Abstract
Alzheimer's disease (AD) is a progressive neurodegenerative disease and in its familial form is associated with mutations in the amyloid precursor protein (APP) and the presenilins (PSs). Much data regarding the interactions of APP, its proteolytic fragments and PS have been generated, expanding our understanding of the roles of these proteins in mechanisms underlying cognitive function and revealing many complex relationships with wide ranging cellular systems. In this review, we examine the multiple interactions of APP and its proteolytic fragments with other neuronal systems in terms of feedback loops and use these relationships to build a map. We highlight the complexity involved in the APP proteolytic system and discuss alternative perspectives on the roles of APP and its proteolytic fragments in dynamic processes associated with disease progression in AD. We highlight areas where data are missing and suggest potential confounding factors. We suggest that a systems biology approach enhances representations of the data and may be more useful in modelling both normal cognition and disease processes.
Introduction
Alzheimer's disease (AD) is a clinico-pathological diagnosis in which progressive neurodegenerative disease is associated with the dementia syndrome. Neuropathologically, AD is generally characterised as including deposits of amyloid beta protein (Aβ) in senile and neuritic plaques [1] and neurofibrillary tangles of paired helical filaments of hyperphosphorylated tau [2,3]. Currently, AD is classified as familial AD, which has an early age of onset and is associated with mutations in either the amyloid precursor protein (APP) or the presenilins (PSs), and sporadic AD, which has a later age of onset and a less clear aetiology [4,5]. Down's syndrome, with an extra copy of APP on chromosome 21, also leads to a form of AD with increasing age. The genetic data associated with familial AD [6] strongly suggest that PS, APP and its proteolytic fragment Aβ are involved in AD disease progression but exactly how they contribute to both normal cognition and dementia is the focus of much debate.
Many non-genetic factors also appear to affect disease initiation and progression, including diet, exercise and education, and many models have been proposed to account for their roles in AD. Population studies (for example, the Medical Research Council Cognitive Function and Aging Study [7]) demonstrate that vascular and AD-associated pathologies are common in people with and without any clinical manifestation of dementia. Coupled with data suggesting that the relationship between amyloid pathology and dementia attenuates with age [8], this suggests that sporadic AD especially has a complex disease aetiology [9].
Many models of disease progression have been proposed to account for the accumulating data on AD. These models form the basis of experimental design and affect how findings are interpreted. Data relevant to disease progression in AD are accumulating from many diverse fields, including neuropsychology, brain imaging, molecular biology and genetics. Integrating these data is becoming harder as expertise is required in so many areas.
The pre-eminent model of the last few decades has been the amyloid cascade hypothesis [10-12], based on genetic data and the role of PS in the proteolysis of APP to release the 39 to 43 residue Aβ. This model proposes that overproduction of Aβ, or an increase in Aβ(1-42) relative to Aβ(1-40), is causal in the disease process. In this model, Aβ, perhaps in a specific aggregation state, is proposed as neurotoxic. An alternative interpretation of data relating to APP, PS and Aβ is the presenilin hypothesis [13], where loss or altered function of PS is proposed as the causal factor in disease progression. Other models suggest different initiating factors, such as cholesterol [14], mitochondria and apoptosis [15], insulin signalling and energy metabolism [16,17], cholinergic insufficiency [18], Ca2+ deregulation [19,20], signalling decoherence [21] or a combination of multiple factors (for example, [22,23]). No single model adequately explains how APP, PS, cholesterol and other factors interact with synaptic plasticity, learning and memory in normal and disease states. This suggests that models currently in use in AD research require re-evaluation.
Recent approaches to integrating the data relevant to AD have involved the use of text mining, bioinformatic databases and network analysis for gene co-expression and protein-protein interactions [24-33]. Studies using automated interrogation of genomic and proteomic databases, such as UNIPROTKB [34], NCBI Entrez Gene [35], and IntAct [36], and text mining in literature databases such as Pubmed [37] have confirmed the involvement of many previously highlighted neurological processes, extended the connectivity between known molecular targets and revealed new targets for investigation (summarised in Table 1).
Table 1.
Overview of protein-protein interaction network studies
| Reference | Selection criteria | Exclusion criteria | Main focus |
|---|---|---|---|
| [25] | Evidence of direct interaction between molecules from literature searches | Metals, non-protein molecules, poorly characterised proteins, specific peptides are included as parent genes | Direct PPI involving APP and associated fragments by domain with reference to APP770 isoform but with application to all isoforms; molecular networks with reference to biological processes |
| [33] | Twelve causative or susceptibility 'seed' genes previously associated with AD; candidate genes were selected due to close proximity to GWAS-identified gene loci associated with AD | Proteins lacking open reading frames; transcription factors, highly glycosylated proteins, extracellular proteins, proteins containing several transmembrane regions; co-expression data were used as a filter | Identification of genes in AD with reference to direct PPI and biological processes |
| [31] | Co-expressed genes that differ between controls and AD | Probe-sets not mapping to any gene or mapping to hypothetical proteins were removed | Variations in transcriptomes of AD suggest similarities with molecular networks associated with CVD and diabetes. Cis-regulatory elements identified in several diseases known to co-occur with AD |
| [30] | Genes with variable expression between an AD and an aging microarray study | Arrays that significantly varied between subjects in each study group; outlier removal, unreliable probe sets defined as being present in three or fewer arrays; control probe sets and probes not associated with known genes | Transcriptional changes between AD and aging highlight possible contributors to disease pathways. Many biological processes are shared between AD and aging. Many novel associations found, including MAPK pathways and unknown proteins. New functional and disease related association for PSEN1 with glial/neuronal interactions; confirms and highlights gamma14.3.3 signalling in AD |
| [32] | Genes with variable expression between human and mouse from brain sample microarray gene expression data sets | Outlier removal, filtered (method not specified) to remove datasets with low interspecies expression or low connectivity correlations; only top 5,000 Human and 3,000 Mouse connected genes included, rest removed to reduce 'noise' | Mouse and human networks are highly similar with expression levels more preserved than connectivity. Significant species difference in: i) co-expression arrays from astroglia and microglia but not neurons; ii) the role of PSEN1 in oligodendrocytes and myelination; and iii) further evidence of species difference in glial cells linked to neuroinflammation in human AD. Neuronal death is a small part of the biological changes associated with AD in this dataset; new transcription factors associated with AD |
| [27] | All AD and related pathways in KEGG database with co-expression data | Genes without corresponding data in the assembled PPI network and proteins with no corresponding genes in the co-expression data | Crosstalk between pathways involved in AD; close relationships between APP and apoptosis, Notch, Wnt pathways and cytokine-cytokine interactions in this dataset, brain areas vary in specific pathway relationships and order of significance |
AD, Alzheimer's disease; APP, amyloid precursor protein; CVD, cardiovascular disease; GWAS, genome-wide association study; KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, mitogen-activated protein kinase; PPI, protein-protein interaction; PSEN, presenilin.
Understanding how APP contributes to synaptic plasticity is important in understanding normal cognition and disease progression in AD. Here, we approach the process of integrating the available data relating to the relationships between APP and its proteolytic fragments and neuronal processes based on a systems biology approach and build a map of relationships. We are not aiming to define protein interactions nor find new targets for investigation, but to highlight relationships in the existing literature that illustrate the complexity involved and how this relates to our current understanding of the role of APP in AD. We highlight areas where data are missing and predict relationships that may confound current disease models.
Building the map
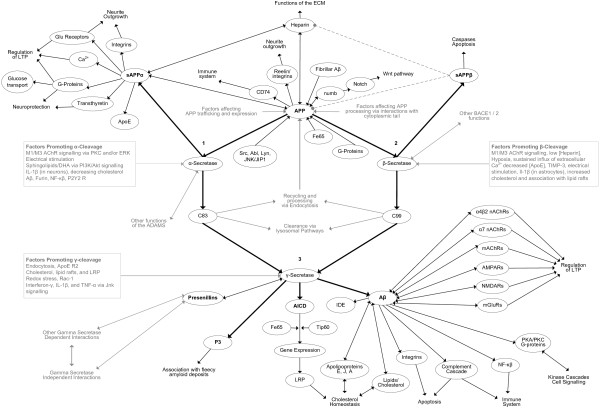
Relationships involving APP and each of its proteolytic fragments were identified from literature searches using relevant key words. Each interaction identified was manually entered into a non-hierarchical network using the mapping software Compendium [38]. This software allowed each molecule or neuronal system to be represented as a node within the map. Each node in the map could contain further networks of nodes, leading to a multi-dimensional organisation of information. These further nodes represented information relevant to the molecule of the parent node and could include notes describing possible relevance, questions raised and links to relevant entries on various bioinformatic databases. The map is not meant to be read as a protein-protein interaction network (PPI), but rather was designed to summarise and collate information relevant to the APP system in a more general way. Physiological outcomes, such as changes in signalling, were included, even though the precise molecular interactions are not clear. Any additional details, such as new biological relationships or new molecular interactions, can be added as they arise. Relationships between molecules are represented by connectors, with annotation if required. Figure 1 shows a limited map built in this way from references cited in the text, with additional references listed in Table 2.
Figure 1.
A limited map of amyloid precursor protein relationships showing molecules/neuronal systems as nodes and interactions between them as arrows. Evidence for each relationship is summarised in Table 2. Numbers are referred to in the text. Aβ, amyloid beta protein; Abl, tyrosine-protein kinase ABL1; AChR, acetylcholine receptor; ADAM, a disintegrin and metalloproteinase domain-containing protein; AICD, APP intracellular domain; Akt, RAC-alpha serine/threonine-protein kinase; AMPAR, AMPA receptor; ApoE, apolipoprotein E; APP, amyloid precursor protein; BACE, beta-site amyloid precursor protein cleaving enzyme; CD74, HLA class II histocompatibility antigen gamma chain; ECM, extracellular matrix; IDE, insulin degrading enzyme; JIP1, C-Jun-amino-terminal kinase-interacting protein 1; JNK, C-Jun amino-terminal kinase; Lyn, tyrosine-protein kinase Lyn; PKA, protein kinase A; PKC, protein kinase C; LRP, low-density lipoprotein receptor; LTP, long-term potentiation; mAChR, muscarinic acetylcholine receptor; mGluR, metabotropic glutamate receptor; nAChR, nicotinic acetylcholine receptor; NMDAR, NMDA receptor; P2Y2, P2Y purinoceptor 2; PI3K, phosphoinositide 3-kinase; sAPP, secreted amyloid precursor protein; Src, proto-oncogene tyrosine-protein kinase Src; TIMP, tissue inhibitor of metalloproteinases; TNF, tumour necrosis factor.
Table 2.
