Abstract
A specialized orchid database, named Orchidstra (URL: http://orchidstra.abrc.sinica.edu.tw), has been constructed to collect, annotate and share genomic information for orchid functional genomics studies. The Orchidaceae is a large family of Angiosperms that exhibits extraordinary biodiversity in terms of both the number of species and their distribution worldwide. Orchids exhibit many unique biological features; however, investigation of these traits is currently constrained due to the limited availability of genomic information. Transcriptome information for five orchid species and one commercial hybrid has been included in the Orchidstra database. Altogether, these comprise >380,000 non-redundant orchid transcript sequences, of which >110,000 are protein-coding genes. Sequences from the transcriptome shotgun assembly (TSA) were obtained either from output reads from next-generation sequencing technologies assembled into contigs, or from conventional cDNA library approaches. An annotation pipeline using Gene Ontology, KEGG and Pfam was built to assign gene descriptions and functional annotation to protein-coding genes. Deep sequencing of small RNA was also performed for Phalaenopsis aphrodite to search for microRNAs (miRNAs), extending the information archived for this species to miRNA annotation, precursors and putative target genes. The P. aphrodite transcriptome information was further used to design probes for an oligonucleotide microarray, and expression profiling analysis was carried out. The intensities of hybridized probes derived from microarray assays of various tissues were incorporated into the database as part of the functional evidence. In the future, the content of the Orchidstra database will be expanded with transcriptome data and genomic information from more orchid species.
Keywords: Annotation, Expression profile, Next-generation sequencing, Orchidaceae, Transcriptome shotgun assembly
Introduction
Orchidaceae, the orchid family, diverged from the Liliaceae and Amaryllidaceae, is the largest family of Angiosperms, with >800 genera and >25,000 species. Continuous identification of new species and molecular markers (from both the chloroplast genome and repetitive sequences), combined with already complex morphological variations, means that systematic orchid classification is a never-ending pursuit that continues to change as new criteria and evidence emerge (Dressler 1993, Pridgeon et al. 1999, Chase et al. 2005, Pridgeon et al. 2005). Orchid family genomes are generally large, and vary 168-fold (1C = 0.33–55.4 pg) overall, indicating great evolutionary diversity (Leitch et al. 2009). The large size and complexity of most orchid genomes tend to hamper genomic approaches to orchid research.
The broad range of biodiversity seen among orchids provides a great opportunity for exploration of the unique and intriguing features that evolved during the adaptation of the family to various environments that are not represented by model organisms such as Arabidopsis and rice. Such features include flower pattern formation, crassulacean acid metabolism (CAM) photosynthesis to assimilate carbon at night, epiphytic habitat with high water and nutrient usage efficiency, unique seed development, symbiosis with mycorrhizae, and many others. Besides their biological novelty, orchids are also of great commercial interest. Taiwan is one of the major orchid-producing and exporting countries in the world. Together with other countries such as Japan, the USA, China, The Netherlands and countries in Southeast Asia, they share a large market of orchid trading and have built an industry of orchid nurseries maintained by advanced greenhouse facilities.
Despite being an important family in the Plantae, genomic information about orchids has been relatively scarce until recently. Rapid advances in DNA sequencing technology known as next-generation sequencing (NGS or massively parallel sequencing) in recent years have led to wide and popular applications in genomic research generating a large volume of sequence information and causing a drop in per-base cost (Wall et al. 2009, Metzker 2010). Abundant orchid sequences including our own research results have been deposited in the GenBank TSA/SRA database (transcriptome shotgun assembly/sequence read archive) since the technology has been available. Another important milestone in recent genomic research is the development of bioinformatic processes, especially for de novo assembly when the reference genome is unavailable. Various strategies, algorithms and software have been developed, resulting in rapid accumulation of sequence data of many non-model organisms in the databases (Surget-Groba and Montoya-Burgos 2010, Su et al. 2011, Zhang et al. 2011). NGS techniques are also applied to the identification and detection of small functional RNA, especially microRNA (miRNA) (Gustafson et al. 2005, Johnson et al. 2007, Simon et al. 2009). In addition, high-throughput sequencing techniques provide an alternative tool for gene expression profiling known as RNaseq (Martin and Wang 2011, Tariq et al. 2011). With developments of the technique, computation and application, NGS has revolutionized modern genomic research with rich information, high pace and low cost.
Several databases specialized to certain orchid species have been previously established with additional functional annotation. OrchidBase (URL: http://http://orchidbase.itps.ncku.edu.tw/est/home2012.aspx) stores sequences of expressed transcripts from three Phalaenopsis species, P. aphrodite, P. equestris and P. bellina, obtained using a combination of conventional Sanger sequencing and the high-throughput sequencing platforms, Roche 454 and Illumina Solexa (Fu et al. 2011). This database offers 8,501 assembled contigs and 76,116 from cDNA libraries built from various tissues, resulting in 84,617 non-redundant transcribed sequences. The sources of the cDNA library covered 11 tissues from the three species (Fu et al. 2011). Another database, OncidiumOrchidGenomeBase (URL: http://predictor.nchu.edu.tw/oogb/) was built specifically for the transcriptome sequences of Oncidium Gower Ramsey (Chang et al. 2011), a commercial hybrid. The authors applied Roche 454 pyrosequencing techniques to obtain sequence data from six tissues, including many flower stages, and generated reads for assembly. This database offers 50,908 assembled contigs and 120,219 singletons that led to the discovery of flowering-associated genes.
Although sequence information is growing in public databases, proper and sufficient annotation is often not associated with the assembled contigs or expressed sequence tags (ESTs) accessible to the public. We hope to promote orchid functional research with rich sequence information in association with functional annotation. Since genomic research often consumes a large amount of resources, focusing on a model orchid species is important for in-depth research. Our research team applied NGS technologies to obtain transcriptome shotgun sequence and developed a streamlined process for de novo assembly followed by the annotation of a potential orchid model species, P. aphrodite (Su et al. 2011).
