Abstract
Plant hormones play important roles as signaling molecules in the regulation of growth and development by controlling the expression of downstream genes. Since the hormone signaling system represents a complex network involving functional cross-talk through the mutual regulation of signaling and metabolism, a comprehensive and integrative analysis of plant hormone concentrations and gene expression is important for a deeper understanding of hormone actions. We have developed a database named Uniformed Viewer for Integrated Omics (UniVIO: http://univio.psc.riken.jp/), which displays hormone-metabolome (hormonome) and transcriptome data in a single formatted (uniformed) heat map. At the present time, hormonome and transcriptome data obtained from 14 organ parts of rice plants at the reproductive stage and seedling shoots of three gibberellin signaling mutants are included in the database. The hormone concentration and gene expression data can be searched by substance name, probe ID, gene locus ID or gene description. A correlation search function has been implemented to enable users to obtain information of correlated substance accumulation and gene expression. In the correlation search, calculation method, range of correlation coefficient and plant samples can be selected freely.
Keywords: Database, Multiomics, Oryza sativa, Plant hormones, Transcriptome
Introduction
Plant hormones are signaling molecules that regulate plant growth and development. Plant hormones function, at least partially, through the regulation of the expression of downstream genes, and the underlying molecular mechanisms are being revealed (Shan et al. 2011, Garg et al. 2012). For instance, cytokinin is perceived by membrane-localized histidine kinases, and the signal is transduced by a His–Asp phosphorelay in a two-component system (Kieber and Schaller 2010, Hwang et al. 2012). Auxin and gibberellin are perceived by nuclear receptors and signal transduction is mediated by the 26S proteasome pathway (Hartweck 2008, Mockaitis and Estelle 2008, Lumba et al. 2010 Hayashi 2012). ABA is perceived by a regulator of type 2C protein phosphatases localized in the nucleocytoplasm; the signal is transmitted by the phosphatase activity and the phosphorylation status of downstream components (Cutler et al. 2010, Umezawa et al. 2010, Qin et al. 2011). Plasma membrane-localized G protein-coupled receptor-type G proteins are also suggested as ABA receptors (Pandey et al. 2009). The spatiotemporal distribution of the signaling systems specifies the situation in which a hormone response can occur. In addition to the signaling systems, hormone concentration and distribution are determinants of hormone action. Hormone metabolism, including biosynthesis, conjugation and degradation, is intricately regulated by feedback loops in space and time to finely control hormone activities.
It has long been known that there are functional interactions between plant hormones, especially between cytokinin and auxin (Loomis and Torrey 1964, Sachs and Thimann 1967), cytokinin and ethylene (Cary et al. 1995), auxin and gibberellin (Weijer 1959), auxin and ABA (Saftner and Wyse 1984), and gibberellin and ABA (Pearce et al. 1987). Current research aims to explain molecular mechanisms of the interactions between plant hormones via the regulation of their signaling and metabolic genes. For instance, the negative regulation of key genes of cytokinin biosynthesis by auxin has been demonstrated to underlie apical dominance in stems (Tanaka et al. 2006, Kitazawa et al. 2008). The cytokinin signaling factor ARR1 directly regulates the expression of the auxin signaling repressor gene SHY2/IAA3 (Taniguchi et al. 2007, Dello Ioio et al. 2008), and auxin regulates a subset of the type-A response regulator genes in Arabidopsis and rice (Müller and Sheen 2008, Zhao et al. 2010, Tsai et al. 2012). Frigerio et al. (2006) showed that auxin up-regulated the expression of gibberellin metabolic genes in Arabidopsis, and several studies highlighted the cross-talk between auxin and gibberellin signaling and metabolism at the molecular level (Ozga et al. 2009, Weston et al. 2009, O’Neill et al. 2010).
Recent omics analyses further illuminate interactions among plant hormones. Comprehensive transcriptome analysis in Arabidopsis outlined the interplay between major plant hormones (Nemhauser et al. 2006, Goda et al. 2008). Transcriptome analysis of cytokinin responses and meta-analysis of public transcriptome data showed that cytokinin regulates various hormone-related genes including auxin signaling AXR3/IAA17, gibberellin synthesis GA4, gibberellin signaling GAI, ethylene synthesis ACC genes and ethylene signaling ERF/AP2 genes (Brenner et al. 2005, Brenner et al. 2012). Transcriptome and hormone-metabolome (hormonome) analyses in rice gibberellin signaling mutants, such as gid1, gid2 and slr1, showed that both hormone contents and expression of gibberellin, cytokinin, ABA and IAA metabolic genes are affected in at least one of the mutants (Kojima et al. 2009). Therefore, to characterize the global interactive network of plant hormone actions, comparative analyses of the hormonome and transcriptome are essential. Although the comprehensive analysis of plant hormones remains difficult because of their varied chemical and structural properties and low concentrations, recent developments in mass spectrometry and solid-phase extraction enabled us to quantify multiple hormone species in small amounts of plant material (Kojima et al. 2009, Kanno et al. 2010, Chen et al. 2011).
In addition to general transcriptome databases such as the co-expression database ATTED-II (Obayashi et al. 2007, Obayashi et al. 2009, Obayashi et al. 2011), several specialist databases provide information related to the hormone network in plants (Mochida and Shinozaki 2011). For example, RiceFOX provides plant hormone profiles of transgenic Arabidopsis plants overexpressing full-length rice cDNAs (T. Sakurai et al. 2011), while the Arabidopsis Hormone Database contains a collection of hormone-related genes (Jiang et al. 2011). Correlation analysis using the web-based tool AtCAST demonstrated correlations between transcriptome data related to hormone responses (Sasaki et al. 2011). However, there is no public platform that provides integrated endogenous plant hormone concentrations and transcriptome data. Here we describe Uniformed Viewer for Integrative Omics (UniVIO), which houses comparative hormonome and transcriptome data analyzed in the same plant samples, and offers useful search functions including correlation search.
Database Contents
UniVIO currently houses contents of four major plant hormones (cytokinin, auxin, gibberellin and ABA) including active forms and their precursors and conjugates (43 compounds in total; Table 1), transcriptome data analyzed in 14 organ parts of rice plants (Fig. 1 and Table 2) and shoots of three gibberellin signaling mutant seedlings (Kojima et al. 2009). Abbreviations of names of substances listed in Table 1 are used hereafter.
Table 1.