Additional references supporting the network diagram in Figure 1
| Factor | Details | Reference |
|---|---|---|
| Akt | Aβ increases Akt phosphorylation in the short term via a mechanism involving α7-nAChR and NMDARs, with phosphorylation levels returning to baseline over the long term | [99] |
| AMPA receptors | AMPA glutamate receptor density is reduced by Aβ oligomers via reduction of CamKII | [100,101] |
| ApoE | Decreased levels of ApoE lead to increased β-cleavage | [102] |
| ApoE promotes polymerisation of Aβ into fibrils and enhances fibrillar Aβ deposition in neuritic plaques. The high affinity binding of Aβ to ApoE reduces ability of ApoE to bind lipids | [103-105] | |
| Ca2+ | sAPPα modulates Ca2+ signalling by activating high conductance K+ channels via a mechanism dependent on cGMP | [106] |
| CD74 | Interacts with APP and reduces expression of Aβ | [107] |
| Cholesterol | Interactions of cholesterol and APP may allow APP to react to cholesterol status of the cell | [108] |
| Membrane cholesterol correlates with β-secretase activity and inhibition of β-secretase activity leads to increased membrane cholesterol levels. Moderate reductions in cholesterol enhance the co-expression of APP and BACE1 and promote the production of Aβ | [109,110] | |
| Aβ binds lipids and has high affinity for cholesterol. Aggregated Aβ(1-40) may affect lipid transport. | [111,112] | |
| Aβ binds 24-hydroxycholesterol and affects membrane choline carriers | ||
| Complement cascade | Aβ activates neuronal complement cascade to induce the membrane attack complex and reduces complement regulatory proteins, increasing complement-mediated cytotoxicity | [113] |
| Dishevelled | Dvl-1 increases sAPPα production mediated via JNK and PKC/MAPK but not via p38 MAPK | [114] |
| Electrophysiology | Hippocampal and cortical electrophysiological processes are modulated by sAPPα | [115] |
| Aβ(1-40) suppresses epileptiform activity in hippocampal neurons | [116] | |
| Fe65 | APP binds Fe65 at the YENPTY sequence with effects on gene transcription, cytoskeleton and cell motility. Binding of Fe65 to APP is dependent on phosphorylation state of Y682; phosphorylation of T668 reduces the binding of Fe65 to YENPTY. Binding of Fe65 reduces Aβ | [117-121] |
| Furin | Furin enhances cleavage to active forms of ADAM10 and ADAM17, leading to enhanced α-cleavage | [122] |
| Glucose/glutamate transport | sAPPα enhances transport of glucose and glutamate in synapses and protects from oxidative stress via a mechanism involving cGMP | [123] |
| Glutamate signalling | sAPPα suppresses NMDA currents rapidly and reversibly at concentrations of approximately 0.011 nM, possibly involving cGMP and a protein phosphatase. Reductions in sAPPα lead to reduced tetanically induced NMDA currents while increased sAPPα increased these currents and enhanced LTP | [124,125] |
| G-protein signalling | Aβ directly increases TNF-α at high levels and at low levels increases TNF-α release by altering GPCR signaling at early stages of disease progression by indirect effects on GPCR kinase 2/5 | [126] |
| Full length and processed APP can potentially interact with G proteins via the cytoplasmic tail and this can be altered by APP mutations around the G protein binding site. This interaction has the potential to alter G-protein signalling with wide ranging effects, including Ca2+ regulation and cell cycle pathways | [127-129] | |
| HDL | Aβ binds ApoA-I, ApoA-II, ApoE and ApoJ; binding modulates Aβ solubility | [130] |
| Heparins | Proteolysis of immature BACE1 to its mature active form is promoted by low concentrations of heparin and inhibited at higher concentrations. Certain heparin derivatives may act as inhibitors of BACE1 and have therapeutic potential | [70,131,132] |
| IL-1β | Enhanced α-cleavage by ADAM10/17 via up-regulation of P2Y2 receptors and may increase levels of ADAM10/17 by approximately threefold | [133,134] |
| Increases expression of APP and β-cleavage in astrocytes | [135] | |
| Insulin degrading enzyme | Aβ competes with insulin for IDE and reduced IDE availability may contribute to dementia | [136] |
| Integrins | sAPPα competes with APP for binding sites on integrin-β-1 and promotes neurite outgrowth | [137] |
| Aβ binds focal adhesion molecules and integrins and modulates integrin/FA signalling pathways involved in cell cycle activation and cell death. The αv integrin subunit is required for Aβ-associated suppression of LTP | [138,139] | |
| Lipids | Binding of Aβ to acidic lipid molecules promotes Aβ aggregation. Aβ binds membrane gangliosides, sphingolipids and cholesterol, which enhance Aβ aggregation | [140-142] |
| Association of APP and BACE1 with lipid rafts increases Aβ | [143] | |
| Aβ endocytosis may also involve lipid rafts | [144] | |
| Sphingolipids enhance α-cleavage via MAPK/ERK signalling | [145] | |
| LTP | Aβ suppresses LTP in hippocampal neurons via a mechanism involving α4β2 nAChRs; Aβ affects cascades downstream of NMDA GluR signaling | [146,147] |
| Muscarinic Ach signaling | Increases in mAChR-M1 and -M3 activation upregulate α-cleavage via PKC activation. Muscarinic upregulation of sAPPα secretion may involve the activation of a Src tyrosine kinase, leading to activation of PKCα and ERK1/2 Increased M2 activation decreases sAPPα secretion | [75,148-151] |
| Inhibition of muscarinic signalling promotes APP processing via β-pathway. Increased M1/M3 signalling promoted β-cleavage via PKC; MEK/ERK and increased expression of BACE1; M2 activation suppressed BACE expression | [152,153] | |
| MAPK/ERK signalling | MAPK cascade may mediate the independent effects of PKC and tyrosine kinase in human astrocytes | [154] |
| Neurite outgrowth | APP enhances neurite outgrowth independently from sAPPα | [155] |
| NF-κB | May reduce expression of BACE1 | [156] |
| NF-κB upregulation by capacitive Ca2+ entry enhances sAPPα release via mAChR signalling | [157] | |
| Nicotinic ACh signalling | Aβ(1-40) and Aβ(1-42) reduced Α4β2 nAChR and α7 nAChR currents. Aβ(1-40) but not Aβ(1-42) increased glutamatergic AMPA. Signalling via Α4β2 nAChR is associated with reduced Aβ | [40,158] |
| Aβ has high affinity for the α7 nAChR and this may be associated with increased Aβ accumulation. Differential effects of Aβ(1-40) and Aβ(1-42) on α7 nAChR as seen by different effects on ACh release and Ca2+ influx. Disruption of signalling by α7 nAChR may be associated with Aβ-mediated increases in pre-synaptic Ca2+ | [39,159,160] | |
| Aβ(1-42) has approximately 5,000-fold greater affinity for α7 nAChR than for Α4β2 nAChR | [161] | |
| NMDA GluR | Aβ promotes endocytosis of NMDARs in cortical neurons with the involvement of protein phosphatase 2B and the tyrosine phosphatase STEP | [162] |
| Nucleotide signaling via P2Y2 receptors | G-protein-coupled purine receptor, P2Y2 enhanced the release of sAPPα in a time- and dose-dependent manner; probably mediated via ADAM10 and ADAM17 | [163] |
| numb | APP binds numb when Y682 is unphosphorylated and inhibits Notch signalling | [164] |
| PKA/CREB | Aβ inhibits PKA via increased persistence of its regulatory subunit PKAIIα, resulting in reduced CREB phosphorylation in response to glutamate | [73] |
| PKC | PKC activators enhance α-cleavage | [165,166] |
| Aβ inhibits PKC | [167] | |
| Reelin | Reelin interacts with APP and Α3/β1-integrins and promotes neurite extension; APP endocytosis is reduced. Reelin signalling opposes the actions of Aβ | [56,168] |
| Src, Abl, Lyn, JNK/JIP1 | These tyrosine kinases bind to APP when phosphorylated at Y682 with affinity increased by phosphorylation of T668. JNK phosphorylation of APP at T668 modulated by JIP1 | [169,170] |
| TIMP-3 | Increases in TIMP-3 led to decreased surface expression of ADAM10 and APP. The production of Aβ and CTF is increased. TIMP-3 appears to promote endocytosis and β-secretase cleavage | [171] |
| Transthyretin | Neuroprotection in transgenic mice over-expressing mutant APP is associated with elevated levels of transthyretin and sAPPα and may be linked to increased proteolysis of Aβ | [172,173] |
Aβ, amyloid beta protein; ACh, acetylcholine; ADAM, a disintegrin and metalloproteinase domain-containing protein; Akt, RAC-alpha serine/threonine-protein kinase; AMPA, 2-amino-3-(5-methyl-3-oxo-1,2- oxazol-4-yl)propanoic acid; ApoE, apolipoprotein E; APP, amyloid precursor protein; CamKII, calcium/calmodulin dependent protein kinase type II; CD74, HLA class II histocompatibility antigen gamma chain; CREB, cyclic adenosine monophosphate response element-binding; CTF, carboxy-terminal fragment; ERK, extracellular signal-regulated kinase; FA, fatty acids; GluR, glutamate receptor; GPCR, G-protein-coupled receptor; HDL, high-density lipoprotein; IDE, insulin degrading enzyme; JIP1, C-Jun-amino-terminal kinase-interacting protein 1; JNK, C-Jun amino-terminal kinase; LTD, long-term depression; LTP, long-term potentiation; Lyn, tyrosine-protein kinase Lyn; MAPK, mitogen-activated protein kinase; mAChR, muscarinic acetylcholine receptor; MEK, ERK activator kinase; nAChR, nicotinic acetylcholine receptor; NMDAR, NMDA receptor; P2Y2, P2Y purinoceptor 2; PKA, protein kinase A; PKC, protein kinase C; sAPP, secreted amyloid precursor protein; Src, proto-oncogene tyrosine-protein kinase Src; TIMP, tissue inhibitor of metalloproteinases.
Unlike more automated network studies such as that by Perreau and colleagues [25] where nodes were included in the network only if there was direct evidence of molecular interaction, or various studies [30-33] where co-expression criteria were applied, no specific criteria were imposed and any relationship found was included. Known differences in peptide behaviour (for example, [39,40]) necessitated separate nodes for Aβ(1-40) and Aβ(1-42). This was extended systematically to include all the peptide fragments, including P3(17-40), P3(17-42), secreted APP (sAPP)α and sAPPβ (the extracellular amino-terminal fragments following α- and β-cleavage, respectively), C31, and so on to take account of any potential behavioural differences. This is in contrast to various network constructions where the various proteolytic fragments are collapsed into an APP parental node, possibly reflecting the UNIPROTKB identifier (P05067) being the same for each peptide. Additionally, different aggregation states of each of the peptides (monomers, dimers, oligomers or fibrils) were considered separately. These additional details have been omitted from Figure 1 for clarity. Since there was little or no evidence for many of the Aβ-type peptides [41], these were collapsed into a separate node (not shown).
The relationships in Figure 1 were analysed manually by identifying feedback loops. Direct feedback loops were defined as relationships between APP, any of its proteolytic fragments and any other molecule or functional change that was represented at more than one point in the map. Possible indirect feedback loops between any neural system, such as neurotransmitter signalling, Ca2+ homeostasis or the inflammation cascade, were also investigated by looking for molecules or functional change in these systems that interact with APP or any of its proteolytic fragments at more than one point in the map.
Following a brief summary of APP cleavage, two examples of the relationships between APP pathways and neuronal processes, looking at the extracellular matrix (ECM) and endocytosis, serve to illustrate the implications of the complexity involved.
The amyloid precursor protein
Overview
APP is a type I transmembrane protein [42] with a large extracellular amino-terminal domain, a transmembrane domain and an intracellular carboxy-terminal domain that resembles a cell surface receptor [43]. It is expressed in several isoforms, with APP695 the main isoform expressed by neurons. APP has a high turnover, with a half life ranging from approximately 1 hour [44-46] to approximately 4 hours [47-49]. In addition to its functions at the cell surface, full length mature APP is also processed via competing pathways that release proteolytic fragments [5,43,50] (recently reviewed in [51]). In effect there are four possible routes: APP may remain functionally active at the cell surface; it may be internalised and recycled or degraded; it may be cleaved via the α-pathway, releasing the sAPPα fragment, leaving the membrane bound C83 carboxy-terminal fragment that is further processed to the P3 fragment; or it may be cleaved via the β-pathway releasing sAPPβ, leaving the membrane bound C99 carboxy-terminal fragment that is further processed to the Aβ fragment.
