Abstract
Background
Gut microbes influence animal health and thus, are potential targets for interventions that slow aging. Live E. coli provides the nematode worm Caenorhabditis elegans with vital micronutrients, such as folates that cannot be synthesized by animals. However, the microbe also limits C. elegans lifespan. Understanding these interactions may shed light on how intestinal microbes influence mammalian aging.
Results
Serendipitously, we isolated an E. coli mutant that slows C. elegans aging. We identified the disrupted gene to be aroD, which is required to synthesize aromatic compounds in the microbe. Adding back aromatic compounds to the media revealed that the increased C. elegans lifespan was caused by decreased availability of para-aminobenzoic acid, a precursor to folate. Consistent with this result, inhibition of folate synthesis by sulfamethoxazole, a sulfonamide, led to a dose-dependent increase in C. elegans lifespan. As expected, these treatments caused a decrease in bacterial and worm folate levels, as measured by mass spectrometry of intact folates. The folate cycle is essential for cellular biosynthesis. However, bacterial proliferation and C. elegans growth and reproduction were unaffected under the conditions that increased lifespan.
Conclusions
In this animal:microbe system, folates are in excess of that required for biosynthesis. This study suggests that microbial folate synthesis is a pharmacologically accessible target to slow animal aging without detrimental effects.
Keywords: aging, microbes, folate, C. elegans, E. coli
Background
The microbial flora found in the gastrointestinal tract influences human metabolism and physiology and is thus likely to impact aging [1,2]. Changes in the microbial flora are associated with obesity [3,4] and microbial metabolism may influence cardiovascular disease [5]. In the C. elegans model, the E. coli foodstuff must be alive, but not necessarily able to divide, for the worm to achieve maximal growth and reproduction [6-8], suggesting that microbial metabolic activity is required for optimal C. elegans nutrition. Dietary restriction of C. elegans by limiting E. coli availability extends lifespan but the mechanisms involved remain unclear [9,10]. Treating E. coli with antibiotics that either stop proliferation or kill the bacteria increases C. elegans lifespan [11,12] and it has been previously suggested the mutants in E. coli genes that disrupt ubiquinone synthesis increase C. elegans lifespan by blocking bacterial respiration [13]. However, slowing or stopping bacterial growth is not a viable starting point to treat healthy microbiota. Lipopolysaccharide structures on the E. coli cell surface explain bacterial strain-specific effects on C. elegans lifespan and interactions with the C. elegans sensory system but do not provide a clear route to slow aging [14]. Here we explore the C. elegans/E. coli interaction further and identify microbial folate synthesis as a specific target to slow animal aging pharmacologically without cost to the microbe or animal.
Results and Discussion
A spontaneous mutation in the E. coli gene aroD increases C. elegans lifespan
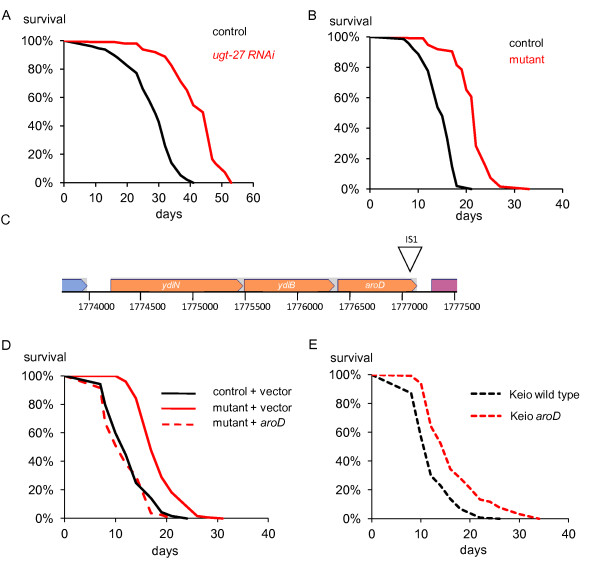
While performing lifespan experiments using RNA interference (RNAi) by feeding, we discovered an E. coli HT115(DE3) RNAi strain, for the C. elegans ugt-27 gene, that causes a substantial (30 to 50%) increase in lifespan of the long-lived daf-2 mutant of C. elegans compared to animals maintained on the HT115(DE3) control (Figure 1A). The strain also extends the lifespan of wild type C. elegans, and a daf-16 mutant lacking the FOXO transcription factor required for daf-2 mutant longevity (Additional file 1). Surprisingly, the lifespan increase persisted once the RNAi plasmid was lost, implicating a spontaneous mutation of the E. coli strain as the causative factor (Figure 1B). Consistent with this conclusion, fresh HT115(DE3) bacteria transformed with the ugt-27 RNAi plasmid had no effect on lifespan (Additional file 2). Unlike previously identified E. coli mutants that result in extended C. elegans lifespan [13,15], this mutant E. coli strain was able to respire but unable to grow on minimal media. This auxotrophy allowed us to perform a plasmid complementation screen leading to the identification of an IS1 transposon insertion in the gene aroD (Figure 1C). Plasmid rescue with aroD confirmed that mutation of this gene increased C. elegans lifespan (Figure 1D). A deletion mutation of aroD from the Keio strain collection also showed an increased lifespan compared to animals fed the control strain [16]. (Additional file 1, Figure 1E), demonstrating that the effect was not allele- or strain-specific.
Figure 1.
Identification of a spontaneous E. coli mutant that extends lifespan. A) Survival curves of rrf-3(pk1426); daf-2(m577) animals at 25°C on the control strain (n = 201): HT115(DE3) containing the empty vector L4440 and a strain, containing the plasmid for the ugt-27 gene, that caused a 50% increase in lifespan (n = 92, P = < 0.0001). B) Survival of rrf-3 (25°C) on the mutant strain from which the ugt-27 has been lost (n = 68) compared with the control strain in which the L4440 empty vector has been lost (n = 62). Increase in lifespan = 49.6% (P = < 0.0001). C) Position of the IS1 transposon insertion at position 1777116 on the E. coli K12 W3110 chromosome [44] (diagram based on EcoCyc.org [45]). As the insertion is at nucleotide 717 of the aroD open reading frame, the allele is designated aroD717::IS1. D) The lifespan effect of the mutant bacteria is rescued by the plasmid containing aroD. glp-4(bn2) animals were raised on the aroD mutant until L4 (15°C) and then transferred to the aroD mutant + pMMB67EH vector (n = 126), aroD mutant + pMMB67EH plasmid containing aroD region (n = 85), wild type bacteria + vector (n = 126) (25°C). E) Worms maintained on the aroD deletion mutant from the Keio collection (n = 128) show an extended lifespan compared to wild type (n = 131). P = < 0.001.