The original Orchidstra database was constructed mainly based on the transcriptomic information including sequences and annotations of P. aphrodite derived from a previous study (Su et al. 2011). After the development of methodology for de novo assembly and annotation of transcriptome information, we expand the applications to the transcriptomic information collected from various orchid species. More genomic data such as miRNA information and expression profiles of P. aphrodite have been continuously generated by us ever since. With more sequence information available including those of multiple orchid species retrieved from the internet database, small RNA data as well as expression profiles, there is a need to update the Orchidstra database for the purposes of studies of comparative genomics and functional genomics. The Orchidstra database is now expanded and reconstructed to integrate complex genomic information.
In order to enrich useful transcriptome resources for researchers interested in orchid functional studies and comparative genomics, we downloaded the TSA/EST data of several orchid species from GenBank to carry out further analysis. Altogether, transcriptomic information from six orchids was collected in the database we built (for illustration, see Supplementary Fig. S1). All of these orchids belong to the Epidendroideae, one of the five subfamilies of Orchidaceae. Epidendroideae is the largest subfamily, with >500 genera and >20,000 species. Most members of this subfamily are tropical epiphytes, some with pseudobulbs (Dressler 1990). Many genera in this subfamily such as Phalaenopsis, Cattleya, Oncidium, Dendrobium and Cymbidium are of major commercial value in the world floral market. However, not all transcriptome sequences of orchid species in GenBank are abundant enough or their library sources cover insufficient representative plant tissues to reach the level of depth for a fair comparison. Genomic information of P. aphrodite is still the most abundant that is available to carry out a thorough analysis.
The main purpose of constructing the Orchidstra database is to share rich genomic information with researchers in order to facilitate molecular biological research, including functional studies of genes, interacting networks and regulatory mechanisms of orchid biology. The moth orchid, P. aphrodite, with abundant genomic information in our database, may serve as a research model system for exploring many interesting and unique biological features in orchids.
Database Contents
Orchid species and library source
Currently, five species and one hybrid of orchids are included in the Orchidstra database (Supplementary Fig. S1). These species represent the orchid species for which the most abundant sequence information is deposited in GenBank. They are P. aphrodite, P. equestris, P. bellina, Erycina pusilla, Dendrobium nobile and Oncidium Gower Ramsey. We generated NGS data for the two of them, P. aphrodite and E. pusilla. Sequences of the other four, P. equestris, P. bellina, D. nobile and Oncidium Gower Ramsey, were downloaded from GenBank in NCBI. A detailed description of the libraries and sequence information is given in the Materials and Methods.
Phalaenopsis aphrodite
The genus Phalaenopsis (also known as moth orchids) belonging to tribe Vandeae contains >60 species and is an important source of breeding parents for commercial hybrids. The Taiwan native P. aphrodite is diploid (2n = 2x = 38) with a genome size estimated as about 1C = 1.4 pg (Lin et al. 2001). The transcriptome sequence of this Phalaenopsis species was obtained by assembly of reads from high-throughput sequencing techniques, namely Roche 454 and Illumina Solexa (Su et al. 2011). Phalaenopsis aphrodite is an epiphyte with a CAM photosynthesis pattern and has large white flowers that are of interest to orchid breeders. Phalaenopsis is indigenous throughout most of Southeast Asia and Australia.
Phalaenopsis equestris
Also native to Taiwan, P. equestris is closely related to P. aphrodite, with a similar geological distribution. The diploid P. equestris (2n = 2x = 38) has a genome size estimated to be 1C = 1.69 pg (Lin et al. 2001). The characteristic large number of small flowers on the stalk of P. equestris is also of interest to breeders and it is often used as a breeding parent. The transcript sequences of P. equestris and P. bellina have been published (Tsai et al. 2006), and were downloaded from NCBI and annotated in-house.
Phalaenopsis bellina
Phalaenopsis bellina (2n = 2x = 38) is one of few Phalaenopsis with fragrance and has a genome size of 1C = 7.52 pg (Lin et al. 2001).
Oncidium Gower Ramsey
Oncidium is a genus with about 330 species under Tribe Cymbidieae, Subtribe Oncidiinae. Oncidium has a genome size of 1C = 0.6–5.73 pg (L. Hanson, I.J. Leitch and M.D. Bennett, unpublished data from the Jodrell Laboratory, Royal Botanic Gardens, Kew, 1999). Oncidium spp. and hybrids are important in the flower market mostly as cut flowers. Oncidium Gower Ramsey is a popular commercial hybrid with a complex breeding background. Sequence information of this hybrid was mainly contributed by the group building the OncidiumOrchidGenomeBase database (Chang et al. 2011) and some ESTs contributed by other researchers.
Erycina pusilla
Erycina is closely related to the genus, Oncidium, and also belongs to the Tribe Cymbidieae, Subtribe Oncidiinae. Erycina has a different morphology and physiology from Phalaenopsis orchids. Erycina is mainly distributed in tropical America and has a diploid (2n = 2x = 10) genome of 1C = 1.5 pg (Chase et al. 2005). Transcriptome information for Erycina was built in the same way as we built it for P. aphrodite. Erycina has the advantage of small plant size, short life cycle and year-round blooming, suggesting its suitability for use as a model orchid.
Dendrobium nobile
The genus Dendrobium belongs to the Tribe Podocjilaeae, Subtribe Dendrobiinae. There are about 1,200 species in the genus Dendrobium, distributed throughout most of Southeast Asia and the Southwest Pacific islands. Dendrobium has a genome size of 1C = 0.75–5.85 pg (Jones et al. 1998). The Dendrobium sequence was obtained from NCBI (Liang et al. 2012). A collection of 15,017 ESTs from the vernalized axillary buds and vegetative tissues of D. nobile were assembled for 9,616 unique gene clusters (Liang et al. 2012). Dendrobium spp. is known to botanists for both its value in the ornamental flower market and its use in traditional herbal medicine.