As can be seen in Figure 1, α- and β-cleavage pathways (numbered 1 and 2, respectively) converge on a further intramembrane γ-cleavage (numbered 3) by the PS-containing γ-secretase complex to release the variable length Aβ and P3 peptides and the variable length APP intracellular domain (AICD). Additional cleavages, including caspase cleavage of the carboxyl terminus at residue D664 of APP695, producing the alternative length carboxy-terminal fragment C31 [51], and cleavage by BACE2 (beta-site amyloid precursor protein cleaving enzyme 2) within the Aβ sequence [52], have been omitted for clarity.
Unprocessed APP is degraded or recycled via the endosomal or lysosomal pathways and may be recycled back to the membrane and processed within approximately 30 minutes [46], with perhaps a third to a half being processed via the cleavage pathways as measured by secreted sAPPα/β [46]. Regulatory relationships promoting expression or cleavage have been collapsed into summary boxes in Figure 1 for simplicity.
Complex feedback pathways involving APP and its proteolytic fragments; the extracellular matrix and endocytosis
The extracellular matrix
APP interacts with itself and its own proteolytic fragments at many points in the network with various functional consequences. At the cell surface, the large amino-terminal domain of full length APP interacts with many components of the ECM, including heparins and heparin sulphate proteoglycans [42], laminin, collagen [53,54] and β-1-integrin [43,55,56], and contributes to early neuronal development, axon and neurite growth, cell adhesion and synaptic plasticity [43,57]. Many of these interactions are now represented in MatrixDB [58], an ECM interaction database.
The amino-terminal domain of APP binds heparin in a pH- and zinc-dependant manner and this binding promotes the formation of APP homodimers [42,59-61]. Depending on various factors, including cell compartment and pH, this dimerisation may influence APP proteolysis via α- or β-cleavages due to conformational changes [59], though this requires further investigation.
The large amino-terminal fragment released by α-cleavage, sAPPα, is a soluble monomer. It retains two heparin binding sites and has been shown to bind heparin as a dimer [60]. The ability of sAPPα to disrupt APP dimerisation at the cell surface is thought to contribute to its neuroprotective actions [62-64] and may partly explain why sAPPα is approximately 100 times more neuroprotective against excitotoxicity, glucose deprivation and the addition of Aβ in hippocampal cultures than sAPPβ, which lacks the second carboxy-terminal heparin binding site [63]. Dementia status has been associated with both reduced sAPPα levels in cerebrospinal fluid [65] and an increased half life of sAPPα [49].
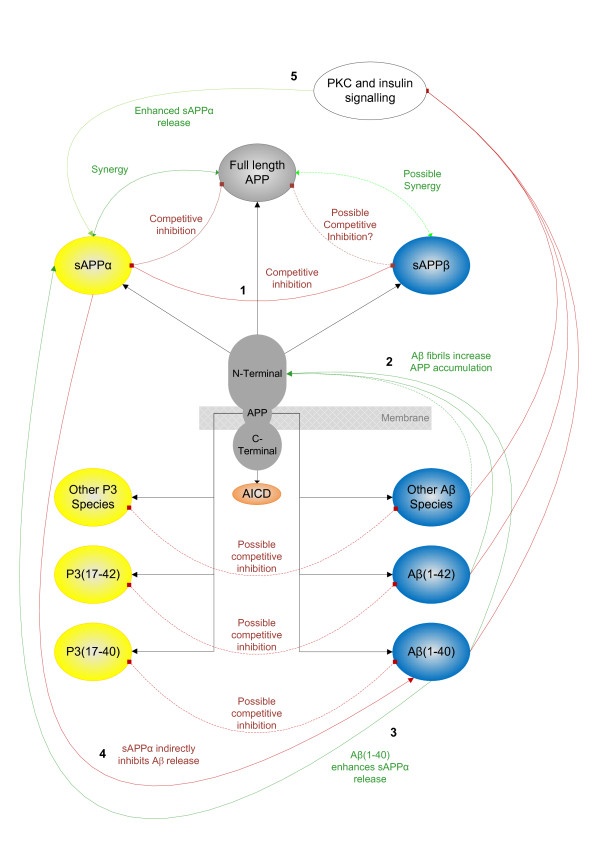
Differing by only 16 carboxy-terminal residues, sAPPα and sAPPβ share a high degree of sequence homology and have the potential to compete with each other for binding sites. Since both sAPPβ and sAPPα are constantly present in the ECM in dynamically changing ratios, we predict that sAPPβ could act as an antagonist of sAPPα/APP (shown in Figure 2 as competitive inhibition (line 1)), with the potential to modulate various interactions between APP and sAPPα and the ECM. We predict, therefore, that any functional outcome from the interactions of the APP pathway will depend on the synergy between APP and its amino-terminal fragments.
Figure 2.
Feedback between amyloid precursor protein and its proteolytic fragments. Inhibition is shown in red, enhancement in green; numbers are referred to in the text. Functionally different aggregation states for the Aβ-type peptides are collapsed into P3 and Aβ nodes for clarity. Aβ, amyloid beta protein; AICD, APP intracellular domain; APP, amyloid precursor protein; PKC, protein kinase C; sAPP, secreted amyloid precursor protein.
Aβ interacts with heparins in the ECM and at high levels may prevent the catabolism of proteoglycans and promote amyloid formation [66]. Reciprocally, heparins modulate many of the interactions involving Aβ. Heparins enhance both nucleation and elongation processes in the aggregation of Aβ [67] and modulate interactions of Aβ; for example, heparins can limit the neurotoxic and pro-inflammatory activity of Aβ in a dose-dependent manner [68]. Additionally, heparins contribute to the uptake of Aβ by a pathway shared with apolipoprotein E [69] and also contribute to the regulation of the β-secretase BACE in a dose-dependent manner, with low concentrations promoting and high concentrations inhibiting the activation of BACE1 [70].
In terms of direct feedback, Aβ may be part of a negative feedback loop involved in the regulation of APP expression and the promotion of α-cleavage. Fibrillar forms of various proteins, including Aβ, interact with amino-terminal APP and increase APP expression [71] (Figure 2, line 2) and Aβ(1-40) may up-regulate expression of α-secretase via integrin receptors and matrix metallo-proteinase-9 activity [72] (Figure 2, line 3). Additionally, Aβ has many indirect negative feedback relationships with α-cleavage - for example, Aβ inhibits protein kinase C [73] and insulin signalling [74], both involved in up-regulation of α-cleavage [75] (Figure 2, line 5) - and additionally may negatively feedback on APP expression and its own production via a pathway involving Fe65 and glycogen synthase kinase 3 [76]. Reciprocally, changes to gene expression by sAPPα via the activation of NF-κβ may attenuate the interactions of Aβ [77] and this can be understood as reciprocal indirect feedback from the α-pathway on the β-pathway (Figure 2, line 4).
The data reviewed above suggest that Aβ may contribute to both direct and indirect feedback loops in the ECM at multiple points, with consequences for the interactions of APP, sAPPα, Aβ and sAPPβ that may ultimately affect cell adhesion, neurite outgrowth and synaptic plasticity. These effects will depend on the specific sequence length, concentration and aggregation state of the Aβ-type peptides, which may dynamically change in response to the neuronal environment.
Endocytosis
Increases in endocytosis, often associated with synaptic activity, lead to both an increase in Aβ [78] and a reduction in α-cleavage and lower secreted sAPPα levels [79]. This may reflect the regulatory role of compartmentation in APP proteolysis, where α-cleavage occurs at the cell surface and Aβ production is associated with factors that promote endocytosis. No studies could be found investigating the role of endocytosis on P3 production, so this cannot be discussed specifically. Endocytosis allows a degree of dissociation between the α- and β-cleavages and γ-cleavage, resulting in levels of sAPPα and sAPPβ that may be independent of Aβ(1-40/42) and P3 levels, respectively. This means that in terms of function, APP processing via the β-pathway has the potential to either: modulate actions of sAPPα via competition with sAPPβ; or modulate the actions of sAPPα and initiate actions mediated by Aβ.
P3 corresponds to Aβ(17-40/42) and we predict that this shared sequence could allow P3 to modulate the interactions of Aβ (shown in Figure 2 as competitive inhibition), especially where those interactions involve the carboxy-terminal amino acids. While no studies can be found that address this particular question directly, various amino-terminal truncated forms of Aβ have been associated with diffuse plaques [80,81] and the Aβ(17-40) fragment, corresponding to P3, has been studied in terms of aggregation [82]. It is clear that amino-truncated Aβ peptides, including P3, have potential for aggregation and can interact with Aβ, but how this affects the physiological behaviour of neuronal systems is not clear. In terms of functional flow through the APP pathway, processing via the α-pathway to release sAPPα and P3 could either: initiate actions of sAPPα; or initiate actions of sAPPα and modulate actions mediated by Aβ.
In terms of modelling these interactions in functional contexts, PPI networks may require a more detailed approach, where each sequence length and aggregation state is represented as an individual node.
The effects of concentration, sequence length aggregation state and affinity
Aβ is produced in a range of sequence lengths [41] and can form monomers, dimers, oligomers and fibrils. The amount of Aβ produced is regulated and can dynamically change in response to many factors, including increased cholesterol [83], increased synaptic activity [84], heparins via activation of BACE1 [70], reduced acetylcholine (Ach) signalling via muscarinic receptors [70], sustained increased cytosolic Ca2+ [85], and hypoxia [86]. Changes in concentration are a well recognised mechanism of regulation in cellular processes; for example, Aβ(1-40) and Aβ(1-42) both promote angiogenesis at nanomolar concentrations but inhibit it at higher micromolar concentrations [87].
Different sequence lengths show different behaviours; for example, Aβ(1-42) is more prone to aggregation than Aβ(1-40) due to a more rigid carboxyl terminus [88]. Aβ monomers appear to have structured and unstructured regions, with differences in aqueous conformations between Aβ(1-40) and Aβ(1-42) [89]. While both Aβ(1-40) and Aβ(1-42) reduced currents via α7 and α4β2 ACh receptors (AChRs), only Aβ(1-40) increased glutamatergic signalling via AMPA receptors [40]. Another study found that the effects of Aβ(1-40) and Aβ(1-42) on α7 AChRs, as measured by ACh release and Ca2+ influx, were different [39]. Additionally, Aβ(1-40) may protect neurons from the effects of Aβ(1-42) by disrupting the aggregation of Aβ(1-42) [90] and this raises the possibility that P3-type peptides may act in similar ways. The functional effects of sequence length are only just beginning to be elucidated and should be expanded to include all the Aβ-type peptides, including P3. In addition to differences in peptide sequence and concentration, aggregation state also modulates function. Aβ oligomers appear to be spherical [91,92] and Aβ fibrils appear as fine fibres [92]. Aβ oligomers, but not monomers or fibrils, enhance lipid release [93] and at low concentrations enhance neuronal differentiation [92] whereas Aβ fibrils, but not monomers or oligomers, may interact with APP and increase its expression [71]. While both soluble and aggregated forms of Aβ(1-40) inhibit long-term potentiation, the fibrillar forms may additionally affect neurotransmitter release [94].
Within a specific cellular compartment, whether one interaction is more likely to occur than another depends on the relative affinities of the reactants involved. While dissociation constants have been measured for some reactions in rodent models (for example, K(i) values of Aβ(1-42) for α7 nicotinic AChRs from rat and guinea pig are 4.1 and 5.0 pM, respectively, and the K(i) of Aβ(1-42) with α4β2 AChRs is approximately 5,000-fold lower at 30 and 23 nM, respectively [95]), we are missing dissociation constants for most interactions in humans.
Taken together, the data discussed above suggest that the interactions of APP and its proteolytic fragments with synaptic systems are complex and involve subtle changes in the relative ratios between APP and all its fragments. Given the complexity of the APP system, multiple neuronal systems have the potential to regulate the expression and proteolysis of APP and could also initiate imbalance within this system, leading to multiple possible disease pathways.
