Abstract
Metastasis remains a significant challenge in treating cancer. A better understanding of the molecular mechanisms underlying metastasis is needed to develop more effective treatments. Here we show that human breast tumor biomarker miR-30c regulates invasion by targeting the cytoskeleton network genes encoding Twinfilin 1 (TWF1) and Vimentin (VIM). Both VIM and TWF1 have been shown to regulate epithelial-to-mesenchymal transition (EMT). Similar to TWF1, VIM also regulates F-actin formation, a key component of cellular transition to a more invasive mesenchymal phenotype. To further characterize the role of the TWF1 pathway in breast cancer, we found that IL-11 is an important target of TWF1 that regulates breast cancer cell invasion and STAT3 phosphorylation. The miR-30c-VIM/TWF1 signaling cascade is also associated with clinical outcome in breast cancer patients.
Keywords: miR-30c, breast tumor invasion, TWF1, VIM, IL-11
Introduction
Metastasis causes 90% of cancer-related mortality and requires better targeting strategies[1]. MicroRNAs (miRNAs) have emerged as important epigenetic regulators of various cellular processes during cancer development and progression. However, the functions, signaling pathways, and relevance of most miRNAs in human cancer are not well characterized. The goal of this study was to characterize signaling pathways for miRNA biomarkers that regulate breast cancer metastasis. Because invasion is the first and rate-limiting step of metastasis, we focused on characterizing the role of a miRNA regulator of invasion.
Human breast cancers can be classified in five intrinsic subtypes (luminal A, luminal B, HER2-enriched, basal-like, and claudin-low) and a normal-like group [2,3]. These subtypes are linked to human mammary epithelial hierarchy, epithelial-to-mesenchymal transition (EMT), breast cancer stem cells (BCSCs), tumor proliferation, treatment response, and metastatic aggressiveness [2,4,5]. Claudin-low and basal-like breast tumors are usually negative for estrogen receptor (ER), progesterone receptor (PR), and HER2 (triple-negative), with high expression of EMT markers, high rates of relapse and metastasis, and poor prognosis [2,4,5]. Given these connections, we speculated that miRNA regulators of metastasis might be associated with different breast cancer subtypes and prognosis.
Our previous work identified the miR-30 family, especially miR-30c, as a breast cancer prognostic biomarker [6]. In addition, miR-30c expression is associated with breast cancer subtypes, such as high miR-30c levels in luminal A tumors and low miR-30c levels in basal-like tumors [6]. MiR-30c regulates EMT and chemo-resistance of breast tumor cells by directly targeting the cytoskeleton gene TWF1 and downstream IL-11 [6]. EMT has been demonstrated to regulate both metastasis and drug resistance [7,8]. We hypothesized that microRNAs and genes that regulate EMT might be critical for both drug resistance and metastasis.
While the actin dynamics regulator TWF1 is involved in cell motility and migration [9], the role of IL-11 in invasion is unknown. Our previous work also identified another cytoskeleton gene, VIM, as a downstream target of miR-30c [6]. However, it was not clear whether VIM is a direct target of miR-30c or if its expression is regulated by miR-30c at the mRNA level. We therefore sought to further characterize the miR-30c pathway and examine whether alteration of miR-30c and the cytoskeleton regulation pathway contributes to breast tumor metastasis or invasion.
Materials and Methods
All experiments were carried out under the approval of Institutional Biosafety Committee, Institutional Review Board, and the Administrative Panel on Laboratory Animal Care of University of Chicago. All experiments were repeated multiple times with three or more experimental replicates.
Real-time PCR
As described [6], total RNAs were extracted from cells or tissues using Trizol (Invitrogen) and followed up by isopropanol precipitation or RNAeasy mini kit (Qiagen). After reverse transcription reactions, real-time PCR for miRNAs/genes were performed using individual miRNA/gene Taqman primers (Applied Biosystems) with a Onestep Real time PCR system (Applied Biosystems). RNU44 and U6 primers were used for miRNA internal controls and GAPDH was used as a housekeeping gene control.