Plant hormones and related compounds analyzed in this study
| Hormone | Class | Abbreviation | Physiological property |
|---|---|---|---|
| Abscisic acid | Abscisic acid | ABA | Active compound |
| Indole-3-acetic acid | Auxin | IAA | Active compound |
| Indole-3-acetyl-l-alanine | Auxin | IAAla | Deactivated compound |
| Indole-3-acetyl-l-isoleucine | Auxin | IAIle | Deactivated compound |
| Indole-3-acetyl-l-leucine | Auxin | IALeu | Deactivated compound |
| Indole-3-acetyl-l-aspartic acid | Auxin | IAAsp | Deactivated compound |
| Indole-3-acetyl-l-tryptophan | Auxin | IATrp | Deactivated compound |
| Indole-3-acetyl-l-phenylalanine | Auxin | IAPhe | Deactivated compound |
| N6-(Δ2-Isopentenyl)adenine | Cytokinin | iP | Active compound |
| N6-(Δ2-Isopentenyl)adenine riboside | Cytokinin | iPR | Intermediate |
| N6-(Δ2-Isopentenyl)adenine ribotides | Cytokinin | iPRPs | Intermediate |
| N6-(Δ2-Isopentenyl)adenine-N7-glucoside | Cytokinin | iP7G | Deactivated compound |
| N6-(Δ2-Isopentenyl)adenine-N9-glucoside | Cytokinin | iP9G | Deactivated compound |
| trans-Zeatin | Cytokinin | tZ | Active compound |
| trans-Zeatin riboside | Cytokinin | tZR | Intermediate |
| trans-Zeatin ribotides | Cytokinin | tZRPs | Intermediate |
| trans-Zeatin-N7-glucoside | Cytokinin | tZ7G | Deactivated compound |
| trans-Zeatin-N9-glucoside | Cytokinin | tZ9G | Deactivated compound |
| trans-Zeatin-O-glucoside | Cytokinin | tZOG | Deactivated compound |
| trans-Zeatin riboside-O-glucoside | Cytokinin | tZROG | Deactivated compound |
| trans-Zeatin ribotide-O-glucosides | Cytokinin | tZRPsOG | Deactivated compound |
| cis-Zeatin | Cytokinin | cZ | Active compound |
| cis-Zeatin riboside | Cytokinin | cZR | Intermediate |
| cis-Zeatin ribotides | Cytokinin | cZRPs | Intermediate |
| cis-Zeatin-O-glucoside | Cytokinin | cZOG | Deactivated compound |
| cis-Zeatin riboside-O-glucoside | Cytokinin | cZROG | Deactivated compound |
| cis-Zeatin ribotide-O-glucosides | Cytokinin | cZRPsOG | Deactivated compound |
| Dihydro-zeatin | Cytokinin | DZ | Active compound |
| Dihydro-zeatin riboside | Cytokinin | DZR | Intermediate |
| Dihydro-zeatin ribotides | Cytokinin | DZRPs | Intermediate |
| Dihydro-zeatin-N9-glucoside | Cytokinin | DZ9G | Deactivated compound |
| Gibberellin A1 | Gibberellin | GA1 | Active compound |
| Gibberellin A3 | Gibberellin | GA3 | Active compound |
| Gibberellin A4 | Gibberellin | GA4 | Active compound |
| Gibberellin A7 | Gibberellin | GA7 | Intermediate |
| Gibberellin A8 | Gibberellin | GA8 | Deactivated compound |
| Gibberellin A9 | Gibberellin | GA9 | Intermediate |
| Gibberellin A12 | Gibberellin | GA12 | Intermediate |
| Gibberellin A19 | Gibberellin | GA19 | Intermediate |
| Gibberellin A20 | Gibberellin | GA20 | Intermediate |
| Gibberellin A24 | Gibberellin | GA24 | Intermediate |
| Gibberellin A44 | Gibberellin | GA44 | Intermediate |
| Gibberellin A53 | Gibberellin | GA53 | Intermediate |
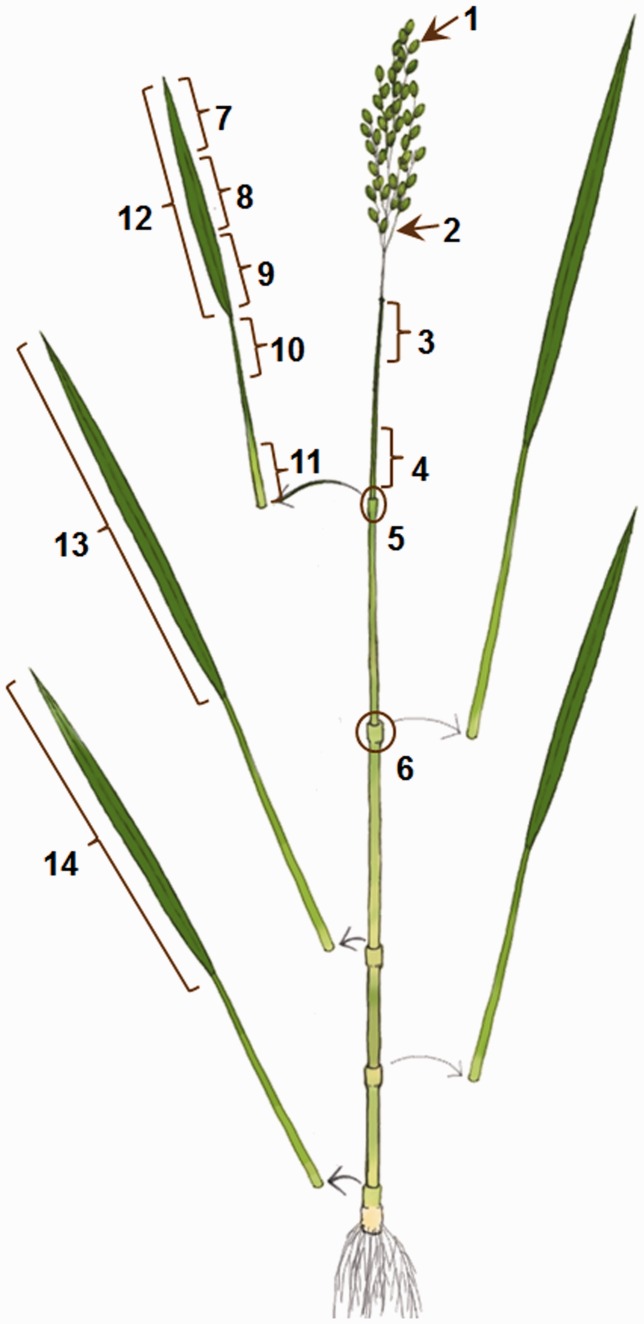
Fig. 1.
Illustration of rice plant organs used for hormone and transcriptome analyses. 1, flowers before anthesis; 2, panicle branches; 3, top part of internode I; 4, basal part of internode I; 5, node I; 6, node II; 7, tip of the blade of the flag leaf; 8, middle part of the blade of the flag leaf; 9, basal part of the blade of the flag leaf; 10, top part of the sheath of the flag leaf; 11, basal part of the sheath of the flag leaf; 12, whole blade of the flag leaf; 13, whole blade of leaf 2 counted down from the flag leaf; 14, whole blade of leaf 4 counted down from the flag leaf. The numbers correspond to the numbers in Table 2.
Table 2.
