Abstract
Nuclear receptor interacting protein (Nrip1), also known as RIP140, is a co-regulator for nuclear receptors that plays an essential role in ovulation by regulating the expression of the epidermal growth factor-like family of growth factors. Although several studies indicate a role for RIP140 in breast cancer, its role in the development of the mammary gland is unclear. By using RIP140-null and RIP140 transgenic mice, we demonstrate that RIP140 is an essential factor for normal mammary gland development and that it functions by mediating oestrogen signalling. RIP140-null mice exhibit minimal ductal elongation with no side-branching, whereas RIP140-overexpressing mice show increased cell proliferation and ductal branching with age. Tissue recombination experiments demonstrate that RIP140 expression is required in both the mammary epithelial and stromal compartments for ductal elongation during puberty and that loss of RIP140 leads to a catastrophic loss of the mammary epithelium, whereas RIP140 overexpression augments the mammary basal cell population and shifts the progenitor/differentiated cell balance within the luminal cell compartment towards the progenitors. For the first time, we present a genome-wide global view of oestrogen receptor-α (ERα) binding events in the developing mammary gland, which unravels 881 ERα binding sites. Unbiased evaluation of several ERα binding sites for RIP140 co-occupancy reveals selectivity and demonstrates that RIP140 acts as a co-regulator with ERα to regulate directly the expression of amphiregulin (Areg), the progesterone receptor (Pgr) and signal transducer and activator of transcription 5a (Stat5a), factors that influence key mitogenic pathways that regulate normal mammary gland development.
Keywords: Amphiregulin, Mammary gland, Mammary progenitors, Oestrogen receptor, Progesterone receptor, RIP140, Mouse
INTRODUCTION
The development of the mouse mammary epithelium is regulated by female reproductive hormones, including oestrogen, progesterone and prolactin, together with their downstream paracrine effectors (Brisken and O’Malley, 2010; McNally and Martin, 2011). At birth, the mammary epithelium is composed of a rudimentary ductal system but at the onset of puberty the increased levels of circulating oestrogen promote the elongation of these ducts. This growth is marked by bulb-like structures called terminal end buds (TEBs), which are located at the distal tips of the growing ducts and are highly proliferative (Hinck and Silberstein, 2005). By 8-10 weeks of age, a well-formed epithelial network fills the mammary fat pad. Secondary and tertiary branching, which is under the influence of progesterone (Lydon et al., 1995), increases the complexity of the ductal tree with each oestrus cycle. During pregnancy, prolactin and progesterone promote the budding and expansion of alveoli (lobules) from the existing ductal network and the cells lining these alveoli synthesise and secrete milk during lactation (Gjorevski and Nelson, 2011).
Oestrogen, bound to oestrogen receptor-α (ERα; Esr1 – Mouse Genome Informatics), promotes ductal elongation during puberty. One of the most rapid changes induced by oestrogen is the induction of Areg, which is an important paracrine mediator required for ductal elongation. Areg knockout (KO) mice phenocopy ER KO mice as ablation of either gene results in severely impaired ductal development during puberty (Bocchinfuso et al., 2000; Mallepell et al., 2006; Ciarloni et al., 2007). Tissue recombination experiments and gene knockout mouse models indicate that Areg generated in the epithelial compartment activates the epidermal growth factor receptor (Egfr) in the stroma, which, as a feedback mechanism, promotes the synthesis of stromal growth-promoting factors (Sternlicht et al., 2005). Thus, signalling cross-talk between the mammary epithelium and stroma plays a crucial role in the development of the mammary gland.
We identified RIP140 as a co-factor for ERα in a breast cancer cell line (Cavaillès et al., 1994; Cavaillès et al., 1995), but subsequently found it interacts with other transcription factors, including many nuclear receptors (L’Horset et al., 1996; Heery et al., 1997). RIP140 is a pleiotropic co-factor that regulates energy homeostasis (Leonardsson et al., 2004; Herzog et al., 2007; Seth et al., 2007) and plays an essential role in ovulation (White et al., 2000; Tullet et al., 2005). Its function seems to vary according to its physiological role, serving as a co-repressor of catabolic gene expression (Christian et al., 2005; Seth et al., 2007) but as a co-activator of inflammatory gene expression (Zschiedrich et al., 2008; Nautiyal et al., 2010). In the ovary, RIP140 is required for the expression of the epidermal growth factor (EGF)-like family of growth factors, including Areg, epiregulin (Ereg) and betacellulin (Btc), all of which are essential for cumulus expansion in response to the luteinizing hormone surge (Tullet et al., 2005; Nautiyal et al., 2010).
Preliminary experiments suggest that RIP140 might have a role in breast cancer. It appears that RIP140 mRNA expression is elevated in ductal carcinoma in situ (Lee et al., 2007; Hannafon et al., 2011) and varies with tumour subtype (Oh et al., 2006; Docquier et al., 2010), with higher RIP140 mRNA levels in luminal-like tumours than in basal-like tumours. This pattern is consistent with the pattern of ERα expression, and earlier work suggests that RIP140 expression is elevated in tamoxifen-resistant breast cancer cells (Chan et al., 1999). Given the potential involvement of RIP140 in breast cancer and its crucial role in the regulation of ovarian expression of Areg, we investigated the role of RIP140 in mammary gland development. Analysis of RIP140 null (RIP140 KO) and overexpressing transgenic (RIP140 Tg) mice indicates that RIP140 is an essential factor required for mammary gland development and that its expression is essential in both the epithelium and the stroma in order to influence glandular development. Furthermore, RIP140 is recruited together with ERα to the promoters/enhancers of a number of regulatory genes, including Areg, Pgr and Stat5a, thereby stimulating their transcription and thus regulating mammary gland development.
MATERIALS AND METHODS
Animals
The generation of RIP140-null animals (RIP140 KO) has been described previously (White et al., 2000). The mice used in this study were backcrossed eight generations to the C57BL/6J background. To generate mice overexpressing RIP140 (RIP140 Tg), human RIP140 (hRIP140; NRIP1 – Human Gene Nomenclature Database) was inserted in FVB/N background using a pCAGGS-hRIP construct (Fritah et al., 2010). All animal studies were carried out in accordance with the UK Home Office guidelines. For tissue recombinant experiments, 3-week-old C57BL/6 and FVB/N wild-type (WT) mice were bought from Harlan Laboratories, UK.
To examine the effect of oestradiol (E2) on mammary gland development, 21-day-old WT and RIP140 KO animals were ovariectomised and implanted with 21-day slow release pellets of E2 or placebo (0.025 mg/pellet, Innovative Research of America, FL, USA). After 21 days, the inguinal glands were excised for whole-mount staining. Uterine weight was taken to analyse the response to the hormone.
Blood collection and hormonal measurements
Mice were anaesthetised with tribromoethanol (Sigma-Aldich, St Louis, MO, USA) by intraperitoneal injection and blood was withdrawn from the heart followed by cervical dislocation. Serum was separated by centrifugation from blood and stored at –20°C until used for oestradiol (E2) and prolactin (PRL) assays. E2 was analysed after diethyl ether extraction using a solid-phase time-resolved fluoroimmunoassay (PerkinElmer, Waltham, MA, USA), adapted for mouse samples (Rulli et al., 2002). PRL was measured using an ELISA immunoassay for mouse PRL (R&D Systems, Minneapolis, MN, USA) (Król et al., 2011).
Mammary gland whole-mounts
Inguinal mammary glands were dissected, spread onto a glass slide and fixed in a mixture of glacial acetic acid and ethanol (1:3) overnight. Tissue was then hydrated and stained in carmine alum stain. Whole-mounts were dehydrated in graded ethanol solutions, followed by clearing in Histoclear (National Diagnostics, Atlanta, GA, USA) and mounting in DPX (VWR Radnor, PA, USA). Digital pictures were taken using a Nikon Eclipse E600 (Nikon, Tokyo, Japan) and images stitched together using Lucia stitching software (Lucia Cytogenetics, Prague, Czech Republic).
Quantitative real-time PCR (Q-PCR)
Total RNA was isolated using TRIzol reagent (Sigma Aldrich) and treated with DNase I (Invitrogen, Carlsbad, CA, USA). RNA was prepared from sorted cells using the Pico Pure RNA Extraction Kit (Applied Biosystems, Warrington, UK) followed by DNase treatment using the RNase-free DNase set (Qiagen, Valencia, CA, USA). cDNA was synthesised using the Super Script First Strand Synthesis Kit (Invitrogen) according to the manufacturer’s instructions. Real-time PCR was performed using Opticon-2 (MJ Research, Watertown, MA, USA) using the SYBR Green Jumpstart Taq Ready Mix (Sigma-Aldrich) and gene-specific primers (supplementary material Table S1). Expression of all genes was normalised against the expression of ribosomal coding gene Rpl7.