Data processing and contents
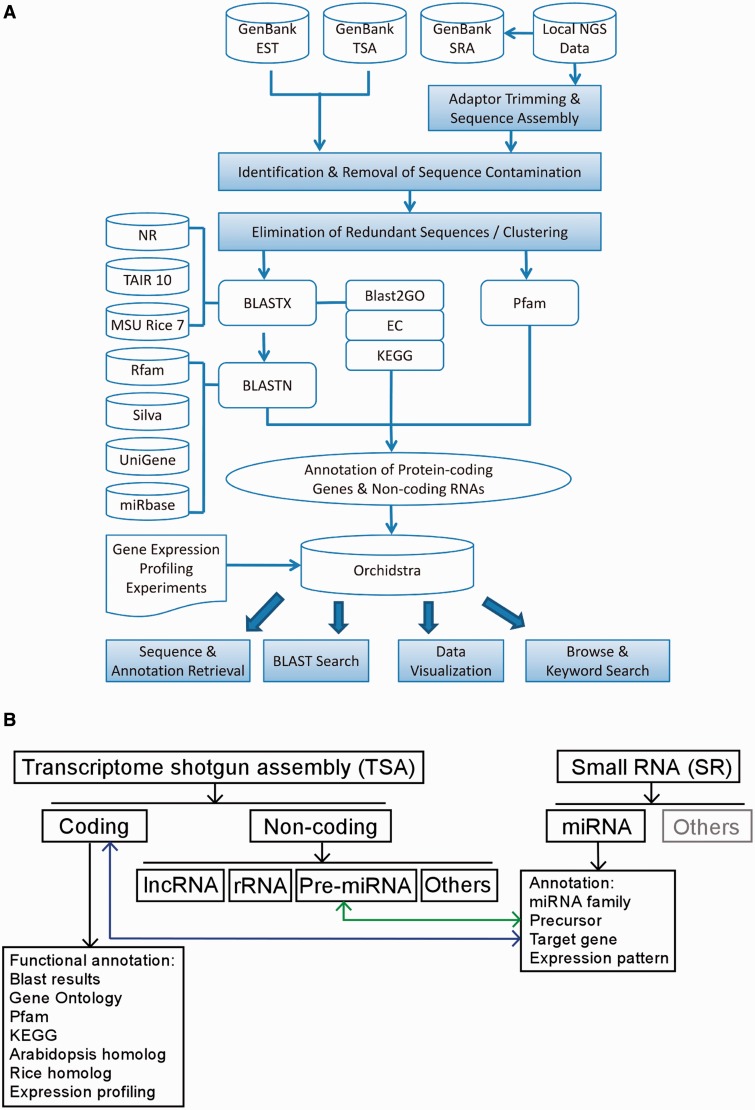
The outline of the analysis process pipeline is shown in a flow chart (Fig. 1A). TSAs of contigs or singletons >200 bp were first searched by BlastX against the NCBI nr database for potential open reading frames and similarity to currently known genes. Altogether, 381,918 non-redundant TSAs were stored in the Orchidstra database and divided into 114,933 protein-coding and 266,985 non-coding TSAs (Table 1). The protein-coding TSAs are annotated with terms identified in Gene Ontology (GO; Gene Ontology Consortium 2013), Pfam (Finn et al. 2010) and KEGG (Kyoto Encyclopedia of Genes and Genomes; Tanabe and Kanehisa 2012) (Table 2). Annotation procedures were performed as described previously (Su et al. 2011). Corresponding homologs to Arabidopsis and rice are also listed, with E-values and identity to indicate sequence similarity.
Fig. 1.
Overview of the information process pipeline for next-generation sequencing data. (A) Flow chart of the sequence data process pipeline (for details see the Materials and Methods). (B) Data content of Phalaenopsis aphrodite in the Orchidstra. Expressed transcripts (TSA for transcriptome shotgun assembly) were cross-linked to miRNA (SR for small RNA) by the target genes and precursors identification.
Table 1.
Statistics of transcriptome shotgun assemblies (TSAs) in the Orchidstra database
| Orchid species | No. of coding TSAs | No. of nc TSAsa | No. of total TSAs | Average lengthb (bp) | N50c (bp) | Tissue sourced | Data sourcee | Expression profiling |
|---|---|---|---|---|---|---|---|---|
| Phalaenopsis aphrodite | 42,573 | 191,233 | 233,806 | 875 | 405 | R, L, S, F, PE, ST, FB, IN, PC | TSA | Yes |
| Erycina pusilla | 31,515 | 51,550 | 83,065 | 783 | 533 | R, L, F, PE | TSA | NA |
| Oncidium Gower Ramsey | 26,786 | 20,900 | 47,686 | 668 | 619 | L, F, PB, IN, FB | TSA, cDNA | NA |
| Dendrobium nobile | 10,515 | 3,302 | 13,817 | 639 | 669 | L, S, AB | cDNA | NA |
| Phalaenopsis equestris | 2,401 | 0 | 2,401 | 631 | 684 | FB | cDNA | NA |
| Phalaenopsis bellina | 1,143 | 0 | 1,143 | 686 | 740 | FB | cDNA | NA |
| Total | 114,933 | 266,985 | 381,918 | 773 | 494 |
a nc TSA, non-coding TSA.
b Average length of the nucleotide sequence of protein-coding TSAs.
c N50 of total assembled TSAs.
d Code for tissue sources: R, root; L, leaf; S, stem; F, open flower; PE, pedicel; ST, stalk; IN, inflorescence; FB, flower bud; PB, pseudobulb; AB, auxiliary bud; PC, protocorm.
e Data source: TSA, transcriptome shotgun assembly sequence; cDNA, reverse transcriptase-mediated cDNA library.
Table 2.
Statistics of functional affiliates in the Orchidstra database
| Orchid species | No. of coding TSAs | No. of TSAs with |
||||
|---|---|---|---|---|---|---|
| Pfam | GO | KEGG | Rice homolog | At homolog | ||
| Phalaenopsis aphrodite | 42,573 | 24,084 | 16,701 | 15,216 | 23,002 | 24,205 |
| Erycina pusilla | 31,515 | 20,731 | 16,229 | 15,932 | 22,686 | 21,833 |
| Oncidium Gower Ramsey | 26,786 | 12,189 | 24,283 | 12,562 | 16,671 | 18,579 |
| Dendrobium nobile | 10,515 | 7,429 | 7,706 | 1,388 | 7,981 | 7,761 |
| Phalaenopsis equestris | 2,401 | 1,667 | 1,335 | 1,675 | 1,852 | 1,805 |
| Phalaenopsis bellina | 1,143 | 839 | 746 | 1,093 | 949 | 934 |
| Total | 114,933 | 66,939 | 67,000 | 47,866 | 73,141 | 75,117 |
Rice homologs were obtained from Blast against MSU Rice Genome Annotation Project Release 7, and At (Arabidopsis thaliana) homologs were from the Arabidopsis TAIR10 release.