Sample information of rice organs and mutants from which plant hormone and transcriptome data housed in UniVIO were obtained
| Nos. | Plant organs | Abbreviations | Biological replicate |
|
|---|---|---|---|---|
| GeneChip | Hormone | |||
| Organs at the heading stage, analyzed in this study | ||||
| 1 | Flowers before anthesis | Flw | 3 | 3 |
| 2 | Panicle branches | PBr | 3 | 3 |
| 3 | Top part of internode I | InN I, top | 3 | 3 |
| 4 | Basal part of internode I | InN I, bsl | 3 | 3 |
| 5 | Node I | Nod I | 3 | 3 |
| 6 | Node II | Nod II | 3 | 3 |
| 7 | Tip of the blade of the flag leaf | FLB, tip | 3 | 3 |
| 8 | Middle part of the blade of the flag leaf | FLB, mid | 3 | 3 |
| 9 | Basal part of the blade of the flag leaf | FLB, bsl | 3 | 3 |
| 10 | Top part of the sheath of the flag leaf | FLS, top | 3 | 3 |
| 11 | Basal part of the sheath of the flag leaf | FLS, bsl | 2 | 3 |
| 12 | Whole blade of the flag leaf | FLB | 1 | 3 |
| 13 | Whole blade of leaf 2 counted down from the flag leaf | LB-2 | 1 | 3 |
| 14 | Whole blade of leaf 4 counted down from the flag leaf | LB-4 | 1 | 3 |
| Gibberellin-related mutants, analyzed by Kojima et al. (2009) | ||||
| Shoot of Taichung 65 (control) | T65 | 3 | 3 | |
| Shoot of gid1-3 mutant | gid1 | 3 | 3 | |
| Shoot of gid2-1 mutant | gid2 | 3 | 3 | |
| Shoot of slr1 mutant | slr1 | 3 | 3 | |
Numbers shown in the first column for organs correspond to the numbers shown in Fig. 1. The abbreviations are those used in UniVIO.
Hormonome and transcriptome analysis
Oryza sativa L. cv. Nipponbare was grown on soil in a greenhouse with irrigation and supplemental artificial light. At the heading stage, tissues were harvested and immediately frozen in liquid nitrogen after measurement of fresh weight. Harvested tissues were stored at –80°C until extraction of plant hormones or total RNA. Plant hormones were extracted, purified and quantified as described previously (Kojima et al. 2009). Microarray analysis was performed using a GeneChip® Rice Genome Array (Affymetrix). Total RNA was extracted from the plant samples using the RNeasy® Mini Kit (QIAGEN). Preparation of labeled target-complementary RNA, subsequent purification and fragmentation were carried out using One-Cycle Target Labeling and Control Reagents (Affymetrix). Double-stranded cDNA was prepared from 5 µg of total RNA. Hybridization, washing, staining and scanning were performed as described in the supplier’s protocol. A 5 µg aliquot of fragmented complementary RNA was used for hybridization. These experiments were conducted according to the manufacturers’ guidelines.
Data processing
Plant hormone contents were normalized by fresh weight and expressed as pmol g FW–1. The microarray data were extracted as CEL files and imported into GeneSpringGX version 11 (Agilent Technologies), followed by summarization using the MAS5 algorithm but no baseline transformation. The summarized microarray data of all probe sets were extracted as raw signal intensities. The hormone contents and signal intensities were averaged over biological replicates (Table 2), and the mean values were included in UniVIO.
Data sources
Microarray data of wild-type organs and gibberellin mutants (Kojima et al. 2009) have been deposited in The National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) database under accession numbers GSE41556 and GSE15046, respectively.
Gene descriptions were obtained from the Rice Annotation Project Databases (http://rapdb.dna.affrc.go.jp/; Itoh et al. 2007, Tanaka et al. 2008) and the MSU Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/; Ouyang et al. 2006). Gene ontologies were obtained from the GO Ontology consortium (http://geneontology.org; Ashburner et al. 2000). A matrix table assigning probe IDs and gene locus IDs was obtained from the Rice Oligonucleotide Array Database (http://www.ricearray.org/index.shtml; Jung et al. 2008).
Database Construction
Concepts and workflow
UniVIO was constructed to present the data in a manner as simple as possible. Since a large number of entries have to be shown in the data viewer, a graphic visualization of the data appeared desirable. On the other hand, a numerical data representation would help users to identify the significance of the data. Therefore, UniVIO presents search results as a heat map table showing hormone contents and gene expression levels.
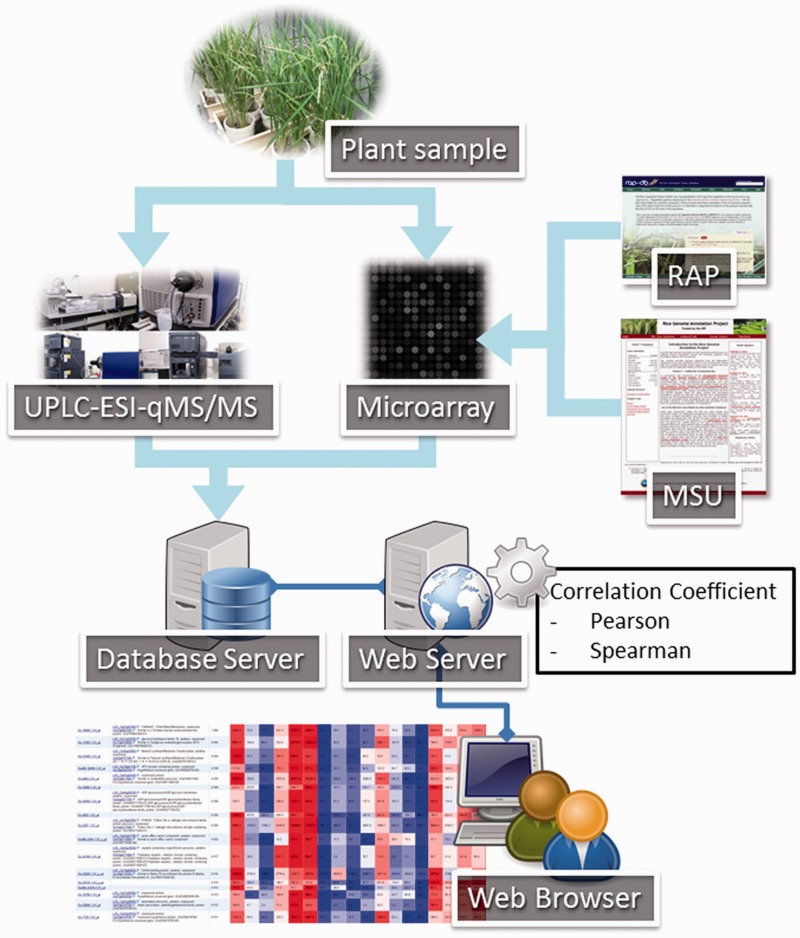
To construct and organize UniVIO, plant hormonome data and transcriptome data were acquired from plant samples as described above. Then, gene annotations were obtained from the two public rice genome databases and combined with the transcriptome data. To let users retrieve the hormonome and transcriptome data using a Web browser, the data were housed in a database server connected to a Web server. Correlation coefficients are computed on the Web server. The data table is controlled on the Web browser only. This workflow is illustrated in Fig. 2.
Fig. 2.
Schematic representation of the workflow to organize UniVIO. All plant samples were collected under the same conditions, and then were used to analyze the concentration of plant hormones and gene expression levels by mass spectrometry and microarray analysis, respectively. The gene annotation information was obtained from the Rice Annotation Project (RAP; http://rapdb.dna.affrc.go.jp/) and the Rice Genome Annotation Project (MSU; http://rice.plantbiology.msu.edu/), and was combined with the transcriptome data. Users retrieve data from the database server through the Web server using Web browsers. Correlation coefficients are computed on the Web server. Data tables are generated in the Web browser only. UPLC, ultra-performance liquid chromatography; ESI, electrospray ionization; qMS/MS, tandem quadrupole mass spectrometry.