Vectors and lentiviral transduction
TWF1 cDNA (ORFeome collection, the University of Chicago) was subcloned into pDEST40 (Invitrogen). VIM and IL-11 expression vectors were purchased from Origene. Luciferase vectors containing the 3’UTR of TWF1 and VIM were obtained from Switchgear Genomics. The miR-30c precursor (AGATACTGTAAACATCCTACACTCTCAGCTGTGGAAAGTAAGAAAGCTGGGAGAAGGCTGTTTACTCTTTCT) was subcloned into a lentiviral gateway vector pFU-attr-PGK-L2G [6] using the gateway entry vector (provided by Dr. Jun Lu at Yale University) and LR clonase II (Invitrogen). The pFU-attr-PGK-L2G vector [6] was developed by cloning the attr site and the PGK promoter into the pFU-L2G vector as used previously [10]. High-titer lentivirus was produced as described [10]. Viable human CD44+ tumorigenic breast cancer cells were isolated from a triple-negative (TN1) patient tumor-derived xenograft model [10] and transduced with lentiviruses at 10 or 20 multiplicities of infection (MOI 10 or 20). The TN1 human-in-mouse breast tumor xenograft model was generated by engrafting the lung metastases (pleural effusion) of a breast tumor patient into NOD/SCID mice as described previously [10].
Cell culture and transfections
Cells were maintained in DMEM (MDA-MB-231, Hek293T with G418) and EMEM (BT-20) with 10% FBS + 1% Penicillin-Streptomycin (P/S). At 100nM (MDA-MB-231) or 50nM (BT-20, Hek293T), miRNAs (Dharmacon, with negative control #4) and siRNAs (Dharmacon, with negative control A) were transfected for 6 hours using Dharmafect (Dharmacon) and repeated on the following day [6]. 3’UTR luciferase vectors and cDNA vectors were co-transfected using Fugene (Roche-Promega), usually twice following each six-hour miRNA transfection.
Cell growth assays
MDA-MB-231 cells (5,000) were plated in 96-well plates in phenol red free media +10% FBS +1%P/S. Calcein AM (BD Biosciences) was added at 4 μg/μl on day 1 and 5 to the wells and incubated for an hour at 37°C. Plates were read at 485/535 nm on a Wallac 1420 plate reader (Perkin Elmer).
Quantitative invasion assays in vitro
MDA-MB-231 cells (80-100k) were plated in serum-free DMEM to the top of 8 um transwell inserts (BD) precoated 1:100 with growth factor-reduced Matrigel (BD) in 24-well companion plates (BD) containing DMEM + 10% FBS (no gradient for controls). After 24 hours, cells were stained for one hour at 37°C in HBSS+ 4 μg/μl calcein AM (BD Biosciences). Cells on the top of inserts were removed using cotton swabs and invaded cells were dissociated in a dissociation buffer (Trevigen) from the bottom side of inserts (shaken for an hour at 37°C) and read at 485/535 nm.
Western blot
MDA-MB-231 cells were lysed and sonicated in RIPA buffer and 100 μg of total protein was loaded onto 4-20% gradient gels for immunoblots with antibodies to TWF1 (Genetex, GTX11439), Vimentin (Abcam, ab61780), STAT3 (Cell Signaling), and β-Actin (Sigma-Aldrich, AC-15). After staining with a fluorescent IRDye-conjugated secondary antibody, blot membranes were directly scanned and analyzed using the Odyssey infrared imaging system (LI-COR).
Luciferase assay
For TWF1-3’UTR, Hek293T cells in 48-well plates were incubated with miR-30c for six hours and then transfected overnight with 3’UTR firefly luciferase and control renilla luciferase vectors (Promega). The firefly luciferase signal was measured 48 hours later and normalized to the renilla luciferase signal using a dual luciferase kit (Promega). For VIM 3’UTR vectors, cells in 96-well plates were co-transfected with 3’UTR vector and miRNA mimics using Dharmafect. Cells were lysed 24 hours later and read using a luciferase assay kit (Switchgear Genomics).