The database we built was named Orchidstra (URL: http://orchidstra.abrc.sinica.edu.tw), a combination of the words ‘orchid’ and ‘orchestra’, to represent the harmonious interplay among collections of genes to bring about the beauty of orchids. Orchidstra is a web-based open-access database with value-added annotations including gene expression profiling as functional evidence. TSA sequences, gene descriptions and functional annotation such as GO, KEGG and Pfam were assigned to protein-coding TSAs.
Structural RNAs including rRNA, tRNA, small nuclear RNA (snRNA) and small nucleolar RNA (snoRNA) were identified and separated from mRNA with Rfam (http://rfam.sanger.ac.uk/) and the Silva database (http://www.arb-silva.de/). Phalaenopsis aphrodite long non-coding RNAs (lncRNAs) that had a high degree of nucleotide sequence homology with many other plant species were identified by multiple sequence alignment and were grouped together in the database. Precursors of miRNA were also discovered in the non-coding TSAs using a small RNA analysis pipeline.
In addition to long expressed transcripts, analysis of deep sequencing results of small RNA for P. aphrodite provides miRNA annotation, precursors and putative target genes for this species. Small RNA was isolated from leaf, root, flower and germinating seeds (protocorm) of P. aphrodite and subjected to Solexa for sequencing. We identified 3,251 P. aphrodite miRNA sequences for 88 publicly known plant miRNA families gathered from GenBank SRA050114. Each miRNA has its own page to display annotation together with the expression level in various tissues, precursors and predicted target genes; if applicable, the relevant internal links to corresponding TSA data (including precursors and target genes) are also provided. Sequences from TSA and small RNA are cross-linked by the identification of precursor and target genes (Fig. 1B).
Gene expression profiling
To enrich functional annotation for P. aphrodite, we included the results of microarray analysis in the transcript information in the Orhcidstra database. The resulting expression profiles generated from hybridization of probes labeled by RNA extracted from various tissues were shown in a heat map pattern to indicate their relative expression level with a color-coded gradient. This information should be useful for determining tissues for amplification of genes of interest by PCR.
Accessory tools for analysis
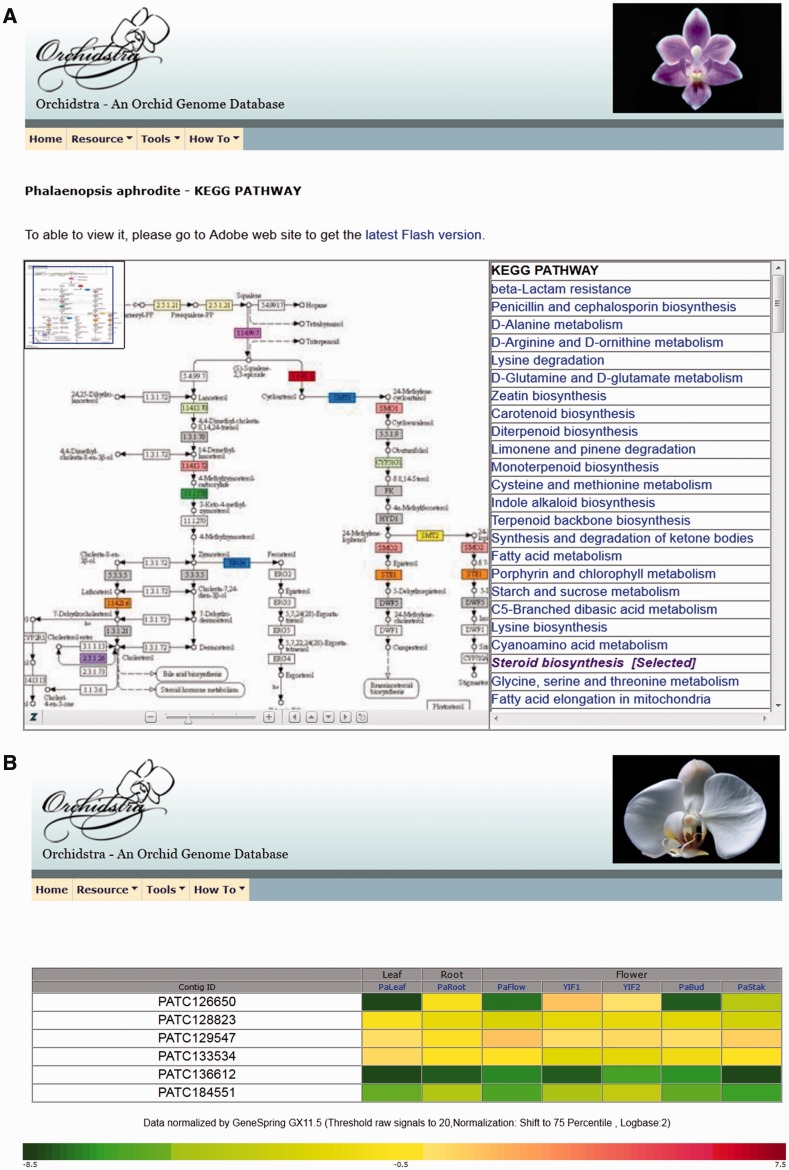
In order to create a user-friendly environment, useful accessory functions are provided on the front page or under the Tool bar (Supplementary Fig. S1). Users have options to initiate the search for a gene of interest. First, a search box on the home page provides a quick choice of species, type of transcripts and keyword. Secondly, an advanced search in the ‘Tools’ provides more detailed criteria for finding the TSA. Thirdly, one can use Blast, also in the ‘Tools’, to find the TSA if the homologous sequence is available. Orchidstra integrates the BLAST utility by providing a web-based interface to search against the local sequence databases of all species in Orchidstra. This function can easily be executed and navigated. In addition, an advanced search function provides improved data search accuracy with input keywords and related information. Users can also browse GO terms or KEGG pathways (EC or K number from the KEGG database, http://www.genome.jp/kegg/) in the database. GO analysis charts and KEGG pathways are also available in graphics (Fig. 2A). Pfam domains can be directly linked to the Sanger Institute Pfam database (http://pfam.sanger.ac.uk/) by the corresponding protein family (PF) number for a more detailed description.