Database implementation and statistical treatment
Since UniVIO is implemented as a typical Web server–client system using general open source software (Linux, Apache, MySQL and Perl), users can query the database via the Internet using a Web browser. The system is compliant with common Web browsers regardless of the operating system, but not with older browsers such as Internet Explorer 7. There is no limit to use; anyone can access all of the data without sign in. To calculate Pearson product–moment correlation coefficients and Spearman’s rank correlation coefficients, the GNU Statistic Library (http://www.gnu.org/software/gsl/) and Statistics::RankCorrelation (http://search.cpan.org/dist/Statistics-RankCorrelation/), respectively, are used. Correlation coefficients are calculated in the Web server on each request (Fig. 2).
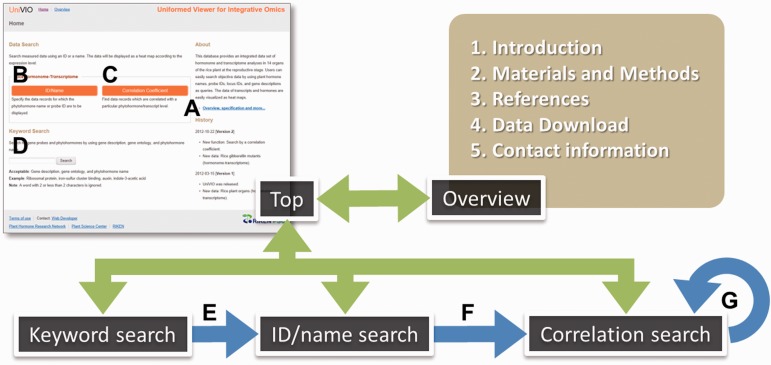
Instructions
The UniVIO website is comprised of five pages (Fig. 3). The top page provides access to other pages: overview, ID/name search, correlation search and keyword search pages (Fig. 3A, B, C, and D, respectively). The overview page provides data information including materials, methods, raw data and contact information. The search pages support corresponding search functions. When users do not have a list of gene(s) and/or hormone(s) to be queries, they may start with a keyword search. A result of a keyword search is shown in the ID/name search page (Fig. 3E). Then, when the users find an interesting ID (gene or plant hormone) in the result, they may go directly on to the correlation search using the interesting ID as a query on the ID/name search page (Fig. 3F). This direct correlation search function is also available on the correlation search page (Fig. 3G). The expected workflow is drawn in Fig. 3. Detailed instructions for the search functions are described in the following sections.
Fig. 3.
Site map of UniVIO. Green arrows indicate mutual accessibility between the top page and other pages comprising the UniVIO website. On the top page, there are links to move to the overview (A), ID/name search (B), correlation search (C) and keyword search (D) pages. Blue arrows indicate functional connections between the keyword search and ID/name search (E), between the ID/name search and correlation search (F), and between corelation searches (G), in a a typical workflow in UniVIO.
ID/name search
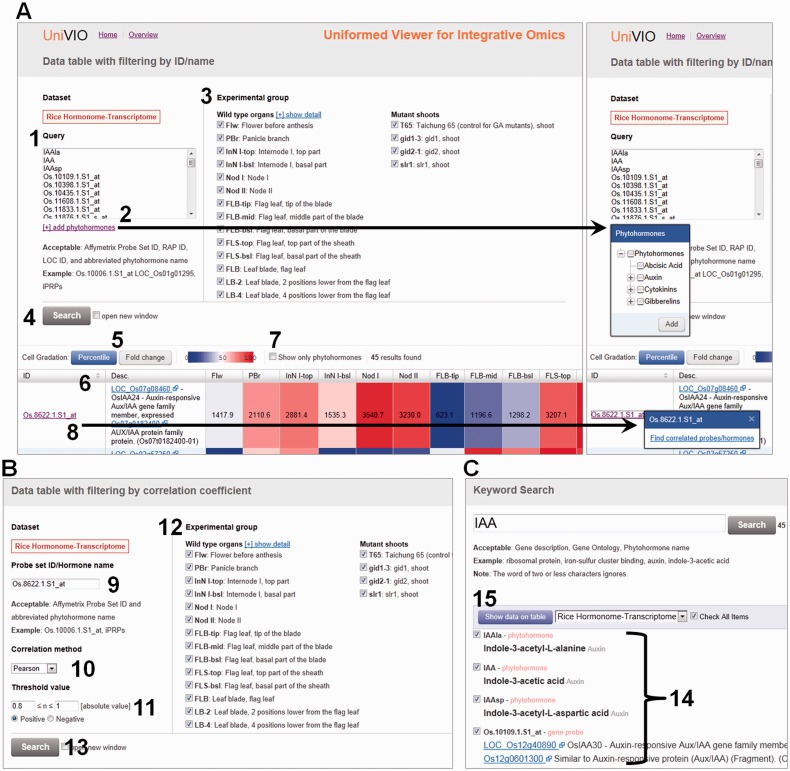
To execute an ID/name search, users move to the search page by clicking a button on the top page (Fig. 3B). UniVIO accepts a single or multiple (up to 1,000) hormone names, gene locus IDs and probe IDs as queries (process 1 in Fig. 4A). Hormone names may be added to a query input form by selecting on a checkbox tree (process 2 in Fig. 4A). Each query can be separated by a space, comma or line break. To focus on particular samples, the plant samples to be shown in a heat map table may be selected using checkboxes (process 3 in Fig. 4A). Color schemes for the heat map can be selected as percentile based or fold change based (process 5 in Fig. 4A). In the former option, red and blue are saturated at 100 and 0 percentiles, respectively; in the latter, the colors are saturated at 3-fold and at one-third, respectively, of the mean values of selected samples. In the current data set of rice organs, microarray analysis was not or was less replicated in some samples (Table 2). It possibly affects the reliability of gene expression levels and correlations for expression patterns. The sample selection function also enable the exclusion of less-replicated samples when users concern this point.
Fig. 4.
Instruction for search functions in UniVIO. Cropped ID/name search (A), correlation search (B) and keyword search (C) pages. 1, query input box for ID/name search; 2, checkbox tree to manage plant hormones; 3, selection of samples to be shown in a heat map table; 4, execution of ID/name search; 5, selection of a coloring method; 6, sorting function by IDs; 7, extraction of plant hormones from the heat map table; 8, direct link to a correlation search result; 9, query input box for correlation search; 10, selection of a method to calculate correlation coefficients; 11, setting a range of correlation coefficients; 12, selection of samples to be used for calculation of correlation coefficients and to be shown in a heat map table; 13, execution of the correlation search; 14, selection of genes and plant hormones shown in a heat map table; 15, execution of ID/name search using probe IDs and plant hormones selected in the keyword search page.