Immunofluorescence and immunohistochemistry
For F-actin staining, cells were plated on microscope slide wells (Millipore), fixed in 4% paraformaldehyde/PBS, and then stained with one unit of fluorescent phalloidin (about 5 μl stock solution at 6.6 μM) (Invitrogen F432) for 20 mins at room temperature and a drop of SlowFade Gold antifade reagent gel with DAPI (Invitrogen) [6]. Tumor and lung samples were fixed in 10% neutral buffered formalin, embedded, and sectioned into 5um-thin tissue sections. After deparaffinization and rehydration, the tissue sections were incubated in antigen retrieval buffer (DAKO, S1699) and heated in steamer at over 97°C for 20 minutes. Immunohistochemical staining was performed using antibodies to GFP (1:100) (Cell Signaling, #2956), hVIM (1:100) (DAKO, M0725), and hTWF1 (1:400) (GENEX, GT111439) at the University of Chicago Immunohistochemistry core facility.
Tumor transplantation in mice and bioluminescence imaging
Human breast cancer cells were dissociated from the TN1 triple-negative patient tumor-derived mouse xenografts labeled with the dual-reporter Luc2-tdTomato as described earlier [10], with 1800 units collagenase III (Worthington) and 100 Kunitz units DNase I (Sigma). DAPI−lineage(H2Kd)−tdTomato+CD44high tumorigenic cells were sorted on FacsAriaII (BD), after staining with anti-CD44-APC, mouse stromal marker H2Kd-biotin and strepavidin-PE-cy7 (BD), and DAPI in HBSS/2% FBS [10]. Sorted tumor cells were transduced with L2G vector control or miR-30c lentiviruses (as described above) for 4 hours and sorted again 16 hours later based on eGFP. Sorted GFP+ cells were then mixed 1:1 with Matrigel (BD) (50 uL tumor cells:50 uL Matrigel, approximately 5,000 cells per site) for transplantation into NOD/SCID mouse mammary fat pads (left 4th or right 4th) [10], with a cohort of mice for 20 tumor injections per sample. Bioluminescence images were acquired for primary tumors starting on day 1 post transplantation until tumors reached 2 cm or dissected lungs and tumors as described [10]. Data was analyzed using LivingImage 4.0 Software (Caliper Life Sciences) and expressed as total photon flux.
Statistical Analysis
For all assays and analyses in vitro, if not specified, student’s t-test was used to evaluate the significant difference or p-values and standard deviations (SD) of mean values were depicted as error bars in figures. For animal studies in vivo, tumor growth curves were analyzed using a linear mixed model in R software [11] with the tumor volume (bioluminescence signals) as a response variable. Lung metastases versus tumor burden signals were analyzed using Wilcoxon rank sum test in R software.
Results
miR-30c regulates invasion of breast cancer cells
Based on miRNA profiling studies of clinical breast tumors [6,12], we previously found that miR-30c was a prognostic marker that associated significantly with breast cancer subtypes, including higher expression levels in luminal A tumors and lower levels in claudin-low and basal-like breast tumors [6]. This observation was consistent with a previous report that miR-30c positively correlates with expression status of hormone receptors ER and PR in primary breast tumors [13,14]. We hypothesized that low expression of miR-30c contributes to metastasis of basal-like and/or claudin-low breast tumors.
To test our hypothesis that miR-30c plays a role in breast tumor progression, we investigated the relevance of miR-30c to the metastatic potential of triple-negative breast tumor xenograft tumor models [10]. Interestingly, the expression level of miR-30c was higher in the M1-derived, and less metastatic, xenograft tumors compared to the more-metastatic M1-parental tumor models (Fig. 1a, p=0.0008). We also examined the expression levels of miR-30c and other miR-30 family members (miR-30a, -30b, -30d, -30a*, -30e, and -30e*) in triple-negative breast cancer cell lines BT-20 and MDA-MB-231. Compared to MDA-MB-231 cells, BT-20 cells were more invasive and expressed lower levels of miR-30c and other members, including miR-30b, miR-30d, and miR-30e (Supplementary Fig. S1a-b).