Folate synthesis is the limiting factor that causes the aroD mutant to increase C. elegans lifespan
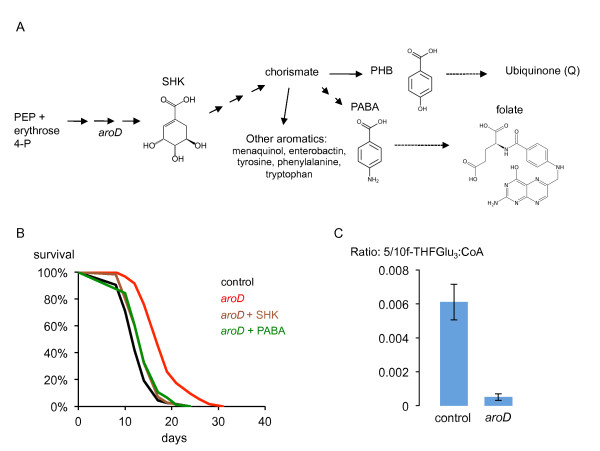
The aroD gene encodes the enzyme 3-dehydroquinate dehydratase, a core component of the shikimic acid pathway that produces chorismate, a precursor to all aromatic compounds in the bacterial cell (Figure 2A) [17,18]. Consistent with the involvement of this pathway, supplementation of the media with shikimic acid causes the lifespan of C. elegans on aroD mutant bacteria to revert to normal (Figure 2B). The aroD mutant can grow on the peptone-based media used in C. elegans studies, so the media must be able to provide either all the essential aromatic compounds needed for growth or the relevant precursors. To test whether the lifespan effect was caused by one of these compounds being present in limiting amounts, we added back compounds known to support growth of aro mutants: the aromatic amino acids, the folate precursor para-aminobenzoic acid (PABA) and the ubiquinone precursor para-hydroxybenzoic acid (PHB) [17]. Of these, only PABA reversed the lifespan increase completely, suggesting that a decrease in bacterial folate synthesis in the aroD mutant is the major cause of the increased C. elegans lifespan (Figures 2B; Additional file 3, Figure A; Additional file 3, Figure C; Additional file 1). PABA supplementation had no effect on C. elegans maintained on the control HT115(DE3) strain or on the extended lifespan of worms on the Q-deficient ubiG mutant bacteria ruling out a toxic effect of PABA (Additional file 3, Figure B). An alternate pathway for ubiquinone synthesis using PABA instead of PHB has been shown in Saccharomyces cerevisiae [19,20] but as PHB has no effect on lifespan (Additional file 3, Figure C) we think it unlikely that ubiquinone synthesis is the limiting factor in the aroD mutant. Folates are needed in all cells for biosynthesis. Generation of purines, pyrimidines, certain amino acids and methyl donors depends on cycling between the various folate species: dihydrofolate (DHF), tetrahydrofolate (THF), 10-formyl THF, 5,10-methenyl THF, 5,10-methylene THF and 5-methyl THF [21]. In addition, over 100 possible folate species result from further derivatization with up to 8 glutamate residues. To understand the effect of the aroD mutation on bacterial folates, we used liquid chromatography coupled to mass spectrometry (LC-MS) to detect individual folate species [21,22]. The most abundant folate species detected in E. coli grown as lawns on NGM agar corresponded to formyl THFGlu3. We found that the aroD mutation in HT115(DE3) caused a large decrease in the detectable levels of this folate and other detectable folate species (Figure 2C, Additional file 4), confirming the effect of the mutation on bacterial folates. Folic acid cannot be taken up directly by E. coli. However, adding back folic acid to the media resulted in a partial suppression of the increase in lifespan, probably because folic acid led to restoration of folate synthesis in the aroD mutant bacteria (Additional file 5). It has been shown that E. coli can use a breakdown product of folic acid to make PABA [23].
Figure 2.
Decreased folate synthesis explained the lifespan increase caused by the aroD mutant. A) Schematic of the shikimic acid and folate synthesis pathways. Solid arrows represent single enzymatic steps. Dashed arrows represent multiple steps. PEP, phosphoenolpyruvate; SHK, shikimic acid; PABA, para-aminobenzoic acid. B) Shikimic acid and PABA reverse the lifespan increase caused by the aroD mutation. glp-4(bn2) animals were raised on the aroD mutant until L4 (15°C) and then transferred to the aroD mutant (n = 116), wild type control (n = 116), aroD + SHK (n = 86), aroD + PABA (n = 76), (25°C). All supplements at 40 μg/ml. aroD + PABA vs aroD, P = < 0.0001; aroD + SHK vs aroD, P = < 0.0001. C) Levels of formylTHFGlu3 as detected by LC-MS are decreased in the aroD mutant compared to the wild type HT115(DE3). Data from two biological replicates.
Pharmacological inhibition of E. coli folate synthesis increases C. elegans lifespan
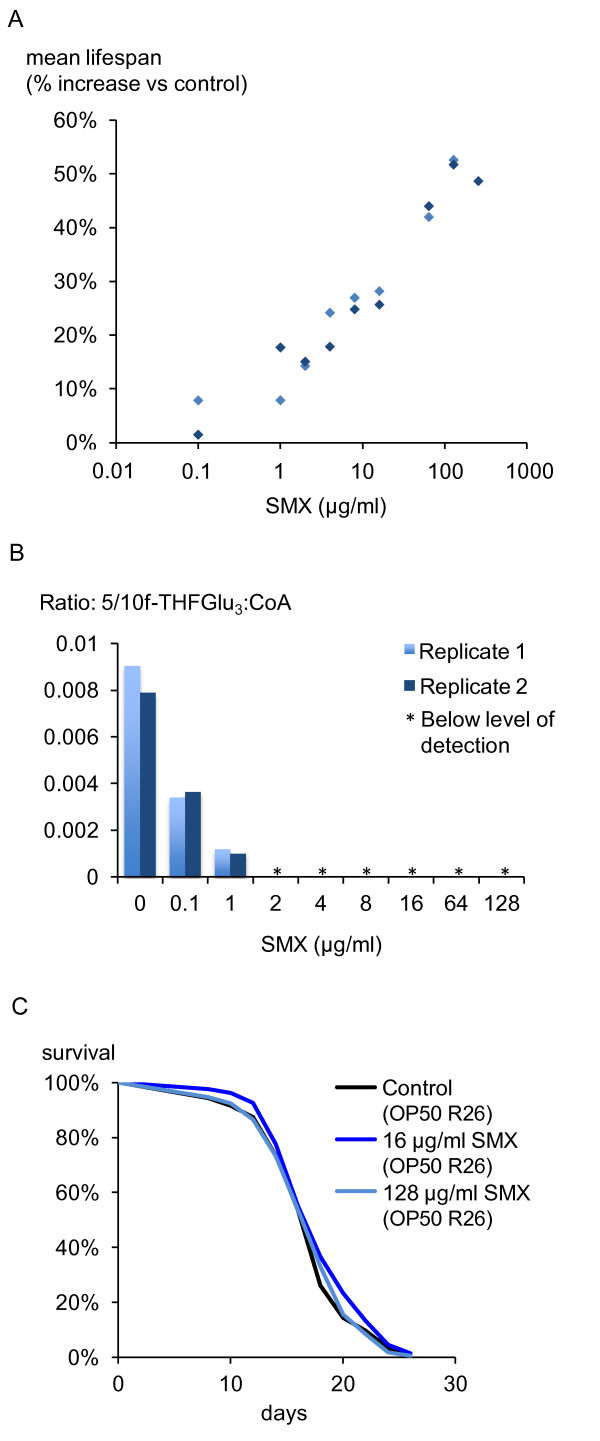
To test further the impact of microbial folate synthesis on C. elegans aging, we employed sulfamethoxazole (SMX), a sulfonamide drug that blocks folate synthesis by competing with PABA for the enzyme dihydropteroate synthase [24]. Addition of SMX to the media caused a dose-dependent increase in the lifespan of worms maintained on E. coli OP50, the strain used in most C. elegans studies (Figure 3A), with 2 μg/ml being the minimal dose that gave a reproducible and statistically significant effect. With increasing drug concentration, the relationship between mean lifespan and log[SMX] is approximately linear until 128 μg/ml, the concentration of SMX that consistently produced the highest increase in mean lifespan (See Additional file 1 for the full data set). This linear relationship suggests a dose response that is pharmacologically amenable. Addition of PABA reverses the increase in lifespan, consistent with folate synthesis being the relevant target of SMX (Additional file 6). To assess the impact of SMX on bacterial folates, we measured formyl THFGlu3. Starting at a concentration of 0.1 μg/ml, SMX reduced the levels of formyl THFGlu3 in OP50 such that at a dose of 2 μg/ml SMX it was effectively below the level of detection (Figure 3B; Additional file 4, Figure B). To determine whether SMX increased lifespan through any direct effects on the worm and/or any non-specific targets in E. coli, we performed lifespan experiments with a sulfonamide-resistant strain of OP50, which contains the multiresistance plasmid R26 encoding a drug-insensitive allele of dihydropteroate synthase [25]. Under these conditions, SMX had no effect on C. elegans lifespan (Figure 3C), demonstrating that SMX acts through inhibition of bacterial folate synthesis to increase C. elegans lifespan.