In a heat map table, data can be sorted by IDs (probe IDs and hormone names) shown in the first column (process 6 in Fig. 4A). To find plant hormones easily in a heat map table, check the box labeled ‘Show only phytohormone’ located just above the heat map table (process 7 in Fig. 4A). Each ID shown in the resulting table is linked to a correlation search using the ID as a query (process 8 in Fig. 4A).
Correlation search
To execute a correlation search, users move to a corresponding search page by clicking a button on the top page (Fig. 3B). UniVIO accepts a single probe ID or hormone name as a query (process 9 in Fig. 4B). Users also may be directed to the results of correlation searches from heat map tables in ID/name searches (Figs. 3F, 4A) and correlation searches (Fig. 3G). To allow free modifications of search conditions, UniVIO provides several options: calculation methods (parametric Pearson product–moment correlation or non-parametric Spearman’s rank correlation), range of correlation coefficients (from –1 to 1) and plant samples (processes 10, 11 and 12 in Fig. 4B). All of these options affect the correlation coefficients. If the number of results is >1,000 (the upper limit in UniVIO), no result is shown. To see results, one has to narrow the range of correlation coefficients. Instructions concerning the resulting heat maps are basically the same as for ID/name searches (see above and Fig. 4A).
Keyword search
Keyword searches can be started from the top page (Fig. 3C). UniVIO accepts a single word of three or more characters, or multiple words as a query, and will search gene descriptions, gene ontologies and hormone names. When a query is submitted from the top page (Fig. 3D), the keyword search page will be displayed on which candidate genes and hormones can be selected (process 14 in Fig. 4C). If the number of candidates exceeds 1,000, they are not shown. To see the candidates, further specify the search conditions by adding or changing keywords. To show a resulting heat map, click ‘show data on table’ (process 15 in Fig. 4C). The results page shown resembles that returned by an ID/name search (see above and Fig. 4A).
Discussion
Among the 43 plant hormone-related compounds analyzed, 13 (ABA, IAA, iPRPs, cZRPs, iPR, tZR, cZR, cZ, cZRPsOG, cZROG, cZOG, GA9 and GA19) were detected in the 14 organ parts analyzed (Fig. 5). Correlative relationships between the hormones and between hormones and hormone-related genes are summarized in Figs. 6, 7 and Supplementary Fig. S1, respectively. Some aspects of these correlations are discussed below. The hormonome and transcriptome data in the gibberellin signaling mutants have been discussed in our previous study (Kojima et al. 2009).
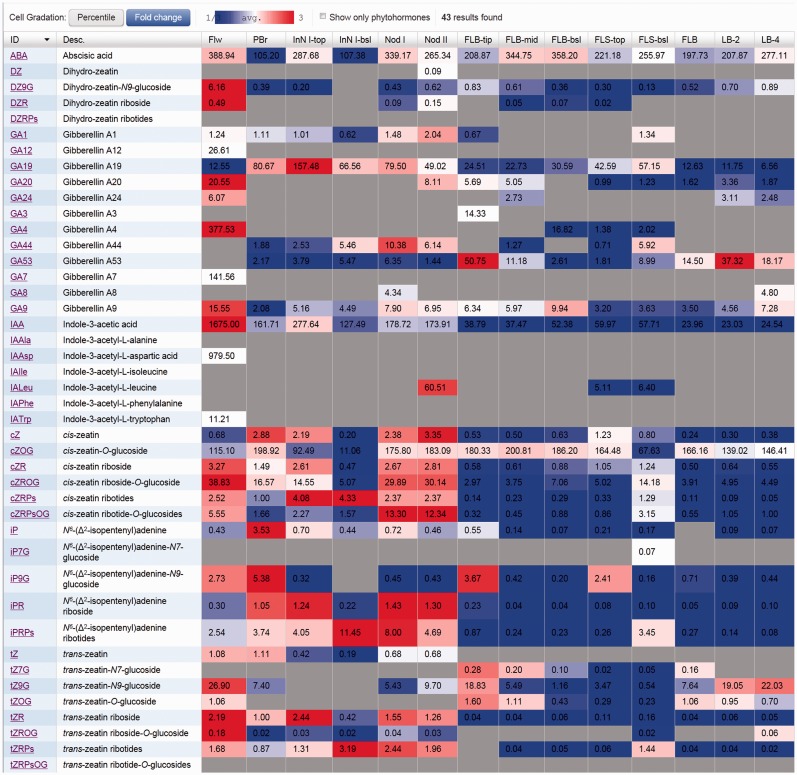
Fig. 5.
Contents of plant hormone-related compounds in various rice organs. The heat map is drawn using UniVIO with fold change coloring. Hormone contents are indicated as means of three biological replicates in units of pmol g–1 FW.
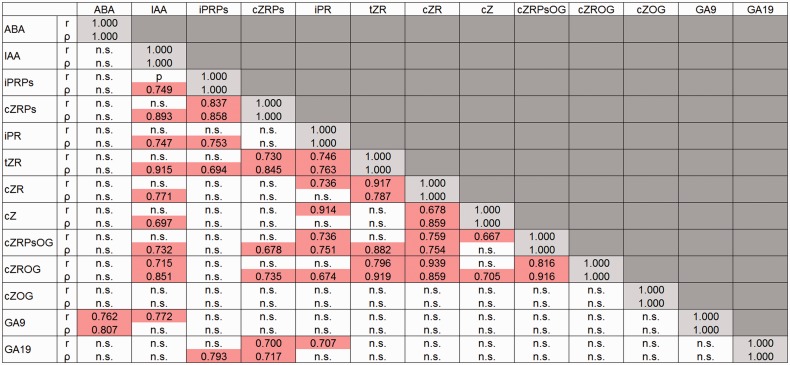
Fig. 6.
Correlations between hormones. Plant hormone-related compounds detected in all organs tested are selected. Pearson product–moment correlation coefficients (upper, r) and Spearman’s rank correlation coefficients (lower, ρ) are indicated and highlighted in red (positive) or blue (negative) where they are significant (α = 0.01). n.s., not significant.
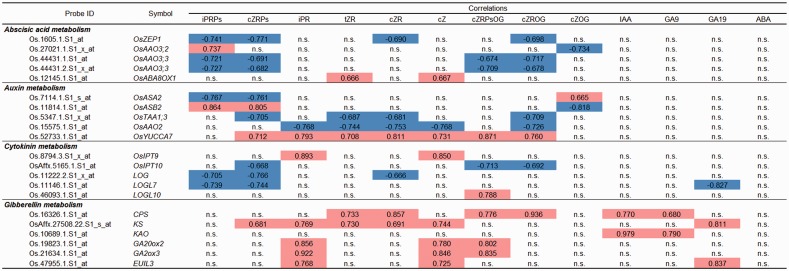
Fig. 7.
Correlations between hormone-related compounds and hormone metabolic genes. Plant hormone-related compounds detected in all organs tested are selected. Microarray probes representing ‘Present’ as flag information in all samples tested are selected. Pearson product–moment correlation coefficients are indicated and highlighted in red (positive) or blue (negative) where they are significant (α = 0.01). n.s., not significant.