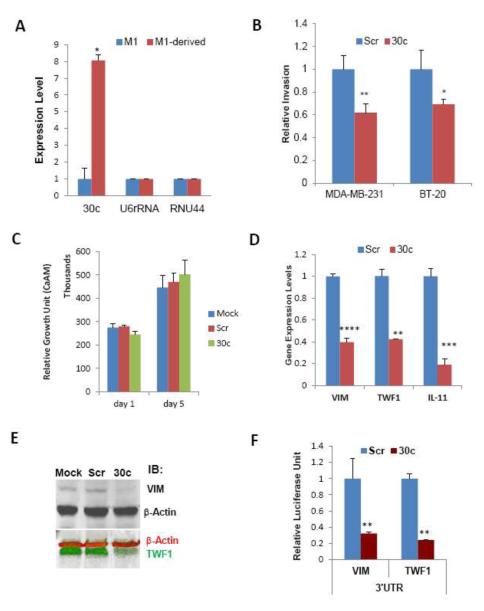
Fig. 1. miR-30c inhibits invasion of breast cancer cells.
aqRT-PCR anlayses of miR-30c expression in triple-negative patient tumor-derived metastatic xenograft model (M1) and M1-derived less metastatic tumor model passaged in mice (M1-less met). U6 and RNU44 were used as internal controls.
bInvasion of MDA-MB-231 and BT-20 cells transiently transfected with oligos of miR-30c or scrambled control (Scr), assessed by transwell invasion assays (n=6, *p<0.05, student’s t-test).
cCell growth analyses on Day 1 and 5 of MDA-MB-231 cells, after transfections of mock, scramble (Scr) and miR-30c (100nM).
dDown-regulated expression levels of VIM, TWF1 and IL-11 36hrs after transfection of miR-30c in MDA-MB-231 cells, measured by real-time PCR analyses. Scr, the scrambled control. ****p<0.00001, **p<0.001, ***p<0.0001 (n=3).
eImmunoblots of VIM (top black band) and TWF1 (bottom green band) along with β-actin controls with protein lysates of MDA-MB-231 cells 36 hrs after transfection of mock control, scrambled (Scr), and miR-30c respectively (full blots see supplementary Fig S2)
fLuciferase reporter activity assays of Hek293T cells co-transfected with scrambled (Scr) or miR-30c and the luciferase vector containing the 3’UTR of VIM or TWF1. **p<0.01 compared to Scr.
We further characterized the role of miR-30c in regulating invasion by modulating its levels in triple-negative breast cancer cells. Upon transient transfection, elevated levels of miR-30c inhibited invasion of both MDA-MB-231 and BT-20 cells in a transwell system in vitro (Fig. 1b). The inhibitory effect of miR-30c on breast cancer invasion was not due to an altered proliferation rate, since MDA-MB-231 tumor cell growth was not significantly affected by miR-30c (Fig. 1c).
TWF1 and VIM are required for miR-30c to regulate breast cancer cell invasion
In a separate study [6], we used microarray analyses (GSE32617) combined with target gene prediction algorithms GeneSet2miRNA [15] to identify TWF1 mRNA, which encodes an actin-binding protein, as a direct downstream target of miR-30c [6]. From these analyses, we also proposed that VIM is a potential direct target of miR-30c [6] but did not further validate it. VIM is a well-known cytoskeleton gene that regulates EMT, invasion, and metastasis. Twinfilin 1 (TWF1), also called protein tyrosine kinase 9 (PTK9), belongs to the actin depolymerizing factor homology (ADF-H) family, along with cofilin and other proteins [16-18]. As cytoskeleton genes often regulate cell motility and EMT, we hypothesized that VIM and TWF1 are both important direct targets of miR-30c in regulating invasion.
We first examined the effect of miR-30c on VIM expression at the mRNA and protein level. Similar to TWF1 and its downstream target IL-11 [6], expression of VIM was down-regulated at both mRNA and protein levels by miR-30c in breast cancer cells, as validated by real-time PCR and western blot (Fig. 1d-e, full immunoblot image in Supplementary Fig. S2a). We then investigated whether VIM is a direct target of miR-30c. The functional interaction between miR-30 and the 3’UTR of VIM was verified in luciferase assays in HEK293T cells, using TWF1 as a positive control (Fig. 1f; see also Supplementary Fig. S2b). Compared to the scrambled control, co-transfected miR-30c suppressed the expression of the luciferase reporter which was located upstream of the wild type 3’UTR of VIM or TWF1 (Fig. 1f).