Western blotting
Protein extracts (30 μg) were resolved by SDS-PAGE (4-12% gels, Invitrogen) and transferred to PVDF membranes (Millipore, Billerica, MA, USA). After blocking in 5% milk in Tris-buffered saline and 0.05% Tween 20 (TBST), blots were probed with primary antibodies and horseradish peroxidase-linked anti-mouse, -rat or -rabbit antibodies (Dako, Cambridge, UK), diluted 1:5000. After three or four washes in TBST, the bound antibodies were visualised using ECL substrate (Pierce, Rockford, IL, USA). The dilutions used for the primary antibodies were: ERα, 1:1000 (MC 200 sc-542, Santa Cruz, CA, USA); Areg, 1:2000 (MAB989, R&D Systems); progesterone receptor, 1:5000 (NCL-PGR-312, Leica, Wetzlar, Germany); cyclin D1, 1:1000 (# 2926, Cell Signaling, Danvers, MA, USA); Gata3, 1:1000 (sc-268, Santa Cruz) and Gapdh, 1:5000 (Mab-374, Millipore).
Immunohistochemistry
Mammary gland tissue was dissected and fixed in neutral buffered formalin for 24 hours, dehydrated as described for whole-mounts and embedded in paraffin. Paraffin sections (5 μm) were mounted on polylysine slides (VWR) and de-waxed before immunostaining using the Vectastain Elite ABC Kit (Vector Laboratories, Burlingame, CA, USA) and ImmPACT DAB Substrate Kit (Vector Laboratories). Primary antibodies were as follows: RIP140 (6D7, in-house monoclonal; 1:500) (Nautiyal et al., 2010), ERα (sc-542, Santa Cruz; 1:1000), Ki67 (Clone TEC-3, M7249, Dako; 1:100) and RANKL (AF462, R&D Systems; 1:100)
Quantification of hyperbudding and Ki67 staining
Epithelial budding in mammary glands derived from 5-week-old mice or tissue recombinants for analysing pubertal growth was quantified by taking whole-mount images (2× magnification) and counting the number of epithelial buds manually. Budding in adult mammary glands was quantified by counting the number of epithelial buds in representative Haematoxylin and Eosin (H&E)-stained sections of the glands (10× magnification). Ki67-positive cells among total epithelial cells were counted manually in representative immunostained tissue sections (50× magnification).
Chromatin immunoprecipitation assay (ChIP)
At least six pairs of mammary gland pads from mice at 5-6 weeks of age were excised, washed with PBS and minced. The tissue was incubated with a solution of Gentle Collagenase/Hyaluronidase (StemCell Technologies, Vancouver, Canada) for 30 minutes at 37°C with agitation. After digestion, cell suspensions were filtered with a 100 μM pore strainer (BD Biosciences, San Jose, CA, USA) in order to eliminate any undigested tissue and were centrifuged for 10 minutes at 2000 rpm (400 g). The pellet, consisting of stromal and epithelial cells, was washed twice with PBS and fixed with 1% formaldehyde [15 minutes at room temperature (RT)]. The cross-linking reaction was stopped by adding 125 mM glycine for 5 minutes at RT. Cells were pelleted and washed again with PBS and resuspended in 10% SDS cell lysis buffer. Homogenate was then sonicated to get DNA fragments of 500 bp length. The ChIP assay was performed as described previously (Nautiyal et al., 2010). Specific proteins were immunoprecipitated with Dynabeads (Invitrogen) using mouse IgG (sc-2025, Santa Cruz), rabbit IgG (sc-2027 Santa Cruz), an ERα antibody (sc-543, Santa Cruz) and a validated RIP140 antibody for ChIP (Nautiyal et al., 2010). Enrichment relative to input was determined by Q-PCR. All Q-PCR reactions were carried out in duplicate. For primer sequences see supplementary material Table S1.
Solexa sequencing and enrichment analysis
ChIP DNA was amplified as described (Schmidt et al., 2009). Sequences were generated by the Illumina Hiseq 2000 genome analyser (using 50 bp reads), and aligned to the Mouse Reference Genome (mm9; July 2007). Enriched regions of the genome were identified by comparing the ChIP sample with input using the MACS peak caller (Zhang et al., 2008) version 1.3.7.1. ChIP-Seq data have been deposited with NCBI GEO under accession number GSE43415.
Motif analysis, heatmaps and genomic distributions of binding events
ChIP-seq data snapshots were generated using the Integrative Genome Viewer IGV 2.1 (www.broadinstitute.org/igv/). Motif analysis was performed through Cistrome (cistrome.org), applying the SeqPos motif tool (He et al., 2010). The genomic distributions of binding sites were analysed using the cis-regulatory element annotation system (CEAS) (Ji et al., 2006). The genes closest to the binding site on both strands were analysed. If the binding region is within a gene, CEAS software indicates whether it is a 5′UTR, a 3′UTR, a coding exon or an intron. Promoter is defined as 3 kb upstream from RefSeq 5′ start. If a binding site is >3 kb away from the RefSeq transcription start site, it is considered to be distal intergenic.
Preparation of mammary cell suspensions
Single mammary cell suspensions were generated by dissecting out the fourth (inguinal) mammary gland from mature 8- to 12-week-old virgin female mice and incubating them overnight in DMEM (GIBCO) supplemented with 1× Gentle Collagenase/Hyaluronidase. Glands were then triturated with a p1000 pipette, washed and then treated with NH4Cl (StemCell Technologies) to lyse red blood cells. After washing, cells were sequentially incubated with 0.25% trypsin (StemCell Technologies) then 5 mg/ml dispase (StemCell Technologies) supplemented with 0.1 mg/ml DNase (Roche, West Sussex, UK), and then filtered through 40 μM mesh (BD Biosciences). Cells at this stage were either seeded into colony-forming assays or prepared for flow cytometry.
Flow cytometry
Single cells suspensions were blocked in 10% normal rat serum (Sigma Aldrich) diluted in Hank’s Balanced Salt Solution supplemented with 10 mM HEPES and 2% foetal bovine serum (FBS) (referred to as HF). Anti-CD45, -CD31, -BP-1 and -Ter119 antibodies (eBioscience, San Diego, CA, USA) were used to label haematopoietic, endothelial and some stromal cells for depletion at a final concentration of 1 μg/ml. Anti-EpCAM-AF647 (1 μg/ml; Biolegend, San Diego, CA, USA) and anti CD49f-AF488 (1:100, Biolegend) were used to identify the mammary epithelial cell populations from the remaining non-depleted stromal cells. Anti CD49b-PE (0.5 μg/ml; Biolegend) was used to further resolve luminal progenitor cells from differentiated cells. All antibodies were diluted in HF and all incubations were performed for 10 minutes on ice. Dead cells were excluded from the analysis by staining with 4′,6-diamino-2-phenylindole (DAPI, Invitrogen). Flow cytometric analysis was performed using a FACS Calibur and an LSRII (both from BD Biosciences) and data were analysed using FlowJo software (Tree Star, Olten, Switzerland). Cell sorting was performed using a FACS Aria (BD Biosciences).
Colony forming assays
Enzymatically dissociated mammary cells were mixed with γ-irradiated NIH3T3 cells in complete Mouse EpiCult-B media (StemCell Technologies). Cell suspensions were seeded in 60-mm cell culture dishes and maintained in a hypoxic incubator (5% oxygen, 5% carbon dioxide) for 7 days at 37°C. Colonies were fixed in acetone:methanol (1:1), stained with Giemsa stain and counted under a low power microscope.
Transplantation of mammary epithelium
Three-week-old WT (C57/BL6/FVB/N) or RIP140 KO/RIP140 Tg females had their endogenous mammary epithelium surgically removed as previously described (Deome et al., 1959). Freshly dissociated and sorted primary mammary epithelial cells derived from WT, RIP140 KO or RIP140 Tg mice were suspended in 25% growth factor-reduced Matrigel (BD Biosciences), diluted in PBS and injected into cleared fat pads. After 6 weeks, some of the animals were sacrificed and the glands stained with carmine or X-gal (see below). A subset of the recipient mice were mated and sacrificed at parturition or at day 15 of pregnancy, depending on genotype.
X-gal staining
Whole-mounts were prepared as described and fixed in acetone for 2 hours, then rinsed in PBS (3×30 minutes). Tissue was incubated in X-gal staining solution [PBS containing 2 mM MgCl2, 0.02% Nonidet P-40, 0.01% sodium deoxycholate, 20 mM K3Fe(CN)6, 20 mM K4Fe(CN)6.3H2O and 1 mg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside] at 37°C, overnight in the dark. The stained whole-mounts were then rinsed in PBS (3×30 minutes) and dehydrated as described above, before mounting in DPX.
Statistics
Data are presented as the mean ± s.e.m. and were analysed by Student’s t-test. P<0.05 was considered to be statistically significant.
RESULTS
RIP140 regulates ductal morphogenesis in the mammary gland
To study the role played by RIP140 in mammary gland development, we prepared mammary gland whole-mounts from 24-day-old, 5-week-old and 12-week-old WT and RIP140 KO mice. These time points represent pre-pubertal, pubertal and mature stages of mammary gland development, respectively. A rudimentary epithelium was observed at 24 days in both WT (Fig. 1A,A′) and RIP140 KO (Fig. 1B,B′) animals with no apparent differences in branching. At 5 weeks, the developing mammary tree had invaded approximately half the fat pad of WT mice with TEBs at the tips of the ducts (Fig. 1C,C′), but this was not apparent in RIP140 KO mice (Fig. 1D,D′). By 12 weeks, the mammary tree had filled the entire fat pad with extensive branching in WT mice (Fig. 1E,E′) but RIP140 KO mice exhibited a minimal ductal network (Fig. 1F,F′), indicating an essential role for RIP140 in mammary gland development. The mammary tree in the RIP140 KO mice did eventually fill the fat pad by 5 months of age, but the ductal network was sparse relative to that in heterozygous and WT mice (supplementary material Fig. S1).
