Abstract
Planarians are flatworms capable of regenerating all body parts. Planarian regeneration requires neoblasts, a population of dividing cells that has been studied for over a century. Neoblast progeny generate new cells of blastemas, which are the regenerative outgrowths at wounds. If the neoblasts comprise a uniform population of cells during regeneration (e.g. they are all uncommitted and pluripotent), then specialization of new cell types should occur in multipotent, non-dividing neoblast progeny cells. By contrast, recent data indicate that some neoblasts express lineage-specific transcription factors during regeneration and in uninjured animals. These observations raise the possibility that an important early step in planarian regeneration is the specialization of neoblasts to produce specified rather than naïve blastema cells.
Keywords: Blastema, Neoblast, Planarian, Regeneration, Stem cell
Introduction
Planarians are freshwater flatworms with the ability to regenerate any missing body part (Reddien and Sánchez Alvarado, 2004). Planarians have a centralized nervous system with brain and nerve cords, a peripheral nervous system, eyes, intestine, epidermis, an excretory system, musculature and many other cell types (Hyman, 1951). Because a small fragment of the planarian body can regenerate an entire animal (Reddien and Sánchez Alvarado, 2004) mechanisms must exist in the adult for the production of all of these cell types and organ systems. These dramatic regenerative abilities, combined with the successful development of molecular tools and resources for planarian research, have helped planarians emerge as a powerful molecular genetic system for the investigation of regeneration (Newmark and Sánchez Alvarado, 2002; Sánchez Alvarado, 2006; Reddien, 2011; Elliott and Sánchez Alvarado, 2012).
The neoblast concept
The cellular source for new tissues in adult injured animals is a central problem in understanding regeneration (Tanaka and Reddien, 2011). Planarian regeneration requires a population of proliferative cells, called neoblasts, which have been studied for over a century (Keller, 1894; Baguñà, 2012). Not only do planarians regenerate, but adult animals also constantly replace aged differentiated cells. This process of tissue turnover, which is essential for viability, also requires neoblasts (Pellettieri and Sánchez Alvarado, 2007). The long-term maintenance of neoblasts as a cell population in adult planarians and the ability of neoblasts to produce differentiated cells suggest that the neoblast population contains stem cells (Fig. 1). Neoblasts are an attractive cell population for study of the cellular basis of regeneration and tissue turnover, and for investigation of stem cell biology, and consequently have recently become the subject of extensive molecular investigation (Shibata et al., 2010; Aboobaker, 2011; Rink, 2012). In this article, I focus on two possible models for how neoblasts produce replacement parts: naïve and specialized neoblast models (see Fig. 2). These models are not entirely mutually exclusive. However, understanding which of these models most closely reflects how cell fate specification occurs in planarian regeneration has important implications for understanding regeneration and the regeneration blastema. First, however, it is important to consider exactly what a neoblast is.
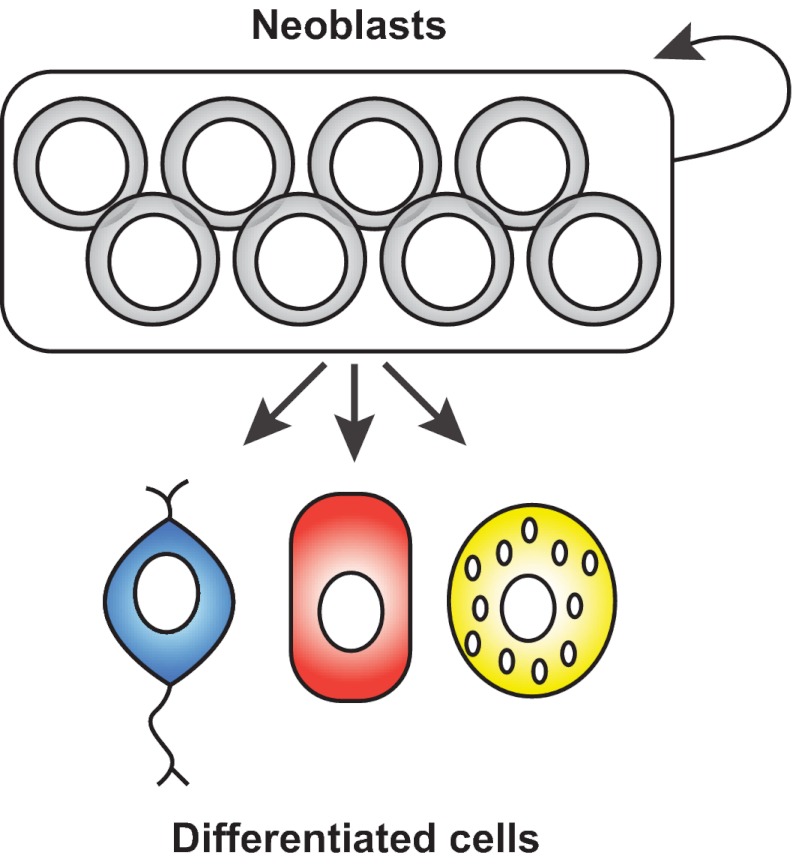
Fig. 1.
Neoblasts are the source of new cells in planarian regeneration. Neoblasts are a population of all adult dividing cells in planarians and collectively produce all adult cell types. Neoblasts are required, as a population, for regeneration and for the replacement of aged cells that occurs during natural tissue turnover.