Figure 3.
Inhibition of bacterial folate synthesis causes an increase in C. elegans lifespan. A) Treatment of OP50 with various concentrations of SMX increases mean C. elegans lifespan by the indicated percentage. Control (n = 102, 191), 0.1 μg/ml SMX (n = 139, 221), 1 μg/ml (n = 154, 229), 2 μg/ml (n = 152, 210), 4 μg/ml (n = 161, 226), 8 μg/ml (n = 146, 224), 16 μg/ml (n = 176, 226), 64 μg/ml (n = 235, 238), 128 μg/ml (n = 229, 230). 256 μg/ml (n = 253). B) SMX treatment decreases levels of formylTHFGlu3 in E. coli OP50 until it becomes undetectable at 2 μg/ml. * = below the level of detection. Two biological replicates are shown. C) The lifespan increase induced by 16 and 128 μg/ml SMX is eliminated when worms are maintained on OP50 containing the R26 plasmid that confers sulfonamide resistance. Control (n = 224), 16 μg/ml SMX (n = 218), 128 μg/ml SMX (n = 215).
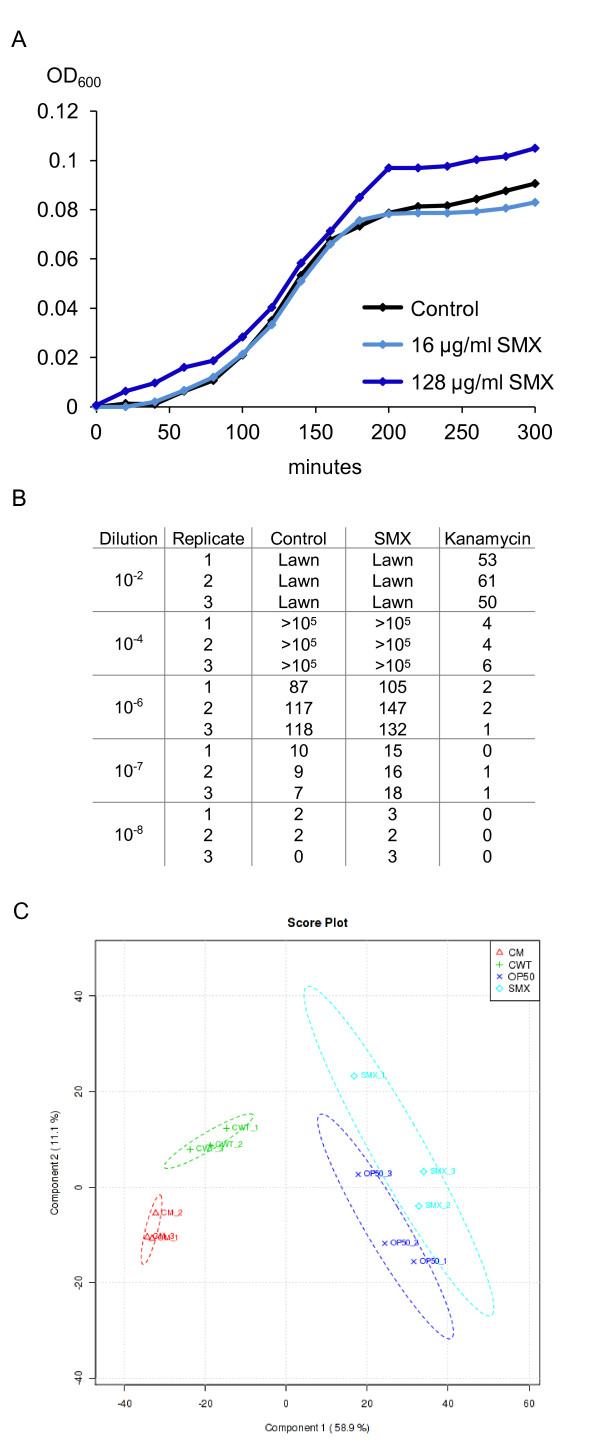
SMX has no effect on bacterial growth or viability
SMX is known to have antibiotic properties. However, we found that in nematode growth media (NGM), concentrations of SMX that extended C. elegans lifespan had no significant effect on E. coli growth in liquid culture (Figure 4A) or on the final size of the E. coli lawn grown on the solid media used to culture worms (Additional file 7). These results suggest that SMX does not extend lifespan by inhibiting E. coli proliferation as suggested for other antibiotics [11]. To test whether SMX-treated E. coli encountered by worms had a decreased ability to proliferate, we tested E. coli lawns for cell viability. In contrast to kanamycin treatment, 128 μg/ml SMX had no effect on the colony-forming ability of OP50 (Figure 4B). Given that the folate cycle is required for cell growth, these results suggest that E. coli can use metabolites from the media to overcome decreased folate synthesis. In agreement with this model, pabA and pabB mutants are reported to be viable and grow normally on rich media [16,26]. Thus, there are conditions that folate synthesis can be inhibited without affecting bacterial growth and it may be that E. coli are adapted for such conditions in the mammalian intestine.
Figure 4.
SMX has little effect on OP50 growth, viability or metabolome. A) in OD600 measurements of bacterial density show that SMX at 16 and 128 μg/ml has no effect on the log-phase growth rate of OP50 in liquid NGM at 37°C. B) Table showing colony forming units from bacteria scraped from lawns on agar plates, concentration of SMX used = 128 μg/ml. C) PLS-DA score plot showing two components that explain 58.9% (x-axis) and a further 11.9% (y-axis) of the variance between the conditions: whole LC-MS data from strains HT115(DE3) (labelled CWT), HT115(DE3)aroD (CM), OP50 (OP50) and OP50 treated with 128 μg/ml of SMX (SMX). Three replicates were performed for each condition.