This complexity has the potential to confound purely empirical studies into the behaviour of the peptides in model systems. Over-expressing Aβ peptides in, for example, mouse models without knowing their concentrations, sequences and aggregation states and the background concentrations, sequences and aggregation states of the other relevant peptides in the experimental system that is being studied means that we do not know if the experimental results obtained are due to the properties of the Aβ peptides themselves or reflect a disrupted balance in a complex system over time, or perhaps both. While this distinction may seem subtle, given the self-organising and self-referencing properties of the human brain, an approach based on the principles in systems biology may better represent the roles of the APP pathway in functional contexts.
Discussion
It is not possible to include the full extent of the known interaction of APP and its proteolytic fragments here and further interactions may yet be discovered. Data relating to the roles of APP and its proteolytic fragments in synaptic plasticity and AD have accumulated from many diverse fields and these need to be placed into context. One way of doing this is to investigate the relationships between APP, its proteolytic fragments and wider neuronal systems by mapping networks of interactions.
It is interesting that the networks generated in the various studies reviewed in Table 1 do not always overlap and different studies highlight different pathways or biological processes, for example, Fe2+ [28], apoptosis [96], or cardiovascular disease/diabetes [31]. Each study has different starting points, inclusion/exclusion criteria and network construction methods, so this lack of agreement is no surprise. It is difficult to assess the degree to which the various starting points, criteria and network construction methods bias results towards an outcome.
The study by Soler-Lopez and colleagues [33] may not model the interactions of full length APP in the membrane adequately, as many of the ECM proteins that might be expected to interact are excluded due to difficulties involved in expressing them in the experimental microarray used. This may shift the focus of their network more towards intracellular interactions. Given the importance of the various interactions of APP with components of the ECM, any study excluding such proteins and proteoglycans could be seriously confounded.
While the study by Perreau and colleagues [25] maps PPIs to specific domains of full length APP, they do not distinguish between the different proteolytic peptides. Additionally, transient interactions and biologically essential post-translational modifications, such as glycosylation and phosphorylation, cannot yet be fully represented in PPI networks [27], severely limiting the modelling of regulation and control in these models.
Studies comparing mouse and human gene expression networks [32] or aging and AD pathways [30] have revealed interesting results. Perhaps most striking is the study revealing a human-specific network for PS function in oligodendrocytes and myelination, with the potential to confound the current approach to modelling AD in rodents [32]. Miller and colleagues [30,32] also show important contributions from both neuronal and glial pathways, perhaps reflecting the contributions of glia to disease pathways found in humans [97,98]. The differences found in networks between glia and neurons could be extended to differentiate between specific neuronal types and future network models may usefully investigate why cholinergic neurons in AD appear more susceptible to degeneration than others.
We do not yet have totally inclusive, gene co-expression or PPI models of APP and related pathways to the level of detail required and this places limitations on their interpretation. Given the limitations, however, network studies do show the usefulness of the systems biology approach in integrating huge volumes of data and a simple search of patents in Google with the terms 'protein interaction network Alzheimer disease' retrieves more than 700 applications based on this approach. Network studies highlight the idea that multiple processes are involved in AD disease pathways, reveal new targets for further investigation and have generated new hypotheses. Network models have the potential to avoid biases inherent in other approaches, such as the emphasis on Aβ in animal models based on the more reductionist amyloid cascade hypothesis, and allow multiple disease pathways to be represented in a single model, perhaps better relating to the processes involved in human disease.
We have suggested that APP and its proteolytic fragments appear to dynamically modulate each other by multiple mechanisms with evidence for both direct and indirect feedback loops between Aβ, sAPPα, sAPPβ and full length APP. Additionally, the fragments derived from γ-cleavage, including the variable length fragments, P3 and Aβ, have the potential to interact with each other and modulate multiple neuronal processes in subtle and dynamic ways. Unfortunately, evidence for the role(s) of P3 is almost entirely missing from the literature and that for sAPPβ is limited. Without this information, we cannot have a full picture of the contributions of the APP proteolytic system to normal neuronal processes and how these change during disease progression.
Given the number and range of reciprocal feedback relationships that APP and its proteolytic fragments are involved in, the effects of perturbing the APP proteolytic system may be better understood as dynamic, synergistic actions involving multiple players. A wide range of neuronal processes have the potential to interact with and regulate APP processing, allowing APP to monitor the current state of the cell. The multiple proteolytic pathways allow APP to assimilate signals from wide-ranging cellular systems and pass these signals on via the ratios of APP fragments. The constant turnover of APP contributes to the ability of neurons to sense the current state of the cell and use this information to set in motion future responses. The potential for subtly different signals both from the α- and β-pathways via alternative peptides and the complex feedback relationships between them described above fits well with the fine control required for integrating neuronal systems.
The predicted synergy between APP and all its proteolytic fragments implies that the ratios of full length APP and all the various fragments, not just Aβ(1-40) and Aβ(1-42), are more important than absolute levels in coordinating neuronal responses. The complexity of the relationships described here has the potential to confound purely empirical investigations into the activities of any one proteolytic fragment studied in isolation. To avoid these predicted confounding effects, experimental approaches should be extended to account for the different concentrations, affinities and aggregation states of each peptide length, and experimental systems should be followed over time for each biological process in a systematic manner. With over 40 Aβ-type peptides [41], this presents a challenge.
Generating useful network models will require a more detailed and comprehensive approach to experimental design, involving a range of experimental controls to account for all the confounding factors. These data will be essential for generating network models that are truly relevant to our understanding of normal and abnormal processes in the context of the human population.
Abbreviations
Aβ: amyloid beta protein; ACh: acetylcholine; AChR: acetylcholine receptor; AD: Alzheimer's disease; APP: amyloid precursor protein; BACE: beta-site amyloid precursor protein cleaving enzyme; ECM: extracellular matrix; IL: interleukin; NF: nuclear factor; PPI: protein-protein interaction; PS: presenilin; sAPP: secreted amyloid precursor protein.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SH wrote the paper in discussion and with contributions from CB.
Contributor Information
Sally Hunter, Email: seh66@medschl.cam.ac.uk.
Carol Brayne, Email: cb105@medschl.cam.ac.uk.
Acknowledgements
This work was supported by funding from the Cambridge and Peterborough CLAHRC.
References
- Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, Brownlee LM, Vogel FS, Hughes JP, van Belle G, Berg L. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41:479–486. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Diagnostic criteria for neuropathologic assessment of Alzheimer's disease. Neurobiol Aging. 1997;18(4 Suppl):S85–88. doi: 10.1016/s0197-4580(97)00062-6. [DOI] [PubMed] [Google Scholar]
- Rossor MN, Fox NC, Freeborough PA, Harvey RJ. Clinical features of sporadic and familial Alzheimer's disease. Neurodegeneration. 1996;5:393–397. doi: 10.1006/neur.1996.0052. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer's disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- Zekanowski C, Religa D, Graff C, Filipek S, Kuźnicki J. Genetic aspects of Alzheimer's disease. Acta Neurobiol Exp (Wars) 2004;64:19–31. doi: 10.55782/ane-2004-1488. [DOI] [PubMed] [Google Scholar]
- Neuropathology Group. Medical Research Council Cognitive Function and Aging Study. Pathological correlates of late-onset dementia in a multicentre, community-based population in England and Wales. Neuropathology Group of the Medical Research Council Cognitive Function and Ageing Study (MRC CFAS) Lancet. 2001;357:169–75. doi: 10.1016/s0140-6736(00)03589-3. [DOI] [PubMed] [Google Scholar]
- Savva GM, Wharton SB, Ince PG, Forster G, Matthews FE, Brayne C. Medical Research Council Cognitive Function and Ageing Study. Age, neuropathology, and dementia. N Engl J Med. 2009;360:2302–2309. doi: 10.1056/NEJMoa0806142. [DOI] [PubMed] [Google Scholar]
- Jellinger KA, Attems J. Neuropathological evaluation of mixed dementia. J Neurol Sci. 2007;257:80–87. doi: 10.1016/j.jns.2007.01.045. [DOI] [PubMed] [Google Scholar]
- Hardy JA, Higgins GA. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer's disease results from the cerebral accumulation and cytotoxicity of amyloid beta-protein. J Alzheimers Dis. 2001;3:75–80. doi: 10.3233/jad-2001-3111. [DOI] [PubMed] [Google Scholar]
- Shen J, Kelleher RJ. The presenilin hypothesis of Alzheimer's disease: evidence for a loss-of-function pathogenic mechanism. Proc Natl Acad Sci USA. 2007;104:403–409. doi: 10.1073/pnas.0608332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudinov AR, Koudinova NV. Cholesterol homeostasis failure as a unifying cause of synaptic degeneration. J Neurol Sci. 2005;229-230:233–240. doi: 10.1016/j.jns.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Eckert A, Keil U, Marques CA, Bonert A, Frey C, Schüssel K, Müller WE. Mitochondrial dysfunction, apoptotic cell death, and Alzheimer's disease. Biochem Pharmacol. 2003;66:1627–1634. doi: 10.1016/S0006-2952(03)00534-3. [DOI] [PubMed] [Google Scholar]
