE | ||||
| NAC | Cold, drought,NaCl, ABA | 20632034 | CATGTG | 17587305 | |
| 19135985 | |||||
| 16924117 | |||||
| WRKY | Bacterial blight | 17986178 | PRE2 | ACGCTGCCG | 17986178 |
| PRE4 | TGCGCTT | ||||
| Oryza sativa subsp. Indica | |||||
| AP2/EREBP | Drought, cold, salinity | 12609047 | CRT/DRE | (G/A)CCGAC | 12609047 |
| 18470484 | |||||
| bZIP | ABA, NaCl, drought | 10636868 | G-box1 | CCACGTGTC | 18236009 |
| 19048288 | ABRE | (C/T)ACGTGGC | 11828032 | ||
| NAC | Cold, drought, NaCl | 17587305 | CATGTG | 17587305 | |
Table 3.
Stress-specific distribution of Arabidopsis thaliana genes in STIFDB2 with predicted binding sites using the STIF algorithm
| Transcription factors | ABA | Alu | Col | CDS | Dehy | Dro | Heat | Light | Iron | NaCl | OS | OxS | UV-B | Wounding |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABRE_ABI3_VP1 | 1 | 0 | 2 | 0 | 0 | 2 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 0 |
| AuxRE_ARF | 299 | 13 | 520 | 17 | 48 | 372 | 32 | 313 | 70 | 473 | 31 | 15 | 54 | 23 |
| C_ABRE_bZIP | 52 | 1 | 105 | 3 | 5 | 86 | 6 | 48 | 12 | 80 | 5 | 4 | 10 | 6 |
| DREB_AP2_EREBP | 230 | 8 | 375 | 23 | 29 | 344 | 12 | 198 | 34 | 362 | 13 | 14 | 29 | 17 |
| GCC_box_AP2_EREBP | 70 | 2 | 112 | 5 | 12 | 98 | 2 | 63 | 11 | 88 | 1 | 6 | 9 | 5 |
| G_ABRE_bZIP | 54 | 0 | 63 | 3 | 1 | 79 | 8 | 39 | 1 | 90 | 7 | 2 | 7 | 3 |
| G_box1_bZIP | 33 | 0 | 46 | 3 | 1 | 51 | 6 | 19 | 6 | 61 | 5 | 1 | 8 | 4 |
| G_box2_bZIP | 234 | 7 | 346 | 9 | 29 | 294 | 26 | 223 | 50 | 356 | 24 | 17 | 38 | 12 |
| G_box_bHLH | 241 | 4 | 361 | 20 | 17 | 369 | 33 | 201 | 33 | 405 | 31 | 10 | 40 | 20 |
| HBE_HB | 48 | 2 | 71 | 2 | 6 | 65 | 5 | 34 | 9 | 81 | 5 | 3 | 11 | 2 |
| HSE1_HSF | 563 | 19 | 892 | 30 | 73 | 728 | 51 | 558 | 106 | 876 | 47 | 34 | 94 | 35 |
| Myb_box1_MYB | 502 | 16 | 832 | 29 | 56 | 691 | 49 | 502 | 114 | 822 | 45 | 33 | 95 | 38 |
| Myb_box2_MYB | 247 | 10 | 396 | 15 | 28 | 315 | 13 | 224 | 51 | 381 | 15 | 14 | 47 | 24 |
| Myb_box3_MYB | 307 | 4 | 470 | 15 | 25 | 395 | 24 | 280 | 53 | 472 | 24 | 21 | 55 | 20 |
| Myb_box4_MYB | 142 | 5 | 242 | 4 | 19 | 172 | 8 | 150 | 24 | 248 | 7 | 9 | 29 | 10 |
| Myb_box5_MYB | 703 | 24 | 1,133 | 41 | 93 | 922 | 63 | 674 | 144 | 1,117 | 58 | 40 | 120 | 47 |
| N_box_bHLH | 163 | 6 | 228 | 13 | 13 | 224 | 12 | 164 | 25 | 261 | 10 | 10 | 26 | 15 |
| Nac_box_NAC | 225 | 6 | 352 | 14 | 25 | 330 | 30 | 227 | 50 | 382 | 24 | 12 | 42 | 16 |
| W_box_WRKY | 464 | 20 | 741 | 24 | 64 | 612 | 47 | 463 | 102 | 716 | 45 | 27 | 84 | 30 |
Alu, aluminum; Col, cold; CDS, cold–drought–salt, Dehy, dehydration; Dro, drought; OS, osmotic stress; OxS, oxidative stress.
Table 4.
Stress-specific distribution of genes of Oryza sativa subsp. japonica genes in STIFDB2 with predicted binding sites using the STIF algorithm
| Transcription factors | ABA | Bacterial blight | Cold | Drought | Heat | Iron | NaCl |
|---|---|---|---|---|---|---|---|
| ABRE_ABI3_VP1 | 30 | 424 | 11 | 87 | 531 | 157 | 33 |
| ABRE_bZIP | 3 | 63 | 0 | 14 | 71 | 15 | 6 |
| DREB_AP2_EREBP | 20 | 260 | 9 | 64 | 289 | 102 | 20 |
| G_box1_bZIP | 1 | 18 | 0 | 2 | 13 | 5 | 1 |
| Nac_box_NAC | 16 | 208 | 5 | 54 | 330 | 97 | 21 |
| OsIRO2_bHLH | 6 | 69 | 2 | 17 | 82 | 21 | 3 |
| PRE2_WRKY | 0 | 1 | 2 | 1 | 7 | 2 | 0 |
| PRE4_WRKY | 2 | 26 | 1 | 2 | 30 | 11 | 0 |
Table 5.
Stress-specific distribution of Oryza sativa subsp. indica genes in STIFDB2 with predicted binding sites using the STIF algorithm
| Transcription factors | Cold | Drought | NaCl |
|---|---|---|---|
| ABRE_bZIP | 26 | 98 | 109 |
| DREB_AP2_EREBP | 181 | 473 | 785 |
| G_box1_bZIP | 3 | 29 | 22 |
| Nac_box_NAC | 132 | 321 | 576 |
Table 6.