Interhormone correlation in wild-type organs
In a current model of cytokinin biosynthesis that is mostly based on studies of Arabidopsis, cZ is thought to originate from the degradation of prenylated tRNA that has been modified by tRNA-isopentenyltransferase (tRNA-IPT). This pathway is distinct from iP and tZ biosynthesis initialized by prenylation of adenosine phosphates (Kakimoto 2001, Takei et al. 2001, Kasahara et al. 2004, Miyawaki et al. 2006). However, cZRPs, cZR, cZ, cZRPsOG and cZROG were positively correlated with iPRPs, iPR and/or tZR (Fig. 6). This result suggests two possibilities. First, there could be a novel cZ biosynthesis pathway linked to iP and tZ biosynthesis in rice, and, secondly, the cZ levels in organs could be controlled by mechanisms other than de novo synthesis. However, cZ was strongly negatively correlated with OsIPT9 which encodes a tRNA-IPT (Supplementary Fig. S1). Regarding the latter possibility, reversible inactivation by O-glucosylation could be a candidate pathway controlling the cZ level. In rice, cZOG was the most abundant cytokinin derivative and it was not correlated with other cytokinins (Figs. 5, 6). Putative cZ-O-glucosyltransferases have been found in cZ-rich plants such as maize and rice, but not in Arabidopsis which does not accumulate significant amounts of cZ derivatives (Veach et al. 2003, Hou et al. 2004, Wang et al. 2011, Kudo et al. 2012). Positive correlation of cytokinins was found with IAA (Fig. 6).
GA9 and GA19 were positively correlated with other hormones: GA9 with ABA and IAA, and GA19 with iPRPs, cZRPs and iPR (Fig. 6). Since GA9 and GA19 are precursors of the bioactive GA4 and GA1, respectively, this may imply different physiological roles for GA4 and GA1 in rice. The synthetic pathway of GA1 is thought to branch from the GA4 synthesis pathway by 13-hydroxylation of GA12 (Kobayashi et al. 2000, Hedden and Thomas 2012). The enzyme responsible for the 13-hydroxylation has not been identified.
Negative correlation was not found in any combinations of the 13 hormone-related compounds when data of all organs were used to calculate correlation coefficients (Fig. 6). However, modification of search conditions, such as selection of samples, allowed us to find negative correlations. For example, negative correlations were detected between tZ and iPRPs (r = –0.750), tZRPs (r = –0.681) or cZRPs (r = –0.881), when flower, panicle branch, and the top and basal part of internode I, node I and node II were selected.
Correlation between hormones and hormone-related genes in wild-type organs
Among cytokinin precursors including ribotides and ribosides, ribotides such as iPRPs and cZRPs showed negative correlation with cytokinin-activating LOG (Os01g0588900) and LOGL7 (Os05g0541200) (Fig. 7). Since iPRPs and cZRPs are substrates for the enzymes LOG, it was not surprising that iPRP and cZRP levels correlated negatively with the activities of the enzyme genes LOG and LOGL7. However, the roles of many LOG family members in rice have not been determined yet, whereas metabolic profiling of multiple log mutants showed that LOG7 makes the largest contribution to the total cytokinin activation on the whole seedling level in Arabidopsis (Tokunaga et al. 2012). The iPRPs and cZRPs showed strongly positive correlation with the gibberellin signaling repressor gene SLR1 (Os03g0707600) (Supplementary Fig. S1). Although Arabidopsis GAI, a homolog of SLR1, was up-regulated by cytokinin (Brenner et al. 2005), SLR1 was not listed as a cytokinin-inducible gene in a recent transcriptome analysis (Garg et al. 2012). While cytokinin derivatives, IAA, GA9 and GA19 showed positive or negative correlations with several IAA, gibbellin and/or ABA metabolic genes, ABA did not show any correlation with any hormone metabolic genes we tested (Fig. 7). Neither did ABA correlate significantly with any hormone signaling genes (Supplementary Fig. S1).
Consistent with our previous analysis (Kojima et al 2009), IAA was present at high levels in flower (Fig. 5). Expression of several IAA biosynthetic genes also appeared high in flowers: OsASA1 (Os.4176.1.S1_at, Os03g0826500), OsASB1 (Os.6876.1.S1_at, Os04g0463500), OsTAA1;4 (Os.13941.1.S1_x_at and Os.13941.1.S1_s_at, Os05g0169300), OsYUCCA4 (Os.22585.1.S1_at, Os01g0224700) and OsYUCCA5 (OsAffx.31989.1.S1_at, Os12g0512000) (Supplementary Fig. S2A). These results are in line with the reported abundance of IAA in anthers and the expression of OsTAA1;4 and OsYUCCA4 at later stages of pollen development (Hirano et al. 2008). IAA–amino acid conjugates were detected in specific organs: IAAsp and IATrp in flowers, and IALeu in the top and basal parts of the sheath of the flag leaf as well as in node II (Fig. 5). Among the IAA–amino acid synthase (GH3) family, OsMGH/OsGH3-8 (Os.11798.1.S1_at, Os07g0592600), which plays an important role in rice floret fertility (Yadav et al. 2011), showed high expression in flowers (Supplementary Fig. S2B). This result is consistent with a previous study (Jain et al. 2006).
Perspective
Obviously, the regulative network of plant hormone metabolism and gene expression plays a pivotal role in the orchestration of physiological and morphological functions in plants. However, because of its complexity, we have only begun to understand this network. Integrative analyses of the plant metabolome and transcriptome have already contributed to studies on metabolic regulation, such as enviromental response (Yamakawa and Hakata 2010, Schlüter et al. 2012) and spatial control (Matsuda et al. 2010, Pick et al. 2011), and on secondary metabolism, such as glucosinolate (Hirai et al. 2004, 2005) and terpenoid (Rischer et al. 2006) matabolism. Thus, integrative omics is a valuable method to figure out the complex network.
There are several web-based platforms to analyze and visualize correlations in rice omics data. For instance, OryzaExpress (http://riceball.lab.nig.ac.jp/oryzaexpress/) integrating public rice omics data provides a tool to visualize the gene co-expression network (Hamada et al. 2011). KaPPA-View4 (http://kpv.kazusa.or.jp/kpv4/) can visualize gene expression, metabolite accumulation and gene–gene and metabolite–metabolite correlation in a matabolic pathway map based on transciptome and metabolome data uploaded by users (N. Sakurai et al. 2011). Our UniVIO provides both a comparative hormonome–transcriptome data set and a web application supporting a correlation search function, enabling exploration of positive or negative correlations between the concentration of a plant hormone-related compound and gene expression level. However, further accumulation of comparative hormonome–transcriptome data concerning organ/tissue development and environmental response, and additional mutants in rice and other model plants is needed to increase the utility of UniVIO. We are going to add new data annually, and Arabidopsis data will be installed within 2 years.
Supplementary data
Supplementary data are available at PCP online.
Funding
This study was supported by the Japan Society for the Promotion of Science [Grants-in-Aid for Scientific Research (B) (grant No. 21370023 to H.S.)].
Supplementary Material
Acknowledgments
We thank T. Kiba, N. Ueda and H. Tokunaga (RIKEN) for their help in sampling rice organs, and S. Oyama (RIKEN) for assistance with the GeneChip experiment. The hormone analysis reported here was supported by the Japan Advanced Plant Science Network.