- Hoyer S. The aging brain. Changes in the neuronal insulin/insulin receptor signal transduction cascade trigger late-onset sporadic Alzheimer disease (SAD). A mini-review. J Neural Transm. 2002;109:991–1002. doi: 10.1007/s007020200082. [DOI] [PubMed] [Google Scholar]
- Hoyer S. Causes and consequences of disturbances of cerebral glucose metabolism in sporadic Alzheimer disease: therapeutic implications. Adv Exp Med Biol. 2004;541:135–152. doi: 10.1007/978-1-4419-8969-7_8. [DOI] [PubMed] [Google Scholar]
- Bartus RT, Dean RL, Beer B, Lippa AS. The cholinergic hypothesis of geriatric memory dysfunction. Science. 1982;217:408–414. doi: 10.1126/science.7046051. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Calcium and neurodegeneration. Aging Cell. 2007;6:337–350. doi: 10.1111/j.1474-9726.2007.00275.x. [DOI] [PubMed] [Google Scholar]
- Bojarski L, Herms J, Kuznicki J. Calcium dysregulation in Alzheimer's disease. Neurochem Int. 2008;52:621–633. doi: 10.1016/j.neuint.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Arendt T. Synaptic plasticity and cell cycle activation in neurons are alternative effector pathways: the 'Dr. Jekyll and Mr. Hyde concept' of Alzheimer's disease or the yin and yang of neuroplasticity. Prog Neurobiol. 2003;71:83–248. doi: 10.1016/j.pneurobio.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Fotuhi M, Hachinski V, Whitehouse PJ. Changing perspectives regarding late-life dementia. Nat Rev Neurol. 2009;5:649–658. doi: 10.1038/nrneurol.2009.175. [DOI] [PubMed] [Google Scholar]
- Brayne C. Research and Alzheimer's disease: an epidemiological perspective. Psychol Med. 1993;23:287–296. doi: 10.1017/S003329170002835X. [DOI] [PubMed] [Google Scholar]
- Yuraszeck TM, Neveu P, Rodriguez-Fernandez M, Robinson A, Kosik KS, Doyle FJ. Vulnerabilities in the tau network and the role of ultrasensitive points in tau pathophysiology. PLoS Comput Biol. 2010;6:e1000997. doi: 10.1371/journal.pcbi.1000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreau VM, Orchard S, Adlard PA, Bellingham SA, Cappai R, Ciccotosto GD, Cowie TF, Crouch PJ, Duce JA, Evin G, Faux NG, Hill AF, Hung YH, James SA, Li QX, Mok SS, Tew DJ, White AR, Bush AI, Hermjakob H, Masters CL. A domain level interaction network of amyloid precursor protein and Abeta of Alzheimer's disease. Proteomics. 2010;10:2377–2395. doi: 10.1002/pmic.200900773. [DOI] [PubMed] [Google Scholar]
- Mewes HW, Wachinger B, Stumpflen V. Perspectives of a systems biology of the synapse: how to transform an indefinite data space into a model? Pharmacopsychiatry. 2010;43(Suppl 1):S2–8. doi: 10.1055/s-0030-1249666. [DOI] [PubMed] [Google Scholar]
- Liu ZP, Wang Y, Zhang XS, Chen L. Identifying dysfunctional crosstalk of pathways in various regions of Alzheimer's disease brains. BMC Syst Biol. 2010;4(Suppl 2):S11. doi: 10.1186/1752-0509-4-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell DB. Towards a unifying, systems biology understanding of large-scale cellular death and destruction caused by poorly liganded iron: Parkinson's, Huntington's, Alzheimer's, prions, bactericides, chemical toxicology and others as examples. Arch Toxicol. 2010;84:825–889. doi: 10.1007/s00204-010-0577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noorbakhsh F, Overall CM, Power C. Deciphering complex mechanisms in neurodegenerative diseases: the advent of systems biology. Trends Neurosci. 2009;32:88–100. doi: 10.1016/j.tins.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Miller JA, Oldham MC, Geschwind DH. A systems level analysis of transcriptional changes in Alzheimer's disease and normal aging. J Neurosci. 2008;28:1410–1420. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray M, Ruan J, Zhang W. Variations in the transcriptome of Alzheimer's disease reveal molecular networks involved in cardiovascular diseases. Genome Biol. 2008;9:R148. doi: 10.1186/gb-2008-9-10-r148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci USA. 2010;107:12698–12703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler-López M, Zanzoni A, Lluís R, Stelzl U, Aloy P. Interactome mapping suggests new mechanistic details underlying Alzheimer's disease. Genome Res. 2011;21:364–376. doi: 10.1101/gr.114280.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNIPROTKB. http://www.uniprot.org/
- NCBI Entrez Gene. http://www.ncbi.nlm.nih.gov/gene
- IntAct. http://ebi.ac.uk/intact/site/
- Pubmed. http://www.ncbi.nlm.nih.gov/pubmed/
- Compendium. http://compendium.open.ac.uk/
- Lee DH, Wang HY. Differential physiologic responses of alpha7 nicotinic acetylcholine receptors to beta-amyloid1-40 and beta-amyloid1-42. J Neurobiol. 2003;55:25–30. doi: 10.1002/neu.10203. [DOI] [PubMed] [Google Scholar]
- Tozaki H, Matsumoto A, Kanno T, Nagai K, Nagata T, Yamamoto S, Nishizaki T. The inhibitory and facilitatory actions of amyloid-beta peptides on nicotinic ACh receptors and AMPA receptors. Biochem Biophys Res Commun. 2002;294:42–45. doi: 10.1016/S0006-291X(02)00429-1. [DOI] [PubMed] [Google Scholar]
- Wang R, Sweeney D, Gandy SE, Sisodia SS. The profile of soluble amyloid beta protein in cultured cell media. Detection and quantification of amyloid beta protein and variants by immunoprecipitation-mass spectrometry. J Biol Chem. 1996;271:31894–31902. doi: 10.1074/jbc.271.50.31894. [DOI] [PubMed] [Google Scholar]
- Bush AI, Pettingell WH Jr, de Paradis M, Tanzi RE, Wasco W. The amyloid beta-protein precursor and its mammalian homologues. Evidence for a zincmodulated heparin-binding superfamily. J Biol Chem. 1994;269:26618–26621. [PubMed] [Google Scholar]
- Turner PR, O'Connor K, Tate WP, Abraham WC. Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog Neurobiol. 2003;70:1–32. doi: 10.1016/S0301-0082(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Perez RG, Soriano S, Hayes JD, Ostaszewski B, Xia W, Selkoe DJ, Chen X, Stokin GB, Koo EH. Mutagenesis identifies new signals for beta-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Abeta42. J Biol Chem. 1999;274:18851–18856. doi: 10.1074/jbc.274.27.18851. [DOI] [PubMed] [Google Scholar]
- Pietrzik CU, Busse T, Merriam DE, Weggen S, Koo EH. The cytoplasmic domain of the LDL receptor-related protein regulates multiple steps in APP processing. EMBO J. 2002;21:5691–5700. doi: 10.1093/emboj/cdf568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo EH, Squazzo SL, Selkoe DJ, Koo CH. Trafficking of cell-surface amyloid beta-protein precursor. I. Secretion, endocytosis and recycling as detected by labeled monoclonal antibody. J Cell Sci. 1996;109:991–998. doi: 10.1242/jcs.109.5.991. [DOI] [PubMed] [Google Scholar]
- Savage MJ, Trusko SP, Howland DS, Pinsker LR, Mistretta S, Reaume AG, Greenberg BD, Siman R, Scott RW. Turnover of amyloid beta-protein in mouse brain and acute reduction of its level by phorbol ester. J Neurosci. 1998;18:1743–1752. doi: 10.1523/JNEUROSCI.18-05-01743.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyckman AW, Confaloni AM, Thinakaran G, Sisodia SS, Moya KL. Post-translational processing and turnover kinetics of presynaptically targeted amyloid precursor superfamily proteins in the central nervous system. J Biol Chem. 1998;273:11100–11106. doi: 10.1074/jbc.273.18.11100. [DOI] [PubMed] [Google Scholar]
- Morales-Corraliza J, Mazzella MJ, Berger JD, Diaz NS, Choi JH, Levy E, Matsuoka Y, Planel E, Mathews PM. In vivo turnover of tau and APP metabolites in the brains of wild-type and Tg2576 mice: greater stability of sAPP in the beta-amyloid depositing mice. PLoS ONE. 2009;4:e7134. doi: 10.1371/journal.pone.0007134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. Cell biology of the amyloid beta-protein precursor and the mechanism of Alzheimer's disease. Annu Rev Cell Biol. 1994;10:373–403. doi: 10.1146/annurev.cb.10.110194.002105. [DOI] [PubMed] [Google Scholar]
- Zhang YW, Thompson R, Zhang H, Xu H. APP processing in Alzheimer's disease. Mol Brain. 2011;4:3. doi: 10.1186/1756-6606-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, He G, Song W. BACE2, as a novel APP theta-secretase, is not responsible for the pathogenesis of Alzheimer's disease in Down syndrome. FASEB J. 2006;20:1369–1376. doi: 10.1096/fj.05-5632com. [DOI] [PubMed] [Google Scholar]
- Beher D, Hesse L, Masters CL, Multhaup G. Regulation of amyloid protein precursor (APP) binding to collagen and mapping of the binding sites on APP and collagen type I. J Biol Chem. 1996;271:1613–1620. doi: 10.1074/jbc.271.3.1613. [DOI] [PubMed] [Google Scholar]
- Breen KC. APP-collagen interaction is mediated by a heparin bridge mechanism. Mol Chem Neuropathol. 1992;16:109–121. doi: 10.1007/BF03159964. [DOI] [PubMed] [Google Scholar]
- Hoe HS, Tran TS, Matsuoka Y, Howell BW, Rebeck GW. DAB1 and Reelin effects on amyloid precursor protein and ApoE receptor 2 trafficking and processing. J Biol Chem. 2006;281:35176–35185. doi: 10.1074/jbc.M602162200. [DOI] [PubMed] [Google Scholar]
- Hoe HS, Lee KJ, Carney RS, Lee J, Markova A, Lee JY, Howell BW, Hyman BT, Pak DT, Bu G, Rebeck GW. Interaction of reelin with amyloid precursor protein promotes neurite outgrowth. J Neurosci. 2009;29:7459–7473. doi: 10.1523/JNEUROSCI.4872-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralle M, Ferreira ST. Structure and functions of the human amyloid precursor protein: the whole is more than the sum of its parts. Prog Neurobiol. 2007;82:11–32. doi: 10.1016/j.pneurobio.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Chautard E, Fatoux-Ardore M, Ballut L, Thierry-Mieg N, Ricard-Blum S. MatrixDB, the extracellular matrix interaction database. Nucleic Acids Res. 2011;39(Database issue):D235–240. doi: 10.1093/nar/gkq830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahms SO, Hoefgen S, Roeser D, Schlott B, Gührs KH, Than ME. Structure and biochemical analysis of the heparin-induced E1 dimer of the amyloid precursor protein. Proc Natl Acad Sci USA. 2010;107:5381–5386. doi: 10.1073/pnas.0911326107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralle M, Oliveira CL, Guerreiro LH, McKinstry WJ, Galatis D, Masters CL, Cappai R, Parker MW, Ramos CH, Torriani I, Ferreira ST. Solution conformation and heparin-induced dimerization of the full-length extracellular domain of the human amyloid precursor protein. J Mol Biol. 2006;357:493–508. doi: 10.1016/j.jmb.2005.12.053. [DOI] [PubMed] [Google Scholar]
- Multhaup G. Amyloid precursor protein and BACE function as oligomers. Neurodegener Dis. 2006;3:270–274. doi: 10.1159/000095266. [DOI] [PubMed] [Google Scholar]