Fig. 1.
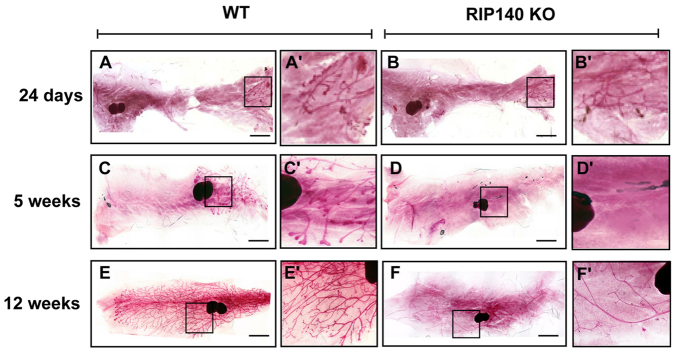
Regulation of mammary gland development by RIP140. (A-F) Mammary gland whole-mounts from representative WT (A,C,E) and RIP140 KO (B,D,F) mice at different stages of development. Whole-mounts were prepared from mice at 24 days (A,B), 5 weeks (C,D) and 12 weeks (E,F) (n=3-5). (A′-F′) Magnified images of the boxed areas in A-F, respectively. Scale bars: 2 mm.
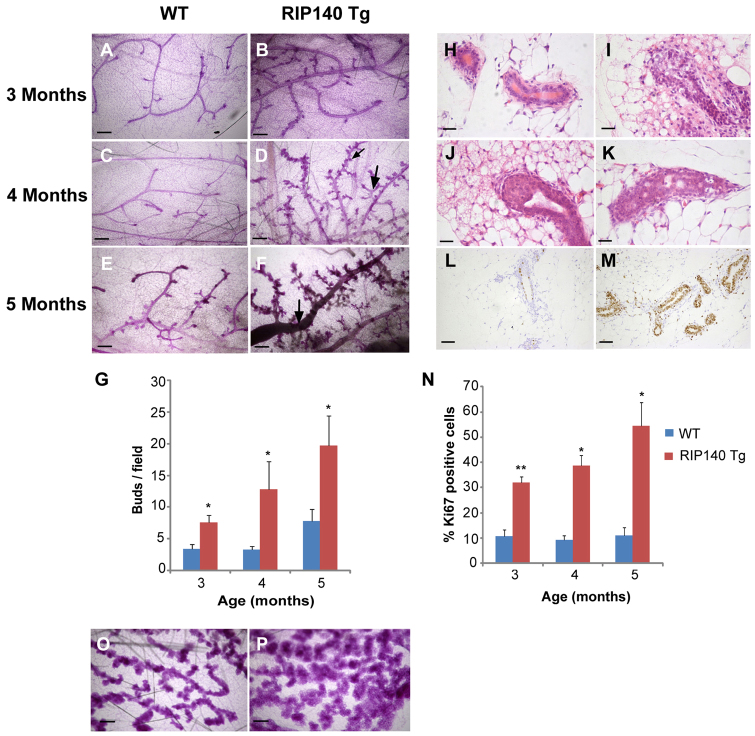
To examine the influence of overexpression of RIP140 (supplementary material Fig. S2A,B), we compared whole-mounts from aged-matched 3-, 4- and 5-month-old RIP140 Tg and WT mice. RIP140 Tg mice (Fig. 2B,D,F) developed numerous alveolar bud-like structures that became more extensive with age and the ductal network became more branched compared with the WT animals (Fig. 2A-G; supplementary material Fig. S3). By 4-5 months, the mammary glands in RIP140 Tg mice contained distended primary ducts (Fig. 2F), areas of hyperplastic growth (Fig. 2I), TEB-like structures (Fig. 2J) and structures with multiple cell layers (Fig. 2K); none of these features was present in the WT mammary epithelium (Fig. 2H). There was a marked increase in the proportion of proliferating (Ki67+) cells in mammary glands from RIP140 Tg compared with WT mice and the level of proliferation in RIP140 Tg mice increased with age (Fig. 2L-N). Thus, we conclude that RIP140 is a rate-limiting factor required for ductal development in the mammary epithelium. Analysis of mice at earlier time points indicated that there was no difference in the development of rudimentary ducts at day 24 (supplementary material Fig. S4A-B′) and that the ducts had progressed to a similar extent at 5 weeks (supplementary material Fig. S4C,D,E) although more buds were evident at this stage (supplementary material Fig. S4C′,D′,F). In addition, we examined the effect of RIP140 overexpression on alveolar development during pregnancy. We observed that RIP140 Tg mice had larger and denser alveolar structures (Fig. 2P) compared with WT mice (Fig. 2O), indicating that RIP140 also influences alveologenesis. Examination of the mammary glands from RIP140 KO mice during pregnancy was not possible because these animals are infertile as a result of defective ovulation.
Fig. 2.
Analysis of mammary glands from RIP140 Tg mice. (A-F) Photomicrographs of mammary glands from WT (A,C,E) and RIP140 Tg mice (B,D,F) at 3 months (A,B), 4 months (C,D) and 5 months (E,F). (D) The large arrow shows more side branching and the small arrow highlights the alveolar buds observed in RIP140 Tg mice. (F) Arrow shows a distended primary duct. (G) Quantification of hyperbudding. (H-K) Normal ducts are present in mammary gland sections from 5-month-old WT (H) mice whereas age-matched RIP140 Tg mice show hyperplastic areas (I), terminal end buds (J) and multi-layered ducts (K). (L,M) Ki67 staining in mammary gland sections from WT (L) and RIP140 Tg (M) mice. (N) Quantification of Ki67 staining at 3, 4 and 5 months for WT and RIP140 Tg mice. (O,P) Photomicrographs of mammary glands from day 15 pregnant WT (O) and RIP140 Tg (P) mice. (n=3). *P<0.05, **P<0.005. Error bars represent s.e.m. Scale bars: 400 μm in A-F,O,P; 50 μm in H-K; 100 μm in L,M.
RIP140 is required for oestrogen signalling in the mammary gland
Given the importance of oestrogen and progesterone signalling in mammary gland development, we next examined the expression of their receptors, a number of their targets and other transcription factors associated with steroid hormone signalling. The expression of ERα mRNA and protein in mammary glands from RIP140 KO mice were similar to those in WT mice (Fig. 3A,B); however, the expression of a number of oestrogen targets, including Areg, Pgr, cyclin D1 (Ccnd1) and Gata3 were markedly reduced at both the mRNA and protein levels in the absence of RIP140. Consistent with these observations, there was a decrease in the expression of genes downstream of progesterone signalling, including Wnt4 and calcitonin (supplementary material Fig. S5A). It is noteworthy that the expression levels of many other genes that we monitored, including the ERα target gene c-Myc (Myc), the transcription factors cJun (Jun), cFos (Fos) and Sp1, the growth factor Igf2 and RANKL (Tnfsf11) were maintained in the RIP140 KO mammary glands (supplementary material Fig. S5A). Thus, expression of only a subset of oestrogen receptor target genes was reduced in the absence of RIP140 in the mammary gland. The reduction in Areg and Pgr expression suggests that oestrogen signalling is impaired in the absence of RIP140. This, together with the observed partially impaired progesterone signalling, provides an explanation for the defect in primary duct formation and side-branching in RIP140 KO mice. Parallel gene expression analysis of RIP140 Tg and WT mice revealed similar levels of ERα, an increase in expression of Areg, Pgr, cyclin D1, Gata3 and a surge in the expression of Igf2 and RANKL in the RIP140 Tg mammary glands (supplementary material Fig. S5B). Immunocytochemical analysis of RANKL expression in mammary gland sections revealed that its expression was maintained in the absence of RIP140. In the RIP140 Tg mice, RANKL expression was more widespread, consistent with the increase in epithelial structures (supplementary material Fig. S6).
Fig. 3.
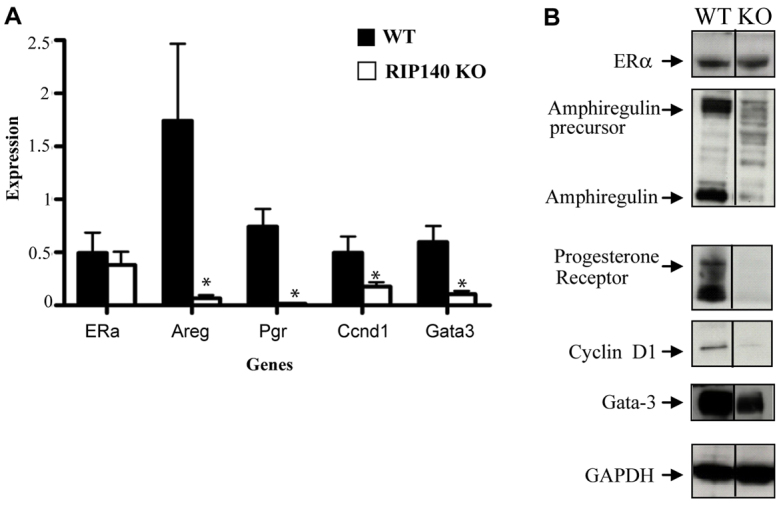
Expression analysis of the mammary glands. Q-PCR analysis (A) and protein expression (western blots) (B) of mammary glands from mature WT and RIP140 KO mice (n=4). Error bars represent s.e.m. *P<0.05.