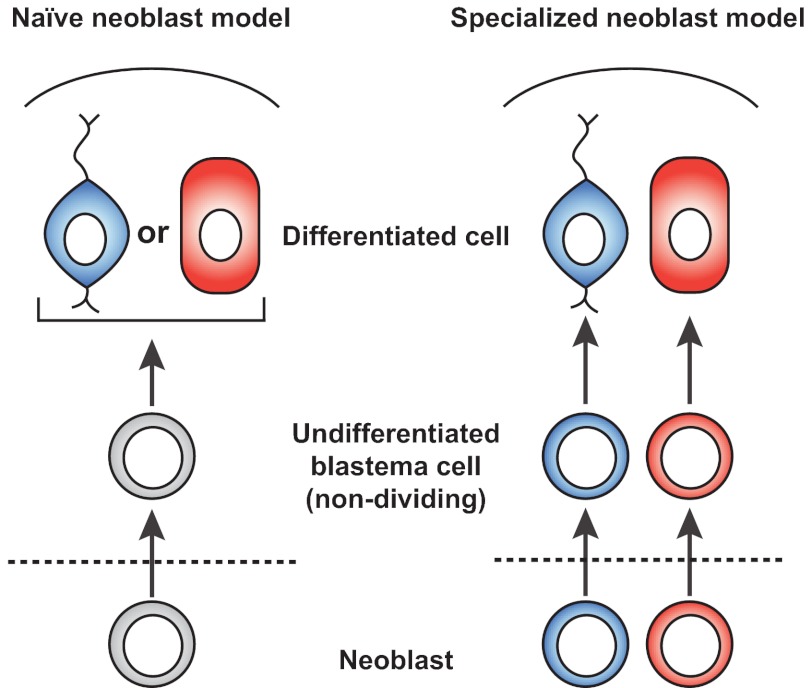
Fig. 2.
Two models for cell fate specification in planarian regeneration. In the naïve neoblast model for regeneration, neoblasts at wound sites (dashed line) produce undifferentiated blastema cells (e.g. for head regeneration) that are non-dividing and multipotent. These blastema cells differentiate according to signals received, for example, based upon their position in the blastema. In the specialized neoblast model for regeneration, the fate of individual neoblasts at wound sites is specified. Undifferentiated blastema cells that are neoblast progeny and non-dividing are therefore specified to adopt particular differentiated cell identities.
The term ‘neoblasts’ refers to a population of cells, and because cell-to-cell differences exist in any population, the criteria that designate a particular cell as a neoblast are necessarily imprecise. Assigning a label (neoblast) to this population is useful for reference to cells with many common features, but is also potentially problematic because of implied cell population uniformity. Cell division is commonly used as a defining neoblast characteristic: all adult dividing cells are neoblasts. Cells in S phase or mitosis (by definition neoblasts) can easily be experimentally labeled and hence visualized with BrdU, by an antibody to a mitosis-specific histone modification, or by RNA probes to genes that are transcribed during S phase (e.g. h2b, pcna, MCM2) (Table 1) (Newmark and Sánchez Alvarado, 2000; Shibata et al., 2010). Cells in G0/G1 that will divide again (and which could therefore be considered neoblasts) are more difficult to experimentally label. Because dividing cells are sensitive to irradiation, acute irradiation sensitivity can also be used as a (perhaps more inclusive) feature of neoblasts (Bardeen and Baetjer, 1904; Dubois, 1949; Reddien et al., 2005a; Hayashi et al., 2006).
Table 1.
Features commonly used to experimentally define neoblasts
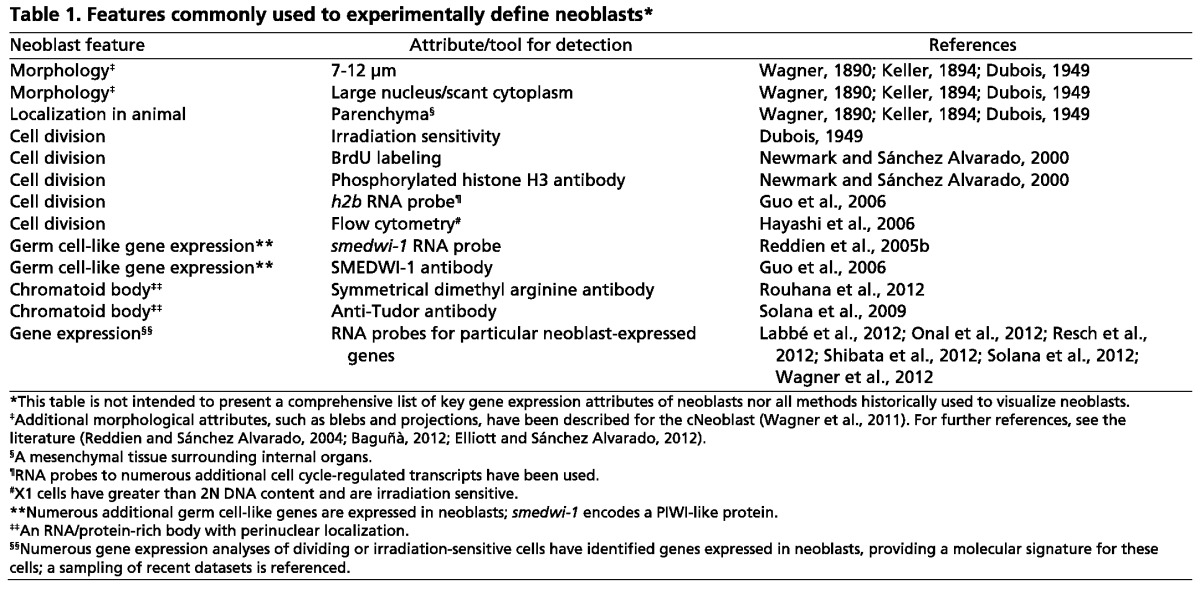
Additional (non-cell cycle-related) features have been associated with cells of the neoblast population (Table 1). Specifically, many acutely irradiation-sensitive cells are small in size (∼7-12 μm in diameter), have a large nucleus and scant cytoplasm, exhibit chromatoid bodies (RNA-protein bodies near nuclei), and reside in the planarian parenchyma – a mesenchymal tissue surrounding organ systems, such as nerve cords, intestine and protonephridia (Hyman, 1951; Reddien and Sánchez Alvarado, 2004; Baguñà, 2012). Neoblasts are scattered broadly throughout the parenchyma, being absent only from the animal head tips and the pharynx – the only two regions that cannot support regeneration in isolation (Reddien and Sánchez Alvarado, 2004).