Orthologs of stress-responsive genes compiled in STIFDB2 from Ensembl Plant BioMart using the Compara pipeline
| Species | Stress- responsive genes in STIFDB2 | Orthologs retrieved using BioMart |
||
|---|---|---|---|---|
| Sorghum (Sorghum bicolor) | Maize (Zea mays) | Soybean (Glycine max) | ||
| Arabidopsis thaliana | 3,150 | 2,515 | 2,506 | 2,519 |
| Oryza sativa subsp. japonica | 1,118 | 909 | 838 | 727 |
| Oryza sativa subsp. indica | 1,716 | 1,155 | 1,055 | 926 |
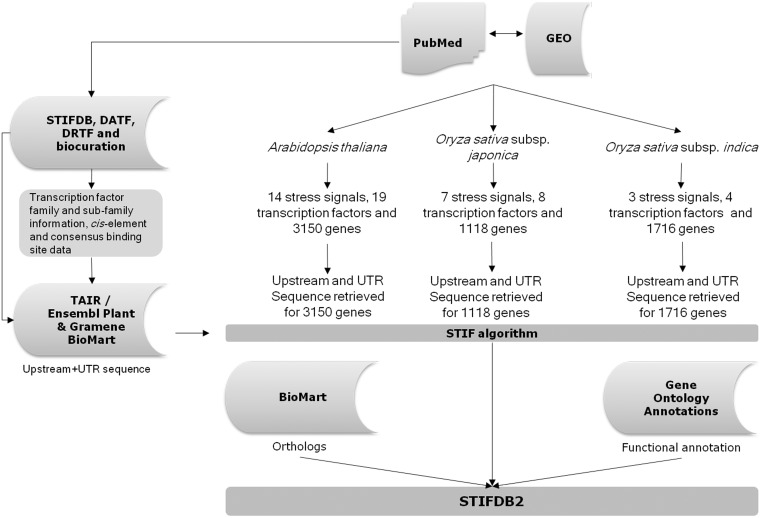
Fig. 1.
Bioinformatics pipeline used to develop STIFDB2.
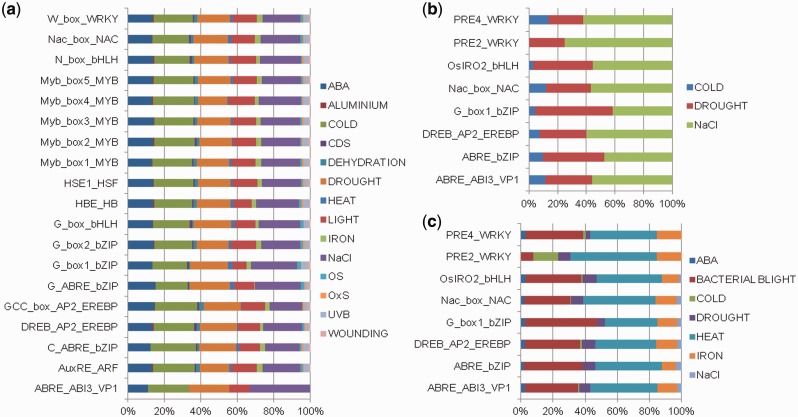
Fig. 2.
Distribution of transcription factor binding sites predicted in the up-regulated stress-responsive genes due to the perturbation of various stress signals in (a) Arabidopsis thaliana (b) Oryza sativa subsp. japonica and (c) Oryza sativa subsp. indica.
Features of STIFDB2
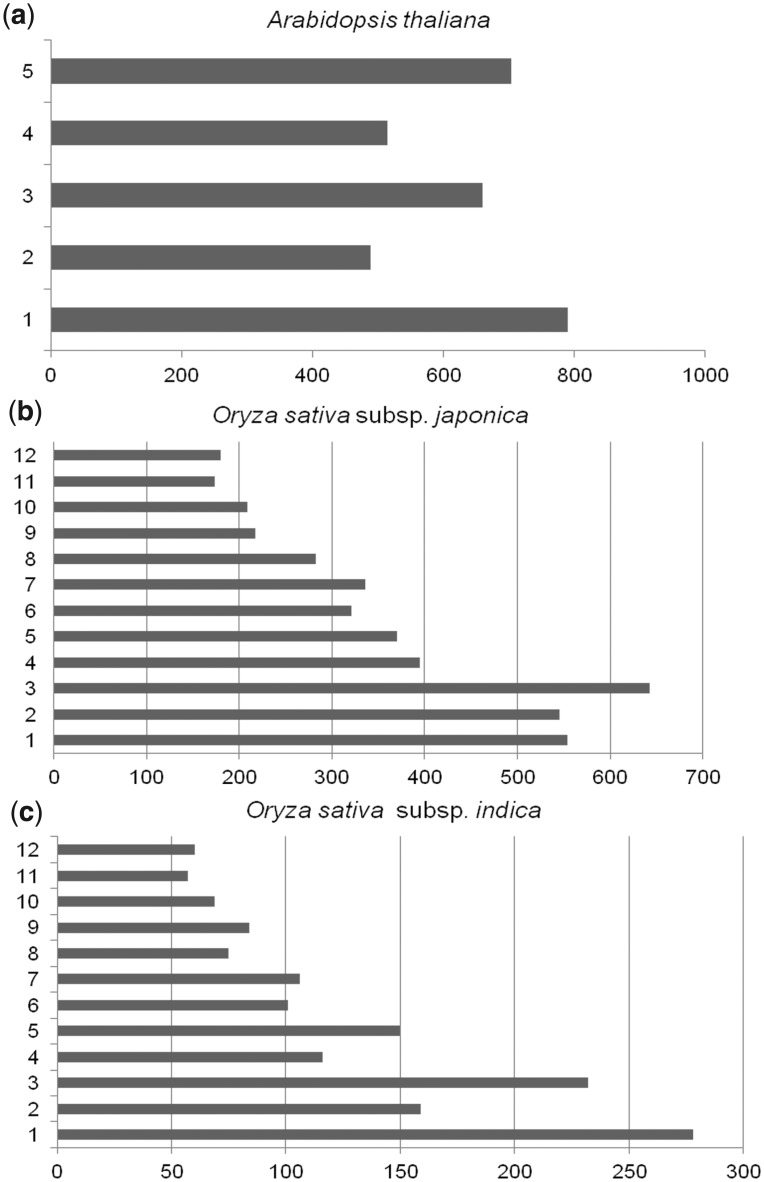
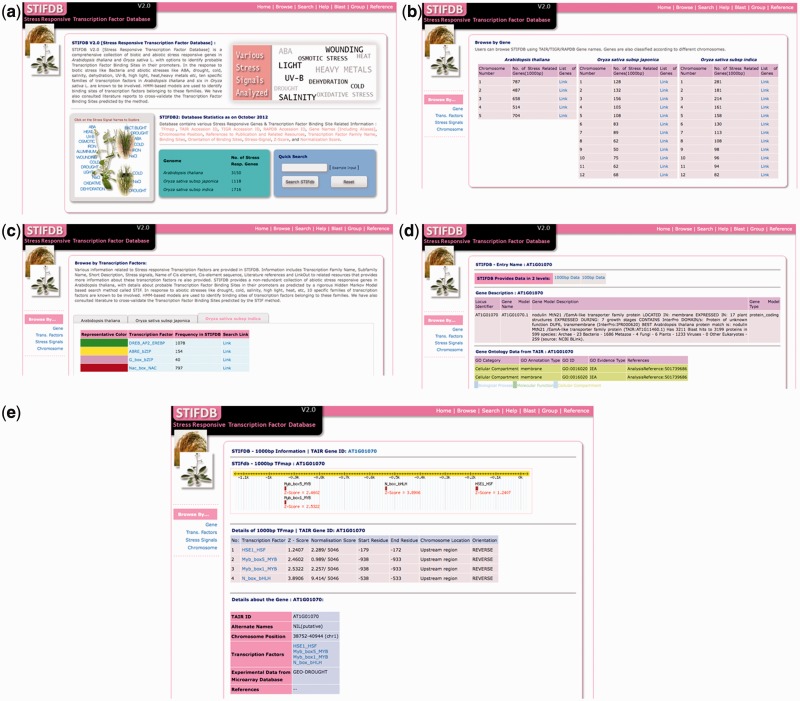
Users can browse and search STIFDB2 using TAIR, TIGR or RAP database identifiers. Genes were also segregated into different chromosomes across three species and can be browsed on the basis of chromosomal location. The data can be browsed using one of the four key data sets: gene, transcription factors, stress signals and chromosome (Fig. 3). Users can also search the database using a variety of keywords including gene descriptions, stress signals, transcription factors and Gene Ontology (GO) annotations. A Basic Local Alignment Search Tool (BLAST) interface is provided to search STIFDB2 using nucleotide sequences. Sequence searches can be performed against the completed set of sequences in STIFDB2 or can be selectively searched against the genome of interest. STIFDB2 also provides ‘TFMap’ that enables users to track the transcription factor of interest visually using a 2D map that consists of the upstream region + untranslated region (UTR) and predicted transcription factors. The database also provides additional information including information about predicted binding sites, chromosomes, various database identifiers and cross-references to other plant databases.
Fig. 3.
Chromosomal distribution of stress-responsive genes in STIFDB2. The x-axis shows the number of stress-up-regulated genes curated from publicly available microarray data and the y-axis shows chromosome numbers.
Applications of STIFDB2