Further evidence to implicate RIP140 in oestrogen signalling in the mammary gland was obtained by examining the ability of oestrogen to rescue mammary gland development in ovariectomised WT and RIP140 KO animals. Ovariectomy was performed at 3 weeks of age and mice were implanted with slow-releasing oestradiol (E2) or placebo pellets for 21 days. Whereas placebo treatment in both genotypes did not promote ductal growth (Fig. 4B,D), E2 treatment rescued ductal development in WT (Fig. 4A) but not in RIP140 KO mice (Fig. 4C), which failed to show any ductal elongation, indicating that RIP140 is essential for oestrogen-induced ductal morphogenesis in the mammary gland at puberty. The efficacy of the E2 pellets in mice was confirmed by examining their uterine response; both genotypes exhibited larger uteri with water retention (supplementary material Fig. S7). Both RIP140 KO and RIP140 Tg mice have a normal oestrus cycle and analysis of circulating levels of oestradiol in both RIP140 KO and RIP140 Tg mice revealed no significant differences in their levels. There was a slight reduction in circulating prolactin in RIP140 KO mice and a corresponding increase in RIP140 Tg mice but the differences were not statistically significant (supplementary material Fig. S8).
Fig. 4.
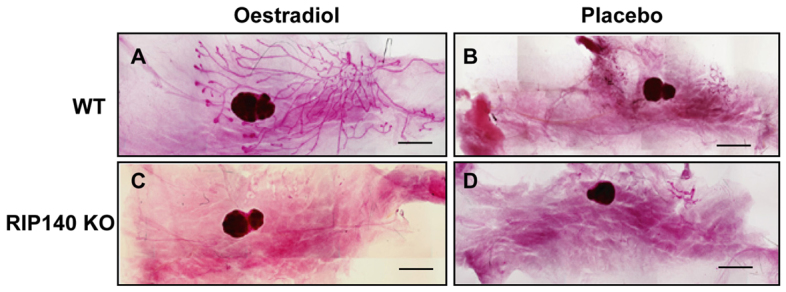
Ductal elongation rescue using slow-release oestrogen pellets. (A-D) Mammary gland whole-mounts from 3-week-old, ovariectomised WT (A,B) and RIP140 KO (C,D) mice which were implanted with 21-day slow release oestradiol (A,C) or placebo (B,D) pellets (n=3). Scale bars: 2 mm.
RIP140 is required in both the epithelial and the stromal compartments for the development of the mammary gland
RIP140 KO mice contain the lacZ gene expression cassette in place of the RIP140 coding exon, which allows β-galactosidase activity to be used as a marker for RIP140 gene expression (White et al., 2000). RIP140 expression depicted by blue staining is localised in both the epithelial and the stromal compartments in the mammary gland (supplementary material Fig. S9). To identify the mammary gland compartment in which RIP140 expression is essential for ductal morphogenesis, tissue recombination experiments were performed. To determine whether RIP140 expression is required in the mammary epithelium, inguinal mammary fat pads of 21-day-old C57BL/6 WT animals were surgically cleared of endogenous epithelium and either maintained as controls (no epithelium injected) or injected with equal numbers of flow-sorted epithelial cells from WT or RIP140 KO donors (supplementary material Fig. S10A,B) in contralateral fat pads. After 6 weeks, an epithelial network was evident in those mice injected with WT epithelium (Fig. 5B) but not in those injected with RIP140 KO epithelium (Fig. 5C) or in control mice (Fig. 5A). The RIP140-null epithelium was unable to repopulate the fat pad even when the number of epithelial cells injected was doubled. Thus, we conclude that RIP140 expression is essential in the epithelium for ductal morphogenesis.
Fig. 5.
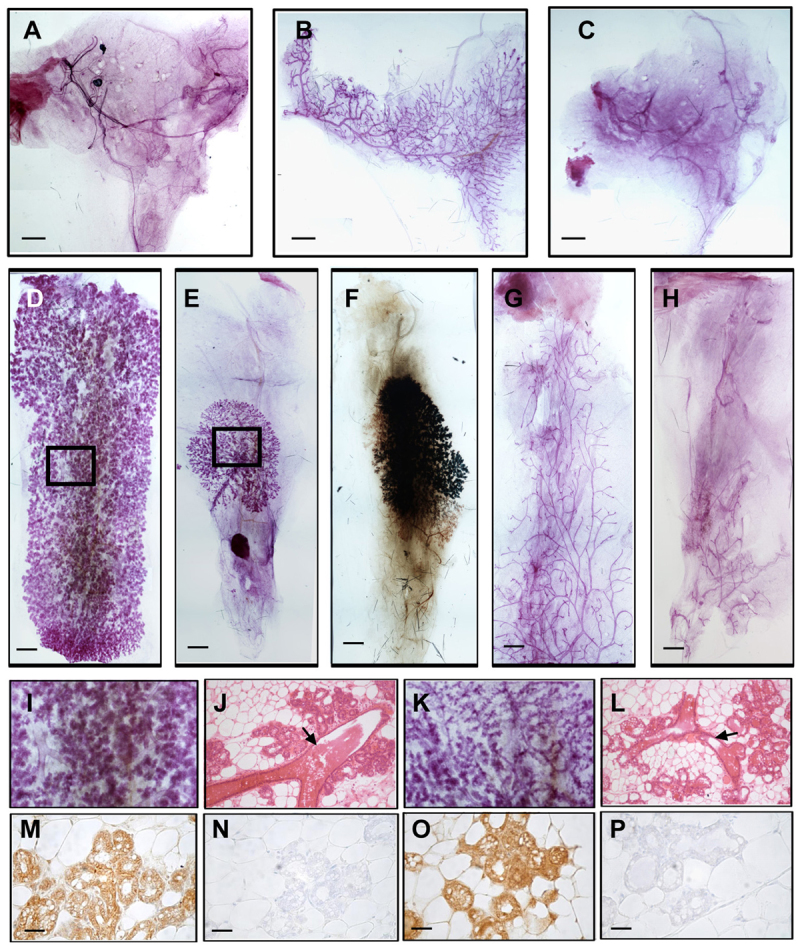
Tissue recombinants in the mammary gland. (A-C) Analysis of pubertal growth. Fat pads cleared of endogenous epithelium from 3-week-old WT mice were either injected with no donor epithelium (A) or injected with epithelium from WT (B) or RIP140 KO (C) donors. Images represent the recipient fat pads after 6 weeks of injections. (D-F,I-L) Analysis of differentiation during pregnancy. A subset of the donor epithelium-injected WT animals were mated and sacrificed at parturition. (D,I,J) Injected WT epithelium shows differentiation throughout the injected fat pad (D) with well formed alveolar structures (I), cross-sections through which show well-formed primary ducts (arrow) and alveolar structures (J). (E,F,K,L) The injected RIP140 KO epithelium shows localised differentiation (E), is positive for X-gal staining (F) but shows scantier and smaller alveolar structures (K) and thinner primary ducts (L, arrow) (n=5). (G,H) Fat pads from 3-week-old RIP140 KO animals cleared of endogenous epithelium, injected with donor epithelium from WT animals showing minimal ductal growth (H) compared with the control animals (G) (n=7). (M-P) RANKL staining in sections obtained from tissue recombinants obtained at parturition. RANKL staining in WT recipients that received WT epithelium (M) with its no primary antibody control (N) and WT recipients that received RIP140 KO epithelium (O) with no primary antibody control (P) (n=3). Scale bars: 1 mm in A-H; 50 μm in M-P.
Next, we investigated the effect of pregnancy on the growth of the tissue recombinants by examining their morphology at parturition. Recombinants in which WT epithelium was transplanted generated differentiated alveolar structures throughout the fat pad (Fig. 5D). Interestingly, recombinants containing RIP140 KO epithelium also had the potential to generate differentiated alveolar outgrowths (Fig. 5E), albeit to a lesser extent than those containing WT epithelium. We were able to confirm that this engraftment was derived from the RIP140 KO epithelium because the outgrowths stained blue as a consequence of positive β-galactosidase activity (Fig. 5F). Thus, although the KO epithelium gave rise to thinner primary ducts and sparse alveolar structures upon transplantation (Fig. 5K,L) compared with WT epithelium (Fig. 5I,J), the lack of mammary gland development observed in the absence of RIP140 could be partially reversed in the presence of pregnancy-associated hormones, indicating that certain pathways are still conserved in RIP140 KO epithelial cells. To evaluate this further, we examined RANKL expression, which is a downstream effector of progesterone and prolactin signalling implicated in alveologenesis. We found that the expression of this mitogen was maintained in the alveolar structures generated in tissue recombinants with RIP140 KO epithelium (Fig. 5O,P), suggesting that a subset of pathways that promote alveologenesis do not require RIP140 expression. To determine whether stromal expression of RIP140 is required in mammary gland development, we injected mammary epithelial cells from WT animals into the cleared fat pads of 3-week-old RIP140 KO and control WT mice. After 6 weeks, control WT recipients showed ductal elongation (Fig. 5G) but none of the RIP140 KO fat pads supported the growth of WT epithelium (Fig. 5H). Thus, the expression of RIP140 is essential in both the epithelial and the stromal compartments to support ductal morphogenesis of the mammary gland.