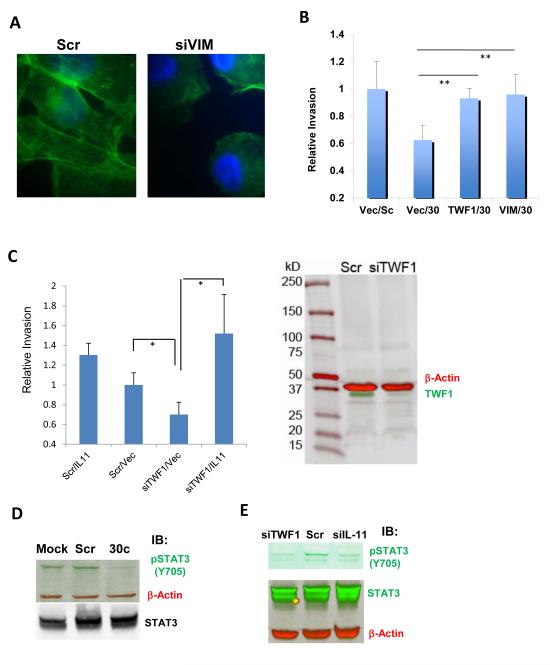
In a subsequent EMT analysis, similar to that of TWF1 knockdown [6], knockdown of VIM expression by siRNAs caused a mesenchymal-to-epithelial-transition (MET) phenotype, with decreased F-actin formation in MDA-MB-231 breast cancer cells (Fig. 2a).
Fig. 2. Cytoskeleton genes important for miR-30c to regulate invasion.
aIimages of F-actin staining of MDA-MB-231 cells inhibited by gene knockdowns mediated by siRNAs of VIM (1000x, green for F-actin staining and blue for DAPI staining). Scr, scrambled control. Cells were harvested 36 hrs after transfections.
bOverexpression of TWF1 or VIM restored the invasion inhibited by miR-30c in MDA-MB-231 cells. **p<0.01 (n=4).
cLeft panel: knockdown of TWF1 by siRNAs (siTWF1) suppressed the invasion of MDA-MB-231 cells and the inhibited invasion was reversed by overexpression of human IL-11 cDNA. Cell invasion were measured 24 hours after two transient transfections for four combinations: scrambled/vector control, scrambled/IL-11, siTWF1/vector, and siTWF1/IL-11. *p<0.05 (n=4). Right panel: Full image of the immunoblots of TWF1 (bottom green band) and beta-actin (top red band) with protein lysates of MDA-MB-231 cells 36hrs after transfections of scrambled (Scr) and siTWF1 respectively.
dImmunoblots of phosphor-STAT3 (Y705) (green band), total STAT3 (black band) and β-actin control (red band) with protein lysates of MDA-MB-231 cells 36 hrs after transfections of mock control, scrambled (Scr), and miR-30c respectively (full blots see supplementary Fis. S3).
eImmunoblots for pSTAT3 (Y705) (upper green band), total STAT3 (lower green band) and the loading control β-actin (red band) with lysates of MDA-MB-231 cells transfected with siTWF1, scrambled control (Scr) and siIL-11 (full blots see supplementary Fis. S3)
More importantly, to determine the importance of the cytoskeleton target genes TWF1 and VIM in miR-30c regulated invasion, we performed functional rescue assays by co-transfecting 3’UTR-deficient cDNA and miR-30c in MDA-MB-231 cells. Overexpressed TWF1 or VIM restored breast cancer cell invasion inhibited by miR-30c (Fig. 2b), suggesting that these two EMT-related cytoskeleton genes are required for miR-30c in regulating invasion.
IL-11 is a relevant downstream target of TWF1 in breast cancer invasion
We previously identified interleukin 11 (IL-11), as a downstream target of TWF1 in regulating drug resistance of breast cancer [6]. We then investigated whether IL-11 is also important for TWF1 signaling in regulating invasion. To determine the role of IL-11 in the pathway, we also performed functional rescue studies by modulating IL-11 expression level. While knockdown of TWF1 mimicked miR-30c phenotype in inhibiting breast cancer cell invasion, overexpression of IL-11 restored the invasiveness of MDA-MB-231 cells inhibited by siTWF1 (Fig. 2c). These data suggest that IL-11 is an important target of TWF1 in the miR-30c signaling pathway that regulates breast cancer cell invasion.