The data compiled in the previous version of STIFDB were utilized for a variety of experimental and computational studies related to abiotic stress response and transcription factor binding site predictions. Briefly, the database can be utilized for identifying putative transcription factor binding sites in the upstream regions/UTRs of stress-upregulated genes curated from gene expression studies. The data can be utilized to study various network features associated with genes up-regulated in the setting of various stresses. These data can also be extrapolated to identify protein–protein interactions amongst transcription factors (manuscript in preparation). STIFDB2 enables the identification of master transcription factors (transcription factors that bind in the up-regulated region of genes perturbed due to diverse stress signals). The database can also be used to study the functional role of highly up-regulated genes using function annotation data integrated in the database (manuscript in preparation). With the inclusion of orthologs, the database can be employed to study evolutionary conservation and cross-genome comparative analyses of abiotic stress-responsive genes across different plant species. Data compiled in STIFDB2 can also be used for the analysis of stress-responsive transcriptional regulatory networks in Arabidopsis or rice genomes.
Analysis of the transcriptional regulatory cascade of the abiotic stress response in A. thaliana using STIFDB2
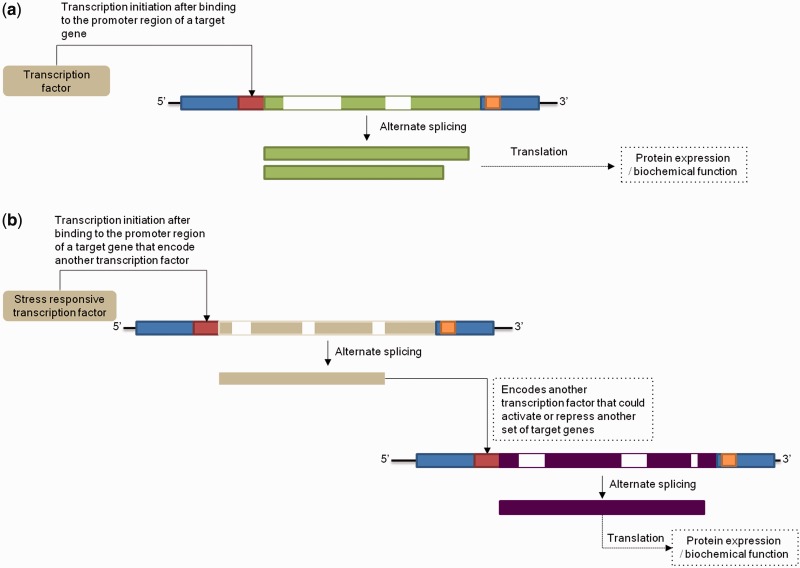
To illustrate the application of STIFDB2, we performed an in-depth analysis of the transcriptional regulatory cascades of 19 stress-responsive transcription factors in A. thaliana. Each transcription factor, inducing expression of another transcription factor due to a stress signal, may act as an activator or repressor of several other genes including those that encode transcription factors. A schematic diagram to depict primary differences between normal transcription factor activity and a transcriptional regulatory cascade event is provided in Fig. 3. The initial list of stress-responsive transcription factors in STIFDB was identified from biocuration followed by genomic data mining. These transcription factors may, in turn, activate other families of transcription factors via a transcriptional regulatory cascade mechanism. The transcriptional regulatory cascade mechanism was studied using the kinetics of regulatory cascades of regulatory networks including developmental gene networks (Davidson et al. 2002. Bolouri and Davidson 2003).