Fig. 2.
Examples of functional annotation in the Orchidstra database. (A) KEGG pathway in graphic view. Steroid biosynthesis is demonstrated here. The EC number within a colored box indicates genes with P. aphrodite identity. (B) Comparison of expression profiles of multiple genes shows tissue expression patterns.
Besides a functional annotation search, Orchidstra provides a visual display of microarray data using a color chart to show relative intensities of signals among tissues. Users can use the function of ‘Expression profile’ under the ‘Tools’ to input multiple sequence contig IDs to generate an integrated expression profile (Fig. 2B).
Cross-species comparison
Many of the genomic databases were comprised of information on a single species as a model organism for research. The Arabidopsis 1001 genome project was intended for whole-genome sequencing of 1,001 Arabidopsis ecotypes (Cao et al. 2011, Schneeberger et al. 2011) and to build a database for comparative purposes (http://www.1001genomes.org/index.html). Another example is the Sol Genomics Network (http://solgenomics.net/) which consists of genomic information on tomato, tobacco, potato, petunia and pepper, all within the Solanaceae family. Information including sequences of genomes and transcriptomes, mutant phenotypes and available lines, quantitiative loci and markers, and many features are present in the Sol Genomics Network for comprehensive analysis (Bombarely et al. 2011).
Orchidstra was designed to integrate information across orchid species and to serve as a general reference source in support of orchid functional genomics research. Since orchids adapt to widely distributed habitats and evolve into a large number of species, it is rational to believe frequent events of gene variation and natural selection may have occurred. Incorporation of transcriptome data from multiple orchid species can broaden the information base available for comparative analysis within and between species, especially in the absence of a reference genome for most orchids.
Due to incomplete transcriptome information of the current database collection including an uneven depth of sequencing efforts and various tissues from where RNA samples were extracted, it is difficult to make an overall comparison to distinguish genes of those commonly shared among plants or uniquely owned by certain species (Table 1).
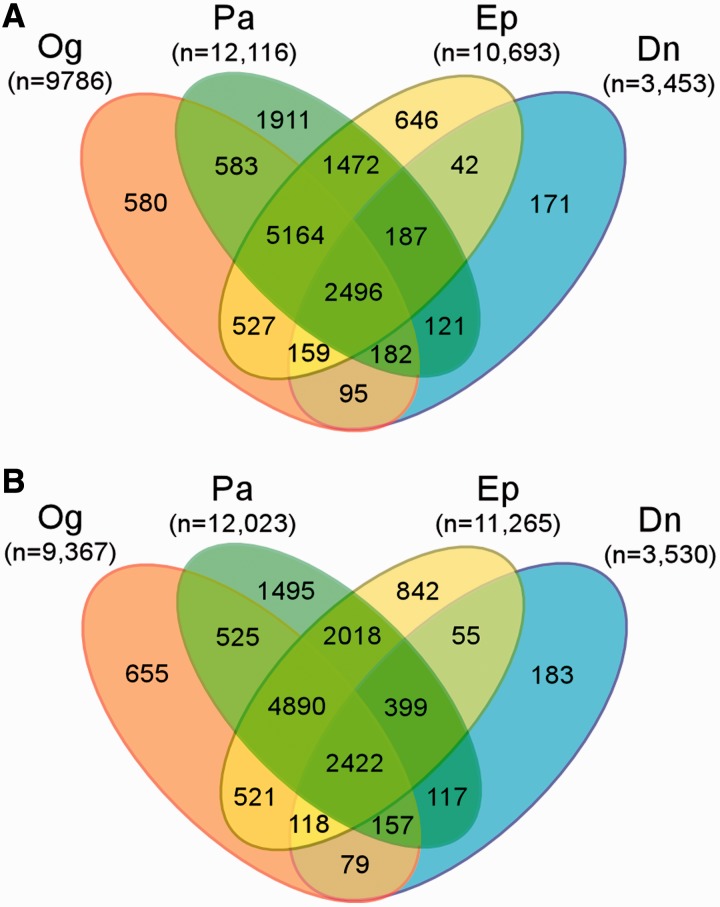
A Venn diagram was plotted for comparing genes in common among orchid species. The number of homologous TSA sequences derived from sequence alignment to Arabidopsis and rice was compared (Fig. 3A, B). Phalaenopsis equestris and P. bellina were excluded from this analysis because of their small sample sizes in TSA number. The number of homologs from D. nobile is also low for comparison, although not excluded. Homologs commonly shared among P. aphrodite, E. pusilla and Oncidium Gower Ramsey (Fig. 3; 2,496 Arabidopsis homologs and 2,422 for rice) make up approximately 20% of the total coding TSAs. These TSAs are probably responsible for the fundamental physiology found across higher plant species. From 12% to 16% of the homologs in P. aphrodite are unique. This may reflect rich sequence information including the number and length of the assembled contigs derived from P. aphrodite. The interspecies variation in number of homologs may reflect a combination of differences in sequencing depth and tissue sources of transcriptome, and genome variations between species.
Fig. 3.
Comparison of Arabidopsis and rice homologs of expressed TSAs among four orchid species in the Orchidstra database. (A) Number of Arabidopsis homologs. (B) Number of rice homologs. The number in brackets indicates the total number of homologs found in the species. Pa, Phalaenopsis aphrodite; Ep, Erycina pusilla; Og, Oncidium Gower Ramsey; and Dn, Dendrobium nobile.