IL-11 and other IL-6 family members are known to mediate activation or phosphorylation of STAT3 in tumor regulation [19,20]. To determine the influence of miR-30c and TWF1 on the IL-11 pathway, we examined phosphorylation of STAT3 at tyrosine 705 (Y705). As expected, both miR-30c and siRNA-mediated knockdown of TWF1 decreased the Y705 phosphorylation of STAT3, similar to the effects of siRNA-mediated knockdown of IL-11 (Fig. 2d-e, full immunoblot images in Supplementary Fig. S3a-d).
miR-30c regulates invasion in vivo
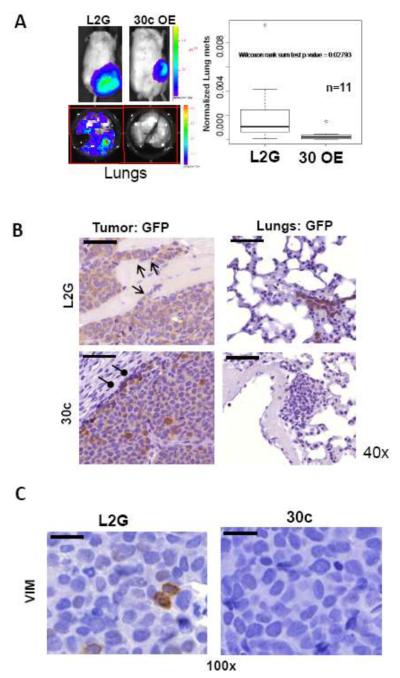
We also utilized bioluminescence imaging to determine the effect of miR-30c on spontaneous lung metastasis in the TN1 L2G-transduced breast tumor model as described in the methods section [10]. MiR-30c-transduced CD44+ breast tumor initiating cells (BTICs) mediated tumor growth with increased miR-30c expression in both CD44+ and CD44− tumor cells (Supplementary Fig. S4a). MiR-30c significantly suppressed lung metastases when normalized to tumor burden signals compared to L2G vector control (the empty vector control without a scrambled miRNA) (Fig. 3a). Immunohistochemical staining for GFP verified that most of the implanted primary tumor cells of the L2G and miR-30c groups were transduced (Fig. 3b). However, L2G tumors displayed more invasive fronts facing the surrounding tissues or stroma compared to the more defined boundaries of miR-30c-expressing tumors (Fig. 3b). As shown by GFP immunostaining of the lung sections, L2G tumors developed GFP+ lung metastases, whereas miR-30c-expressing tumors did not generate GFP+ metastases, although a few GFP-negative metastatic lesions were found in the lungs without co-expression of miR-30c (Fig. 3b).
Fig. 3. miR-30c regulates metastasis of breast tumor in vivo.
aLung metastasis analyses. Left panels: representative bioluminescence images of labeled breast tumors (top) and lung metastases (bottom). Right panel: a box plot of lung metastasis index between vector control group and miR-30c-overexpressing group (n=11, p<0.05, Wilcoxon rank sum test). Lung metastases were assessed by bioluminescence after lungs dissected from mice at the end point and were presented by normalized metastasis index, the total flux from the lungs divided by the total flux from the mammary tumors.
bImages of immunohistochemistry staining against GFP for both L2G-vector-transduced and 30c-overexpressing primary breast tumor sections as well as respective mouse lung sections (40x). The invasive fronts of L2G tumors were shown with open arrows and the defined or contained boundary of 30c-expressing tumors were pointed by oval-arrows. GFP+ metastatic lesions were absent in the lungs of mice bearing 30c-overexpressing tumors. Scale bars=50 μm in all images.
cReduced immunohistochemistry staining against human VIM in 30c-overexpressing primary breast tumor sections, compared to L2G-vector-transduced tumor sections (100x). Scale bars=20 μm in the images
The reduction of VIM expression at the protein level was confirmed in miR-30c-expressing tumors by immunohistochemical staining (Fig. 3c), similar to the reduction of TWF1 by miR-30c observed both in lung metastases (Supplementary Fig. S4b) and in primary tumors [6]. These studies convincingly demonstrate that miR-30c regulates the downstream targets TWF1 and VIM and their signaling pathways in vivo.