Both the aroD mutation and SMX treatment have minor effects on metabolism
To examine the effects of SMX on broader metabolism, we re-analyzed the metabolite data from the LC-MS analysis of E. coli folates. We performed a global comparison between the aroD mutant, the HT115(DE3) wild type, OP50 and OP50 treated with 128 μg/ml SMX. A total of 1,539 features were detected, including common metabolites such as ATP, NADPH and acetyl CoA. Partial least squares discriminant analysis (PLS-DA) of the data (see Methods) shows that both the SMX and the aroD mutation have metabolite profiles very similar to their respective controls, having a smaller effect than the difference between the OP50 and HT115 control strains (Figure 4C). This analysis supports the hypothesis that the inhibition of folate synthesis under conditions that result in increased lifespan has only a minor effect on whole cell metabolism.
SMX leads to a decrease in C. elegans folate levels without adverse effects
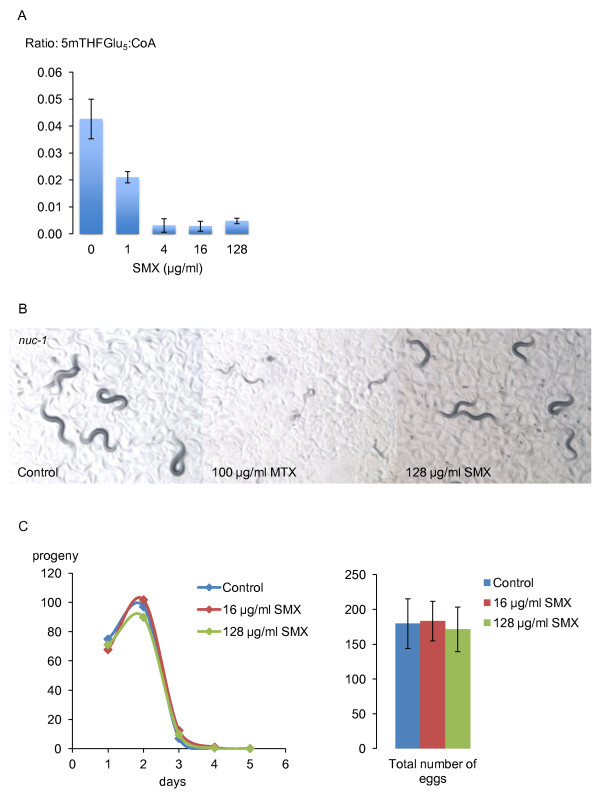
All animals must obtain folate from their food or intestinal microbes so inhibition of bacterial folate synthesis would be expected to decrease C. elegans folate levels. 5-methylTHFGlu5 was the most abundant folate species we could detect in worms. SMX decreased levels of this folate substantially but detectable levels remained (Figure 5A, Additional file 4, Methods). To test whether this decrease in folate levels led to a functional deficiency, we examined nuc-1 mutants, which are sensitized to methotrexate, an inhibitor of dihydrofolate reductase. Concentrations of methotrexate that have no effect on the wild-type animals cause nuc-1 mutants to produce sterile and uncoordinated progeny [27] (Figure 5B). However, SMX had no effect on nuc-1 mutants (Figure 5B), demonstrating that animals maintain a functional folate cycle. To test folate sufficiency further, we examined developmental rate and fecundity in the presence of SMX. We found that SMX had no effect on the time taken for animals to reach reproductive age or their subsequent brood size (Figure 5C). This result suggests that SMX does not impact the biosynthetic capability of the folate cycle. Further, this result confirms the drug does not interfere with C. elegans nutrition, or cause dietary restriction in the usual sense. However, it remains possible that life extension in C. elegans by dietary restriction via limitation of E. coli, and inhibition of microbial folate synthesis involve some common mechanisms.
Figure 5.
Effects of SMX on C. elegans folates and folate-dependent functions. A) SMX causes a dose-dependent decrease in C. elegans 5-methylTHFGlu5 levels. Data shown are from three biological replicates. B) Methotrexate induces slowed development, sterility and uncoordination in nuc-1 mutant worms but 128 μg/ml SMX has no effect. Pictures were taken three days after egg-laying. C) 16 and 128 μg/ml SMX have no effect on development time, egg-laying schedule or total brood size of N2 worms raised at 25°C. Control (number of animals = 15), 16 μg/ml SMX (n = 18), 128 μg/ml SMX (n = 17). Error bars are ± standard deviation.
Our data show that, via inhibition of bacterial folate synthesis, it is possible to slow animal aging with a minimal effect on bacterial growth. Such an intervention may allow manipulation in the mammalian gut without large disruption of the microbiota. Intriguingly, an early study showed that administration of a sulfonamide (sulfadiazine) extends the lifespan of rodents [28,29]. Moreover, as in the present study, this effect was reversed by the addition of PABA, implicating microbial folate synthesis as the target. Several species of bacteria, including E. coli, excrete folates, suggesting that they produce more than they need [30]. In our system, C. elegans is solely dependent on bacterial folate, and yet we can achieve a positive effect on lifespan without compromising healthy development, suggesting that the C. elegans requirement for folate is much lower than is available to them from E. coli. Interestingly, when added to high-folate rat food, the sulfonamide succinyl sulfathiazole, inhibited bacterial folate synthesis but had only a minor effect on rat liver folate levels [31], raising the possiblity that we can reduce folate specifically in the gut bacteria, without folate restricting the animal.
Conclusions
Genetic or pharmacological inhibition of E. coli folate synthesis leads to an increase in C. elegans lifespan without causing detrimental effects on either the microbe or the animal. Whether a decrease in folate acts on a process in the microbe or directly in the animal or both to extend lifespan remains to be determined. However, the identification of bacterial folate synthesis as a target suggests that eliminating excessive microbial folate in the gut microbe environment could be a route to slow aging.
Methods
Strains
C. elegans strains used in this study are GA303 rrf-3(pk 1426); daf-2(m577) [32], GR1307 daf-16(mgDf50) [33], N2 (wild type), NL2099 rrf-3(pk1426) [34] and SS104 glp-4(bn2) [35].
All E. coli strains used in this study are listed in Table 1.
Table 1.
E. coli strains
| Strains | Genotype/Relevant characteristics | Source |
|---|---|---|
| OP50 | OP50 ura | [37] |
| RNAi control strain | HT115(DE3) W3110 rnc14::Tn10 (DE3 PlacUV5-T7 polymerase) | [46] |
| aroD mutant | HT115(DE3) aroD717::IS1 | This study |
| Keio collection WT | BW25113 | [16] |
| Keio collection aroD | BW25113 aroD | [16,36], This study |
| OP50 Su resistant | OP50 R26 | This study; [47] |
| aroD mutant/ugt-27 RNAi | HT115(DE3)aroD717::IS1/L4440(ugt-27) | This study; [38] |
| RNAi control + vector | HT115(DE3)/pMMB67EH | This study |
| aroD mutant + vector | HT115(DE3)aroD717::IS1/pMMB67EH | This study |
| mutant + aroD | HT115(DE3)aroD717::IS1/pMMB67EH[aroD] | This study |
| ubiG mutant | GD1 ubiG | [15] |
| ubiG mutant + rescue | GD1 ubiG/pAHG(ubiG+) | [15] |
OP50 R26 was made in this study by mating with C600 R26 and selecting with SMX on minimal media containing uracil. As the Keio aroD mutant was reported to be a mixture of mutant and other strains [36], a colony was isolated and confirmed to be mutant by PCR.