- Gralle M, Botelho MG, Wouters FS. Neuroprotective secreted amyloid precursor protein acts by disrupting amyloid precursor protein dimers. J Biol Chem. 2009;284:15016–15025. doi: 10.1074/jbc.M808755200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Sopher BL, Rydel RE, Begley JG, Pham DG, Martin GM, Fox M, Mattson MP. Increased activity-regulating and neuroprotective efficacy of alpha-secretase-derived secreted amyloid precursor protein conferred by a C-terminal heparin-binding domain. J Neurochem. 1996;67:1882–1896. doi: 10.1046/j.1471-4159.1996.67051882.x. [DOI] [PubMed] [Google Scholar]
- Copanaki E, Chang S, Vlachos A, Tschäpe JA, Müller UC, Kögel D, Deller T. sAPPalpha antagonizes dendritic degeneration and neuron death triggered by proteasomal stress. Mol Cell Neurosci. 2010;44:386–393. doi: 10.1016/j.mcn.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Sennvik K, Fastbom J, Blomberg M, Wahlund LO, Winblad B, Benedikz E. Levels of alpha- and beta-secretase cleaved amyloid precursor protein in the cerebrospinal fluid of Alzheimer's disease patients. Neurosci Lett. 2000;278:169–172. doi: 10.1016/S0304-3940(99)00929-5. [DOI] [PubMed] [Google Scholar]
- Bame KJ, Danda J, Hassall A, Tumova S. Abeta(1-40) prevents heparanasecatalyzed degradation of heparan sulfate glycosaminoglycans and proteoglycans in vitro. A role for heparan sulfate proteoglycan turnover in Alzheimer's disease. J Biol Chem. 1997;272:17005–17011. doi: 10.1074/jbc.272.27.17005. [DOI] [PubMed] [Google Scholar]
- Klajnert B, Cortijo-Arellano M, Bryszewska M, Cladera J. Influence of heparin and dendrimers on the aggregation of two amyloid peptides related to Alzheimer's and prion diseases. Biochem Biophys Res Commun. 2006;339:577–582. doi: 10.1016/j.bbrc.2005.11.053. [DOI] [PubMed] [Google Scholar]
- Bergamaschini L, Donarini C, Rossi E, De Luigi A, Vergani C, De Simoni MG. Heparin attenuates cytotoxic and inflammatory activity of Alzheimer amyloid-beta in vitro. Neurobiol Aging. 2002;23:531–536. doi: 10.1016/S0197-4580(02)00003-9. [DOI] [PubMed] [Google Scholar]
- Winkler K, Scharnagl H, Tisljar U, Hoschützky H, Friedrich I, Hoffmann MM, Hüttinger M, Wieland H, März W. Competition of Abeta amyloid peptide and apolipoprotein E for receptor-mediated endocytosis. J Lipid Res. 1999;40:447–455. [PubMed] [Google Scholar]
- Beckman M, Holsinger RM, Small DH. Heparin activates beta-secretase (BACE1) of Alzheimer's disease and increases autocatalysis of the enzyme. Biochemistry. 2006;45:6703–6714. doi: 10.1021/bi052498t. [DOI] [PubMed] [Google Scholar]
- White AR, Maher F, Brazier MW, Jobling MF, Thyer J, Stewart LR, Thompson A, Gibson R, Masters CL, Multhaup G, Beyreuther K, Barrow CJ, Collins SJ, Cappai R. Diverse fibrillar peptides directly bind the Alzheimer's amyloid precursor protein and amyloid precursor-like protein 2 resulting in cellular accumulation. Brain Res. 2003;966:231–244. doi: 10.1016/S0006-8993(02)04173-2. [DOI] [PubMed] [Google Scholar]
- Talamagas AA, Efthimiopoulos S, Tsilibary EC, Figueiredo-Pereira ME, Tzinia AK. Abeta(1-40)-induced secretion of matrix metalloproteinase-9 results in sAPPalpha release by association with cell surface APP. Neurobiol Dis. 2007;28:304–315. doi: 10.1016/j.nbd.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Vitolo OV, Sant'Angelo A, Costanzo V, Battaglia F, Arancio O, Shelanski M. Amyloid beta -peptide inhibition of the PKA/CREB pathway and long-term potentiation: reversibility by drugs that enhance cAMP signaling. Proc Natl Acad Sci USA. 2002;99:13217–13221. doi: 10.1073/pnas.172504199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling X, Martins RN, Racchi M, Craft S, Helmerhorst E. Amyloid beta antagonizes insulin promoted secretion of the amyloid beta protein precursor. J Alzheimers Dis. 2002;4:369–374. doi: 10.3233/jad-2002-4504. [DOI] [PubMed] [Google Scholar]
- Canet-Aviles RM, Anderton M, Hooper NM, Turner AJ, Vaughan PF. Muscarine enhances soluble amyloid precursor protein secretion in human neuroblastoma SH-SY5Y by a pathway dependent on protein kinase C(alpha), src-tyrosine kinase and extracellular signal-regulated kinase but not phospholipase C. Brain Res Mol Brain Res. 2002;102:62–72. doi: 10.1016/s0169-328x(02)00184-5. [DOI] [PubMed] [Google Scholar]
- Henriques AG, Vieira SI, da Cruz e Silva EF, da Cruz e Silva OA. Abeta hinders nuclear targeting of AICD and Fe65 in primary neuronal cultures. J Mol Neurosci. 2009;39:248–255. doi: 10.1007/s12031-009-9192-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barger SW, Mattson MP. Participation of gene expression in the protection against amyloid beta-peptide toxicity by the beta-amyloid precursor protein. Ann N Y Acad Sci. 1996;777:303–309. doi: 10.1111/j.1749-6632.1996.tb34437.x. [DOI] [PubMed] [Google Scholar]
- Cirrito JR, Kang JE, Lee J, Stewart FR, Verges DK, Silverio LM, Bu G, Mennerick S, Holtzman DM. Endocytosis is required for synaptic activity-dependent release of amyloid-beta in vivo. Neuron. 2008;58:42–51. doi: 10.1016/j.neuron.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöbel S, Neumann S, Hertweck M, Dislich B, Kuhn PH, Kremmer E, Seed B, Baumeister R, Haass C, Lichtenthaler SF. A novel sorting nexin modulates endocytic trafficking and alpha-secretase cleavage of the amyloid precursor protein. J Biol Chem. 2008;283:14257–14268. doi: 10.1074/jbc.M801531200. [DOI] [PubMed] [Google Scholar]
- Thal DR, Sassin I, Schultz C, Haass C, Braak E, Braak H. Fleecy amyloid deposits in the internal layers of the human entorhinal cortex are comprised of N-terminal truncated fragments of Abeta. J Neuropathol Exp Neurol. 1999;58:210–216. doi: 10.1097/00005072-199902000-00010. [DOI] [PubMed] [Google Scholar]
- Miravalle L, Calero M, Takao M, Roher AE, Ghetti B, Vidal R. Amino-terminally truncated Abeta peptide species are the main component of cotton wool plaques. Biochemistry. 2005;44:10810–10821. doi: 10.1021/bi0508237. [DOI] [PubMed] [Google Scholar]
- Zhao JH, Liu HL, Liu YF, Lin HY, Fang HW, Ho Y, Tsai WB. Molecular dynamics simulations to investigate the aggregation behaviors of the Abeta(17-42) oligomers. J Biomol Struct Dyn. 2009;26:481–490. doi: 10.1080/07391102.2009.10507263. [DOI] [PubMed] [Google Scholar]
- Cordy JM, Hooper NM, Turner AJ. The involvement of lipid rafts in Alzheimer's disease. Mol Membr Biol. 2006;23:111–122. doi: 10.1080/09687860500496417. [DOI] [PubMed] [Google Scholar]
- Kamenetz F, Tomita T, Hsieh H, Seabrook G, Borchelt D, Iwatsubo T, Sisodia S, Malinow R. APP processing and synaptic function. Neuron. 2003;37:925–937. doi: 10.1016/S0896-6273(03)00124-7. [DOI] [PubMed] [Google Scholar]
- Pierrot N, Ghisdal P, Caumont AS, Octave JN. Intraneuronal amyloid-beta1-42 production triggered by sustained increase of cytosolic calcium concentration induces neuronal death. J Neurochem. 2004;88:1140–1150. doi: 10.1046/j.1471-4159.2003.02227.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhou K, Wang R, Cui J, Lipton SA, Liao FF, Xu H, Zhang YW. Hypoxia-inducible factor 1alpha (HIF-1alpha)-mediated hypoxia increases BACE1 expression and beta-amyloid generation. J Biol Chem. 2007;282:10873–10880. doi: 10.1074/jbc.M608856200. [DOI] [PubMed] [Google Scholar]
- Cantara S, Donnini S, Morbidelli L, Giachetti A, Schulz R, Memo M, Ziche M. Physiological levels of amyloid peptides stimulate the angiogenic response through FGF-2. FASEB J. 2004;18:1943–1945. doi: 10.1096/fj.04-2114fje. [DOI] [PubMed] [Google Scholar]
- Yan Y, Wang C. Abeta42 is more rigid than Abeta40 at the C terminus: implications for Abeta aggregation and toxicity. J Mol Biol. 2006;364:853–862. doi: 10.1016/j.jmb.2006.09.046. [DOI] [PubMed] [Google Scholar]
- Sgourakis NG, Yan Y, McCallum SA, Wang C, Garcia AE. The Alzheimer's peptides Abeta40 and 42 adopt distinct conformations in water: a combined MD/NMR study. J Mol Biol. 2007;368:1448–1457. doi: 10.1016/j.jmb.2007.02.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou K, Kim D, Kakio A, Byun K, Gong JS, Kim J, Kim M, Sawamura N, Nishimoto S, Matsuzaki K, Lee B, Yanagisawa K, Michikawa M. Amyloid beta-protein (Abeta)1-40 protects neurons from damage induced by Abeta1-42 in culture and in rat brain. J Neurochem. 2003;87:609–619. doi: 10.1046/j.1471-4159.2003.02018.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Matsubara E, Maeda S, Minagawa H, Takashima A, Maruyama W, Michikawa M, Yanagisawa K. A ganglioside-induced toxic soluble Abeta assembly. Its enhanced formation from Abeta bearing the Arctic mutation. J Biol Chem. 2007;282:2646–2655. doi: 10.1074/jbc.M606202200. [DOI] [PubMed] [Google Scholar]
- Heo C, Chang KA, Choi HS, Kim HS, Kim S, Liew H, Kim JA, Yu E, Ma J, Suh YH. Effects of the monomeric, oligomeric, and fibrillar Abeta42 peptides on the proliferation and differentiation of adult neural stem cells from subventricular zone. J Neurochem. 2007;102:493–500. doi: 10.1111/j.1471-4159.2007.04499.x. [DOI] [PubMed] [Google Scholar]
- Michikawa M, Gong JS, Fan QW, Sawamura N, Yanagisawa K. A novel action of alzheimer's amyloid beta-protein (Abeta): oligomeric Abeta promotes lipid release. J Neurosci. 2001;21:7226–7235. doi: 10.1523/JNEUROSCI.21-18-07226.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid AW, Freir DB, Herron CE. Inhibition of LTP in vivo by beta-amyloid peptide in different conformational states. Brain Res. 2008;1197:135–142. doi: 10.1016/j.brainres.2007.11.056. [DOI] [PubMed] [Google Scholar]
- Wang HY, Lee DH, Davis CB, Shank RP. Amyloid peptide Abeta(1-42) binds selectively and with picomolar affinity to alpha7 nicotinic acetylcholine receptors. J Neurochem. 2000;75:1155–1161. doi: 10.1046/j.1471-4159.2000.0751155.x. [DOI] [PubMed] [Google Scholar]
- Alberghina L, Colangelo AM. The modular systems biology approach to investigate the control of apoptosis in Alzheimer's disease neurodegeneration. BMC Neurosci. 2006;7(Suppl 1):S2. doi: 10.1186/1471-2202-7-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JE, Ince PG, Haynes LJ, Theaker R, Gelsthorpe C, Baxter L, Forster G, Lace GL, Shaw PJ, Matthews FE, Savva GM, Brayne C, Wharton SB. MRC Cognitive Function and Ageing Neuropathology Study Group. Population variation in oxidative stress and astrocyte DNA damage in relation to Alzheimer-type pathology in the ageing brain. Neuropathol Appl Neurobiol. 2010;36:25–40. doi: 10.1111/j.1365-2990.2009.01030.x. [DOI] [PubMed] [Google Scholar]
- Simpson JE, Ince PG, Lace G, Forster G, Shaw PJ, Matthews F, Savva G, Brayne C, Wharton SB. MRC Cognitive Function and Ageing Neuropathology Study Group. Astrocyte phenotype in relation to Alzheimer-type pathology in the ageing brain. Neurobiol Aging. 2010;31:578–590. doi: 10.1016/j.neurobiolaging.2008.05.015. [DOI] [PubMed] [Google Scholar]