The importance of cytoskeleton genes in a clinically relevant signaling pathway
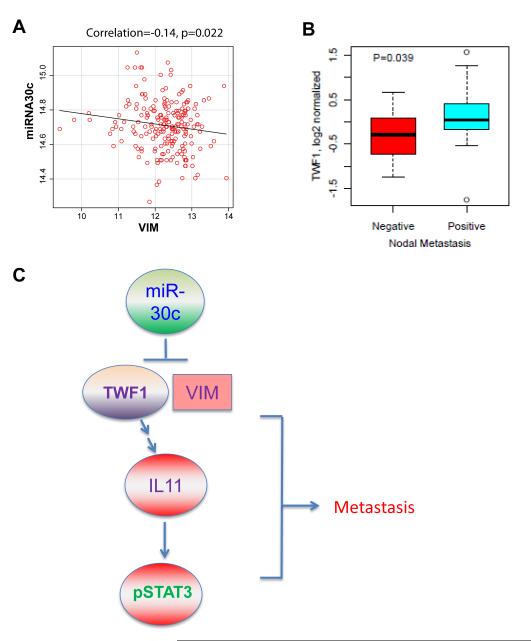
To validate the importance of the cytoskeleton signaling transduction pathway in clinical tumors, we previously demonstrated an inverse association of miR-30c with its target gene IL-11 [6]. Based on the combined breast tumor dataset with matched mRNA and miRNA arrays deposited as GSE22220 and GSE22216 respectively [12], we also observed a negative correlation between miR-30c and VIM (Fig. 4a) (n=210, p=0.022) in the current study.
Fig. 4. Clinical relevance of the miR-30c signaling pathway.
aScatter plots between VIM single gene expression and the expression of miR-30c in GSE22220 and GSE22216 (n=210). Pearson correlation was estimated in each comparison and plots were drawn using the scatterplot function found in the Car package (http://cran.r-project.org).
bBox plots of TWF1 expression compared between nodal metastasis positive (n=30) versus nodal-metastasis-negative (n=13) clinical breast tumors (UC set). A student’s t-test was used to calculate p values.
cSignaling pathway scheme of miR30-TWF1(VIM)-IL-11-pSTAT3 regulation of breast cancer metastasis
Regarding the clinical relevance of miR-30c targets to tumor metastasis in the UC breast tumor set we profiled previously [6], we noted that TWF1 expression is associated with more aggressive nodal metastasis status (n=43, p=0.039) (Fig. 4b). Another report also showed that TWF1 regulates both drug sensitivity and cell migration in lymphoma progression [9]. In addition, relapse-free analyses with or without adjustment for nodal status, ER, grade, treatment, or tumor stage, demonstrate that VIM is strongly associated with clinical breast cancer outcomes in the UC breast tumor set, for which VIM is a poor prognostic marker (Supplementary Table S1).
The above data suggest a signaling network that links miR-30c and downstream targets VIM, TWF1, and IL-11 (Fig. 4c), as well as the clinical importance of miR-30c in regulation of these targets in human breast cancer.
Discussion
The function of a miRNA depends on the network of genes that it regulates. The current studies demonstrate that, in addition to its role in regulating chemo-resistance of breast tumor cells [6], miR-30c also directs a signaling cascade that regulates tumor cell invasion. Our studies therefore suggest that two important arms driving tumor progression, metastasis and therapy resistance, are mechanistically connected.
MiR-30c is a member of the miR-30 family of miRNAs. These family members share a common seeding sequence located near the 5’ end, but differ in the compensatory sequences located near the 3’ end. This difference in compensatory sequences among the family members allows for miRNAs from the same family to target different genes and pathways [21]. For the miR-30 family, miR-30a, miR-30b, and miR-30d play a demonstrated role in cancer cell invasion. In contrast to the role of miR-30c as a tumor-suppressor in our study, miR-30d and miR-30b have been demonstrated to behave as oncomirs in melanoma and ovarian cancer [22,23]. This difference in behavior is not unusual and has been shown to also occur for the well-studied miR-200 family [24]. Recently, miR-30a was shown to target VIM in breast cancer invasion [25].