Transcriptional cascades were reported to play a mechanistic role in the abiotic stress response in Arabidopsis (Nover et al. 2001, Guo et al. 2008, Vandepoele et al. 2009). Several high-throughput experimental and computational studies (Chen and Zhu 2004, Shinozaki and Yamaguchi-Shinozaki 2007, Vandepoele et al. 2009, Moreno-Risueno et al. 2010, Cramer et al. 2011, Less et al. 2011, Krasensky and Jonak 2012, Walley and Dehesh 2012) were focused on the identification of transcriptional regulatory networks perturbed due to individual stress signals. A global survey of transcriptional regulatory cascades driven by 19 different stress-responsive transcription factors and 14 stresses were not reported elsewhere. Deconvoluting the role of a primary transcription factor in the regulation of one or more secondary transcription factors is challenging in A. thaliana due to various factors including paucity of the data. STIFDB2 offers a solution to this problem by providing a large compendium of stress signals, stress-responsive transcription factors, putative target binding sites predicted using the STIF algorithm and upstream regions of stress-responsive genes curated from GEO. We utilized the data from STIFDB2 to investigate the transcriptional regulatory cascade network underlying abiotic stress responses in A. thaliana.
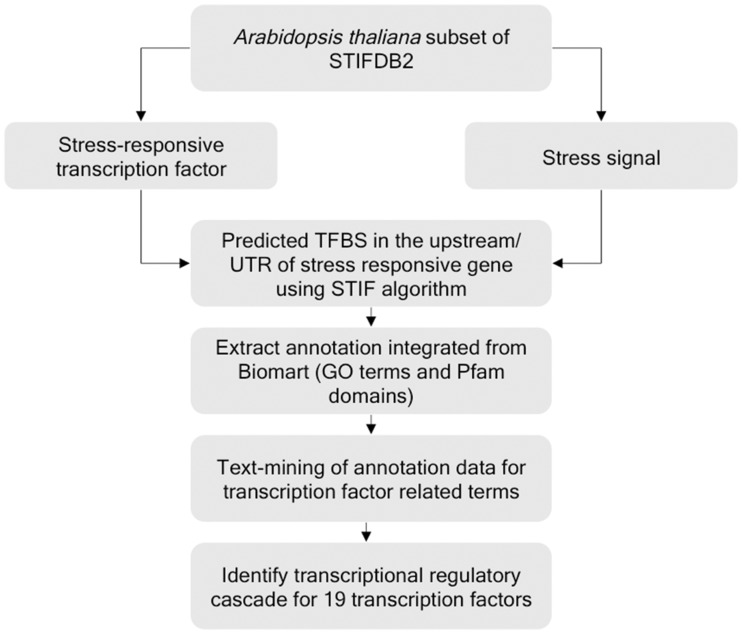
To understand the transcriptional regulatory cascades of transcription factors, associated with various stress conditions, we have used an analytical pipeline that performs text mining of annotation data (such as GO terms, protein domain information and Pfam2GO annotations). First, we grouped the STIF prediction results by transcription factors and retrieved the annotation data [gene description from TAIR, GO terms (molecular function subset) and protein domains for genes annotated with various stress signals]. For each transcription factor, genes that were predicted with their corresponding target binding sites were retrieved from STIFDB2. We performed targeted text mining in the annotation for GO terms in the ‘molecular function’ category pertaining to trancription factors or transcription factor activity. We used the term ‘sequence-specific DNA binding transcription factor activity’ (GO:0003700) as a primary filter. Terms from the neighborhood of GO:0003700 consisting of ‘DNA binding’- or ‘transcription’-related terms (see Table 7) were also used for filtering. Once the genes were identified, we further scanned them for protein domains annotated with transcription factors using Pfam2GO annotations and Pfam domain associations, leading to 90 PFAM domains and GO term association data from Pfam2GO (Supplementary Table S1).
Table 7.
GO terms used for target text mining of stress-responsive genes to find transcriptional regulatory cascades
| GO identifier | GO term |
|---|---|
| GO:0003700 | Sequence-specific DNA binding transcription factor activity |
| GO:0001073 | DNA binding transcription antitermination factor activity |
| GO:0001199 | Metal ion-regulated sequence-specific DNA binding transcription factor activity |
| GO:0001130 | Sequence-specific DNA binding bacterial-type RNA polymerase transcription factor activity |
| GO:0001142 | Sequence-specific DNA binding mitochondrial RNA polymerase transcription factor activity |
| GO:0001167 | Sequence-specific DNA binding RNA polymerase I transcription factor activity |
| GO:0000981 | Sequence-specific DNA binding RNA polymerase II transcription factor activity |
| GO:0001034 | Sequence-specific DNA binding RNA polymerase III transcription factor activity |
| GO:0001011 | Sequence-specific DNA binding RNA polymerase recruiting transcription factor activity |
| GO:0001010 | Sequence-specific DNA binding transcription factor recruiting transcription factor activity |
| GO:0000975 | Regulatory region DNA binding |
| GO:0043565 | Sequence-specific DNA binding |
| GO:0000975 | Regulatory region DNA binding |
| GO:0043565 | Sequence-specific DNA binding |
| GO:0043566 | Structure-specific DNA binding |
A summary of transcription factors perturbed by stress-responsive transcription factors is provided in Table 8. We noted that ABRE_ABI3_VP1 has predicted binding sites in five different genes in Arabidopsis. One of these genes (AT3G16770; ethylene-responsive transcription factor RAP2-3; AtEBP) was annotated with GO terms (DNA binding molecular_function, sequence-specific DNA binding transcription factor activity) and encodes a Pfam domain Apetala 2 (PF00847; AP2 that belongs to a large family of transcription factors). In addition to ABRE_ABI3_VP1, consensus binding regions for six other cis-elements were also predicted in the upstream region + UTR of AtEBP, confirming the importance of this gene in the stress response. ABRE_ABI3_VP1 was primarily considered as a transcription factor responsive to ABA-related stress. Several independent studies have reported that ABA and other plant hormones could participate in the same signaling cascades (Beaudoin et al. 2000, Tuteja 2007). Functional analysis of AtEBP conferred its role in abiotic stress response pathways in A. thaliana (Buttner and Singh 1997, Ogawa et al. 2005). Several interesting stress-responsive transcription-based regulatory cascades were observed and may reveal more interesting biological connections that can be studied using functional genomics and regulatory network analyses.
Table 8.