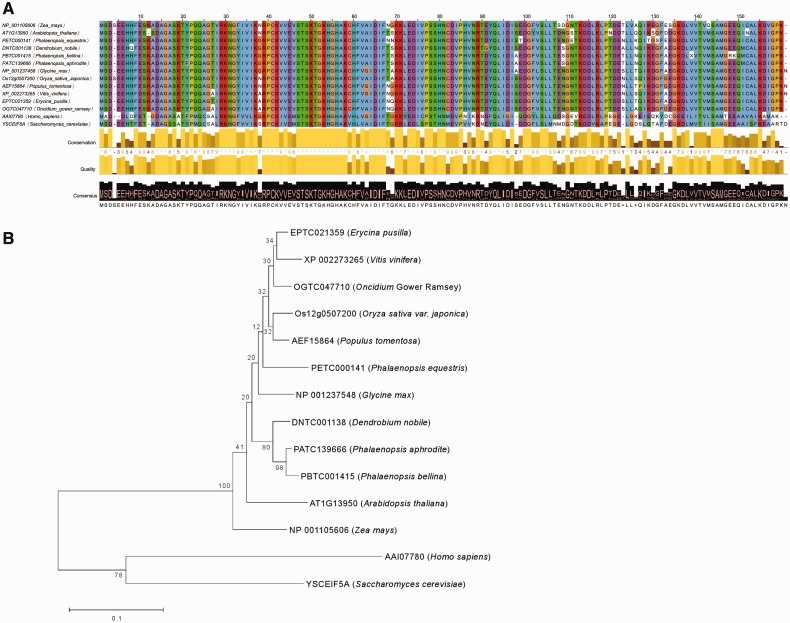
We selected the eukaryotic translation initiation factor 5A (EIF5A) gene as an example to demonstrate the usage of the Orchidstra database for phylogeny analysis. The amino acid identities of EIF5A between plants we selected are high, ranging from 84% to 97% (Fig. 4A). Phylogeny analysis indicated that these EIF5A orthologs are highly conserved among plant species and that sequence diversity exists among orchids (Fig. 4B).
Fig. 4.
Sequence comparisons of EIF5A genes. (A) Multiple sequence alignment reveals high amino acid identity of EIF5A between plant species. (B) Phylogenetic analysis of EIF5A from various species. Contig id is used for EIF5A of orchid species that can be found in the Orchidstra database, while other species are given a GenBank id.
Conclusions and Future Implementation
Orchid biology research has gained momentum in recent years due to significant value of the commercial market and the unique biological features exhibited by orchids attracting researcher’s interest. Technological developments such as molecular tools and availability of genomic sequence information have also contributed to the research progress. Whether to facilitate biotechnological development for horticultural purposes or for fundamental research purposes, easy access to rich genomic information about the orchid family will facilitate in-depth research on the molecular function and regulation of genes as well as areas of evolutionary and ecological interest. NGS technology has successfully reduced the cost and effort required for fast accumulation of sequence information. Here, we built an informatics data processing pipeline to assemble reads into contigs and functionally annotate genes from de novo organisms effectively and constructed a database, Orchidstra, which can serve as a gateway to access information about the abundance of genes expressed in orchid species.
In the future, we intend to generate and collect more sequence information from other orchid species especially in different subfamilies. Since there are a great number of species in the orchid family, it will be intriguing to understand the phylogeny of gene families during the course of their evolution. Providing more transcript sequences from species in different orchid subfamilies should be helpful in promoting orchid comparative genomic studies, and a comprehensive orchid genomic information database should facilitate molecular research on gene functions and regulatory mechanisms of the many interesting biological features of orchids.
Materials and Methods
Library construction and data sources
Phalaenopsis aphrodite
Tissue source: vegetative (roots, stem, leaf); reproductive (stalk, flower buds, young inflorescences, flowers of full blossom and senescence); germinating seed (protocorm formation, protocorm development and seedling formation)
Library construction and data source: Su et al. (2011)
Sequencing techniques: Illumia Solexa and Roche 454 platform
NCBI accession numbers: SRA030409, SRA050114; TSA: JI626343–JI831113
Sequences obtained: 246,242 contigs (from SRA030409) and 22,829,317 unique reads (from SRA050114)
Phalaenopsis equestris
Tissue source: flower bud
Library construction and data source: Tsai et al. (2006)
Sequencing technique: Sanger sequencing
NCBI accession numbers: CK855526–CK857579, CB031751–CB035289, BU744268–BU744277, CK901119
Sequences obtained: 2,455 ESTs (after data clean up)
Phalaenopsis bellina
Tissue source: flower bud
Library construction and data source: Tsai et al. (2006)
Sequencing technique: Sanger sequencing
NCBI accession numbers: CK857580–CK859399, CO742089–CO742627
Sequence obtained: 1,208 ESTs (after data clean up)
Erycina pusilla
Tissue source: leaf, root, flower, pedicel
Library construction and data source: unpublished data
Sequencing technique: Illumia Solexa and Roche 454 platform
NCBI accession number: SRA037585.1
Sequences obtained: 88,203 contigs
Dendrobium nobile
Tissue source: vegetative (auxiliary bud) and seedling (leaf and stem)
Library construction and data source: Liang et al. (2012)
Sequencing technique: Sanger sequencing
NCBI accession numbers: HO189246–HO204626, JQ063042, JQ063043, JQ063457, JQ063458, JQ063459, JQ063460, AY608889, DQ462460, DQ462469, EF535598, EF535599, GR410230, GR410231, GU357498, GU382674, GU382675, HQ388352
Sequences obtained: 15,398 ESTs
Oncidium Gower Ramsey
Tissue source: leaf, pseudobulbs, young inflorescences, inflorescences, flower buds, mature flowers
Library construction and data source: Chang et al. (2011)
Sequencing technique: Roche 454 and Sanger sequencing
NCBI accession numbers: HS521830–HS524732 and JL898334–JL943742; AF276233, AF276234, AF276235, AF276236, AF276237, AY196350, AY496865, AY940147, AY940148, AY953937, AY953938, AY953939, AY973631, AY973632, AY973633, AY973634, AY974325, AY974326, AY974327, DQ289592, DQ289593, DQ289594, DQ289595, DQ302727, EF570111, EF570112, EF570113, EF570114, EF570115, EF570116, EU130454, EU130455, EU130456, EU130457, EU130458, EU130459, EU130460, EU583501, EU583502, FJ237035, FJ237036, FJ237037, FJ237038, FJ237040, FJ348573, FJ618566, FJ618567, FJ859988, FJ859989, FJ859990, FJ859991, FJ859993, FJ859994, FJ859995, FJ859996, HM140840, HM140841, HM140842, HM140843, HM140844, HM140845, HM140846, HM140847, HM146076, HM146077, HQ585983, HQ585984, HQ591455
Sequences obtained: 48,380 contigs
Data processing—sequence analysis and functional annotation
Raw data were obtained from three sources: local NGS data for whole transcriptomes and small RNA transcriptomes, GenBank ESTs (Benson et al. 2012) and GenBank TSAs (for the process pipeline, see Fig. 1A). After assembly, sequences with high similarity to sequences of potential ‘contaminants’ such as bacteria, viruses and chloroplasts were removed prior to annotation. Blast2GO was incorporated into the autoannotation pipeline for functional annotation and possible pathway analysis (Gotz et al. 2008). After annotation procedures, description of the best BLAST (Altschul et al. 1990) hit, the GO terms (Gene Ontology Consortium 2013), Pfam domains (http://pfam.sanger.ac.uk/) (Finn et al. 2010), enzyme codes and corresponding KEGG pathway (http://www.genome.jp/kegg/) (Tanabe and Kanehisa 2012) were assigned to every protein-coding TSA that meets the specified threshold. Non-coding RNAs were annotated according to the similarity search of the public database for non-coding RNA families and structured RNA elements including Rfam (http://rfam.sanger.ac.uk/), Silva (http://www.arb-silva.de/) and miRBase (http://www.mirbase.org/) (Griffiths-Jones et al. 2008). Non-coding RNAs showing a high degree of similarity between various species were identified using UniGene data. The sequence annotation information and data from microarray experiments were included in the database. The protein-coding TSAs and non-coding RNAs in Orchidstra can be accessed and analyzed by various online tools and services, including a variety of keyword search or browse options, visualization of microarray profiling of various tissues and a BLAST server for database search. The sequences and annotations can be retrieved directly from the database.