Glossary
Abbreviations
- tRNA-IPT
tRNA-isopentenyltransferase
- UniVIO
Uniformed Viewer for Integrative Omics.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Ramireddy E, Heyl A, Schmülling T. Gene regulation by cytokinin in Arabidopsis. Front. Plant Sci. 2012;3:8. doi: 10.3389/fpls.2012.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T. Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J. 2005;44:314–333. doi: 10.1111/j.1365-313X.2005.02530.x. [DOI] [PubMed] [Google Scholar]
- Cary AJ, Liu W, Howell SH. Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyl elongation in Arabidopsis thaliana seedlings. Plant Physiol. 1995;107:1075–1082. doi: 10.1104/pp.107.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ML, Huang YQ, Liu JQ, Yuan BF, Feng YQ. Highly sensitive profiling assay of acidic plant hormones using a novel mass probe by capillary electrophoresis–time of flight-mass spectrometry. J. Chromatogr. B. 2011;879:938–944. doi: 10.1016/j.jchromb.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Cutler S, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, et al. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322:1380–1384. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- Frigerio M, Alabadí D, Pérez-Gómez J, García-Cárcel L, Phillips AL, Hedden P, et al. Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 2006;142:553–563. doi: 10.1104/pp.106.084871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R, Tyagi AK, Jain M. Microarray analysis reveals overlapping and specific transcriptional responses to different plant hormones in rice. Plant Signal. Behav. 2012;7:1–6. doi: 10.4161/psb.20910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, et al. The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 2008;55:526–542. doi: 10.1111/j.0960-7412.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- Hamada K, Hongo K, Suwabe K, Shimizu A, Nagayama T, Abe R, et al. OryzaExpress: an integrated database of gene expression networks and omics annotations in rice. Plant Cell Physiol. 2011;52:220–229. doi: 10.1093/pcp/pcq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartweck LM. Gibberellin signaling. Planta. 2008;229:1–13. doi: 10.1007/s00425-008-0830-1. [DOI] [PubMed] [Google Scholar]
- Hayashi K. The interaction and integration of auxin signaling components. Plant Cell Physiol. 2012;53:965–975. doi: 10.1093/pcp/pcs035. [DOI] [PubMed] [Google Scholar]
- Hedden P, Thomas SG. Gibberellin biosynthesis and its regulation. Biochem. J. 2012;444:11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Klein M, Fujikawa Y, Yano M, Goodenowe DB, Yamazaki Y, et al. Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J. Biol. Chem. 2005;280:25590–25595. doi: 10.1074/jbc.M502332200. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, et al. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2004;101:10205–10210. doi: 10.1073/pnas.0403218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Aya K, Hobo T, Sakakibara H, Kojima M, et al. Comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes in microspore/pollen and tapetum of rice. Plant Cell Physiol. 2008;49:1429–1450. doi: 10.1093/pcp/pcn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou B, Lim EK, Higgins GS, Bowles DJ. N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J. Biol. Chem. 2004;279:47822–47832. doi: 10.1074/jbc.M409569200. [DOI] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B. Cytokinin signaling networks. Annu. Rev. Plant Biol. 2012;63:353–380. doi: 10.1146/annurev-arplant-042811-105503. [DOI] [PubMed] [Google Scholar]
- Itoh T, Tanaka T, Barrero RA, Yamasaki C, Fujii Y, Hilton PB, et al. Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res. 2007;17:175–183. doi: 10.1101/gr.5509507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Kaur N, Tyagi AK, Khurana JP. The auxin-responsive GH3 gene family in rice (Oryza sativa). Funct. Integr. Genomics. 2006;6:36–46. doi: 10.1007/s10142-005-0142-5. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Liu X, Peng Z, Wan Y, Ji Y, He W, et al. AHD2.0: an updated version of Arabidopsis Hormone Database for plant systematic studies. Nucleic Acids Res. 2011;39:D1123–D1129. doi: 10.1093/nar/gkq1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Dardick C, Bartley LE, Cao P, Phetsom J, Canlas P, et al. Refinement of light-responsive transcript li lists using rice oligonucleotide arrays: evaluation of gene-redundancy. PLoS One. 2008;3:e3337. doi: 10.1371/journal.pone.0003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltransferases. Plant Cell Physiol. 2001;42:677–685. doi: 10.1093/pcp/pce112. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Jikumaru Y, Hanada A, Nambara E, Abrams SR, Kamiya Y, et al. Comprehensive hormone profiling in developing Arabidopsis seeds: examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol. 2010;51:1988–2001. doi: 10.1093/pcp/pcq158. [DOI] [PubMed] [Google Scholar]
- Kasahara H, Takei K, Ueda N, Hishiyama S, Yamaya T, Kamiya Y, et al. Distinct isoprenoid origins of cis- and trans-zeatin biosyntheses in Arabidopsis. J. Biol. Chem. 2004;279:14049–14054. doi: 10.1074/jbc.M314195200. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE. The perception of cytokinin: a story 50 years in the making. Plant Physiol. 2010;154:487–492. doi: 10.1104/pp.110.161596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa D, Miyazawa Y, Fujii N, Hoshino A, Iida S, Nitasaka E, et al. The gravity-regulated growth of axillary buds is mediated by a mechanism different from decapitation-induced release. Plant Cell Physiol. 2008;49:891–900. doi: 10.1093/pcp/pcn063. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, MacMillan J, Phinney B, Gaskin P, Spray CR, Hedden P. Gibberellin biosynthesis: metabolic evidence for three steps in the early 13-hydroxylation pathway of rice. Phytochemistry. 2000;55:317–321. doi: 10.1016/s0031-9422(00)00265-x. [DOI] [PubMed] [Google Scholar]
- Kojima M, Kamada-Nobusada T, Komatsu H, Takei K, Kuroha T, Mizutani M, et al. Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography–tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol. 2009;50:1201–1214. doi: 10.1093/pcp/pcp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo T, Makita N, Kojima M, Tokunaga H, Sakakibara H. Cytokinin activity of cis-zeatin and phenotypic alterations induced by overexpression of putative cis-zeatin-O-glucosyltransferase in rice. Plant Physiol. 2012;160:319–331. doi: 10.1104/pp.112.196733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis RS, Torrey JG. Chemical control of vascular cambium initiation in isolated radish roots. Proc. Natl Acad. Sci. USA. 1964;52:3–11. doi: 10.1073/pnas.52.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumba S, Cutler S, McCourt P. Plant nuclear hormone receptors: a role for small molecules in protein–protein interactions. Annu. Rev. Cell Dev. Biol. 2010;26:445–469. doi: 10.1146/annurev-cellbio-100109-103956. [DOI] [PubMed] [Google Scholar]