Summary of the transcription factors identified using the targeted annotation mining approach
| Arabidopsis stress-responsive transcription factors in STIFDB2 | Target genes in STIFDB | GO term hitsa with transcription factor annotation | Pfam domain hitsb with transcription factor annotation |
|---|---|---|---|
| ABRE_ABI3_VP1 | 5 | 1 | 1 |
| AuxRE_ARF | 1,405 | 117 | 40 |
| C_ABRE_bZIP | 248 | 27 | 8 |
| DREB_AP2_EREBP | 952 | 82 | 31 |
| GCC_box_AP2_EREBP | 299 | 12 | 4 |
| G_ABRE_bZIP | 173 | 16 | 4 |
| G_box1_bZIP | 111 | 6 | 2 |
| G_box2_bZIP | 1,020 | 60 | 18 |
| G_box_bHLH | 931 | 87 | 28 |
| HBE_HB | 188 | 23 | 8 |
| HSE1_HSF | 2,472 | 197 | 74 |
| Myb_box1_MYB | 2,318 | 176 | 68 |
| Myb_box2_MYB | 1,050 | 103 | 34 |
| Myb_box3_MYB | 1,269 | 108 | 38 |
| Myb_box4_MYB | 659 | 58 | 21 |
| Myb_box5_MYB | 3,113 | 250 | 92 |
| N_box_bHLH | 683 | 47 | 15 |
| Nac_box_NAC | 1,020 | 81 | 30 |
| W_box_WRKY | 2,077 | 167 | 64 |
a GO terms used to filter genes associated with transcription factor/transcription factor activities are provided in Table 7.
bPfam domains used to define the functional role pertaining to transcription factor/transcription factor activity are filtered from Pfam2GO annotation (see Supplementary Table S1).
In summary, our analysis indicates that varying number of transcription factors could be modulated by the main set of 19 stress-responsive factors. Extensive experimental validation will be required to elucidate the complex patterns of gene regulation via the regulatory network cascade. The data summarized for each stress signal [see Supplementary File (.xls)] and data for 19 different transcription factors (labeled using the transcription factor name and given in Table 8) could be used for designing such experiments including ChIP-seq experiments to understand stress regulation pathways including mechanisms of response, adaptation and tolerance.
Discussion
Plants are commercially important crops, and it is important to understand the basic mechanism of the natural stress response and improve upon their stress response for better productivity. It will also be crucial to know the transcription factors that control and help with combating stress using stress tolerance pathways and adaptive mechanisms. STIFDB2 is a large compendium of curated stress signals, stress-responsive genes and stress-responsive transcription factors, along with information on putative binding sites where the stress-responsive transcription factors are predicted to be bound to the upstream regions and UTRs of stress-responsive genes. STIFDB2 will be a data-centric platform for performing analyses pertaining to stress response, tolerance and adaptation.
The data compiled in STIFDB can be categorized into three classes: (i) data extracted from biocuration; (ii) prediction results; and (iii) annotation data compiled from primary databases. Here, the first class of data refers to a list of Arabidopsis and rice stresses and stress-associated transcription factors curated from the literature. These data points were used as a query to find stress-responsive gene expression studies in Arabidopsis and rice transcriptomes. For each study, we curated corresponding up-regulated gene lists from the literature. After defining the stress-centric gene list, primary databases were mined to retrieve sequences, upstream regions, UTRs and annotations. Thus, STIFDB2 has multiple layers of biological data types integrated for stress gene analyses. The limitation of such knowledge-based approaches is the availability and reliability of validated binding sites and annotation of all genomes, including plant genomes. In this specific case, the underlying algorithm ‘STIF’ uses a Hidden Markov model (HMM). However, the models were generated using published consensus sites curated from the literature. The predictive approach in STIFDB2 is limited to pattern searching, and each binding site is provided with a z-score to indicate the strength of prediction. This enables the users to filter out different sites using the z-score threshold. The data, compiled in STIFDB2, can be used to answer biologically relevant questions on the stress regulome. For example, the data can be examined to identify the functional repertoire and molecular pathways associated with stress-responsive genes and various stress signals in A. thaliana (manuscript submitted) and rice (manuscript in preparation). The data can also be utilized to find whether the stress-responsive genes are GC-rich or GC-poor classes of plant genes (Carels and Bernardi 2000). Such analyses can also performed on the type of stress to understand the role of GC content and stress responses. Transcriptomic diversity of stress-responsive genes has been reported (Carels and Berbnardi 2000, Duque 2011, Mastrangelo et al 2012, Syed et al. 2012), and detailed analysis of the transcriptomic plasticity, due to alternative splicing (Reddy et al. 2012) of genes up-regulated due to various stress signals, can be performed using the data integrated in STIFDB2. Recent studies have indicated that RNA-binding proteins may play a crucial role in the stress response in plants (Lorkovic 2009, Duque 2011, Nakaminami et al. 2012). The annotation data integrated in the database can be utilized to identify genes involved in RNA-binding proteins. Another interesting avenue where data in STIFDB2 can be utilized is in the analysis of transcriptional regulatory cascades of abiotic and biotic stress responses in plants. We performed an extensive analysis with the curated transcription factor information, a rich set of annotation data in STIFDB2, and the results were discussed in the application section. In a similar manner, the database can be used for both experimental and computational studies pertaining to plant stress response, tolerance and adaptation.
Materials and Methods
The content of STIFDB2 was generated using a bioinformatics pipeline consisting of three modules as follows: biocuration and genomic data mining; prediction of stress-responsive transcription factor binding sites; and data integration for developing a database and web-based platform for the analysis of stress-responsive genes and the stress regulome in plants. The analytic pipeline used to develop STIFDB2 is provided below (Fig. 4).
Fig. 5.
Schematic diagram of the targeted text mining approach used to find transcriptional regulatory cascades mediated by stress-responsive genes from Arabidopsis thaliana in STIFDB2.
Fig. 6.
Features of STIFDB2. (a) Front-page of STIFDB2. (b) Browse by chromosome in STIFDB2. (c) Browse by transcription factor page. (d) An intermediate page showing annotations and the link to access the 1,000 or 100 bp prediction results. (e) STIFDB2 page for the gene coding for the nodulin MtN21-like transporter family protein (AT1G01070) in Arabidopsis thaliana.
Fig. 4.
Schematic diagram of (a) normal transcription factor activity and (b) transcriptional regulatory cascade network. Blue rectangles indicate untranslated regions, red rectangles indicate promoter regions, exon regions are highlighted using green, creme and violet color. Introns are defined using a white region interspersed between exons, and polyadenylation sites in the 3′ untranslated regions are colored in orange.