Furthermore, many genes are commonly expressed in cells that have these features (morphology, localization, division, irradiation sensitivity), and the expression of such genes has been used to label a cell as a neoblast (Shibata et al., 1999; Reddien et al., 2005b; Salvetti et al., 2005; Guo et al., 2006; Palakodeti et al., 2008; Solana et al., 2009; Fernandéz-Taboada et al., 2010; Rouhana et al., 2010; Shibata et al., 2010; Labbé et al., 2012; Onal et al., 2012; Resch et al., 2012; Shibata et al., 2012; Solana et al., 2012; Wagner et al., 2012). Many of the genes that are expressed broadly, if not uniformly, in the neoblast population encode proteins similar to those found in germ cells in other organisms, suggesting a germ cell-like character of cells throughout the neoblast population. The presence of abundant mRNA for the gene smedwi-1 (which encodes a PIWI-like protein) is perhaps the most commonly used means to label a cell as a neoblast at present (Reddien et al., 2005b). All cells in S phase are smedwi-1+, but not all smedwi-1+ cells are in S phase (Wagner et al., 2011). However, all smedwi-1+ cells are eliminated within ∼24 hours following irradiation, consistent with the possibility that smedwi-1 mRNA labels all dividing cells and cells that will divide, and no further cells (Reddien et al., 2005b; Eisenhoffer et al., 2008).
Despite these defining characteristics, the field would benefit from a more precise definition of neoblasts (or cells within the neoblast population) based on the function and potential of cells with particular molecular and morphological characteristics. With this caveat in mind, the term neoblast as used in this article will refer to cells that are smedwi-1+ and acutely irradiation sensitive (lost within 24 hours following irradiation); cells labeled with putative S-phase markers will also be referred to as neoblasts.
Naïve versus specialized neoblast models for planarian regeneration
Neoblasts respond to wounds with stereotypic cell division and migration responses (Baguñà, 1976; Saló and Baguñà, 1984; Wenemoser and Reddien, 2010; Guedelhoefer and Sánchez Alvarado, 2012). The local production of non-dividing neoblast progeny cells at wounds results in the formation of a regenerative outgrowth called a blastema. The blastema produces some of the replacement parts needed in regeneration. In the case of head regeneration, proliferation is an early response to injury, and the blastema is largely devoid of dividing cells after the first 2 days of regeneration initiation (Pedersen, 1972; Saló and Baguñà, 1984; Wenemoser and Reddien, 2010). Over the next 1 to 2 weeks following amputation, a new head is fully regenerated.
At what step in regeneration are cells specified to make appropriate replacement cell types? Here, I consider two possible models that address this question (Fig. 2). (1) In the ‘naïve neoblast model’, neoblasts produce non-dividing, multipotent blastema cells. Cells in the neoblast population are essentially all the same, responding like drones to wounds by simply migrating and dividing, producing the blastema cells. The action would then happen in blastema cells, with these multipotent and naïve postmitotic cells adopting appropriate identities based on the external signals that they receive; for example, as a consequence of their position in a blastema. (2) In the ‘specialized neoblast model’, neoblasts involved in regeneration have different fates. These specialized neoblasts produce different lineage-committed and non-dividing blastema cells. Which differentiated cells are ultimately generated by the different neoblasts is thus predetermined. The specialization of neoblasts could occur before or after injury. It is also possible that some regenerative lineages follow a naïve neoblast model and others a specialized neoblast model.
The naïve neoblast model
The capacity of fragments of planarians from any region containing neoblasts to regenerate suggests that the attribute of pluripotency (defined here as the capacity to generate somatic lineages spanning embryonic germ layers) is widespread, if not uniform, in the neoblast population (Randolph, 1897). Although the possibility of neoblast heterogeneity has been entertained (Reddien and Sánchez Alvarado, 2004; Baguñà, 2012), the paucity of evidence for cell-to-cell differences in the fate of neoblasts historically led to their consideration largely as a uniform cell population. If the neoblasts are a homogenous population of cells with respect to potential and specification during regeneration and tissue turnover (e.g. if every neoblast is pluripotent and not specified), some version of the naïve neoblast model must be true. The specification process, or adoption of cell fate, would then occur in non-dividing neoblast progeny cells in the blastema. Little evidence, however, presently exists to support the possibility that fate specification occurs in non-dividing blastema cells and not in the neoblasts. Splitting a head blastema in two early in regeneration (within the first 3 days) results in the regeneration of two heads, rather than two half heads, which is consistent with plasticity in the fates of early blastema cells (Morgan, 1902). However, following the fates of individual cells in these experiments would be necessary for clear interpretation. For example, the contribution of neoblasts versus neoblast progeny to new cell production after blastema splitting is unclear. If the naïve neoblast model is correct for most or all lineages, the patterning mechanisms that are active in the blastema are likely to be crucial for specifying fate based, for example, on blastema cell position.
The specialized neoblast model
Despite their similarity in appearance, different subtypes of cells could in principle exist in the neoblast population. There are many precedents for progenitor cell populations in other organisms that contain numerous cell types with different properties or lineage potential (Spangrude et al., 1988; Chao et al., 2008; Biressi and Rando, 2010). Furthermore, blastema cells in amphibians were long considered to be a multipotent progenitor type, but it is now known that there is lineage restriction in the cells of the blastema in Xenopus and axolotl, as well as in zebrafish (Gargioli and Slack, 2004; Kragl et al., 2009; Knopf et al., 2011; Tanaka and Reddien, 2011; Tu and Johnson, 2011).
The irradiation-sensitive adult planarian cell population also includes germ cells (even in asexual animals), so this cell population is already known to contain at least two cell types (Sato et al., 2006; Handberg-Thorsager and Saló, 2007; Wang et al., 2007). In principle, multiple other neoblast cell types could exist in regeneration and/or tissue turnover. A long-standing question, therefore, has been whether a pluripotent cell type exists in the neoblast population, or whether regeneration is accomplished instead by the collective action of multiple, lineage-restricted cell types (Fig. 3A). Two approaches involving clonal analyses of single cells from the neoblast population have demonstrated that some neoblasts are indeed pluripotent (Fig. 3B) (Wagner et al., 2011). First, irradiation was used to eliminate most – but not all – neoblasts. Sparse, surviving neoblasts produced clones, or colonies, of descendant cells. These colonies produced neurons and intestine cells, as well as multiple other known neoblast descendent cell types. Second, transplantation of individual neoblasts revealed that some could produce neoblast colonies and restore tissue turnover and regenerative capacity to lethally irradiated animals. These data indicate that at least some neoblasts are pluripotent (neoblasts with these attributes are referred to as clonogenic neoblasts, or cNeoblasts) (Wagner et al., 2011). Furthermore, the cNeoblast attributes fulfill the criteria for a stem cell: the capacity for renewal and differentiation.