- Matsuda F, Hirai MY, Sasaki E, Akiyama K, Yonekura-Sakakibara K, Provart NJ, et al. AtMetExpress development: a phytochemical atlas of Arabidopsis development. Plant Physiol. 2010;152:566–578. doi: 10.1104/pp.109.148031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K, Tarkowski P, Matsumoto-Kitano M, Kato T, Sato S, et al. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl Acad. Sci. USA. 2006;103:16598–16603. doi: 10.1073/pnas.0603522103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Shinozaki K. Advances in omics and bioinformatics tools for systems analyses of plant functions. Plant Cell Physiol. 2011;52:2017–2038. doi: 10.1093/pcp/pcr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M. Auxin receptors and plant development: a new signaling paradigm. Annu. Rev. Cell Dev. Biol. 2008;24:55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature. 2008;453:1094–1097. doi: 10.1038/nature06943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K. ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 2009;37:D987–D991. doi: 10.1093/nar/gkn807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, et al. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 2007;35:D863–D869. doi: 10.1093/nar/gkl783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Nishida K, Kasahara K, Kinoshita K. ATTED-II updates: condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol. 2011;52:213–219. doi: 10.1093/pcp/pcq203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill DP, Davidson SE, Clarke VC, Yamauchi Y, Yamaguchi S, Kamiya Y, et al. Regulation of the gibberellin pathway by auxin and DELLA proteins. Planta. 2010;232:1141–1149. doi: 10.1007/s00425-010-1248-0. [DOI] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga JA, Reinecke DM, Ayele BT, Ngo P, Nadeau C, Wickramarathna AD. Developmental and hormonal regulation of gibberellin biosynthesis and catabolism in pea fruit. Plant Physiol. 2009;150:448–462. doi: 10.1104/pp.108.132027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Nelson DC, Assmann SM. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell. 2009;136:136–148. doi: 10.1016/j.cell.2008.12.026. [DOI] [PubMed] [Google Scholar]
- Pearce D, Pharis RP, Rajasekaran K, Mullins MG. Effects of chilling and ABA on [3H]gibberellin A4 metabolism in somatic embryos of grape (Vitis vinifera L.×V. rupestris Scheele) Plant Physiol. 1987;84:381–385. doi: 10.1104/pp.84.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick TR, Bräutigam A, Schlüter U, Denton AK, Colmsee C, Scholz U, et al. Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. Plant Cell. 2011;23:4208–4220. doi: 10.1105/tpc.111.090324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Shinozaki K, Yamaguchi-Shinozaki K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011;52:1569–1582. doi: 10.1093/pcp/pcr106. [DOI] [PubMed] [Google Scholar]
- Rischer H, Oresic M, Seppänen-Laakso T, Katajamaa M, Lammertyn F, Ardiles-Diaz W, et al. Gene-to-metabolite networks for terpenoid indole alkaloid biosynthesis in Catharanthus roseus cells. Proc. Natl Acad. Sci. USA. 2006;103:5614–5619. doi: 10.1073/pnas.0601027103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs T, Thimann KV. The role of auxins and cytokinins in the release of buds from dominance. Amer. J. Bot. 1967;54:136–144. [Google Scholar]
- Saftner RA, Wyse RE. Effect of plant hormones on sucrose uptake by sugar beet root tissue discs. Plant Physiol. 1984;74:951–955. doi: 10.1104/pp.74.4.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai N, Ara T, Ogata Y, Sano R, Ohno T, Sugiyama K, et al. KaPPA-View4: a metabolic pathway database for representation and analysis of correlation networks of gene co-expression and metabolite co-accumulation and omics data. Nucleic Acids Res. 2011;39:D677–D684. doi: 10.1093/nar/gkq989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Kondou Y, Akiyama K, Kurotani A, Higuchi M, Ichikawa T, et al. RiceFOX: a database of Arabidopsis mutant lines overexpressing rice full-length cDNA that contains a wide range of trait information to facilitate analysis of gene function. Plant Cell Physiol. 2011;52:265–273. doi: 10.1093/pcp/pcq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E, Takahashi C, Asami T, Shimada Y. AtCAST, a tool for exploring gene expression similarities among DNA microarray experiments using networks. Plant Cell Physiol. 2011;52:169–180. doi: 10.1093/pcp/pcq185. [DOI] [PubMed] [Google Scholar]
- Schlüter U, Mascher M, Colmsee C, Scholz U, Bräutigam A, Fahnenstich H, et al. Maize source leaf adaptation to nitrogen deficiency affects not only nitrogen and carbon metabolism but also control of phosphate homeostasis. Plant Physiol. 2012;160:1384–1406. doi: 10.1104/pp.112.204420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Yan J, Xie D. Comparison of phytohormone signaling mechanisms. Curr. Opin. Plant Biol. 2011;15:84–91. doi: 10.1016/j.pbi.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Sugiyama T. Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 2001;276:26405–26410. doi: 10.1074/jbc.M102130200. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Antonio BA, Kikuchi S, Matsumoto T, Nagamura Y, Numa H, et al. The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res. 2008;36:D1028–D1033. doi: 10.1093/nar/gkm978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 2006;45:1028–1036. doi: 10.1111/j.1365-313X.2006.02656.x. [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A. ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol. 2007;48:263–277. doi: 10.1093/pcp/pcl063. [DOI] [PubMed] [Google Scholar]
- Tokunaga H, Kojima M, Kuroha T, Ishida T, Sugimoto K, Kiba T, et al. Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. 2012;69:355–365. doi: 10.1111/j.1365-313X.2011.04795.x. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Weir N, Hill K, Zhang W, Kim HJ, Shiu SH, et al. Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 2012;158:1666–1684. doi: 10.1104/pp.111.192765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, et al. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 2010;51:1821–1839. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veach YK, Martin RC, Mok DWS, Malbeck J, Vankova R, Mok MC. O-Glucosylation of cis-zeatin in maize. Characterization of genes, enzymes, and endogenous cytokinins. Plant Physiol. 2003;131:1374–1380. doi: 10.1104/pp.017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ma XM, Kojima M, Sakakibara H, Hou BK. N-glucosyltransferase UGT76C2 is involved in cytokinin homeostasis and cytokinin response in Arabidopsis thaliana. Plant Cell Physiol. 2011;52:2200–2213. doi: 10.1093/pcp/pcr152. [DOI] [PubMed] [Google Scholar]
- Weijer J. Interaction of gibberellic acid and indoleacetic acid in Impatiens. Science. 1959;129:896–897. doi: 10.1126/science.129.3353.896-a. [DOI] [PubMed] [Google Scholar]
- Weston DE, Reid JB, Ross JJ. Auxin regulation of gibberellin biosynthesis in the roots of pea (Pisum sativum) Funct. Plant Biol. 2009;36:362–369. doi: 10.1071/FP08301. [DOI] [PubMed] [Google Scholar]
- Yadav SR, Khanday I, Majhi BB, Veluthambi K, Vijayraghavan U. Auxin-responsive OsMGH3, a common downstream target of OsMADS1 and OsMADS6, controls rice floret fertility. Plant Cell Physiol. 2011;52:2123–2135. doi: 10.1093/pcp/pcr142. [DOI] [PubMed] [Google Scholar]
- Yamakawa H, Hakata M. Atlas of rice grain filling-related metabolism under high temperature: joint analysis of metabolome and transcriptome demonstrated inhibition of starch accumulation and induction of amino acid accumulation. Plant Cell Physiol. 2010;51:795–809. doi: 10.1093/pcp/pcq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, et al. Hormonal control of the shoot stem-cell niche. Nature. 2010;465:1089–1092. doi: 10.1038/nature09126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.