Culture conditions
NGM was prepared as described [37] using 2.5 g/l soy peptone (Product number P6713, Sigma-Aldrich Corp., Saint Louis, MO, USA) and 20 g/l high-purity agar (Sigma, Product number 05038). High purity agar is used because standard agar can give batch-to-batch variation in the aroD effect on lifespan, probably due to contaminating aromatic compounds. Plates were supplemented with the indicated compounds. For the kanamycin treatment of bacteria, 80 μl of 10 mM kanamycin was added after 24 hours of bacterial growth as described [11]. All compounds were from Sigma-Aldrich.
Lifespan analysis
Unless indicated differently in Additional file 1, survival analyses were performed by the following method: eggs were prepared by bleaching adults to remove all microbes, and then placed onto plates containing either aroD mutant bacteria or, where relevant, SMX-treated OP50 plates. Animals were raised at 15°C until adulthood due to the temperature sensitivity of mutant phenotypes. Gravid adults were used to lay eggs onto fresh aroD mutant or SMX-treated OP50 plates. At L3/L4 these animals were transferred to 25°C and larvae of equivalent stage were put onto at least 5 plates of 25 worms for each condition. Animals were transferred to fresh plates after 7 and 14 days and scored for survival every 2 or 3 days. Lifespan data were analysed by JMP statistical software (SAS Institute Inc., Cary, NC, USA). Where relevant, statistical significance was determined using the Log-Rank and Wilcoxon tests of fitting to the Kaplan-Meier survival model.
Characterisation of the life-extending effect of the mutant E. coli strain
All lifespan experiments conducted in this study are summarized in Additional file 1. The E. coli mutant was discovered because it extended the lifespan of rrf-3(pk1426); daf-2(m577) mutants. We then tested wild type C. elegans (N2), rrf-3(pk1426) mutants, daf-16(mgDf50) mutants and temperature-sensitive sterile mutants glp-4(bn2), shifted from 15°C to 25°C at L4. The mutant bacteria extended lifespan of all C. elegans strains. The effect at 25°C was stronger than at 20°C. To test whether the mutant bacteria exerted its effect during adulthood, we shifted animals on mutant bacteria onto wild type bacteria, and vice versa, just before the beginning of adulthood as L4 larvae. Shifting from mutant bacteria to wild type bacteria caused C. elegans to have a wild type lifespan. Shifting in the other direction from wild type to mutant increased lifespan but took several days to have an effect, probably because residual wild type bacteria chemically complement the mutant bacteria with secreted PABA. These experiments imply that the effect of the bacteria on lifespan is exerted during adulthood. In all subsequent experiments, animals were raised on the mutant bacteria and then transferred to the experimental conditions at the L4 or young adult stage. Although the mutant occurred in a strain from the Ahringer RNAi library [38], the mutation occurred during culture in our laboratory. We have tested the original ugt-27 strain in the Ahringer library and it does not have the mutation.
Complementation screen to identify aroD
To identify the mutated gene we took advantage of the inability of the mutant bacterial strain to grow on minimal media plates. We partially digested the genomic DNA of the control strain using the four-base cutter BfuCI that leaves BamHI compatible ends and ligated the fragments in a BamHI-digested pMMB67EH, a low copy IncQ plasmid [39]. We transformed the ligation mixture into the mutant strain and screened for large colonies on minimal media plates. Untransformed dead bacteria provided enough nutrients to support the growth of small colonies of transformed mutant bacteria, allowing estimation of the numbers screened. Colonies that were clearly larger than their neighbors were picked, grown up and the plasmid isolated by mini-prep. These plasmids were retested by transformation into the mutant bacteria to test for enhanced growth on minimal media. From over 6,000 colonies, 6 independent plasmids passed this second round of screening. Sequencing of these plasmids revealed that one contained the gene folC, one contained folD, three contained aroK and one contained aroD/ydiB. This latter plasmid had the largest effect on mutant growth. The other plasmids appeared to have a positive effect on growth of both mutant and control strains. PCR and sequencing of the aroD region identified an IS1 transposon inserted in the aroD gene in the mutant strain but not in the control (Figure 1C). Composition of minimal media (based on ref [40]) is as follows: 15 g/l agar, 2 g/l D-glucose, 2.17 g/l Na2HPO4, 1.35 g/l KH2PO4, 0.5 g/l (NH4)2SO4, 0.01 g/l Ca(NO3)2, 0.005 g/l thiamine, 0.088 g/l adenine, 0.044 g/l arginine, 0.11 g/l asparagine, 0.088 g/l cysteine, 0.11 g/l glutamatic acid, 0.088 g/l histidine, 0.066 g/l isoleucine, 0.133 g/l leucine, 0.066 g/l lysine, 0.088 g/l methionine, 0.066 g/l phenylalanine, 0.422 g/l serine, 0.221 g/l threonine, 0.088 g/l tryptophan, 0.044 g/l tyrosine, 0.088 g/l uracil, 0.166 g/l valine. The composition of 1% trace element solution is 5 g/l EDTA, 0.5 g/l FeCl3, 0.05 g/l ZnO, 0.01 g/l CuCl2, 0.01 g/l CoCl2.6H2O, 0.01 g/l H3BO3.
Folate analysis
E. coli extraction
Bacterial lawns that had been incubated at 25°C were scraped from NGM agar plates with M9 solution. The final volume of the solution multiplied by the OD600 of the solution diluted 1:5 gave a measure of the amount of material. The samples were concentrated by centrifugation, washed into microcentrifuge tubes and centrifuged again. The pellets were snap frozen in liquid nitrogen and resuspended in a volume of cold 80% methanol: 20% folate extraction buffer (FEB - 50 mM HEPES, 50 mM CHES, 0.5% w/v ascorbic acid, 0.2 M DTT, pH 7.85 with NaOH) in proportion to bacterial content (0.0375 × OD600 × original solution volume). Samples were spiked with 2:1,000 1 mg/ml methotrexateGlu6 as an internal standard and were sonicated on ice using a probe sonicator, centrifuged for five minutes in a cooled microcentrifuge at full speed and the supernatants were kept for analysis.
C. elegans extraction
Synchronized worms at the first day of adulthood incubated at 25°C were washed from 9 cm plates with M9 and allowed to settle. The supernatant was removed and the worms were washed with M9 and allowed to settle again to remove any remaining bacteria. Worms were then transferred to microcentrifuge tubes, gently centrifuged, the volume of pellets estimated. Worms were then washed twice into FEB and left in a total of twice the pellet volume. Proteinase K was added to a final concentration of 0.5 mg/ml and animals were then shaken vigorous at 37°C for 90 minutes. An equal volume of ice cold methanol spiked with 1:1,000 1 mg/ml methotrexateGlu6 was added, vortexed and centrifuged at 4°C as above.