Biocuration and genomic data mining of stress-responsive transcription factors and stress-responsive genes
Information pertaining to stress-responsive transcription factors and transcription factor binding sites of consensus nucleotide regions was curated from the literature, as explained in the previous version of STIFDB (Sundar et al. 2008, Shameer et al. 2009). The initial list of stress-responsive transcription factors was identified, and the consensus sequences were retrieved and used to generate HMMs for prediction of transcription factor binding sites using the STIF algorithm. The compendium of stress-responsive genes was mined from GEO by consulting the corresponding genes from the literature retrieved from PubMed. A gene was considered as stress responsive in STIFDB2 when it was reported to be differentially up-regulated in a perturbation experiment using one of the stress signals in the stress signal library. The data mining approach was performed to filter stress-responsive genes from three different plant species as follows: A. thaliana, O. sativa subsp. japonica and O. sativa subsp. indica from public expression data sets. The transcription factors identified from biocuration were named using a convention that consists of the name of the cis-element followed by the transcription family name or subfamily. For example: Myb_box1 consists of the name of the cis-element ‘Myb_box1’ and the name of the transcription factor family ‘Myb’. Biotic and abiotic stress-responsive genes up-regulated (in at least two replicates) in microarray experiments with a ≥2.5-fold expression change were considered as candidates for STIFDB2. Gene sequences for 100 and 1,000 bp with their 5′ UTR were extracted using Ensembl Plant BioMart (Guberman et al. 2011, Kinsella et al. 2011) using the Arabidopsis Information Resource (TAIR Version 10) database for Arabidopsis (Lamesch et al. 2012), Rice Genome Annotation Project MSU/TIGR (Ouyang et al. 2007) for O. sativa subsp. indica, and Rice Annotation Project (RAP-DB) (Ohyanagi et al. 2006) for O. sativa subsp. japonica. Various annotation data (GO annotations, gene descriptions, etc.) and ortholog information were retrieved using Ensembl plant and Gramene BioMart.
Prediction of stress-responsive transcription factor binding sites
The STIF algorithm was used to identify putative transcription factor binding sites in the upstream regions and UTRs of stress-responsive genes retrieved from publicly available microarray data. Briefly, the STIF algorithm encodes the consensus binding site data as an HMM model (Suppelementary Fig. S1) and performs a sensitive sequence search in both the forward and reverse direction. The program gives the following output: transcription factor, z-score, normalization score, start and end of the predicted binding site, chromosomal location (upstream region or UTR depends on the location of the predicted binding site) and orientation of the strand (forward or reverse). We predicted stress-responsive transcription factors at two levels. The first level surveyed 1,000 bp + UTRs and the second level surveyed 100 bp + UTRs for putative binding sites. The detailed background on the STIF algorithm is available elsewhere [see Sundar et al. (2008) and Shameer et al. (2009) for a detailed description of the STIF algorithm and scoring method].
Data integration
Data retrieved from biocuration (transcription factor, cis-elements, binding site information and library of stress signals), genomic data mining (genes differentially expressed due to plant stress) and STIF prediction (transcription factors in the upstream regions + UTRs of stress-up-regulated genes) were integrated in STIFDB2. STIFDB2 has a large number of unique associations of stress signals, stress-responsive genes and transcription factor binding sites predicted using the STIF algorithm and targeted for plant stress regulome studies. These data sets were compiled as a database using a new interface that enables searching, browsing and retrieval of various information via a user-friendly web interface. STIFDB was developed on a MySQL backend. The web interface of STIFDB was developed using HTML and JavaScript. Perl-CGI programs were used for the development of search, query and retrieval system. Programs for searching putative binding sites and for performing STIF prediction were coded in Perl. Stress profiles have been created for each gene that indicates the associated stress signals. We have also integrated GO associations, gene descriptions from TAIR (Lamesch et al. 2012), Rice Annotation Project (RAP) database (Ouynag et al. 2007), and transcription factor-related information from Database of Arabidopsis Transcription Factors (DATF) (Guo et al. 2005) and Database of Rice Transcrption Factor (DRTF) (Gao et al. 2006).
Conclusion
Understanding the molecular pathways and regulatory networks which influence various facets of stress-responsive events in plants is crucial for developing stress-tolerant or stress-adaptive varieties of plants. Such information will also help to understand the metabolic, physiological and cellular mechanisms implicated in such processes. STIFDB2, as an expanded resource that includes two additional genomes (O. sativa subsp. japonica and O. sativa subsp. indica), seven new stress signals (aluminum, bacterial blight, heat, iron, osmotic stress, UV-B and wounding) and 3,355 genes, will be an ideal information resource for plant biologists and computational biologists to perform in-depth analyses. The database provides 40,217 data points, and the data can be used to find putative RNA-binding proteins, transcriptional regulatory cascades, transcriptomic diversity and GC content of stress-responsive genes. We envisage that, analogously to its previous version, STIFDB2 will be widely accepted by the community and aid in unraveling novel aspects of stress response in plants.
Supplementary data
Supplementary data are available at PCP online.
Funding
The authors thank University of Agricultural Sciences (Bangalore) and National Centre for Biological Sciences, Tata Institute of Fundamental Research for infrastructural support. MN acknowledges University Grant Commission, New Delhi, India, for the Senior Research Fellowship during the course of this research work.