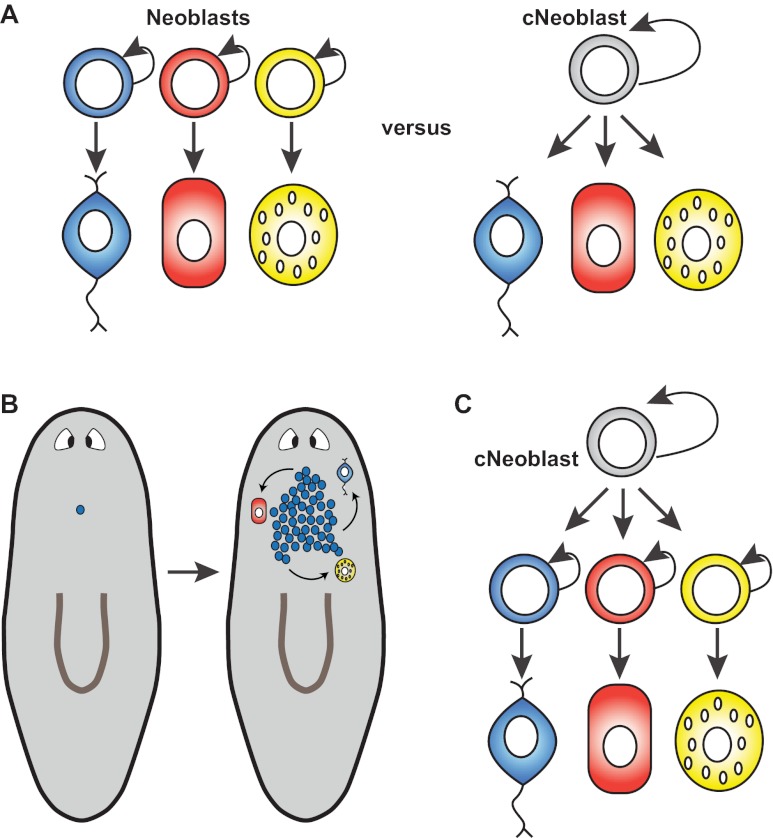
Fig. 3.
Pluripotency in the neoblast population and the cNeoblast. (A) Pluripotency could be accomplished by the collective action of different neoblasts or by individual cells within the neoblast population. Recent data demonstrate that individual neoblasts (termed clonogenic neoblasts, or cNeoblasts) can be pluripotent, generating tissues spanning germ layers and restoring regenerative ability to lethally irradiated animals that lack other sources of cell division (Wagner et al., 2011). (B) A single cell (blue) from the neoblast population can produce colonies of neoblasts that differentiate to produce neurons, intestine and other cell types (Wagner et al., 2011). Colonies can be produced by isolated neoblasts that survive irradiation or by transplantation. (C) The percentage of neoblasts with cNeoblast attributes is unknown. Therefore, cNeoblasts could in principle produce other neoblasts that are more restricted in potential (as depicted) or cNeoblasts might directly differentiate (as depicted in A, right panel).
The neoblast transplantation experiments described above cannot, however, establish whether some neoblasts are non-pluripotent, because the failure of some transplanted cells to generate colonies might have been for technical reasons. Therefore, whether all neoblasts are pluripotent and display cNeoblast attributes or whether cNeoblasts make other neoblasts that are more restricted in their lineage potential remains an important and unresolved problem (Fig. 3A,C). cNeoblast-derived colonies form predominantly on the ventral side of animals (Wagner et al., 2011); indeed, sublethal irradiation results in the preferential presence of neoblasts ventrally (Salvetti et al., 2009). However, whether these results are explained by cNeoblasts being a ventral subpopulation of the neoblasts, or are due to technical reasons regarding the irradiation sensitivity of neoblasts in different locations/cell cycle stages, is unknown. Regardless, the capacity of small fragments of planarians, from many locations, to regenerate suggests that cNeoblasts are widespread and not exceptionally rare.
Dividing planarian cells, with greater than 2N DNA content, can be isolated by DNA labeling and flow cytometry. Cells isolated in this way are called X1 cells (Hayashi et al., 2006). Gene expression analyses, using qPCR, of individual X1 cells have revealed some gene expression heterogeneity (Hayashi et al., 2010; Shibata et al., 2012). Some of the gene expression heterogeneity observed in neoblasts, such as for cell cycle genes that will be expressed at particular cell cycle stages, is expected, but additional gene expression heterogeneity was observed for genes encoding RNA-binding proteins (e.g. for the vasa-like gene vlgA) and for a myosin heavy chain gene. p53 is also expressed in a minority of dividing planarian cells in vivo (Pearson and Sánchez Alvarado, 2010). These findings support the possibility that the neoblast population is heterogeneous.
Recent studies of eye regeneration identified the expression of eye-specific transcription factors in neoblasts in vivo (Lapan and Reddien, 2011) (Fig. 4A). The planarian eye comprises two main cell types: photoreceptor neurons and pigmented optic cup cells. Trails of eye progenitors extend from regenerating eyes in head blastemas to the base of the blastema near the approximate amputation plane (Lapan and Reddien, 2011). Eye trail cells express differentiation markers as they near the eye: tyrosinase, which encodes an enzyme involved in melanin production, is expressed strongly in some trail cells near the eye and weakly in trail cells further away. Separate trails of progenitors exist for the optic cup and the photoreceptor neurons. dlx (distalless family) and sp6-9 encode transcription factors and are expressed together with other eye transcription factor-encoding genes six1/2 (sine oculis family), eya (eyes absent) and ovo in all cells of the regenerative optic cup trail (Lapan and Reddien, 2011; Lapan and Reddien, 2012). The transcription factor otxA is expressed with six1/2, eya and ovo in photoreceptor trail cells (Lapan and Reddien, 2011). Eye trail cells (expressing eye transcription factors) furthest from the regenerating eye that express smedwi-1 and S-phase-associated h2b can be labeled with BrdU and are irradiation sensitive (Lapan and Reddien, 2011). By these criteria, these distal eye progenitor cells are neoblasts. Aside from the expression of a set of eye transcription factors, these cells look like any other neoblast. Whether these smedwi-1+ eye progenitors will undergo renewing divisions or will immediately differentiate once specified is unknown. These data support a model in which a subset of neoblasts is specialized during regeneration to make eye cells (Fig. 4A).