Q-PCR analysis of FACS-sorted mammary gland cells for expression of human RIP140 (hRIP140) (supplementary material Fig. S11A,B) indicated that RIP140 was overexpressed in both stroma and epithelium (luminal and basal cells). The effect of RIP140 overexpression in stroma and epithelium was examined in tissue recombinants during pubertal growth and differentiation during pregnancy. Whereas the control fat pads in which the endogenous epithelium was surgically removed did not show any ductal network after 6 weeks (supplementary material Fig. S11C), all the other tissue recombinants injected with sorted epithelium (supplementary material Fig. S10C,D), namely WT recipients injected with WT epithelium (supplementary material Fig. S11D), WT recipients injected with epithelium derived from RIP140 Tg glands (supplementary material Fig. S11E) and RIP140 Tg recipients injected with WT epithelium (supplementary material Fig. S11F), demonstrated ductal growth after 6 weeks. Quantification of the number of terminal end buds in each of the tissue recombinants revealed that both the RIP140 Tg epithelium and RIP140 Tg stroma promoted the formation of more terminal end buds compared with the WT (supplementary material Fig. S11G). Because RIP140 Tg mice do not usually survive through late pregnancy, a subset of tissue recombinants were analysed at day 15 of pregnancy. No morphological differences were evident in alveolar structures in WT mice that received WT (supplementary material Fig. S11H-H″) or RIP140 Tg epithelium (supplementary material Fig. S11I-I″); however, recombinants that received RIP140 Tg epithelium exhibited slightly denser arrangement of alveolar structures compared with WT controls. Interestingly, after 15 days of pregnancy, RIP140 Tg fat pads that received WT epithelium were distinct from WT recipients (supplementary material Fig. S11J-J″) and cross-sections revealed that they resembled lactating alveolar structures (supplementary material Fig. S11J″), albeit to a lesser extent than intact RIP140 Tg mammary glands at this stage (supplementary material Fig. S11K-K″). Thus, we conclude that RIP140 overexpression in both the epithelium and the stroma contribute to the generation of densely arranged alveoli.
RIP140 regulates mammary cell populations
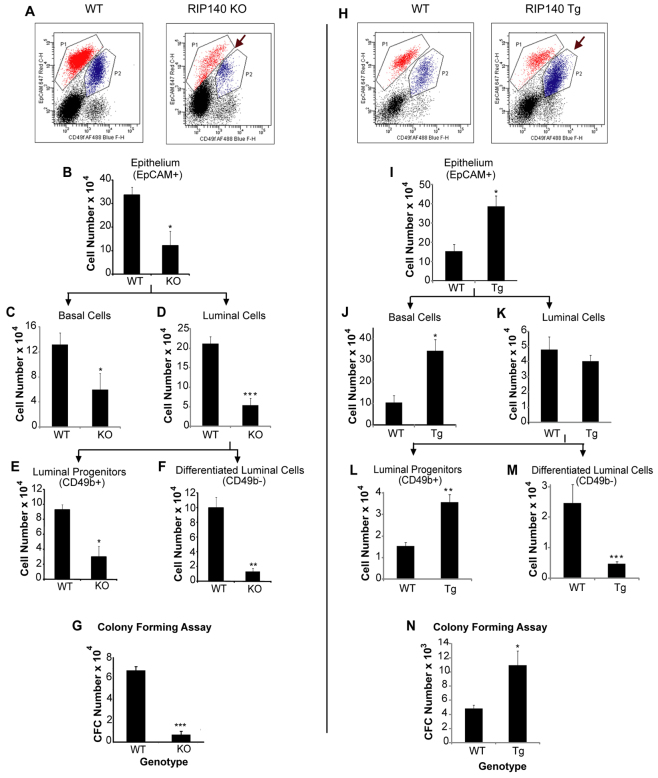
To examine the influence of RIP140 on the development and maintenance of subtypes of mammary cells, we used flow cytometry to quantify the different cell populations in WT, RIP140 KO and RIP140 Tg mice. Mammary glands from mature WT (C57BL/6)/RIP140 KO and WT (FVB/N) /RIP140 Tg mice were dissected and dissociated into single-cell suspensions and immunostained to identify haematopoietic cells, endothelial cells and a fraction of the stromal cells so they could be excluded from the subsequent analysis. The remaining cells (termed Lin–) were also stained to detect expression of EpCAM, CD49f (Itga6 – Mouse Genome Informatics) and CD49b (Itga2 – Mouse Genome Informatics). Staining with this panel of markers permits resolution of stromal (EpCAM–) and epithelial (EpCAM+) cell populations. Epithelial cell populations were further resolved into basal (EpCAMlowCD49fhigh) and luminal (EpCAMhighCD49flow) cells. Within the luminal cell population, CD49b marker was used to resolve the differentiated luminal cells (CD49b–) from the luminal progenitors (CD49b+) (Shehata et al., 2012). We observed that although there was no difference in the total number of stromal cells between the WT and RIP140 KO mice, the total number of epithelial cells was lower in RIP140 KO mice (Fig. 6A,B; supplementary material Fig. S12). Within the epithelium there was a reduction in both the basal and luminal cell populations in the RIP140 KO glands (Fig. 6C,D). Within the luminal compartment, both the differentiated and progenitor populations were reduced in the RIP140 KO mammary glands (Fig. 6E,F). Analysis of mammary cell populations from RIP140 Tg/WT mice revealed an increase in the epithelial cell numbers (Fig. 6I) but, interestingly, this increase was only due to an enhanced basal cell population (Fig. 6J,H). However, when the luminal cell population (Fig. 6K) was further resolved into luminal progenitors and differentiated cells there was an increase in the progenitor cell number (Fig. 6L) and a reduction in the number of differentiated cells (Fig. 6M) in the RIP140 Tg compared with the WT mammary glands. Thus, RIP140 is a key determinant in maintaining cell identity in the mammary epithelium.
Fig. 6.
Analysis of the distribution of mammary gland cell populations in adult virgin WT (C57BL/6)/RIP140 KO and WT (FVB/N)/RIP140 Tg mice. (A,H) CD49f/EpCAM dot plots showing stromal (black), luminal (red, P1) and basal (blue, P2) cell populations in WT/RIP140 KO (A) and WT/RIP140 Tg (H) mice. (A) Arrow indicates reduced luminal and basal cell populations in RIP140 KO mice. (H) Arrow indicates enhanced basal cell population in the RIP140 Tg compared with WT. (B-F) Total number of epithelial cells (B), basal cells (C), luminal cells (D), luminal progenitors (E) and differentiated luminal cells (F) per gland in WT/RIP140 KO mice (n=3). (I-M) Total number of epithelial cells (I), basal cells (J), luminal cells (K), luminal progenitors (L) and differentiated luminal cells (M) per gland in WT/RIP140 Tg mice (n=4). *P<0.05, **P<0.005, ***P<0.0001. (G,N) Absolute number of colony-forming cells (CFCs) per mammary gland in WT/RIP140 KO (G) and WT/RIP140 Tg mice (N) (n=3). *P<0.05, ***P<0.0005. Error bars represent s.e.m.
We also performed in vitro colony forming cell (CFC) assays in order to validate the observation that there is a decreased number of progenitors in the RIP140 KO mice and an increased number in the RIP140 Tg mice compared with WT mice. Mammary glands isolated from RIP140 KO mice contain ∼90% fewer CFCs than age-matched WT mammary glands (Fig. 6G), whereas the RIP140 Tg (Fig. 6N) glands demonstrated an approximate doubling of the number of CFCs compared with WT.