Supplementary Material
Glossary
Abbreviations
- GEO
Gene Expression Omnibus
- GO
gene ontology
- HMM
Hidden Markov model
- STIF
Stress-responsive Transcription Factor
- UTR
untranslated region.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal PK, Jha B. Transcription factors in plants and ABA dependent and independent abiotic stress signaling. Biol. Plant. 2010;54:201–212. [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Atkinson NJ, Urwin PE. The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 2012;63:3523–3543. doi: 10.1093/jxb/ers100. [DOI] [PubMed] [Google Scholar]
- Babitha KC, Ramu SV, Pruthvi V, Mahesh P, Nataraja KN, Udayakumar M. Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 2012 doi: 10.1007/s11248-012-9645-8. (in press) [DOI] [PubMed] [Google Scholar]
- Baena-Gonzalez E. Energy signaling in the regulation of gene expression during stress. Mol. Plant. 2010;3:300–313. doi: 10.1093/mp/ssp113. [DOI] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, et al. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin N, Serizet C, Gosti F, Giraudat J. Interactions between abscisic acid and ethylene signaling cascades. Plant Cell. 2000;12:1103–1115. doi: 10.1105/tpc.12.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolouri H, Davidson EH. Transcriptional regulatory cascades in development: initial rates, not steady state, determine network kinetics. Proc. Natl Acad. Sci. USA. 2003;100:9371–9376. doi: 10.1073/pnas.1533293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner M, Singh KB. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl Acad. Sci. USA. 1997;94:5961–5966. doi: 10.1073/pnas.94.11.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyk ML. Computational prediction of transcription-factor binding site locations. Genome Biol. 2003;5:201. doi: 10.1186/gb-2003-5-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carels N, Bernardi G. Two classes of genes in plants. Genetics. 2000;154:1819–1825. doi: 10.1093/genetics/154.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves MM, Flexas J, Pinheiro C. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann. Bot. 2009;103:551–560. doi: 10.1093/aob/mcn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WJ, Zhu T. Networks of transcription factors with roles in environmental stress response. Trends Plant Sci. 2004;9:591–596. doi: 10.1016/j.tplants.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Cheong YH, Chang HS, Gupta R, Wang X, Zhu T, Luan S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol, 2002;129:661–677. doi: 10.1104/pp.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Schumaker K, Zhu JK. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 2004;55:225–236. doi: 10.1093/jxb/erh005. [DOI] [PubMed] [Google Scholar]
- Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K. Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 2011;11:163. doi: 10.1186/1471-2229-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, et al. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Duque P. A role for SR proteins in plant stress responses. Plant Signal. Behav. 2011;6:49–54. doi: 10.4161/psb.6.1.14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO. 2012. Statistical Yearbook. [Google Scholar]
- Fernandez-Suarez XM, Birney E. Advanced genomic data mining. PLoS Comput. Biol. 2008;4:e1000121. doi: 10.1371/journal.pcbi.1000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury D, Jefferies S, Kuchel H, Langridge P. Genetic and genomic tools to improve drought tolerance in wheat. J. Exp. Bot. 2010;61:3211–3222. doi: 10.1093/jxb/erq152. [DOI] [PubMed] [Google Scholar]
- Gao G, Zhong Y, Guo A, Zhu Q, Tang W, Zheng W, et al. DRTF: a database of rice transcription factors. Bioinformatics. 2006;22:1286–1287. doi: 10.1093/bioinformatics/btl107. [DOI] [PubMed] [Google Scholar]
- Georgii E, Salojarvi J, Brosche M, Kangasjarvi J, Kaski S. Targeted retrieval of gene expression measurements using regulatory models. Bioinformatics. 2012;28:2349–2356. doi: 10.1093/bioinformatics/bts361. [DOI] [PubMed] [Google Scholar]
- Guberman JM, Ai J, Arnaiz O, Baran J, Blake A, Baldock R, et al. BioMart Central Portal: an open database network for the biological community. Database (Oxford) 2011;2011:bar041. doi: 10.1093/database/bar041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, He K, Liu D, Bai S, Gu X, Wei L, et al. DATF: a database of Arabidopsis transcription factors. Bioinformatics. 2005;21:2568–2569. doi: 10.1093/bioinformatics/bti334. [DOI] [PubMed] [Google Scholar]
- Guo J, Wu J, Ji Q, Wang C, Luo L, Yuan Y, et al. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genomics. 2008;35:105–118. doi: 10.1016/S1673-8527(08)60016-8. [DOI] [PubMed] [Google Scholar]
- Harb A, Pereira A. Screening Arabidopsis genotypes for drought stress resistance. Methods Mol. Biol. 2011;678:191–198. doi: 10.1007/978-1-60761-682-5_14. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 2010;61:1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- Hu Y, Hines LM, Weng H, Zuo D, Rivera M, Richardson A, et al. Analysis of genomic and proteomic data using advanced literature mining. J. Proteome Res. 2003;2:405–412. doi: 10.1021/pr0340227. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Jayasekaran K, Kim KN, Vivekanandan M, Shin JS, Ok SH. Novel calcium-binding GTPase (AtCBG) involved in ABA-mediated salt stress signaling in Arabidopsis. Plant Cell Rep. 2006;25:1255–1262. doi: 10.1007/s00299-006-0195-5. [DOI] [PubMed] [Google Scholar]
- Jonak C, Nakagami H, Hirt H. Heavy metal stress. Activation of distinct mitogen-activated protein kinase pathways by copper and cadmium. Plant Physiol. 2004;136:3276–3283. doi: 10.1104/pp.104.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HG, Kim J, Kim B, Jeong H, Choi SH, Kim EK, et al. Overexpression of FTL1/DDF1, an AP2 transcription factor, enhances tolerance to cold, drought, and heat stresses in Arabidopsis thaliana. Plant Sci. 2011;180:634–641. doi: 10.1016/j.plantsci.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Kapushesky M, Emam I, Holloway E, Kurnosov P, Zorin A, Malone J, et al. Gene expression atlas at the European bioinformatics institute. Nucleic Acids Res. 2010;38:D690–698. doi: 10.1093/nar/gkp936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Peschke F, Berendzen KW, Harter K, Wanke D. Prerequisites, performance and profits of transcriptional profiling the abiotic stress response. Biochim. Biophys. Acta. 2012;1819:166–175. doi: 10.1016/j.bbagrm.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- Kinsella RJ, Kahari A, Haider S, Zamora J, Proctor G, Spudich G, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasensky J, Jonak C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012;63:1593–1608. doi: 10.1093/jxb/err460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Less H, Angelovici R, Tzin V, Galili G. Coordinated gene networks regulating Arabidopsis plant metabolism in response to various stresses and nutritional cues. Plant Cell. 2011;23:1264–1271. doi: 10.1105/tpc.110.082867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14:836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 2007;51:897–909. doi: 10.1111/j.1365-313X.2007.03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic ZJ. Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 2009;14:229–236. doi: 10.1016/j.tplants.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Arch. Biochem. Biophys. 2005;444:139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Mantri NL, Ford R, Coram TE, Pang ECK. Evidence of unique and shared responses to major biotic and abiotic stresses in chickpea. Environ. Exp. Bot. 2010;69:286–292. [Google Scholar]