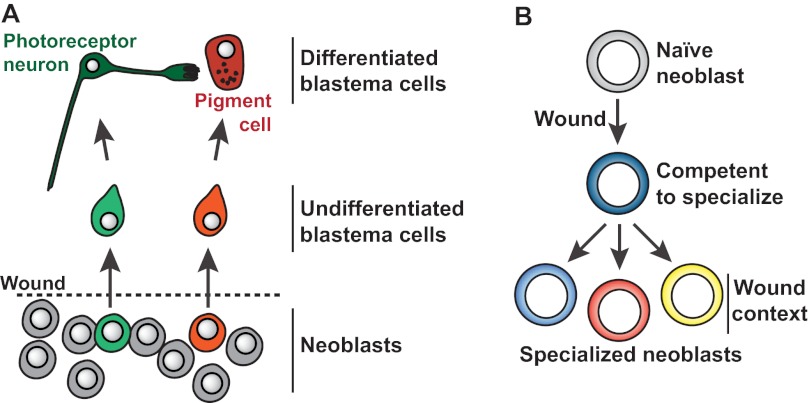
Fig. 4.
Neoblast specialization. (A) Eye transcription factors (e.g. ovo) are expressed in a subset of neoblasts at wound sites. Additional transcription factors distinguish the pigment cup cell lineage (dlx and sp6-9) and the photoreceptor eye lineage (otxA). Specialized neoblasts produce particular differentiated cells. Modified from Lapan and Reddien (Lapan and Reddien, 2011). (B) Model for production of specialized neoblasts during regeneration. Whereas specialized neoblasts do exist in uninjured animals (see text), neoblasts that previously did not express lineage-specific transcription factors (e.g. for eye regeneration) can be induced to do so at wound sites after injury. Injuries can induce the expression of the transcription factor gene runt-1 in neoblasts, with runt-1 expression required for the production of eye progenitors at injuries that remove the head (Wenemoser et al., 2012). Decisions regarding specialization paths in neoblasts are proposed to be regulated by the wound context (i.e. by processes that specify the identity of the missing tissue). It is unknown whether specialized neoblasts undergo any self-renewing cell divisions or immediately differentiate.
Eye regeneration is not the only context in which specialized neoblasts have been observed in vivo. Recent studies of regeneration of the planarian kidney-like excretory system, the protonephridia, also demonstrate neoblast specialization (Scimone et al., 2011). The protonephridia is made up of multiple cell types, including flame cells (filtration cells), tubule cells and tubule-associated cells. six1/2-2 (a second planarian six1/2 family member, distinct from the six1/2 gene expressed in the eye) and POU2/3 encode transcription factors required for protonephridia regeneration (Scimone et al., 2011). These transcription factors are expressed together in neoblasts (smedwi-1+, h2b+, irradiation sensitive) during regeneration. Non-dividing, neoblast-descendant cells expressing these transcription factors initiate expression of differentiation markers for the protonephridia. Therefore, these data suggest that protonephridia cell fates are specified in neoblasts. As is the case for the eye, whether these neoblasts expressing protonephridia-associated transcription factors undergo further division (e.g. self-renewal) or immediately differentiate once specified is unknown.
Specialized neoblasts exist in unamputated animals
In addition to producing new cells during regeneration, planarians constantly produce new cells for replacement of aging differentiated cells for tissue turnover (Pellettieri and Sánchez Alvarado, 2007). Are specialized neoblasts present only during regeneration or also during tissue turnover? Insight into this question recently emerged from a study of the eye transcription factor-encoding ovo gene (Lapan and Reddien, 2012). ovo is specifically expressed in the planarian eye and eye progenitors, and nowhere else in the animal. In uninjured animals, ovo is expressed outside of the eye in smedwi-1+, h2b+, irradiation-sensitive cells (neoblasts) and ovo is required for the long-term maintenance of eyes during tissue turnover (Lapan and Reddien, 2012). These data demonstrate the presence of specialized neoblasts, even in the absence of injury, raising the possibility that specialized neoblasts are a general feature of the neoblast population in uninjured animals. Whether other specialized neoblasts exist in unamputated animals, expressing markers for tissues other than the eye, has yet to be determined.
Production of specialized neoblasts at wound sites
Do specialized neoblasts involved in regeneration come from specialized neoblasts that exist prior to injury, or are previously naïve neoblasts induced to express lineage-specific transcription factors at wound sites? The eye-related neoblasts present in uninjured animals only exist near the head (Lapan and Reddien, 2012). Nonetheless, tail fragments that initially lack any ovo+ cells (i.e. lacking eye-specialized neoblasts) display the rapid appearance of ovo+ cells near wounds, indicating that the major source of eye-specialized neoblasts in regeneration is neoblasts that did not previously show the expression of eye transcription factors. These observations suggest the existence of multipotent (non-specialized) neoblasts that can adopt specialized fates as an early step in regeneration.