HPLC-MS analysis
We used methodology based on previous literature [21,22]. For HPLC, a C18 reversed phase column (Waters Acquity BEH, 100 mm × 2.1 mm (Waters Corporation, Milford, MA, USA)) was used with dimethylhexylamine as an ion-pairing reagent as in reference [21]. The mobile phase consisted of (A) methanol/water (5:95, v/v) with 5 mM dimethylhexylamine, pH 8, and (B) methanol with 5 mM dimethylhexylamine, at a flow rate of 0.2 mL/min. A linear gradient from 22% B to 80% B over nine minutes was followed by a one minute isocratic hold at 80% B. The column was then re-equilibrated for one minute at 22% B. The injection volume was 10 μl. The mass analysis was by negative mode electrospray ionisation time-of-flight (negative ESI TOF) utilizing a Q-TOF Premier instrument (Waters Corporation) calibrated with sodium formate and with dynamic correction from a leucine encephalin lockspray. Sampling cone voltage was -35 V and capillary voltage was -2,500 V. The following standards (from Schircks (Schircks Laboratories, Jona, Switzerland)) were used: 5-formylTHF (folinic acid), folic acid, 5-formylTHFGlu3, methotrexate, methotrexateGlu6. The elution times and masses were consistent between runs and corresponded to the published literature [21,22]. We fragmented the signal corresponding to 5-methylTHFGlu5 in the C. elegans sample using MS-MS and it showed the expected products. Conditions that resulted in decreased levels of signal for 5-methylTHFGlu5 were accompanied by similar changes in the levels of signal for the mass corresponding to 5-methylTHFGlu4. The peaks obtained by selecting the specific mass were integrated using MassLynx software (Waters Corporation) and used as an indication of quantity. The peaks generated by known metabolites were also integrated and the peak corresponding to coenzyme A was chosen for use in normalization because it was large and there was little variation between samples.
Metabolomic PLS-DA
Raw LC-MS data were preprocessed with XCMS [41], a Bioconductor package for R [42]. Sample-wise normalization was done experimentally, by ensuring that all samples had a similar concentration of biological material, and feature intensity was normalized with a log transform. Both normalization and further statistical analysis, including the PLS-DA plot, were performed with MetaboAnalyst [43].
Bacterial growth rate measurements
Liquid NGM media was aliquoted into sterile conical flasks. SMX was added to the appropriate samples and sterile water added to the control samples. OP50 was then seeded from overnight culture into each flask, and initial OD600 measured for each sample using liquid media as a blank. The samples were then placed on a shaker set to 200 rpm at 37°C, and OD600 measured every 20 minutes for 300 minutes or until growth had reached a plateau.
To measure bacterial growth on solid media, bacteria were seeded, left at room temperature for 48 hours and then transferred to 25°C for a further 24 hours. A total of 1.5 ml of M9 buffer was then added to each plate and, using a glass spreader, the bacterial lawn was scraped from the plate. The M9-containing bacteria were then pipetted into a microfuge tube. These samples were diluted 1:5 in M9 buffer for final OD600 measurements. This measurement was multiplied by the final volume of M9 to provide a relative measure of lawn density.
Development and fecundity measurements
N2 worms were cultured individually from eggs at 25°C on the appropriate media. From the beginning of adulthood, animals were transferred to fresh plates every 24 hours until egg-laying stopped. The progeny from each plate was allowed to develop for two days and then counted.
Abbreviations
CHES: N-Cyclohexyl-2-aminoethanesulfonic acid; DHF: dihydrofolate; ESI TOF: electrospray ionisation time-of-flight; FEB: folate extraction buffer; Glu: glutamate; HEPES: 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid; HPLC: high pressure liquid chromatography; HPLC-MS: high pressure liquid chromatography coupled to mass spectrometry; LC-MS: liquid chromatography coupled to mass spectrometry; MS-MS: tandem mass spectrometry; NGM: nematode growth medium; OD600: optical density at absorbance 600 nm; PABA: para-aminobenzoic acid; PEP: phosphoenolpyruvate; PHB: para-hydroxybenzoic acid; PLS-DA: partial least squares discriminant analysis; Q-TOF: quadrupole time-of-flight; RNAi: RNA interference; SHK: shikimic acid; SMX: sulfamethoxazole; THF: tetrahydrofolate.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
The study was conceived by DW with contributions from DG. The experiments were designed by DW, BV and JW. The experiments were performed and analyzed by BV, DW, IF, DPD, JJ, ZB and EH. Further analysis was performed by GC. Method development was carried out by GC, NO, DPD, BV, DW and RE. The manuscript was written by DW with contributions from DG, BV and GC. Correspondence and requests for material should be addressed to DW: david.weinkove@durham.ac.uk.
All authors have read and approved the final manuscript.
Supplementary Material
Summary of all lifespan experiments conducted in this study. File showing individual experiments, number of animals scored as dead, number censored and, where relevant, percent increase in lifespan between mutant and control, and P-values from Log-Rank and Wilcoxon tests of the Kaplan-Meier survival model.
Spontaneous E. coli mutant rather than RNAi plasmid extends C. elegans lifespan. A) A new RNAi strain containing the ugt-27 plasmid has no effect on lifespan. Survival of rrf-3 worms (20°C) on the control HT115(DE3) strain with the L4440 plasmid, (control, n = 61), the original ugt-27 strain (mutant, n = 98) and a new strain consisting of HT115(DE3) transformed with the ugt-27 plasmid (new, n = 50). Difference between mutant and control, 29.3% (P = < 0.0001).
PABA supplementation has no toxic effect. A) PABA supplementation reverses the lifespan extension on the mutant bacteria (aroD + PABA, n = 85, aroD, n = 124), but has no effect on the control bacteria (control + PABA, n = 112, control, n = 84). B) Addition of PABA has no effect on ubiG bacteria (ubiG + PABA, n = 90, ubiG, n = 76) or ubiG- + rescue plasmid (ubiG:pAHG + PABA, n = 116, ubiG:pAHG, n = 118). C) PHB supplementation had no effect of lifespan of C. elegans maintained on either the mutant (aroD, n = 161, aroD + 25 μM PHB, n = 129, aroD + 250 μM, n = 105) or control bacteria (control, n = 173, control + 25 μM PHB, n = 121, control + 250 μM PHB, n = 130). See Additional file 1 for a full listing of all lifespan data in this study.
Relevant traces from the HPLC/MS analysis from E. coli. A) Traces from HT115(DE3) and HT115(DE3)aroD of m/z = -730.244 corresponding to the negative ion of formylTHFGlu3. B) Traces from formylTHFGlu3 show that this species becomes undetectable in OP50 with increasing concentrations of SMX. C) Traces from C. elegans lysates corresponding to the negative ion of 5-methylTHFGlu5, with 0, 0.1 and 1 μg/ml SMX.
Effect of media supplementation with folate on C. elegans lifespan on aroD and ubiG mutants. (A) glp-4(bn2) animals were raised on the aroD mutant until L4 (15°C) and then transferred (25°C) to the aroD mutant (n = 100), control (n = 126), aroD + 1 × folate (n = 101), aroD + 2 × folate (n = 149). aroD vs aroD + 1 × folate, P = < 0.0001; aroD + 1 × folate vs aroD + 2 × folate, P = 0.13 (Log Rank), P = 0.055 (Wilcoxon). (B) glp-4 worms were raised on E. coli OP50 (15°C) and transferred (25°C) at L4 to ubiG (n = 145), ubiG + pAHG (ubiG+ rescue plasmid), GD1 + 1 × folate (n = 145), GD1+ 2 × folate (n = 146). (C) Traces from the HPLC/MS analysis of the wild type (HT115(DE3)) and aroD mutant (HT115(DE3)aroD) bacteria. Peaks shown are m/z = 730.25 and 548.67 corresponding to formylTHFGlu3 and the methotrexateGlu6 spiked standard respectively with increasing folic acid supplementation (1 × folate is equal to 294 μM folic acid).