- Mastrangelo AM, Marone D, Laido G, De Leonardis AM, De Vita P. Alternative splicing: enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 2012;185–186:40–49. doi: 10.1016/j.plantsci.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Mirouze M, Paszkowski J. Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 2011;14:267–274. doi: 10.1016/j.pbi.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Mishra AK, Agarwal S, Jain CK, Rani V. High GC content: critical parameter for predicting stress regulated miRNAs in Arabidopsis thaliana. Bioinformation. 2009;4:151–154. doi: 10.6026/97320630004151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Busch W, Benfey PN. Omics meet networks—using systems approaches to infer regulatory networks in plants. Curr. Opin. Plant Biol. 2010;13:126–131. doi: 10.1016/j.pbi.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami K, Matsui A, Shinozaki K, Seki M. RNA regulation in plant abiotic stress responses. Biochim. Biophys. Acta. 2012;1819:149–153. doi: 10.1016/j.bbagrm.2011.07.015. [DOI] [PubMed] [Google Scholar]
- Newton AC, Johnson S, Gregory PJ. Implications of climate change for diseases, crop yields and food security. Euphytica. 2011;179:3–18. [Google Scholar]
- Nover L, Bharti K, Doring P, Mishra SK, Ganguli A, Scharf KD. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones. 2001;6:177–189. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Pan L, Kawai-Yamada M, Yu LH, Yamamura S, Koyama T, et al. Functional analysis of Arabidopsis ethylene-responsive element binding protein conferring resistance to Bax and abiotic stress-induced plant cell death. Plant Physiol. 2005;138:1436–1445. doi: 10.1104/pp.105.063586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyanagi H, Tanaka T, Sakai H, Shigemoto Y, Yamaguchi K, Habara T, et al. The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res. 2006;34:D741–D744. doi: 10.1093/nar/gkj094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktem HA, Eyidogan F, Selcuk F, Oz MT, Teixeira da Silva JA, Yucel M. Revealing response of plants to biotic and abiotic stresses with microarray technology. Genes Genomes Genomics. 2008;2:1–35. [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Farne A, et al. ArrayExpress—a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Rodriguez P, Riano-Pachon DM, Correa LG, Rensing SA, Kersten B, Mueller-Roeber B. PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010;38:D822–D827. doi: 10.1093/nar/gkp805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Rogers MF, Richardson DN, Hamilton M, Ben-Hur A. Deciphering the plant splicing code: experimental and computational approaches for predicting alternative splicing and splicing regulatory elements. Front. Plant Sci. 2012;3:18. doi: 10.3389/fpls.2012.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SJ, Tucker EJ, Tester M. Genetic analysis of abiotic stress tolerance in crops. Curr. Opin. Plant Biol. 2011;14:232–239. doi: 10.1016/j.pbi.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Sanghera GS, Wani SH, Hussain W, Singh NB. Engineering cold stress tolerance in crop plants. Curr. Genomics. 2011;12:30–43. doi: 10.2174/138920211794520178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shameer K, Ambika S, Varghese SM, Karaba N, Udayakumar M, Sowdhamini R. STIFDB–Arabidopsis Stress-responsive Transcription Factor DataBase. Int. J. Plant Genomics. 2009;2009:583429. doi: 10.1155/2009/583429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000;3:217–223. [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Singh K, Foley RC, Onate-Sanchez L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002;5:430–436. doi: 10.1016/s1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- Sinha AK, Jaggi M, Raghuram B, Tuteja N. Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal. Behav. 2011;6:196–203. doi: 10.4161/psb.6.2.14701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P, Gregory PJ, van Vuuren D, Obersteiner M, Havlik P, Rounsevell M, et al. Competition for land. Philos. Trans. R. Soc. B: Biol. Sci. 2010;365:2941–2957. doi: 10.1098/rstb.2010.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner W, Youens-Clark K, Staines D, Ware D. GrameneMart: the BioMart data portal for the Gramene project. Database (Oxford) 2012;2012:bar056. doi: 10.1093/database/bar056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N, Sopory SK, Kavi Kishor PB. Deciphering the regulatory mechanisms of abiotic stress tolerance in plants by genomic approaches. Gene. 2007;388:1–13. doi: 10.1016/j.gene.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Sucaet Y, Deva T. Evolution and applications of plant pathway resources and databases. Brief. Bioinform. 2011;12:530–544. doi: 10.1093/bib/bbq083. [DOI] [PubMed] [Google Scholar]
- Sundar AS, Varghese SM, Shameer K, Karaba N, Udayakumar M, Sowdhamini R. STIF: identification of stress-upregulated transcription factor binding sites in Arabidopsis thaliana. Bioinformation. 2008;2:431–437. doi: 10.6026/97320630002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed NH, Kalyna M, Marquez Y, Barta A, Brown JW. Alternative splicing in plants—coming of age. Trends Plant Sci. 2012;17:616–623. doi: 10.1016/j.tplants.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Matsuoka M. Genetic approaches to crop improvement: responding to environmental and population changes. Nat. Rev. Genet. 2008;9:444–457. doi: 10.1038/nrg2342. [DOI] [PubMed] [Google Scholar]
- Tran LS, Mochida K. Identification and prediction of abiotic stress-responsive transcription factors involved in abiotic stress signaling in soybean. Plant Signal. Behav. 2010;5:255–257. doi: 10.4161/psb.5.3.10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner WR, Oppenheimer M, Wilcove DS. A force to fight global warming. Nature. 2009;462:278–279. doi: 10.1038/462278a. [DOI] [PubMed] [Google Scholar]
- Tuteja N. Abscisic acid and abiotic stress signaling. Plant Signal. Behav. 2007;2:135–138. doi: 10.4161/psb.2.3.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Ichimura K, Mizoguchi T, Peck SC, Zhu T, Wang X, et al. Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J. 2002;21:6483–6493. doi: 10.1093/emboj/cdf646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell. 1993;5:1529–1539. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Quimbaya M, Casneuf T, De Veylder L, Van de Peer Y. Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol. 2009;150:535–546. doi: 10.1104/pp.109.136028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Zeng L, et al. Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol. 2005;139:822–835. doi: 10.1104/pp.105.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley JW, Dehesh K. Molecular mechanisms regulating rapid stress signaling networks in Arabidopsis. J. Integr. Plant Biol. 2010;52:354–359. doi: 10.1111/j.1744-7909.2010.00940.x. [DOI] [PubMed] [Google Scholar]
- Warren GJ. Cold stress: manipulating freezing tolerance in plants. Curr. Biol. 1998;8:R514–516. doi: 10.1016/s0960-9822(07)00335-1. [DOI] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14(Suppl):S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhu JK. Abiotic stress signal transduction in plants: molecular and genetic perspectives. Physiol. Plant. 2001;112:152–166. doi: 10.1034/j.1399-3054.2001.1120202.x. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Zhao W, Wang J, He X, Huang X, Jiao Y, Dai M, et al. BGI-RIS: an integrated information resource and comparative analysis workbench for rice genomics. Nucleic Acids Res. 2004;32:D377–D382. doi: 10.1093/nar/gkh085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sumazin P, Rajbhandari P, Califano A. A systems biology approach to transcription factor binding site prediction. PLoS One. 2010;5:e9878. doi: 10.1371/journal.pone.0009878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 2001;4:401–406. doi: 10.1016/s1369-5266(00)00192-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.