The transcription factor runt-1 is rapidly induced (within 3 hours following amputation) at planarian wounds (Sandmann et al., 2011; Wenemoser et al., 2012), and expression is within neoblasts (Wenemoser et al., 2012). However, only a subset of neoblasts in wounded planarians (neoblasts near wounds) express runt-1, demonstrating that injury induces gene expression heterogeneity in the neoblasts. runt-1 is required for normal eye regeneration (Sandmann et al., 2011; Wenemoser et al., 2012) and for the presence of sp6-9+ optic cup progenitor trails (Wenemoser et al., 2012). runt-1 does not display any expression specificity for particular regenerative contexts, but is generically induced by many or all injuries. Thus, runt-1 activation is not sufficient to trigger eye regeneration. Moreover, additional transcription factors (ap2 and pax6), which are not associated with either eye or protonephridia regeneration, are also activated in subsets of runt-1+ neoblasts at wounds. Therefore, transcription factors specifying eye and protonephridia fate, as well as those encoded by ap2 and pax6, are all expressed in neoblasts during regeneration. This supports the model that the neoblast population is heterogeneous and that multiple lineages follow a specialized neoblast model during regeneration. runt-1 is required for the formation of normal numbers of several types of specialized neoblasts (eye and ap2+ neoblasts). These data raise the possibility that wounds promote neoblast specialization (possibly mediated by runt-1 activation for at least some neoblasts), with the tissue context of the injury determining which specialization paths neoblast cells adopt (Fig. 4B).
Animals deficient for the Wnt pathway inhibitor APC [APC(RNAi)] regenerate tails in place of heads following head amputation; however, some brain tissue regenerates in these anterior blastemas (Evans et al., 2011; Iglesias et al., 2011). Experiments using cell cycle-inhibitory drugs explored the formation of these neurons in APC(RNAi) anterior blastemas (Evans et al., 2011). Hydroxyurea treatment, to disrupt S-phase progression, 15 hours prior to amputation did not block all anterior neural differentiation following head amputation (Evans et al., 2011). By contrast, mitosis-blocking colchicine treatment 2 hours prior to amputation did block differentiation. These experiments suggest that some neoblasts that have progressed through S phase but not yet divided are capable of responding to wound signals to differentiate appropriately without undergoing further division. Further exploration of this possibility is an important future direction.
Conclusions
The expression of several transcription factors in subsets of neoblasts during regeneration and tissue turnover supports the specialized neoblast hypothesis. This entails that the fates of non-dividing blastema cells (for at least some lineages) are set in their neoblast parents. This hypothesis focuses attention on the responses of neoblasts to wounds for understanding the origins of replacement cells in regeneration. This hypothesis also provides a view of planarian regeneration in which there might not exist a naïve, multipotent blastema cell population, but instead a blastema that comprises a patchwork of lineage-committed cells. In this view, blastema-patterning mechanisms during regeneration would be involved in the spatial organization of specified cells rather than in the fate specification of blastema cells. There is an ongoing need for new differentiated cells in normal adult planarian life, and low levels of specialized neoblasts appear to exist in unamputated animals (Lapan and Reddien, 2012). Following amputation, many newly differentiated cells appear rapidly and, accordingly, the numbers of specialized neoblasts increase robustly at wounds. Therefore, wounding might induce the specialization of neoblasts for regeneration.
There are many important and unresolved questions related to the specialized neoblast model described above. First, exactly how can we best experimentally define a cell as a neoblast? Second, are specialized neoblasts and/or their postmitotic progeny blastema cells irreversibly determined? No experiment has yet clearly tested whether specialized neoblasts or neoblast-descendant blastema cells can change their fates (e.g. in response to blastema injury). Third, because few regenerative lineages have been investigated, whether specialization of neoblasts occurs for the regeneration of most or all target tissues is unknown. It is possible that a combination of specialized neoblasts and naïve neoblasts produces blastema cells that together replace missing cells. Fourth, what proportions of neoblasts in unamputated animals are specialized versus non-specialized? Fifth, what degree of lineage hierarchy exists in the neoblast population? Are there multiple cell division steps from the cNeoblast to differentiation (similar to the hierarchical lineage of hematopoiesis), or can cNeoblasts directly become specialized? Finally, do specialized neoblasts divide and renew during the production of target tissues in regeneration and tissue turnover, or, by contrast, are specialized neoblasts simply a very transient state between a dividing pluripotent cell and a differentiated cell?
The presence of specialized neoblasts opens many important new avenues for investigation of planarian regeneration. For example, how do wound contexts regulate the formation of specialized neoblast types that are tailored to the identity of missing tissues? What molecular mechanisms underlie neoblast specialization and are similar progenitor specialization mechanisms active in the regeneration and repair of tissues in other organisms? Recent expansion of cellular and molecular tools for planarian research has enabled the initial forays described here. The development of additional cell-marking tools for lineage analysis will be an important future direction for characterizing described and additional lineages. Finally, it will be interesting to compare the molecular nature and regenerative paths of specialized neoblasts with those of progenitor cells involved in regeneration in other organisms. For example, the blastemas of amphibians and fish were long thought to involve a multipotent cell population, but now appear to be more mosaic. Xenopus spinal cord, notochord and muscle and axolotl Schwann cell, muscle, dermis and cartilage lineages all appear to be regenerated from cells within their respective lineages prior to amputation (Gargioli and Slack, 2004; Kragl et al., 2009). Therefore, Xenopus tail and axolotl limb blastemas would be expected to contain many different lineage-committed cells rather than a uniform population of multipotent cells. Similarly, lineage restriction in osteoblasts and in several neural crest-derived lineages in the regeneration of zebrafish fins also suggests that zebrafish blastemas might comprise a mosaic patchwork of lineage-committed cells (Knopf et al., 2011; Tu and Johnson, 2011).
Neoblasts have long attracted interest because of the remarkable regenerative potential that this cell population promotes. The initial insights into the cellular steps in the transition from pluripotency to a specialized progenitor cell state set the stage for further molecular dissection of regeneration.
Acknowledgments
I thank all members of the P.W.R. laboratory for comments and discussion. Particular thanks to Sylvain Lapan, Lucila Scimone and Mansi Srivastava for work cited and discussions related to planarian eye and protonephridia regeneration; Danielle Wenemoser for work cited and discussions related to neoblast wound responses; and Dan Wagner and Irving Wang for work cited and discussions related to the pluripotency of neoblasts at the levels of a population and individual cells.
Footnotes
Funding
I acknowledge support from the Howard Hughes Medical Institute; the National Institutes of Health; and the Keck Foundation. Deposited in PMC for release after 12 months.