RIP140 is recruited to the promoters and enhancers of several oestrogen-regulated genes along with ERα
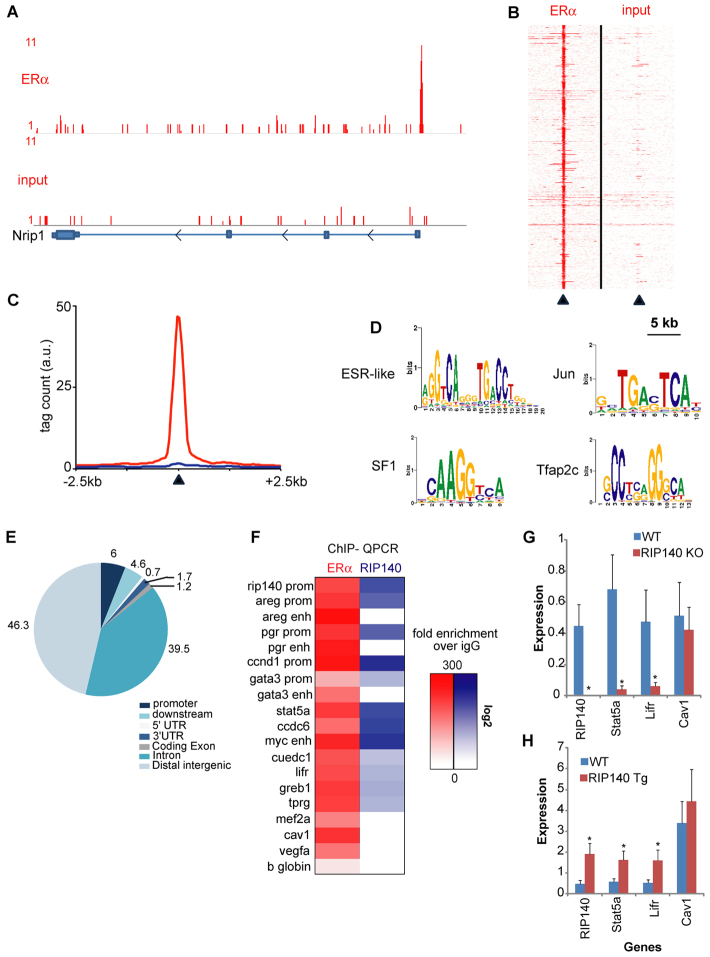
Given that our preliminary studies indicate that oestrogen signalling is impaired in RIP140-null mice leading to a reduction in the expression of several key oestrogen-regulated genes, we explored ERα and RIP140 binding to chromatin in mammary glands from 6-week-old mice in which mammary epithelial development is underway. Using ChIP-seq analysis, we were able to obtain high quality data for ERα-chromatin interactions, which enabled an unbiased Q-PCR-based analysis of RIP140 binding events by interrogating the newly found ERα binding events for RIP140 presence. ERα-chromatin interactions were readily detected compared with input negative control as exemplified at the RIP140 locus (Fig. 7A). The 881 high quality chromatin interaction sites found for ERα are shown as a heatmap visualisation (Fig. 7B) and quantified as a 2D graph (Fig. 7C). Of these, 458 peaks were detected within 20 kb of gene promoters (supplementary material Table S2), which is the genomic distance within which most ERα-mediated gene regulation takes place (Fullwood et al., 2009). Next, the observed chromatin interaction patterns were analysed for enriched motifs. As expected, ESR-like motifs as well as SF1, Jun (Gaub et al., 1990) and TFAP2 (Woodfield et al., 2007) motifs were observed (Fig. 7D). We found only 6% of chromatin interactions of ERα at the promoters; the vast majority of binding events were found at enhancers and introns (Fig. 7E). Previously described ERα-responsive promoters as well as some newly identified ERα-associated enhancers were subsequently tested for RIP140 co-occupancy using ChIP-Q-PCR (Fig. 7F). We found that RIP140 was recruited with ERα on its own promoter (Fig. 7A,F) as well as promoters of the genes encoding several mammary gland mitogens and regulators, including Areg, Pgr, Ccnd1 and Gata3. Whereas RIP140 recruitment was detected on distal enhancer elements of several genes, e.g. Stat5a, Myc, Lifr and Greb1, it was absent from several other proximal ERα binding sites, e.g. Cav1 and Vegfa, as well as enhancer regions for Areg, Pgr and Gata3 (Fig. 7F). Thus, although a sizeable number of ERα binding sites were co-occupied by RIP140 there were other sites for which RIP140 recruitment was not found, indicating selectivity in RIP140 binding. These data are consistent with the gene expression of Areg, Pgr, Ccnd1 and Gata3 in RIP140 KO and RIP140 Tg mice (Fig. 3; supplementary material Fig. S5). Interestingly, although we found that RIP140 was recruited with ERα on the Myc enhancer, Myc expression was unaltered in both RIP140 KO and RIP140 Tg mammary tissues, indicating that either this site is not regulating Myc transcription or that RIP140 binding is dispensable for regulation from this site. To validate further the role of RIP140 in gene regulation on some of the newly identified sites, we compared gene expression in RIP140 KO, RIP140 Tg and WT mice. Expression of Stat5a, RIP140 and Lifr was modulated by RIP140 whereas Cav1 expression was maintained in both the RIP140 KO and RIP140 Tg, indicating that it is not a RIP140 target despite ERα recruitment (Fig. 7G,H). Selective binding might be determined by the architecture (transcription factor binding elements) of the DNA sequence but, importantly, RIP140 together with ERα regulates the transcription of mitogenic signals emanating from the mammary gland.
Fig. 7.
Chromatin interactions of ERα and RIP140 in the mouse mammary gland. (A) Genome browser snapshot of ERα binding events (top) over input control (bottom) at the RIP140 promoter region. Tag count and genomic coordinates are indicated. (B) Heatmap visualisation of raw read counts for ERα ChIP-seq (left) or input control (right) of all 881 identified binding events. A 5-kb window around the top of the peak (arrowhead) is shown. (C) Quantification of signal intensities of binding events as shown in B, depicting ERα (red) and input control (blue) at a 2.5-kb window around the top of the peak region (indicated by arrowhead). (D) Top-enriched motifs for ERα-chromatin binding events. (E) Genomic distributions of ERα binding event, related to the most proximal gene. (F) Heatmap visualization of ERα (red) and RIP140 (blue) binding events as confirmed by Q-PCR. Fold enrichment over IgG control is calculated and shown in a heatmap on log scale. (G,H) Q-PCR analysis of RIP140 expression and other ERα targets in WT/RIP140 KO (G) and WT/RIP140 Tg (H) mammary glands (n=4). *P<0.05. Error bars represent s.e.m.
DISCUSSION
In this article, we demonstrate that RIP140 is an important mediator of oestrogen-induced mammary epithelial cell proliferation as loss of RIP140 severely impairs the formation of TEBs and the ductal elongation and branching morphogenesis that occurs during puberty. This phenotype mirrors that observed in mammary glands in ER KO mice (Mallepell et al., 2006) and thus supports a role for RIP140 in oestrogen-mediated ductal development. This phenomenon appears to be intrinsic to the mammary epithelial cells and is not due to systemic effects, such as disruption of the hypothalamic pituitary axis, because tissue recombination experiments clearly demonstrate that RIP140 KO epithelium is unable to repopulate the normal mammary fat pad, even in a WT host animal. This observation is in agreement with our previous studies that demonstrate that infertility in the RIP140 KO females could be reversed by transplanting WT ovaries, indicating an intact hypothalamic pituitary axis (Leonardsson et al., 2002).
The molecular mechanism by which RIP140 appears to function in the mammary gland is not via transcriptional regulation of ERα expression; instead, RIP140 appears to modulate the expression of several ERα target genes, such as Areg, Pgr, Ccnd1 and Stat5a, by functioning as a co-factor that is recruited with ERα to their promoters/enhancers. When RIP140 expression is ablated, there is loss of expression of Areg and Pgr and this results in profound developmental defects because these factors are essential for subsequent mammary development (Lydon et al., 1995; Ismail et al., 2002; Ciarloni et al., 2007). In contrast to the impaired mammary gland development observed in the RIP140 KO mice, overexpression of RIP140 results in increased side branching and development of alveolar bud-like structures along with increased cell division, which in some cases, results in epithelial hyperplasia and the formation of TEB-like structures in older (e.g. 5-month-old) transgenic animals. The observed hyperplastic structures can be described as simple ductal hyperplasia and are potential tumour precursors (Medina, 1976); however, we were unable to extend these studies to test this directly as RIP140 animals die prematurely because of heart defects (Fritah et al., 2010).
Tissue recombination experiments also demonstrated that RIP140 expression is required in the mammary stroma in order for ductal development to occur. The requirement of RIP140 indicates that although there are similarities in the phenotype of ER KO and RIP140 KO animals, there are also important differences as ERα expression is not required in the stroma for normal mammary development (Mallepell et al., 2006). A comparison of the distribution of ERα and RIP140 shows that, despite there being an overlap in their expression in the body cells and luminal cells in the TEBs and mature ducts, respectively, RIP140 is expressed in cap cells and the basal cells, which are devoid of ERα (supplementary material Fig. S13). Gene expression analysis of purified subsets of mouse mammary cell populations (Shehata et al., 2012) reveals that several nuclear receptors and transcription factors with which RIP140 is known to act as a co-regulator have a differential distribution within these subpopulations (supplementary material Table S3; Fig. S14). As RIP140 is expressed both in the stroma and epithelium, as are several of its potential nuclear receptor partners, it is conceivable that RIP140 could drive transcription of gene targets through several nuclear receptors, including ERα. If so, this could explain why although ERα expression is required only in the epithelium, the expression of RIP140 is essential both in the mammary epithelium and the stroma. It is somewhat surprising that the RIP140 KO epithelium, which failed to show pubertal ductal elongation, could still undergo alveolar differentiation; however, the alveoli generated were smaller and sparser than those obtained with WT controls, indicating that RIP140 is important for the generation of factors that influence cell proliferation and maintenance during pregnancy. The observation that expression levels of RANKL in tissue recombinants derived from RIP140 KO epithelium are comparable to those in the WT indicates that certain pathways that drive alveolar differentiation (Fata et al., 2000; Fernandez-Valdivia et al., 2009) are conserved in the RIP140 KO epithelium.