- Abbott JJ, Howlett DR, Francis PT, Williams RJ. Abeta(1-42) modulation of Akt phosphorylation via alpha7 nAChR and NMDA receptors. Neurobiol Aging. 2008;29:992–1001. doi: 10.1016/j.neurobiolaging.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Gu Z, Liu W, Yan Z. {beta}-Amyloid impairs AMPA receptor trafficking and function by reducing Ca2+/calmodulin-dependent protein kinase II synaptic distribution. J Biol Chem. 2009;284:10639–10649. doi: 10.1074/jbc.M806508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Watson JB, Xie CW. Amyloid beta prevents activation of calcium/calmodulin-dependent protein kinase II and AMPA receptor phosphorylation during hippocampal long-term potentiation. J Neurophysiol. 2004;92:2853–2858. doi: 10.1152/jn.00485.2004. [DOI] [PubMed] [Google Scholar]
- Dodart JC, Bales KR, Johnstone EM, Little SP, Paul SM. Apolipoprotein E alters the processing of the beta-amyloid precursor protein in APP(V717F) transgenic mice. Brain Res. 2002;955:191–199. doi: 10.1016/S0006-8993(02)03437-6. [DOI] [PubMed] [Google Scholar]
- Zlokovic BV, Yamada S, Holtzman D, Ghiso J, Frangione B. Clearance of amyloid beta-peptide from brain: transport or metabolism? Nat Med. 2000;6:718–719. doi: 10.1038/77397. [DOI] [PubMed] [Google Scholar]
- Holtzman DM, Fagan AM, Mackey B, Tenkova T, Sartorius L, Paul SM, Bales K, Ashe KH, Irizarry MC, Hyman BT. Apolipoprotein E facilitates neuritic and cerebrovascular plaque formation in an Alzheimer's disease model. Ann Neurol. 2000;47:739–747. doi: 10.1002/1531-8249(200006)47:6<739::AID-ANA6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Tamamizu-Kato S, Cohen JK, Drake CB, Kosaraju MG, Drury J, Narayanaswami V. Interaction with amyloid beta peptide compromises the lipid binding function of apolipoprotein E. Biochemistry. 2008;47:5225–5234. doi: 10.1021/bi702097s. [DOI] [PubMed] [Google Scholar]
- Furukawa K, Barger SW, Blalock EM, Mattson MP. Activation of K+ channels and suppression of neuronal activity by secreted beta-amyloid-precursor protein. Nature. 1996;379:74–78. doi: 10.1038/379074a0. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Matsuda Y, D'Adamio L. CD74 interacts with APP and suppresses the production of Abeta. Mol Neurodegener. 2009;4:41. doi: 10.1186/1750-1326-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beel AJ, Mobley CK, Kim HJ, Tian F, Hadziselimovic A, Jap B, Prestegard JH, Sanders CR. Structural studies of the transmembrane C-terminal domain of the amyloid precursor protein (APP): does APP function as a cholesterol sensor? Biochemistry. 2008;47:9428–9446. doi: 10.1021/bi800993c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WW, Todd S, Coulson DT, Irvine GB, Passmore AP, McGuinness B, McConville M, Craig D, Johnston JA. A novel reciprocal and biphasic relationship between membrane cholesterol and beta-secretase activity in SH-SY5Y cells and in human platelets. J Neurochem. 2009;108:341–349. doi: 10.1111/j.1471-4159.2008.05753.x. [DOI] [PubMed] [Google Scholar]
- Abad-Rodriguez J, Ledesma MD, Craessaerts K, Perga S, Medina M, Delacourte A, Dingwall C, De Strooper B, Dotti CG. Neuronal membrane cholesterol loss enhances amyloid peptide generation. J Cell Biol. 2004;167:953–960. doi: 10.1083/jcb.200404149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avdulov NA, Chochina SV, Igbavboa U, Warden CS, Vassiliev AV, Wood WG. Lipid binding to amyloid beta-peptide aggregates: preferential binding of cholesterol as compared with phosphatidylcholine and fatty acids. J Neurochem. 1997;69:1746–1752. doi: 10.1046/j.1471-4159.1997.69041746.x. [DOI] [PubMed] [Google Scholar]
- Kristofiková Z, Kopecký V Jr, Hofbauerová K, Hovorková P, Rípová D. Complex of amyloid beta peptides with 24-hydroxycholesterol and its effect on hemicholinium-3 sensitive carriers. Neurochem Res. 2008;33:412–421. doi: 10.1007/s11064-007-9443-5. [DOI] [PubMed] [Google Scholar]
- Shen Y, Sullivan T, Lee CM, Meri S, Shiosaki K, Lin CW. Induced expression of neuronal membrane attack complex and cell death by Alzheimer's beta-amyloid peptide. Brain Res. 1998;796:187–197. doi: 10.1016/S0006-8993(98)00346-1. [DOI] [PubMed] [Google Scholar]
- Mudher A, Chapman S, Richardson J, Asuni A, Gibb G, Pollard C, Killick R, Iqbal T, Raymond L, Varndell I, Sheppard P, Makoff A, Gower E, Soden PE, Lewis P, Murphy M, Golde TE, Rupniak HT, Anderton BH, Lovestone S. Dishevelled regulates the metabolism of amyloid precursor protein via protein kinase C/mitogen-activated protein kinase and c-Jun terminal kinase. J Neurosci. 2001;21:4987–4995. doi: 10.1523/JNEUROSCI.21-14-04987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Alavez M, Chan SL, Mattson MP, Criado JR. Electrophysiological and cerebrovascular effects of the alpha-secretase-derived form of amyloid precursor protein in young and middle-aged rats. Brain Res. 2007;1131:112–117. doi: 10.1016/j.brainres.2006.10.074. [DOI] [PubMed] [Google Scholar]
- Sepulveda FJ, Opazo C, Aguayo LG. Alzheimer beta-amyloid blocks epileptiform activity in hippocampal neurons. Mol Cell Neurosci. 2009;41:420–428. doi: 10.1016/j.mcn.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Sabo SL, Ikin AF, Buxbaum JD, Greengard P. The Alzheimer amyloid precursor protein (APP) and FE65, an APP-binding protein, regulate cell movement. J Cell Biol. 2001;153:1403–1414. doi: 10.1083/jcb.153.7.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberly WT, Zheng JB, Guénette SY, Selkoe DJ. The intracellular domain of the beta-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J Biol Chem. 2001;276:40288–40292. doi: 10.1074/jbc.C100447200. [DOI] [PubMed] [Google Scholar]
- Ando K, Iijima KI, Elliott JI, Kirino Y, Suzuki T. Phosphorylation-dependent regulation of the interaction of amyloid precursor protein with Fe65 affects the production of beta-amyloid. J Biol Chem. 2001;276:40353–40361. doi: 10.1074/jbc.M104059200. [DOI] [PubMed] [Google Scholar]
- Santiard-Baron D, Langui D, Delehedde M, Delatour B, Schombert B, Touchet N, Tremp G, Paul MF, Blanchard V, Sergeant N, Delacourte A, Duyckaerts C, Pradier L, Mercken L. Expression of human FE65 in amyloid precursor protein transgenic mice is associated with a reduction in beta-amyloid load. J Neurochem. 2005;93:330–338. doi: 10.1111/j.1471-4159.2005.03026.x. [DOI] [PubMed] [Google Scholar]
- Zhou D, Zambrano N, Russo T, D'Adamio L. Phosphorylation of a tyrosine in the amyloid-beta protein precursor intracellular domain inhibits Fe65 binding and signaling. J Alzheimers Dis. 2009;16:301–307. doi: 10.3233/JAD-2009-0970. [DOI] [PubMed] [Google Scholar]
- Hwang EM, Kim SK, Sohn JH, Lee JY, Kim Y, Kim YS, Mook-Jung I. Furin is an endogenous regulator of alpha-secretase associated APP processing. Biochem Biophys Res Commun. 2006;349:654–659. doi: 10.1016/j.bbrc.2006.08.077. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Guo ZH, Geiger JD. Secreted form of amyloid precursor protein enhances basal glucose and glutamate transport and protects against oxidative impairment of glucose and glutamate transport in synaptosomes by a cyclic GMP-mediated mechanism. J Neurochem. 1999;73:532–537. doi: 10.1046/j.1471-4159.1999.0730532.x. [DOI] [PubMed] [Google Scholar]
- Furukawa K, Mattson MP. Secreted amyloid precursor protein alpha selectively suppresses N-methyl-D-aspartate currents in hippocampal neurons: involvement of cyclic GMP. Neuroscience. 1998;83:429–438. doi: 10.1016/s0306-4522(97)00398-9. [DOI] [PubMed] [Google Scholar]
- Taylor CJ, Ireland DR, Ballagh I, Bourne K, Marechal NM, Turner PR, Bilkey DK, Tate WP, Abraham WC. Endogenous secreted amyloid precursor protein-alpha regulates hippocampal NMDA receptor function, long-term potentiation and spatial memory. Neurobiol Dis. 2008;31:250–260. doi: 10.1016/j.nbd.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Suo Z, Wu M, Citron BA, Wong GT, Festoff BW. Abnormality of G-protein-coupled receptor kinases at prodromal and early stages of Alzheimer's disease: an association with early beta-amyloid accumulation. J Neurosci. 2004;24:3444–3452. doi: 10.1523/JNEUROSCI.4856-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto I, Okamoto T, Matsuura Y, Takahashi S, Okamoto T, Murayama Y, Ogata E. Alzheimer amyloid protein precursor complexes with brain GTP-binding protein G(o) Nature. 1993;362:75–79. doi: 10.1038/362075a0. [DOI] [PubMed] [Google Scholar]
- Okamoto T, Takeda S, Murayama Y, Ogata E, Nishimoto I. Ligand-dependent G protein coupling function of amyloid transmembrane precursor. J Biol Chem. 1995;270:4205–4208. doi: 10.1074/jbc.270.9.4205. [DOI] [PubMed] [Google Scholar]
- Shaked GM, Chauv S, Ubhi K, Hansen LA, Masliah E. Interactions between the amyloid precursor protein C-terminal domain and G proteins mediate calcium dysregulation and amyloid beta toxicity in Alzheimer's disease. FEBS J. 2009;276:2736–2751. doi: 10.1111/j.1742-4658.2009.06997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudinov AR, Berezov TT, Kumar A, Koudinova NV. Alzheimer's amyloid beta interaction with normal human plasma high density lipoprotein: association with apolipoprotein and lipids. Clin Chim Acta. 1998;270:75–84. doi: 10.1016/S0009-8981(97)00207-6. [DOI] [PubMed] [Google Scholar]
- Patey SJ, Edwards EA, Yates EA, Turnbull JE. Heparin derivatives as inhibitors of BACE-1, the Alzheimer's beta-secretase, with reduced activity against factor Xa and other proteases. J Med Chem. 2006;49:6129–6132. doi: 10.1021/jm051221o. [DOI] [PubMed] [Google Scholar]
- Patey SJ, Edwards EA, Yates EA, Turnbull JE. Engineered heparins: novel beta-secretase inhibitors as potential Alzheimer's disease therapeutics. Neurodegener Dis. 2008;5:197–199. doi: 10.1159/000113701. [DOI] [PubMed] [Google Scholar]
- Kong Q, Peterson TS, Baker O, Stanley E, Camden J, Seye CI, Erb L, Simonyi A, Wood WG, Sun GY, Weisman GA. Interleukin-1beta enhances nucleotideinduced and alpha-secretase-dependent amyloid precursor protein processing in rat primary cortical neurons via up-regulation of the P2Y(2) receptor. J Neurochem. 2009;109:1300–1310. doi: 10.1111/j.1471-4159.2009.06048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachida Y, Nakagawa K, Saito T, Saido TC, Honda T, Saito Y, Murayama S, Endo T, Sakaguchi G, Kato A, Kitazume S, Hashimoto Y. Interleukin-1 beta up-regulates TACE to enhance alpha-cleavage of APP in neurons: resulting decrease in Abeta production. J Neurochem. 2008;104:1387–1393. doi: 10.1111/j.1471-4159.2007.05127.x. [DOI] [PubMed] [Google Scholar]