Competing interests statement
The author declares no competing financial interests.
References
- Aboobaker A. A. (2011). Planarian stem cells: a simple paradigm for regeneration. Trends Cell Biol. 21, 304–311 [DOI] [PubMed] [Google Scholar]
- Baguñà J. (1976). Mitosis in the intact and regenerating planarian Dugesia mediterranea n.sp. II. Mitotic studies during regeneration, and a possible mechanism of blastema formation. J. Exp. Zool. 195, 65–79 [Google Scholar]
- Baguñà J. (2012). The planarian neoblast: the rambling history of its origin and some current black boxes. Int. J. Dev. Biol. 56, 19–37 [DOI] [PubMed] [Google Scholar]
- Bardeen C. R., Baetjer F. H. (1904). The inhibitive action of the Roentgen rays on regeneration in planarians. J. Exp. Zool. 1, 191–195 [Google Scholar]
- Biressi S., Rando T. A. (2010). Heterogeneity in the muscle satellite cell population. Semin. Cell Dev. Biol. 21, 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao M. P., Seita J., Weissman I. L. (2008). Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb. Symp. Quant. Biol. 73, 439–449 [DOI] [PubMed] [Google Scholar]
- Dubois F. (1949). Contribution á l ’ètude de la migration des cellules de règènèration chez les Planaires dulcicoles. Bull. Biol. Fr. Belg. 83, 213–283 [Google Scholar]
- Eisenhoffer G. T., Kang H., Sánchez Alvarado A. (2008). Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell 3, 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott S. A., Sánchez Alvarado A. (2012). The history and enduring contributions of planarians to the study of animal regeneration. WIREs Dev. Biol. 10.1002/wdev.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. J., Owlarn S., Tejada Romero B., Chen C., Aboobaker A. A. (2011). Combining classical and molecular approaches elaborates on the complexity of mechanisms underpinning anterior regeneration. PLoS ONE 6, e27927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandéz-Taboada E., Moritz S., Zeuschner D., Stehling M., Schöler H. R., Saló E., Gentile L. (2010). Smed-SmB, a member of the LSm protein superfamily, is essential for chromatoid body organization and planarian stem cell proliferation. Development 137, 1055–1065 [DOI] [PubMed] [Google Scholar]
- Gargioli C., Slack J. M. (2004). Cell lineage tracing during Xenopus tail regeneration. Development 131, 2669–2679 [DOI] [PubMed] [Google Scholar]
- Guedelhoefer O. C., 4th, Sánchez Alvarado A. (2012). Amputation induces stem cell mobilization to sites of injury during planarian regeneration. Development 139, 3510–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Peters A. H., Newmark P. A. (2006). A Bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell 11, 159–169 [DOI] [PubMed] [Google Scholar]
- Handberg-Thorsager M., Saló E. (2007). The planarian nanos-like gene Smednos is expressed in germline and eye precursor cells during development and regeneration. Dev. Genes Evol. 217, 403–411 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Asami M., Higuchi S., Shibata N., Agata K. (2006). Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev. Growth Differ. 48, 371–380 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Shibata N., Okumura R., Kudome T., Nishimura O., Tarui H., Agata K. (2010). Single-cell gene profiling of planarian stem cells using fluorescent activated cell sorting and its ‘index sorting’ function for stem cell research. Dev. Growth Differ. 52, 131–144 [DOI] [PubMed] [Google Scholar]
- Hyman L. H. (1951). The Invertebrates: Platyhelminthes and Rhynchocoela the Acoelomate Bilateria. New York, NY: McGraw-Hill; [Google Scholar]
- Iglesias M., Almuedo-Castillo M., Aboobaker A. A., Saló E. (2011). Early planarian brain regeneration is independent of blastema polarity mediated by the Wnt/β-catenin pathway. Dev. Biol. 358, 68–78 [DOI] [PubMed] [Google Scholar]
- Keller J. (1894). Die ungeschlechtliche Fortpflanzungder Süsswasser-Turbellarien. Jen. Zeit. Naturw. 28, 370–407 [Google Scholar]
- Knopf F., Hammond C., Chekuru A., Kurth T., Hans S., Weber C. W., Mahatma G., Fisher S., Brand M., Schulte-Merker S., et al. (2011). Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev. Cell 20, 713–724 [DOI] [PubMed] [Google Scholar]
- Kragl M., Knapp D., Nacu E., Khattak S., Maden M., Epperlein H. H., Tanaka E. M. (2009). Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460, 60–65 [DOI] [PubMed] [Google Scholar]
- Labbé R. M., Irimia M., Currie K. W., Lin A., Zhu S. J., Brown D. D., Ross E. J., Voisin V., Bader G. D., Blencowe B. J., et al. (2012). A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem Cells 30, 1734–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan S. W., Reddien P. W. (2011). dlx and sp6-9 control optic cup regeneration in a prototypic eye. PLoS Genet. 7, e1002226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan S. W., Reddien P. W. (2012). Transcriptome analysis of the planarian eye identifies ovo as a specific regulator of eye regeneration. Cell Rep. 2, 294–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T. H. (1902). Growth and regeneration in Planaria lugubris. Arch. Ent. Mech. Org. 13, 179–212 [Google Scholar]
- Newmark P. A., Sánchez Alvarado A. (2000). Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev. Biol. 220, 142–153 [DOI] [PubMed] [Google Scholar]
- Newmark P. A., Sánchez Alvarado A. (2002). Not your father’s planarian: a classic model enters the era of functional genomics. Nat. Rev. Genet. 3, 210–219 [DOI] [PubMed] [Google Scholar]
- Onal P., Grün D., Adamidi C., Rybak A., Solana J., Mastrobuoni G., Wang Y., Rahn H. P., Chen W., Kempa S., et al. (2012). Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells. EMBO J. 31, 2755–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palakodeti D., Smielewska M., Lu Y. C., Yeo G. W., Graveley B. R. (2008). The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA 14, 1174–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson B. J., Sánchez Alvarado A. (2010). A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development 137, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K. J. (1972). Studies on regeneration blastemas of the planarian Dugesia tigrina with special reference to differentiation of the muscle-connective tissue filament system. Arch. Entw. Org. 169, 134–169 [DOI] [PubMed] [Google Scholar]