PABA reverses the lifespan increase caused by 16 μg/ml and 128 μg/ml SMX. Lifespan curves showing Control (n = 102), 16 μg/ml SMX (n = 176), 128 μg/ml SMX (n = 229), 16 μg/ml SMX + 250 μM PABA (n = 160), 128 μg/ml SMX + 250 μM PABA (n = 217).
Lawn density of plates treated with various concentrations of SMX. Relative bacterial content of lawns from the mean values from 10 plates per conditions (see Methods). Error bars are ± standard deviations. Student's t-test values (Control vs 8 μg/ml SMX: P = 0.02, Control vs 16 μg/ml: P = 0.04, Control vs 64 μg/ml: P = 0.12, Control vs 128 μg/ml: P = 0.04).
Contributor Information
Bhupinder Virk, Email: b.k.virk@durham.ac.uk.
Gonçalo Correia, Email: g.d.s.correia@durham.ac.uk.
David P Dixon, Email: d.p.dixon@dunelm.org.uk.
Inna Feyst, Email: inna_feyst@yahoo.de.
Jie Jia, Email: jie.jia@durham.ac.uk.
Nikolin Oberleitner, Email: nikolin.oberleitner@gmx.at.
Zoe Briggs, Email: zoe_briggs@hotmail.co.uk.
Emily Hodge, Email: emi-xx@hotmail.co.uk.
Robert Edwards, Email: robert.edwards@fera.gsi.gov.uk.
John Ward, Email: j.ward@ucl.ac.uk.
David Gems, Email: david.gems@ucl.ac.uk.
David Weinkove, Email: david.weinkove@durham.ac.uk.
Acknowledgements
We thank Andrea Bender for preliminary data. We thank Peter Stokes and Jackie Mosely for the LC-MS service. We thank Filipe Cabreiro, Catherine Au, Eugene Schuster, Ryan Doonan, Philippa Almack and Marjanne Bourgois for sharing unpublished data, and Catherine Clarke for providing ubiG strains and the respiration protocol. We thank Gary Sharples for sharing strains and primers, and Ivana Bjedov, Kate Bowers, Catherine Clarke, Filipe Cabreiro, Nigel Robinson and Elizabeth Veal for useful discussions. This work was supported by the Wellcome Trust (DW, DG), BBSRC (DW, BV, JJ), the Biochemical Society (GC), IAESTE (IF, NO), ERASMUS (GC) and the Durham Biophysical Sciences Institute (GC). Some strains were obtained from the Caenorhabditis Genetics Center, which is supported by the National Institutes of Health National Center for Research Resources.
References
- Ottaviani E, Ventura N, Mandrioli M, Candela M, Franchini A, Franceschi C. Gut microbiota as a candidate for lifespan extension: an ecological/evolutionary perspective targeted on living organisms as metaorganisms. Biogerontology. 2011;12:599–609. doi: 10.1007/s10522-011-9352-5. [DOI] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai F, Coyle WJ. The microbiome and obesity: is obesity linked to our gut flora? Curr Gastroenterol Rep. 2009;11:307–313. doi: 10.1007/s11894-009-0045-z. [DOI] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk NJ, Kozak E, Conley CA. Chemically defined medium and Caenorhabditis elegans. BMC Biotechnol. 2003;3:19. doi: 10.1186/1472-6750-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houthoofd K, Braeckman BP, Lenaerts I, Brys K, De Vreese A, Van Eygen S, Vanfleteren JR. Axenic growth up-regulates mass-specific metabolic rate, stress resistance, and extends life span in Caenorhabditis elegans. Exp Gerontol. 2002;37:1371–1378. doi: 10.1016/S0531-5565(02)00173-0. [DOI] [PubMed] [Google Scholar]
- Lenaerts I, Walker GA, Van Hoorebeke L, Gems D, Vanfleteren JR. Dietary restriction of Caenorhabditis elegans by axenic culture reflects nutritional requirement for constituents provided by metabolically active microbes. J Gerontol A Biol Sci Med Sci. 2008;63:242–252. doi: 10.1093/gerona/63.3.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G, Houthoofd K, Vanfleteren JR, Gems D. Dietary restriction in C. elegans: from rate-of-living effects to nutrient sensing pathways. Mech Ageing Dev. 2005;126:929–937. doi: 10.1016/j.mad.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Riddle DL. Genetic, behavioral and environmental determinants of male longevity in Caenorhabditis elegans. Genetics. 2000;154:1597–1610. doi: 10.1093/genetics/154.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R, Lunceford AL, Bixler T, Dang P, Lee W, Furukawa S, Larsen PL, Clarke CF. Altered bacterial metabolism, not coenzyme Q content, is responsible for the lifespan extension in Caenorhabditis elegans fed an Escherichia coli diet lacking coenzyme Q. Aging Cell. 2008;7:291–304. doi: 10.1111/j.1474-9726.2008.00378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier W, Adilov B, Regenass M, Alcedo J. A neuromedin U receptor acts with the sensory system to modulate food type-dependent effects on C. elegans lifespan. PLoS Biol. 2010;8:e1000376. doi: 10.1371/journal.pbio.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PL, Clarke CF. Extension of life-span in Caenorhabditis elegans by a diet lacking coenzyme Q. Science. 2002;295:120–123. doi: 10.1126/science.1064653. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittard J, Wallace BJ. Distribution and function of genes concerned with aromatic biosynthesis in Escherichia coli. J Bacteriol. 1966;91:1494–1508. doi: 10.1128/jb.91.4.1494-1508.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinghorn JR, Schweizer M, Giles NH, Kushner SR. The cloning and analysis of the aroD gene of E. coli K-12. Gene. 1981;14:73–80. doi: 10.1016/0378-1119(81)90149-9. [DOI] [PubMed] [Google Scholar]
- Marbois B, Xie LX, Choi S, Hirano K, Hyman K, Clarke CF. para-Aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae. J Biol Chem. 2010;285:27827–27838. doi: 10.1074/jbc.M110.151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrel F, Hamelin O, Douki T, Kieffer-Jaquinod S, Muhlenhoff U, Ozeir M, Lill R, Fontecave M. Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem Biol. 2010;17:449–459. doi: 10.1016/j.chembiol.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Garratt LC, Ortori CA, Tucker GA, Sablitzky F, Bennett MJ, Barrett DA. Comprehensive metabolic profiling of mono- and polyglutamated folates and their precursors in plant and animal tissue using liquid chromatography/negative ion electrospray ionisation tandem mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:2390–2398. doi: 10.1002/rcm.2074. [DOI] [PubMed] [Google Scholar]
- Lu W, Kwon YK, Rabinowitz JD. Isotope ratio-based profiling of microbial folates. J Am Soc Mass Spectrom. 2007;18:898–909. doi: 10.1016/j.jasms.2007.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter EL, Jager L, Gardner L, Hall CC, Willis S, Green JM. Escherichia coli abg genes enable uptake and cleavage of the folate catabolite p-aminobenzoyl-glutamate. J Bacteriol. 2007;189:3329–3334. doi: 10.1128/JB.01940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JM, Nichols BP, Matthews RG. In: Escherichia coli and Salmonella. Neidhardt FC, editor. Vol. 1. ASM Press; 1996. Folate Biosynthesis, Reduction and Polyglutamylation; pp. 665–673. [Google Scholar]
- Stanisich VA, Bennett PM, Oritz JM. A molecular analysis of transductional marker rescue involving P-group plasmids in Pseudomonas aeruginosa. Mol Gen Genet. 1976;143:333–337. doi: 10.1007/BF00269412. [DOI] [PubMed] [Google Scholar]