Mammary stem cells and certain progenitor cells within the mammary epithelium do not express ERα or Pgr (Asselin-Labat et al., 2006), but yet are highly sensitive to oestrogen and progesterone (Asselin-Labat et al., 2010; Joshi et al., 2010). It is now emerging that steroid hormones, in addition to having direct effects on relatively mature steroid receptor-expressing progenitor cells, also strongly influence mammary stem and steroid hormone-negative progenitor cells via paracrine interactions that are mediated by the RANK ligands Wnt4 and Areg (Brisken et al., 2000; Ciarloni et al., 2007; Beleut et al., 2010). Thus, the loss of expression of some of these paracrine factors in RIP140 KO mice probably accounts for the reduction in the number of stem and progenitor cells observed in the mammary glands of these mice. Similarly, the surge in RANKL expression in the RIP140 Tg glands, along with other factors, including Stat5 (Furth et al., 2011), could account for the increased number of luminal progenitors and basal cells. It is noteworthy that although expression of RANKL and Igf2 was unaltered in RIP140 KO mice, their expression was greatly amplified in RIP140 Tg animals compared with the WT mice, indicating that this might be a consequence of indirect effects of RIP140 overexpression.
Several co-factors, such as steroid receptor co-activator 1 (SRC-1; Ncoa1 – Mouse Genome Informatics), steroid receptor co-activator 3 (SRC-3; Ncoa3 – Mouse Genome Informatics), CBP/p300 interacting transactivator 1 (Cited1) and mediator-1 (Med1), have been shown to play a role in mammary gland development and breast cancer (Han et al., 2006; Howlin et al., 2006; Jiang et al., 2010), and the binding profiles of these co-factors identify specific transcriptional networks that influence clinical outcome in breast cancer (Zwart et al., 2011). Here, we demonstrate that RIP140 is a co-factor for ERα and that it regulates ductal formation during mammary gland development, but it might also potentially act as a co-factor for other nuclear receptors and transcription factors. RIP140 is a known co-factor for multiple other nuclear receptors, including PPARg (PPARG), farnesoid X receptor (FXR; NR1H4) and estrogen-related receptors (Christian et al., 2006), all of which have been implicated in breast cancer progression (Fenner and Elstner, 2005; Riggins et al., 2010; Giordano et al., 2011). Further analysis of the mechanisms of action of RIP140 in breast cancer will provide new insights and might point towards potential pathways for the development of new strategies for breast cancer treatment.
Supplementary Material
Acknowledgments
We thank Dr Amel Saadi, Dr Mona Shehata and Rajshekhar Girradi for valuable technical advice for performing the mammary gland tissue recombinants and FACS analysis. We also thank Mr Reece Williams and Ms Maeve Troy for their support in the animal facility. In addition we thank Ron Kerkhoven, Arno Velds and Marja Nieuwland for Solexa sequencing. We are grateful to Prof. Daniel Medina for expert advice on the mammary hyperplastic structures found in the RIP140 Tg mice; and Drs Simak Ali and Laki Buluwela for critical reading of the manuscript.
Footnotes
Funding
This work was supported by a Wellcome Trust grant [079200/Z/06/Z to J.N., J.H.S., A.P. and M.G.P.]; a Biotechnology and Biological Sciences Research Council grant [BB/C504327/1 to M.R.M. and M.G.P.]; and the Genesis Research Trust. J.S. is supported by Cancer Research UK, The University of Cambridge and Hutchison Whampoa Limited. Deposited in PMC for immediate release.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
J.N. and M.G.P. conceived the project. J.N., J.H.S., M.P., W.Z., J.S. and M.G.P. designed the research. J.N., J.H.S., M.R.M., O.O., A.P., X.A., W.Z., N.W. and J.S. carried out the research. J.S. critically reviewed the manuscript. J.N. and M.G.P. wrote the manuscript.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.085720/-/DC1
References
- Asselin-Labat M. L., Shackleton M., Stingl J., Vaillant F., Forrest N. C., Eaves C. J., Visvader J. E., Lindeman G. J. (2006). Steroid hormone receptor status of mouse mammary stem cells. J. Natl. Cancer Inst. 98, 1011–1014 [DOI] [PubMed] [Google Scholar]
- Asselin-Labat M. L., Vaillant F., Sheridan J. M., Pal B., Wu D., Simpson E. R., Yasuda H., Smyth G. K., Martin T. J., Lindeman G. J., et al. (2010). Control of mammary stem cell function by steroid hormone signalling. Nature 465, 798–802 [DOI] [PubMed] [Google Scholar]
- Beleut M., Rajaram R. D., Caikovski M., Ayyanan A., Germano D., Choi Y., Schneider P., Brisken C. (2010). Two distinct mechanisms underlie progesterone-induced proliferation in the mammary gland. Proc. Natl. Acad. Sci. USA 107, 2989–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchinfuso W. P., Lindzey J. K., Hewitt S. C., Clark J. A., Myers P. H., Cooper R., Korach K. S. (2000). Induction of mammary gland development in estrogen receptor-alpha knockout mice. Endocrinology 141, 2982–2994 [DOI] [PubMed] [Google Scholar]
- Brisken C., O’Malley B. (2010). Hormone action in the mammary gland. Cold Spring Harb. Perspect. Biol. 2, a003178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisken C., Heineman A., Chavarria T., Elenbaas B., Tan J., Dey S. K., McMahon J. A., McMahon A. P., Weinberg R. A. (2000). Essential function of Wnt-4 in mammary gland development downstream of progesterone signaling. Genes Dev. 14, 650–654 [PMC free article] [PubMed] [Google Scholar]
- Cavaillès V., Dauvois S., Danielian P. S., Parker M. G. (1994). Interaction of proteins with transcriptionally active estrogen receptors. Proc. Natl. Acad. Sci. USA 91, 10009–10013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaillès V., Dauvois S., L’Horset F., Lopez G., Hoare S., Kushner P. J., Parker M. G. (1995). Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 14, 3741–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. M., Lykkesfeldt A. E., Parker M. G., Dowsett M. (1999). Expression of nuclear receptor interacting proteins TIF-1, SUG-1, receptor interacting protein 140, and corepressor SMRT in tamoxifen-resistant breast cancer. Clin. Cancer Res. 5, 3460–3467 [PubMed] [Google Scholar]
- Christian M., Kiskinis E., Debevec D., Leonardsson G., White R., Parker M. G. (2005). RIP140-targeted repression of gene expression in adipocytes. Mol. Cell. Biol. 25, 9383–9391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M., White R., Parker M. G. (2006). Metabolic regulation by the nuclear receptor corepressor RIP140. Trends Endocrinol. Metab. 17, 243–250 [DOI] [PubMed] [Google Scholar]
- Ciarloni L., Mallepell S., Brisken C. (2007). Amphiregulin is an essential mediator of estrogen receptor alpha function in mammary gland development. Proc. Natl. Acad. Sci. USA 104, 5455–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deome K. B., Faulkin L. J., Jr, Bern H. A., Blair P. B. (1959). Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 19, 515–520 [PubMed] [Google Scholar]
- Docquier A., Harmand P. O., Fritsch S., Chanrion M., Darbon J. M., Cavaillès V. (2010). The transcriptional coregulator RIP140 represses E2F1 activity and discriminates breast cancer subtypes. Clin. Cancer Res. 16, 2959–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fata J. E., Kong Y. Y., Li J., Sasaki T., Irie-Sasaki J., Moorehead R. A., Elliott R., Scully S., Voura E. B., Lacey D. L., et al. (2000). The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell 103, 41–50 [DOI] [PubMed] [Google Scholar]
- Fenner M. H., Elstner E. (2005). Peroxisome proliferator-activated receptor-gamma ligands for the treatment of breast cancer. Expert Opin. Investig. Drugs 14, 557–568 [DOI] [PubMed] [Google Scholar]
- Fernandez-Valdivia R., Mukherjee A., Ying Y., Li J., Paquet M., DeMayo F. J., Lydon J. P. (2009). The RANKL signaling axis is sufficient to elicit ductal side-branching and alveologenesis in the mammary gland of the virgin mouse. Dev. Biol. 328, 127–139 [DOI] [PubMed] [Google Scholar]
- Fritah A., Steel J. H., Nichol D., Parker N., Williams S., Price A., Strauss L., Ryder T. A., Mobberley M. A., Poutanen M., et al. (2010). Elevated expression of the metabolic regulator receptor-interacting protein 140 results in cardiac hypertrophy and impaired cardiac function. Cardiovasc. Res. 86, 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood M. J., Liu M. H., Pan Y. F., Liu J., Xu H., Mohamed Y. B., Orlov Y. L., Velkov S., Ho A., Mei P. H., et al. (2009). An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462, 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth P. A., Nakles R. E., Millman S., Diaz-Cruz E. S., Cabrera M. C. (2011). Signal transducer and activator of transcription 5 as a key signaling pathway in normal mammary gland developmental biology and breast cancer. Breast Cancer Res. 13, 220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaub M. P., Bellard M., Scheuer I., Chambon P., Sassone-Corsi P. (1990). Activation of the ovalbumin gene by the estrogen receptor involves the fos-jun complex. Cell 63, 1267–1276 [DOI] [PubMed] [Google Scholar]