- Pellettieri J., Sánchez Alvarado A. (2007). Cell turnover and adult tissue homeostasis: from humans to planarians. Annu. Rev. Genet. 41, 83–105 [DOI] [PubMed] [Google Scholar]
- Randolph H. (1897). Observations and experiments on regeneration in planarians. Arch. Entw. Mech. Org. 5, 352–372 [Google Scholar]
- Reddien P. W. (2011). Constitutive gene expression and the specification of tissue identity in adult planarian biology. Trends Genet. 27, 277–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W., Sánchez Alvarado A. (2004). Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 20, 725–757 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Bermange A. L., Murfitt K. J., Jennings J. R., Sánchez Alvarado A. (2005a). Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell 8, 635–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W., Oviedo N. J., Jennings J. R., Jenkin J. C., Sánchez Alvarado A. (2005b). SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327–1330 [DOI] [PubMed] [Google Scholar]
- Resch A. M., Palakodeti D., Lu Y. C., Horowitz M., Graveley B. R. (2012). Transcriptome analysis reveals strain-specific and conserved stemness genes in Schmidtea mediterranea. PLoS ONE 7, e34447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J. C. (2012). Stem cell systems and regeneration in planaria. Dev. Genes Evol. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L., Shibata N., Nishimura O., Agata K. (2010). Different requirements for conserved post-transcriptional regulators in planarian regeneration and stem cell maintenance. Dev. Biol. 341, 429–443 [DOI] [PubMed] [Google Scholar]
- Rouhana L., Vieira A. P., Roberts-Galbraith R. H., Newmark P. A. (2012). PRMT5 and the role of symmetrical dimethylarginine in chromatoid bodies of planarian stem cells. Development 139, 1083–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saló E., Baguñà J. (1984). Regeneration and pattern formation in planarians. I. The pattern of mitosis in anterior and posterior regeneration in Dugesia (G) tigrina, and a new proposal for blastema formation. J. Embryol. Exp. Morphol. 83, 63–80 [PubMed] [Google Scholar]
- Salvetti A., Rossi L., Lena A., Batistoni R., Deri P., Rainaldi G., Locci M. T., Evangelista M., Gremigni V. (2005). DjPum, a homologue of Drosophila Pumilio, is essential to planarian stem cell maintenance. Development 132, 1863–1874 [DOI] [PubMed] [Google Scholar]
- Salvetti A., Rossi L., Bonuccelli L., Lena A., Pugliesi C., Rainaldi G., Evangelista M., Gremigni V. (2009). Adult stem cell plasticity: neoblast repopulation in non-lethally irradiated planarians. Dev. Biol. 328, 305–314 [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A. (2006). Planarian regeneration: its end is its beginning. Cell 124, 241–245 [DOI] [PubMed] [Google Scholar]
- Sandmann T., Vogg M. C., Owlarn S., Boutros M., Bartscherer K. (2011). The head-regeneration transcriptome of the planarian Schmidtea mediterranea. Genome Biol. 12, R76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Shibata N., Orii H., Amikura R., Sakurai T., Agata K., Kobayashi S., Watanabe K. (2006). Identification and origin of the germline stem cells as revealed by the expression of nanos-related gene in planarians. Dev. Growth Differ. 48, 615–628 [DOI] [PubMed] [Google Scholar]
- Scimone M. L., Srivastava M., Bell G. W., Reddien P. W. (2011). A regulatory program for excretory system regeneration in planarians. Development 138, 4387–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N., Umesono Y., Orii H., Sakurai T., Watanabe K., Agata K. (1999). Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev. Biol. 206, 73–87 [DOI] [PubMed] [Google Scholar]
- Shibata N., Rouhana L., Agata K. (2010). Cellular and molecular dissection of pluripotent adult somatic stem cells in planarians. Dev. Growth Differ. 52, 27–41 [DOI] [PubMed] [Google Scholar]
- Shibata N., Hayashi T., Fukumura R., Fujii J., Kudome-Takamatsu T., Nishimura O., Sano S., Son F., Suzuki N., Araki R., et al. (2012). Comprehensive gene expression analyses in pluripotent stem cells of a planarian, Dugesia japonica. Int. J. Dev. Biol. 56, 93–102 [DOI] [PubMed] [Google Scholar]
- Solana J., Lasko P., Romero R. (2009). Spoltud-1 is a chromatoid body component required for planarian long-term stem cell self-renewal. Dev. Biol. 328, 410–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J., Kao D., Mihaylova Y., Jaber-Hijazi F., Malla S., Wilson R., Aboobaker A. (2012). Defining the molecular profile of planarian pluripotent stem cells using a combinatorial RNAseq, RNA interference and irradiation approach. Genome Biol. 13, R19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangrude G. J., Heimfeld S., Weissman I. L. (1988). Purification and characterization of mouse hematopoietic stem cells. Science 241, 58–62 [DOI] [PubMed] [Google Scholar]
- Tanaka E. M., Reddien P. W. (2011). The cellular basis for animal regeneration. Dev. Cell 21, 172–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S., Johnson S. L. (2011). Fate restriction in the growing and regenerating zebrafish fin. Dev. Cell 20, 725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner F. (1890). Zur Kenntnis der ungeschlechtlichen Fortpflanzung von Microstoma nebst allegemeinen Bemerkungen über Teilung und Knospung im Tierreich. Zool. Jahrb. 4, 349–423 [Google Scholar]
- Wagner D. E., Wang I. E., Reddien P. W. (2011). Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D. E., Ho J. J., Reddien P. W. (2012). Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell 10, 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zayas R. M., Guo T., Newmark P. A. (2007). Nanos function is essential for development and regeneration of planarian germ cells. Proc. Natl. Acad. Sci. USA 104, 5901–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D., Reddien P. W. (2010). Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev. Biol. 344, 979–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D., Lapan S. W., Wilkinson A. W., Bell G. W., Reddien P. W. (2012). A molecular wound response program associated with regeneration initiation in planarians. Genes Dev. 26, 988–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]