Database construction
Nucleotide sequences in fasta format, blast results, annotations including GO results, KEGG results, Pfam and homolog search results were stored. To access these genomic resources in a user-friendly and accurate way, a customized database was designed and built, Orchidstra. Orchidstra is a web application built using three-tier architecture. It runs with the Apache Web server and MySQL database in Linux OS. PHP and JavaScript scripts were used to create the user interface coupled with MySQL, a relational database management system. The URL address of the Orchidstra database is http://orchidstra.abrc.sinica.edu.tw. Current features include resource browsing and online tools such as searching, blasting and linking to corresponding Pfam, GO and KEGG.
Expression profiling
Expression profiles of P. aphrodite were analyzed using a custom-made orchid microarray. This microarray was designed based on the transcriptome sequences we obtained from high-throughput sequencing and printed on glass slides by Agilent eArray and the SurePrint platform (Agilent). The first version of the orchid biochip featured 67,038 probes from 43,662 annotated genes and two popular orchid viruses, Ondontoglossum ringspot virus (ORSV) and Cymbidium mosaic virus (CymMV). RNAs isolated from root, leaf, full-bloom flowers, flower buds, stalk, young inflorescences with scale leaves, and small buds were labeled and hybridized to the microarray. Signals detected were globally normalized and compared using GeneSpring GX7.3 software (Agilent). The intensities of the signals were displayed on a color-coded chart for the convenience of viewers.
Phylogenetic analysis
The amino acid sequence of the EIF5A gene used in the phylogeny analysis was downloaded from the Orchidstra database or NCBI GenBank. The sequence was aligned by Mega5 (Tamura et al. 2011) with the Neighbor–Joining method (Saitou and Nei 1987). The result of the phylogeny tree was evaluated by bootstrap resampling of 1,000 replicates (Felsenstein 1985) and the evolutionary distances were computed using the Poisson correction method.
System requirements
The Orchidstra database is supported by the latest versions of Microsoft Internet Explorer, Mozilla Firefox and Apple Safari. In order to experience the Orchidstra website fully, we suggest that you upgrade to a recent browser such as Microsoft Internet Explorer 8 or later, Mozilla Firefox 9 and Apple Safari 5. We recommend that the best resolution for browsing is 1,024 × 768. In addition, JavaScript is used on this website, thus enabling Javascript 1.2 or later is recommended.
Definition of terms
In this manuscript, TSA is used to describe expressed transcript sequences from in-house-generated or downloaded reads including ESTs and NGS reads (SRA) that were assembled into unique and contiguous sequences. In the Orchidstra database, a TC (for transcript contig) is initialed and placed in front of the identification number. The authors wish to clarify that the TC here does not stand for ‘tentative consensus’ as usually recognized for the clustering procedure of UniGene or Gene index formation. Methods used in our de novo assembly procedure for assembling transcriptomic shotgun reads is different from clustering as described previously in the Materials and Methods. For example, in P. aphrodite, PATC was used to prefix the id number of assembled transcripts and, in E. pucilla, EPTC was used. Small RNA (SR) is used to describe short sequences derived from small RNA deep sequencing after sequencing data are cleaned up. For example, PASR represents small RNA id in P. aphrodite.
We defined the protein-coding TSAs as the transcript assemblies which matched against the NCBI nr database with an E-value of ≤1e-10, as well as the non-coding transcripts with an E-value of >1e-10. Relative degrees of homology from Blast results were applied to assign gene identity for orchid TSAs using terms such as homolog, similar to, weakly similar to and putative protein (Su et al. 2011).
Homologs to Arabidopsis and rice were defined when the TSAs were submitted to BlastX against the protein databases of Arabidopsis TAIR10 (Lamesch et al. 2012) and MSU Rice Genome Annotation Project Release 7 (Ouyang et al. 2007) with an E-value ≤1e-20.
Supplementary data
Supplementary data are available at PCP online.
Disclaimer
The authors accept no liability for the accuracy of the sequence or expression information. Validation by users is strongly encouraged.
Funding
This work was supported by Academia Sinica [under the Development Program of Industrialization for Agricultural Biotechnology (http://dpiab.sinica.edu.tw/index_en.php), (grant No. 098S0311)]; the National Science Council.
Supplementary Material
Acknowledgements
The authors would like to express their gratitude to Dr. Tsai-Mu Shen of the National Chia-Yi University, for providing plant material of native Phalaenopsis aphrodite collected from Da-Wu Mountain in Taiwan, and Dr. Chang Chen from the National Chung Hsing University and Dr. Choun-Sea Lin from Academia Sinica for providing the Erycina pusilla materials. High-throughput sequencing work was performed by the NGS core facility in Academia Sinica led by Dr. Mei-Yeh Lu. Dr. Tzyy-Jen Chiou and Dr. Ho-Ming Chen, both from Academia Sinica, provided precious advice related to small RNA (especially miRNA and tasiRNA).