- Sheng JG, Ito K, Skinner RD, Mrak RE, Rovnaghi CR, Van Eldik LJ, Griffin WS. In vivo and in vitro evidence supporting a role for the inflammatory cytokine interleukin-1 as a driving force in Alzheimer pathogenesis. Neurobiol Aging. 1996;17:761–766. doi: 10.1016/0197-4580(96)00104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez A, Morelli L, Cresto JC, Castaño EM. Degradation of soluble amyloid beta-peptides 1-40, 1-42, and the Dutch variant 1-40Q by insulin degrading enzyme from Alzheimer disease and control brains. Neurochem Res. 2000;25:247–255. doi: 10.1023/A:1007527721160. [DOI] [PubMed] [Google Scholar]
- Young-Pearse TL, Chen AC, Chang R, Marquez C, Selkoe DJ. Secreted APP regulates the function of full-length APP in neurite outgrowth through interaction with integrin beta1. Neural Dev. 2008;3:15. doi: 10.1186/1749-8104-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caltagarone J, Jing Z, Bowser R. Focal adhesions regulate Abeta signaling and cell death in Alzheimer's disease. Biochim Biophys Acta. 2007;1772:438–445. doi: 10.1016/j.bbadis.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Klyubin I, Wright S, Griswold-Prenner I, Rowan MJ, Anwyl R. Alpha v integrins mediate beta-amyloid induced inhibition of long-term potentiation. Neurobiol Aging. 2008;29:1485–1493. doi: 10.1016/j.neurobiolaging.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Terzi E, Holzemann G, Seelig J. Interaction of Alzheimer beta-amyloid peptide(1-40) with lipid membranes. Biochemistry. 1997;36:14845–14852. doi: 10.1021/bi971843e. [DOI] [PubMed] [Google Scholar]
- Hertel C, Terzi E, Hauser N, Jakob-Rotne R, Seelig J, Kemp JA. Inhibition of the electrostatic interaction between beta-amyloid peptide and membranes prevents beta-amyloid-induced toxicity. Proc Natl Acad Sci USA. 1997;94:9412–9416. doi: 10.1073/pnas.94.17.9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier Y, Zarandi M, Penke B. Amyloid beta-peptide interactions with neuronal and glial cell plasma membrane: binding sites and implications for Alzheimer's disease. J Pept Sci. 2004;10:229–248. doi: 10.1002/psc.573. [DOI] [PubMed] [Google Scholar]
- Cordy JM, Hussain I, Dingwall C, Hooper NM, Turner AJ. Exclusively targeting beta-secretase to lipid rafts by GPI-anchor addition up-regulates beta-site processing of the amyloid precursor protein. Proc Natl Acad Sci USA. 2003;100:11735–11740. doi: 10.1073/pnas.1635130100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra L, Mohamed A, Ma V, Kar S, de Chaves EP. Internalization of beta-amyloid peptide by primary neurons in the absence of apolipoprotein E. J Biol Chem. 2007;282:35722–35732. doi: 10.1074/jbc.M701823200. [DOI] [PubMed] [Google Scholar]
- Sawamura N, Ko M, Yu W, Zou K, Hanada K, Suzuki T, Gong JS, Yanagisawa K, Michikawa M. Modulation of amyloid precursor protein cleavage by cellular sphingolipids. J Biol Chem. 2004;279:11984–11991. doi: 10.1074/jbc.M309832200. [DOI] [PubMed] [Google Scholar]
- Wu MN, He YX, Guo F, Qi JS. Alpha4beta2 nicotinic acetylcholine receptors are required for the amyloid beta protein-induced suppression of long-term potentiation in rat hippocampal CA1 region in vivo. Brain Res Bull. 2008;77:84–90. doi: 10.1016/j.brainresbull.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Yamin G. NMDA receptor-dependent signaling pathways that underlie amyloid beta-protein disruption of LTP in the hippocampus. J Neurosci Res. 2009;87:1729–1736. doi: 10.1002/jnr.21998. [DOI] [PubMed] [Google Scholar]
- Caccamo A, Oddo S, Billings LM, Green KN, Martinez-Coria H, Fisher A, LaFerla FM. M1 receptors play a central role in modulating AD-like pathology in transgenic mice. Neuron. 2006;49:671–682. doi: 10.1016/j.neuron.2006.01.020. [DOI] [PubMed] [Google Scholar]
- Fisher A. M1 muscarinic agonists target major hallmarks of Alzheimer's disease - the pivotal role of brain M1 receptors. Neurodegener Dis. 2008;5:237–240. doi: 10.1159/000113712. [DOI] [PubMed] [Google Scholar]
- Nitsch RM, Slack BE, Farber SA, Schulz JG, Deng M, Kim C, Borghesani PR, Korver W, Wurtman RJ, Growdon JH. Regulation of proteolytic processing of the amyloid beta-protein precursor of Alzheimer's disease in transfected cell lines and in brain slices. J Neural Transm Suppl. 1994;44:21–27. doi: 10.1007/978-3-7091-9350-1_2. [DOI] [PubMed] [Google Scholar]
- Farber SA, Nitsch RM, Schulz JG, Wurtman RJ. Regulated secretion of beta-amyloid precursor protein in rat brain. J Neurosci. 1995;15:7442–7451. doi: 10.1523/JNEUROSCI.15-11-07442.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskowsky W, Schliebs R. Muscarinic acetylcholine receptor inhibition in transgenic Alzheimer-like Tg2576 mice by scopolamine favours the amyloidogenic route of processing of amyloid precursor protein. Int J Dev Neurosci. 2006;24:149–156. doi: 10.1016/j.ijdevneu.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Zuchner T, Perez-Polo JR, Schliebs R. Beta-secretase BACE1 is differentially controlled through muscarinic acetylcholine receptor signaling. J Neurosci Res. 2004;77:250–257. doi: 10.1002/jnr.20152. [DOI] [PubMed] [Google Scholar]
- Kim C, Jang CH, Bang JH, Jung MW, Joo I, Kim SU, Mook-Jung I. Amyloid precursor protein processing is separately regulated by protein kinase C and tyrosine kinase in human astrocytes. Neurosci Lett. 2002;324:185–188. doi: 10.1016/S0304-3940(02)00217-3. [DOI] [PubMed] [Google Scholar]
- Qiu WQ, Ferreira A, Miller C, Koo EH, Selkoe DJ. Cell-surface beta-amyloid precursor protein stimulates neurite outgrowth of hippocampal neurons in an isoform-dependent manner. J Neurosci. 1995;15:2157–2167. doi: 10.1523/JNEUROSCI.15-03-02157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossner S, Sastre M, Bourne K, Lichtenthaler SF. Transcriptional and translational regulation of BACE1 expression - implications for Alzheimer's disease. Prog Neurobiol. 2006;79:95–111. doi: 10.1016/j.pneurobio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Choi S, Kim JH, Roh EJ, Ko MJ, Jung JE, Kim HJ. Nuclear factor-kappaB activated by capacitative Ca2+ entry enhances muscarinic receptor-mediated soluble amyloid precursor protein (sAPPalpha) release in SH-SY5Y cells. J Biol Chem. 2006;281:12722–12728. doi: 10.1074/jbc.M601018200. [DOI] [PubMed] [Google Scholar]
- Kihara T, Shimohama S, Urushitani M, Sawada H, Kimura J, Kume T, Maeda T, Akaike A. Stimulation of alpha4beta2 nicotinic acetylcholine receptors inhibits beta-amyloid toxicity. Brain Res. 1998;792:331–334. doi: 10.1016/S0006-8993(98)00138-3. [DOI] [PubMed] [Google Scholar]
- Nagele RG, D'Andrea MR, Anderson WJ, Wang HY. Intracellular accumulation of beta-amyloid(1-42) in neurons is facilitated by the alpha 7 nicotinic acetylcholine receptor in Alzheimer's disease. Neuroscience. 2002;110:199–211. doi: 10.1016/S0306-4522(01)00460-2. [DOI] [PubMed] [Google Scholar]
- Dougherty JJ, Wu J, Nichols RA. Beta-amyloid regulation of presynaptic nicotinic receptors in rat hippocampus and neocortex. J Neurosci. 2003;23:6740–6747. doi: 10.1523/JNEUROSCI.23-17-06740.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Lee DH, D'Andrea MR, Peterson PA, Shank RP, Reitz AB. beta- Amyloid(1-42) binds to alpha7 nicotinic acetylcholine receptor with high affinity. Implications for Alzheimer's disease pathology. J Biol Chem. 2000;275:5626–5632. doi: 10.1074/jbc.275.8.5626. [DOI] [PubMed] [Google Scholar]
- Snyder EM, Nong Y, Almeida CG, Paul S, Moran T, Choi EY, Nairn AC, Salter MW, Lombroso PJ, Gouras GK, Greengard P. Regulation of NMDA receptor trafficking by amyloid-beta. Nat Neurosci. 2005;8:1051–1058. doi: 10.1038/nn1503. [DOI] [PubMed] [Google Scholar]
- Camden JM, Schrader AM, Camden RE, González FA, Erb L, Seye CI, Weisman GA. P2Y2 nucleotide receptors enhance alpha-secretase-dependent amyloid precursor protein processing. J Biol Chem. 2005;280:18696–18702. doi: 10.1074/jbc.M500219200. [DOI] [PubMed] [Google Scholar]
- Roncarati R, Sestan N, Scheinfeld MH, Berechid BE, Lopez PA, Meucci O, McGlade JC, Rakic P, D'Adamio L. The gamma-secretase-generated intracellular domain of beta-amyloid precursor protein binds Numb and inhibits Notch signaling. Proc Natl Acad Sci USA. 2002;99:7102–7107. doi: 10.1073/pnas.102192599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etcheberrigaray R, Tan M, Dewachter I, Kuipéri C, Van der Auwera I, Wera S, Qiao L, Bank B, Nelson TJ, Kozikowski AP, Van Leuven F, Alkon DL. Therapeutic effects of PKC activators in Alzheimer's disease transgenic mice. Proc Natl Acad Sci USA. 2004;101:11141–11146. doi: 10.1073/pnas.0403921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossner S, Mendla K, Schliebs R, Bigl V. Protein kinase Calpha and beta1 isoforms are regulators of alpha-secretory proteolytic processing of amyloid precursor protein in vivo. Eur J Neurosci. 2001;13:1644–1648. doi: 10.1046/j.0953-816x.2001.01525.x. [DOI] [PubMed] [Google Scholar]
- Favit A, Grimaldi M, Nelson TJ, Alkon DL. Alzheimer's-specific effects of soluble beta-amyloid on protein kinase C-alpha and -gamma degradation in human fibroblasts. Proc Natl Acad Sci USA. 1998;95:5562–5567. doi: 10.1073/pnas.95.10.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durakoglugil MS, Chen Y, White CL, Kavalali ET, Herz J. Reelin signaling antagonizes beta-amyloid at the synapse. Proc Natl Acad Sci USA. 2009;106:15938–15943. doi: 10.1073/pnas.0908176106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayev R, Zhou D, D'Adamio L. The interactome of the amyloid beta precursor protein family members is shaped by phosphorylation of their intracellular domains. Mol Neurodegener. 2009;4:28. doi: 10.1186/1750-1326-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinfeld MH, Ghersi E, Davies P, D'Adamio L. Amyloid beta protein precursor is phosphorylated by JNK-1 independent of, yet facilitated by, JNK-interacting protein (JIP)-1. J Biol Chem. 2003;278:42058–42063. doi: 10.1074/jbc.M304853200. [DOI] [PubMed] [Google Scholar]
- Hoe HS, Cooper MJ, Burns MP, Lewis PA, van der Brug M, Chakraborty G, Cartagena CM, Pak DT, Cookson MR, Rebeck GW. The metalloprotease inhibitor TIMP-3 regulates amyloid precursor protein and apolipoprotein E receptor proteolysis. J Neurosci. 2007;27:10895–10905. doi: 10.1523/JNEUROSCI.3135-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein TD, Johnson JA. Lack of neurodegeneration in transgenic mice overexpressing mutant amyloid precursor protein is associated with increased levels of transthyretin and the activation of cell survival pathways. J Neurosci. 2002;22:7380–7388. doi: 10.1523/JNEUROSCI.22-17-07380.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R, Ferreira-da-Silva F, Saraiva MJ, Cardoso I. Transthyretin protects against A-beta peptide toxicity by proteolytic cleavage of the peptide: a mechanism sensitive to the Kunitz protease inhibitor. PLoS ONE. 2008;3:e2899. doi: 10.1371/journal.pone.0002899. [DOI] [PMC free article] [PubMed] [Google Scholar]