- Joyce AR, Reed JL, White A, Edwards R, Osterman A, Baba T, Mori H, Lesely SA, Palsson BO, Agarwalla S. Experimental and computational assessment of conditionally essential genes in Escherichia coli. J Bacteriol. 2006;188:8259–8271. doi: 10.1128/JB.00740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsell D. Effects of sulfonamides on ageing in rats. Experientia. 1962;XVIII/10:476–478. http://www.springerlink.com/content/y14w4u11850u7jq5/ [Google Scholar]
- Hackmann C. Observations on influenceability of age phenomena in experimental animals by peroral administration of combinations of 2-(p-aminobenzolsulfonamide)-pyrimidin. Munch Med Wochenschr. 1958;100:1814–1817. [PubMed] [Google Scholar]
- Iwai K, Kobashi M, Fujisawa H. Occurrence of Crithidia factors and folic acid in various bacteria. J Bacteriol. 1970;104:197–201. doi: 10.1128/jb.104.1.197-201.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepehr E, Peace RW, Storey KB, Jee P, Lampi BJ, Brooks SP. Folate derived from cecal bacterial fermentation does not increase liver folate stores in 28-d folate-depleted male Sprague-Dawley rats. J Nutr. 2003;133:1347–1354. doi: 10.1093/jn/133.5.1347. [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Piper MD, Thomas JH, Patel DS, Selman C, Withers DJ, Thornton JM, Partridge L, Gems D. Evolutionary conservation of regulated longevity assurance mechanisms. Genome Biol. 2007;8(7):R132. doi: 10.1186/gb-2007-8-7-r132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389(6654):994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12(15):1317–1319. doi: 10.1016/S0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Beanan MJ, Strome S. Characterization of a germ-line proliferation mutation in C. elegans. Development. 1992;116(3):755–766. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Nakahigashi K, Nakamichi T, Yoshino M, Takai Y, Touda Y, Furubayashi A, Kinjyo S, Dose H, Hasegawa M, Datsenko KA, Nakayashiki T, Tomita M, Wanner BL, Mori H. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol Syst Biol. 2009;5:335. doi: 10.1038/msb.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Furste JP, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Poole RK, Williams HD, Downie JA, Gibson F. Mutations affecting the cytochrome d-containing oxidase complex of Escherichia coli K12: identification and mapping of a fourth locus, cydD. J Gen Microbiol. 1989;135:1865–1874. doi: 10.1099/00221287-135-7-1865. [DOI] [PubMed] [Google Scholar]
- Tautenhahn R, Bottcher C, Neumann S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics. 2008;9:504. doi: 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. 2012. http://www.R-project.org/ ISBN 3-900051-07-0.
- Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37(Web Server):W652–660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol. 2006;2:2006.0007. doi: 10.1038/msb4100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Bonavides-Martinez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, Peralta-Gil M, Santos-Zavaleta A, Shearer AG, Karp PD. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37(Database):D464–470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/S0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Villarroel R, Hedges RW, Maenhaut R, Leemans J, Engler G, Van Montagu M, Schell J. Heteroduplex analysis of P-plasmid evolution: the role of insertion and deletion of transposable elements. Mol Gen Genet. 1983;189:390–399. doi: 10.1007/BF00325900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of all lifespan experiments conducted in this study. File showing individual experiments, number of animals scored as dead, number censored and, where relevant, percent increase in lifespan between mutant and control, and P-values from Log-Rank and Wilcoxon tests of the Kaplan-Meier survival model.
Spontaneous E. coli mutant rather than RNAi plasmid extends C. elegans lifespan. A) A new RNAi strain containing the ugt-27 plasmid has no effect on lifespan. Survival of rrf-3 worms (20°C) on the control HT115(DE3) strain with the L4440 plasmid, (control, n = 61), the original ugt-27 strain (mutant, n = 98) and a new strain consisting of HT115(DE3) transformed with the ugt-27 plasmid (new, n = 50). Difference between mutant and control, 29.3% (P = < 0.0001).
PABA supplementation has no toxic effect. A) PABA supplementation reverses the lifespan extension on the mutant bacteria (aroD + PABA, n = 85, aroD, n = 124), but has no effect on the control bacteria (control + PABA, n = 112, control, n = 84). B) Addition of PABA has no effect on ubiG bacteria (ubiG + PABA, n = 90, ubiG, n = 76) or ubiG- + rescue plasmid (ubiG:pAHG + PABA, n = 116, ubiG:pAHG, n = 118). C) PHB supplementation had no effect of lifespan of C. elegans maintained on either the mutant (aroD, n = 161, aroD + 25 μM PHB, n = 129, aroD + 250 μM, n = 105) or control bacteria (control, n = 173, control + 25 μM PHB, n = 121, control + 250 μM PHB, n = 130). See Additional file 1 for a full listing of all lifespan data in this study.
Relevant traces from the HPLC/MS analysis from E. coli. A) Traces from HT115(DE3) and HT115(DE3)aroD of m/z = -730.244 corresponding to the negative ion of formylTHFGlu3. B) Traces from formylTHFGlu3 show that this species becomes undetectable in OP50 with increasing concentrations of SMX. C) Traces from C. elegans lysates corresponding to the negative ion of 5-methylTHFGlu5, with 0, 0.1 and 1 μg/ml SMX.
Effect of media supplementation with folate on C. elegans lifespan on aroD and ubiG mutants. (A) glp-4(bn2) animals were raised on the aroD mutant until L4 (15°C) and then transferred (25°C) to the aroD mutant (n = 100), control (n = 126), aroD + 1 × folate (n = 101), aroD + 2 × folate (n = 149). aroD vs aroD + 1 × folate, P = < 0.0001; aroD + 1 × folate vs aroD + 2 × folate, P = 0.13 (Log Rank), P = 0.055 (Wilcoxon). (B) glp-4 worms were raised on E. coli OP50 (15°C) and transferred (25°C) at L4 to ubiG (n = 145), ubiG + pAHG (ubiG+ rescue plasmid), GD1 + 1 × folate (n = 145), GD1+ 2 × folate (n = 146). (C) Traces from the HPLC/MS analysis of the wild type (HT115(DE3)) and aroD mutant (HT115(DE3)aroD) bacteria. Peaks shown are m/z = 730.25 and 548.67 corresponding to formylTHFGlu3 and the methotrexateGlu6 spiked standard respectively with increasing folic acid supplementation (1 × folate is equal to 294 μM folic acid).
PABA reverses the lifespan increase caused by 16 μg/ml and 128 μg/ml SMX. Lifespan curves showing Control (n = 102), 16 μg/ml SMX (n = 176), 128 μg/ml SMX (n = 229), 16 μg/ml SMX + 250 μM PABA (n = 160), 128 μg/ml SMX + 250 μM PABA (n = 217).
Lawn density of plates treated with various concentrations of SMX. Relative bacterial content of lawns from the mean values from 10 plates per conditions (see Methods). Error bars are ± standard deviations. Student's t-test values (Control vs 8 μg/ml SMX: P = 0.02, Control vs 16 μg/ml: P = 0.04, Control vs 64 μg/ml: P = 0.12, Control vs 128 μg/ml: P = 0.04).