Glossary
Abbreviations
- BLAST
basic local alignment search tool
- CAM
crassulacean acid metabolism
- EIF5A
eukaryotic translation initiation factor 5A
- EST
expressed sequence tag
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- miRNA
microRNA
- NGS
next-generation sequencing
- SRA
sequence read archive
- TSA
transcriptome shotgun assembly.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2012;40:D48–D53. doi: 10.1093/nar/gkr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, et al. The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res. 2011;39:D1149–D1155. doi: 10.1093/nar/gkq866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Gunther T, Bender S, Fitz J, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 2011;43:956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- Chang YY, Chu YW, Chen CW, Leu WM, Hsu HF, Yang CH. Characterization of Oncidium ‘Gower Ramsey’ transcriptomes using 454 GS-FLX pyrosequencing and their application to the identification of genes associated with flowering time. Plant Cell Physiol. 2011;52:1532–1545. doi: 10.1093/pcp/pcr101. [DOI] [PubMed] [Google Scholar]
- Chase MW, Hanson L, Albert VA, Whitten WM, Williams NH. Life history evolution and genome size in subtribe Oncidiinae (Orchidaceae) Ann. Bot. 2005;95:191–199. doi: 10.1093/aob/mci012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler RL. Cambridge, MA: Harvard University Press; 1990. The Orchids, Natural History and Classification. [Google Scholar]
- Dressler RL. Cambridge, UK: Cambridge University Press; 1993. Phylogeny and Classification of the Orchid Family. [Google Scholar]
- Felsenstein J. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu CH, Chen YW, Hsiao YY, Pan ZJ, Liu ZJ, Huang YM, et al. OrchidBase: A collection of sequences of transcriptome derived from orchids. Plant Cell Physiol. 2011;52:238–243. doi: 10.1093/pcp/pcq201. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium. Gene Ontology annotations and resources. Nucleic Acids Res. 2013;41:D530–D535. doi: 10.1093/nar/gks1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson AM, Allen E, Givan S, Smith D, Carrington JC, Kasschau KD. ASRP: the Arabidopsis Small RNA Project Database. Nucleic Acids Res. 2005;33:D637–640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Bowman L, Adai AT, Vance V, Sundaresan V. CSRDB: a small RNA integrated database and browser resource for cereals. Nucleic Acids Res. 2007;35:D829–D833. doi: 10.1093/nar/gkl991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WE, Kuehnle AR, Arumuganathan K. Nuclear DNA content of 26 orchid (Orchidaceae) genera with emphasis on Dendrobium. Ann. Bot. 1998;82:189–194. [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch IJ, Kahandawala I, Suda J, Hanson L, Ingrouille MJ, Chase MW, et al. Genome size diversity in orchids: consequences and evolution. Ann. Bot. 2009;104:469–481. doi: 10.1093/aob/mcp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Ye QS, Li RH, Leng JY, Li MR, Wang XJ, et al. Transcriptional regulations on the low-temperature-induced floral transition in an Orchidaceae species, Dendrobium nobile: an expressed sequence tags analysis. Comp. Funct. Genomics. 2012;2012:757801. doi: 10.1155/2012/757801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Lee HC, Chen WH, Chen CC, Kao YY, Fu YM, et al. Nuclear DNA contents of Phalaenopsis sp and Doritis pulcherrima. J. Amer. Soc. Hortic. Sci. 2001;126:195–199. [Google Scholar]
- Martin JA, Wang Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 2011;12:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies—the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN. Oxford: Oxford University Press; 1999. Genera Orchidacearum Vol. 1 General Introduction, Apostasioideae, Cypripedioideae. [Google Scholar]
- Pridgeon, A.M., Cribb, P.J., Chase, M.W. and Rasmussen, F.N., eds. (2005) Genera Orchidacearum: Epidendroideae (part one). Oxford University Press, Oxford.
- Saitou N, Nei M. The Neighbor–Joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Ott F, Klein JD, Wang X, Lanz C, et al. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc. Natl Acad. Sci. USA. 2011;108:10249–10254. doi: 10.1073/pnas.1107739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon SA, Zhai J, Nandety RS, McCormick KP, Zeng J, Mejia D, et al. Short-read sequencing technologies for transcriptional analyses. Annu. Rev. Plant Biol. 2009;60:305–333. doi: 10.1146/annurev.arplant.043008.092032. [DOI] [PubMed] [Google Scholar]
- Su CL, Chao YT, Alex Chang YC, Chen WC, Chen CY, Lee AY, et al. De novo assembly of expressed transcripts and global analysis of the Phalaenopsis aphrodite transcriptome. Plant Cell Physiol. 2011;52:1501–1514. doi: 10.1093/pcp/pcr097. [DOI] [PubMed] [Google Scholar]
- Surget-Groba Y, Montoya-Burgos JI. Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Res. 2010;20:1432–1440. doi: 10.1101/gr.103846.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe M, Kanehisa M. Using the KEGG database resource. Curr. Protoc. Bioinformatics. 2012 doi: 10.1002/0471250953.bi0112s38. Chapter 1: Unit1 12. [DOI] [PubMed] [Google Scholar]
- Tariq MA, Kim HJ, Jejelowo O, Pourmand N. Whole-transcriptome RNaseq analysis from minute amount of total RNA. Nucleic Acids Res. 2011;39:e120. doi: 10.1093/nar/gkr547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WC, Hsiao YY, Lee SH, Tung CW, Wang DP, Wang HC, et al. Expression analysis of the ESTs derived from the flower buds of Phalaenopsis equestris. Plant Sci. 2006;170:426–432. [Google Scholar]
- Wall PK, Leebens-Mack J, Chanderbali AS, Barakat A, Wolcott E, Liang H, et al. Comparison of next generation sequencing technologies for transcriptome characterization. BMC Genomics. 2009;10:347. doi: 10.1186/1471-2164-10-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Chen J, Yang Y, Tang Y, Shang J, Shen B. A practical comparison of de novo genome assembly software tools for next-generation sequencing technologies. PLoS One. 2011;6:e17915. doi: 10.1371/journal.pone.0017915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.