- Giordano C., Catalano S., Panza S., Vizza D., Barone I., Bonofiglio D., Gelsomino L., Rizza P., Fuqua S. A., Andò S. (2011). Farnesoid X receptor inhibits tamoxifen-resistant MCF-7 breast cancer cell growth through downregulation of HER2 expression. Oncogene 30, 4129–4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjorevski N., Nelson C. M. (2011). Integrated morphodynamic signalling of the mammary gland. Nat. Rev. Mol. Cell Biol. 12, 581–593 [DOI] [PubMed] [Google Scholar]
- Han S. J., DeMayo F. J., Xu J., Tsai S. Y., Tsai M. J., O’Malley B. W. (2006). Steroid receptor coactivator (SRC)-1 and SRC-3 differentially modulate tissue-specific activation functions of the progesterone receptor. Mol. Endocrinol. 20, 45–55 [DOI] [PubMed] [Google Scholar]
- Hannafon B. N., Sebastiani P., de las Morenas A., Lu J., Rosenberg C. L. (2011). Expression of microRNA and their gene targets are dysregulated in preinvasive breast cancer. Breast Cancer Res. 13, R24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H. H., Meyer C. A., Shin H., Bailey S. T., Wei G., Wang Q., Zhang Y., Xu K., Ni M., Lupien M., et al. (2010). Nucleosome dynamics define transcriptional enhancers. Nat. Genet. 42, 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery D. M., Kalkhoven E., Hoare S., Parker M. G. (1997). A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387, 733–736 [DOI] [PubMed] [Google Scholar]
- Herzog B., Hallberg M., Seth A., Woods A., White R., Parker M. G. (2007). The nuclear receptor cofactor, receptor-interacting protein 140, is required for the regulation of hepatic lipid and glucose metabolism by liver X receptor. Mol. Endocrinol. 21, 2687–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinck L., Silberstein G. B. (2005). Key stages in mammary gland development: the mammary end bud as a motile organ. Breast Cancer Res. 7, 245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlin J., McBryan J., Napoletano S., Lambe T., McArdle E., Shioda T., Martin F. (2006). CITED1 homozygous null mice display aberrant pubertal mammary ductal morphogenesis. Oncogene 25, 1532–1542 [DOI] [PubMed] [Google Scholar]
- Ismail P. M., Li J., DeMayo F. J., O’Malley B. W., Lydon J. P. (2002). A novel LacZ reporter mouse reveals complex regulation of the progesterone receptor promoter during mammary gland development. Mol. Endocrinol. 16, 2475–2489 [DOI] [PubMed] [Google Scholar]
- Ji X., Li W., Song J., Wei L., Liu X. S. (2006). CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 34, W551–W554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Hu Q., Ito M., Meyer S., Waltz S., Khan S., Roeder R. G., Zhang X. (2010). Key roles for MED1 LxxLL motifs in pubertal mammary gland development and luminal-cell differentiation. Proc. Natl. Acad. Sci. USA 107, 6765–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi P. A., Jackson H. W., Beristain A. G., Di Grappa M. A., Mote P. A., Clarke C. L., Stingl J., Waterhouse P. D., Khokha R. (2010). Progesterone induces adult mammary stem cell expansion. Nature 465, 803–807 [DOI] [PubMed] [Google Scholar]
- Król E., Martin S. A., Huhtaniemi I. T., Douglas A., Speakman J. R. (2011). Negative correlation between milk production and brown adipose tissue gene expression in lactating mice. J. Exp. Biol. 214, 4160–4170 [DOI] [PubMed] [Google Scholar]
- L’Horset F., Dauvois S., Heery D. M., Cavaillès V., Parker M. G. (1996). RIP-140 interacts with multiple nuclear receptors by means of two distinct sites. Mol. Cell. Biol. 16, 6029–6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Medina D., Tsimelzon A., Mohsin S. K., Mao S., Wu Y., Allred D. C. (2007). Alterations of gene expression in the development of early hyperplastic precursors of breast cancer. Am. J. Pathol. 171, 252–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardsson G., Jacobs M. A., White R., Jeffery R., Poulsom R., Milligan S., Parker M. (2002). Embryo transfer experiments and ovarian transplantation identify the ovary as the only site in which nuclear receptor interacting protein 1/RIP140 action is crucial for female fertility. Endocrinology 143, 700–707 [DOI] [PubMed] [Google Scholar]
- Leonardsson G., Steel J. H., Christian M., Pocock V., Milligan S., Bell J., So P. W., Medina-Gomez G., Vidal-Puig A., White R., et al. (2004). Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc. Natl. Acad. Sci. USA 101, 8437–8442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydon J. P., DeMayo F. J., Funk C. R., Mani S. K., Hughes A. R., Montgomery C. A., Jr, Shyamala G., Conneely O. M., O’Malley B. W. (1995). Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 9, 2266–2278 [DOI] [PubMed] [Google Scholar]
- Mallepell S., Krust A., Chambon P., Brisken C. (2006). Paracrine signaling through the epithelial estrogen receptor alpha is required for proliferation and morphogenesis in the mammary gland. Proc. Natl. Acad. Sci. USA 103, 2196–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally S., Martin F. (2011). Molecular regulators of pubertal mammary gland development. Ann. Med. 43, 212–234 [DOI] [PubMed] [Google Scholar]
- Medina D. (1976). Preneoplastic lesions in murine mammary cancer. Cancer Res. 36, 2589–2595 [PubMed] [Google Scholar]
- Nautiyal J., Steel J. H., Rosell M. M., Nikolopoulou E., Lee K., Demayo F. J., White R., Richards J. S., Parker M. G. (2010). The nuclear receptor cofactor receptor-interacting protein 140 is a positive regulator of amphiregulin expression and cumulus cell-oocyte complex expansion in the mouse ovary. Endocrinology 151, 2923–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D. S., Troester M. A., Usary J., Hu Z., He X., Fan C., Wu J., Carey L. A., Perou C. M. (2006). Estrogen-regulated genes predict survival in hormone receptor-positive breast cancers. J. Clin. Oncol. 24, 1656–1664 [DOI] [PubMed] [Google Scholar]
- Riggins R. B., Mazzotta M. M., Maniya O. Z., Clarke R. (2010). Orphan nuclear receptors in breast cancer pathogenesis and therapeutic response. Endocr. Relat. Cancer 17, R213–R231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulli S. B., Kuorelahti A., Karaer O., Pelliniemi L. J., Poutanen M., Huhtaniemi I. (2002). Reproductive disturbances, pituitary lactotrope adenomas, and mammary gland tumors in transgenic female mice producing high levels of human chorionic gonadotropin. Endocrinology 143, 4084–4095 [DOI] [PubMed] [Google Scholar]
- Schmidt D., Wilson M. D., Spyrou C., Brown G. D., Hadfield J., Odom D. T. (2009). ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 48, 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A., Steel J. H., Nichol D., Pocock V., Kumaran M. K., Fritah A., Mobberley M., Ryder T. A., Rowlerson A., Scott J., et al. (2007). The transcriptional corepressor RIP140 regulates oxidative metabolism in skeletal muscle. Cell Metab. 6, 236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M., Teschendorff A., Sharp G., Novcic N., Russell A., Avril S., Prater M., Eirew P., Caldas C., Watson C. J., et al. (2012). Phenotypic and functional characterization of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 14, R134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht M. D., Sunnarborg S. W., Kouros-Mehr H., Yu Y., Lee D. C., Werb Z. (2005). Mammary ductal morphogenesis requires paracrine activation of stromal EGFR via ADAM17-dependent shedding of epithelial amphiregulin. Development 132, 3923–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet J. M., Pocock V., Steel J. H., White R., Milligan S., Parker M. G. (2005). Multiple signaling defects in the absence of RIP140 impair both cumulus expansion and follicle rupture. Endocrinology 146, 4127–4137 [DOI] [PubMed] [Google Scholar]
- White R., Leonardsson G., Rosewell I., Ann Jacobs M., Milligan S., Parker M. (2000). The nuclear receptor co-repressor nrip1 (RIP140) is essential for female fertility. Nat. Med. 6, 1368–1374 [DOI] [PubMed] [Google Scholar]
- Woodfield G. W., Horan A. D., Chen Y., Weigel R. J. (2007). TFAP2C controls hormone response in breast cancer cells through multiple pathways of estrogen signaling. Cancer Res. 67, 8439–8443 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zschiedrich I., Hardeland U., Krones-Herzig A., Berriel Diaz M., Vegiopoulos A., Müggenburg J., Sombroek D., Hofmann T. G., Zawatzky R., Yu X., et al. (2008). Coactivator function of RIP140 for NFkappaB/RelA-dependent cytokine gene expression. Blood 112, 264–276 [DOI] [PubMed] [Google Scholar]
- Zwart W., Theodorou V., Kok M., Canisius S., Linn S., Carroll J. S. (2011). Oestrogen receptor-co-factor-chromatin specificity in the transcriptional regulation of breast cancer. EMBO J. 30, 4764–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.