Our work has demonstrated that while VIM is also a target of miR-30c in regulating invasion, it is not the only target. Notably, TWF1 plays a more prominent role as a downstream target of the miR-30c signaling pathway in invasion. We have demonstrated that the miR-30c/TWF1 pathway, instead of miR-30a, regulates both invasion and chemo-resistance [6]. These differing phenotypes between miR-30c and miR-30a are likely due to differential preferences of target genes and pathways.
Combined with the role of the miR-30c pathway in regulating chemo-resistance [6], the miRNA-cytoskeleton regulation network of miR-30c and its target genes VIM, TWF1, and IL-11 represents a molecular mechanism that links invasion, EMT, and chemo-resistance. Interestingly, the inflammatory cytokine IL-11 plays an autonomous role in tumor cell invasion independent of stromal cells and IL-11 is an indispensable downstream target of the cytoskeleton protein TWF1 in its invasion-regulating function in addition to its chemo-sensitivity regulation [6].
Importantly, the identification of this novel invasion-regulating pathway (miR-30c-VIM/TWF1-IL-11-pSTAT3), with relevance to chemo-resistance in breast cancer, will expedite the development of targeting strategies to prevent and treat breast tumor progression.
Supplementary Material
ACKNOWLEDGMENTS
We are very thankful to Dr. Jun Lu at Yale University for providing miR-30c precursor gene vector, Dr. Seo Young Park for data analyses, and Dr. John Kokontis for helping review and edit our manuscript. We appreciate the experimental support of several core facilities, including the animal facility, optical imaging core facility, flow cytometry facility, integrated microscopy core facility, DNA sequencing facility, functional genomics facility, and IHC core facility at the University of Chicago. We specifically acknowledge Ryan Duggan, James Cao, David Leclerc, Marianne Greene, Terri Li, Xin Jiang, Shirley Bond, Jaejung Kim, Hui Zheng, Andrew Gusev, Dalong Qian, and Yohei Shimono for technical help and support.
Funding This study was supported in part by The University of Chicago Women’s Board (J.B.), National Institutes of Health (NIH) T90 Regenerative Medicine Training Program DK070103-05, Department of Defense Breast Cancer Research Program W81XWH-09-1-0331, Paul Calabresi K12 Award 1K12CA139160-02, NCI K99 CA160638-01A1, Chicago Fellows Program at the University of Chicago, and the University of Chicago Clinical and Translational Science Award (UL1 RR024999) (H.L.), funds from the Sociedad Española de Oncología Médica (SEOM) (A.P.), the Breast SPORE at University of North Carolina 5-P50-CA58223-17 (A.P. and C.M.P.), The University of Chicago Cancer Research Center Pilot Fund, BSD Imaging Research Institute Pilot Research Projects Using Animal Imaging, UCMC/Northshore Collaborative Funds, a Carole and Gordon Segal Grant (G.L.G.), and the Virginia and D. K. Ludwig Fund (G.L.G. and H.L.), NIH Grants U54 CA 126524 and P01 CA139490 (M.F.C.), the Breast Cancer Research Foundation (M.F.C., O.I.O. and C.M.P), and the University of Chicago Cancer Center Support Grant CA 014599.
Footnotes
AUTHOR CONTRIBUTIONS J.B., Y-F.C., C.N., A.P., K.Y., R.D., S.H., K.E.S., and H.L. designed and performed experiments, and analyzed data. S.Y.P. and D. H. performed biostatistical analyses for animal work and association studies. C.M.P., O.I.O., M.F.C., and G.L.G designed the project. J.B. and H.L. wrote the manuscript. All authors contributed to the editing of the manuscript.
Conflicts of interest: Authors of this manuscript have no conflict of interest.
References
- 1.Sporn MB. The war on cancer. Lancet. 1996;347(9012):1377–1381. doi: 10.1016/s0140-6736(96)91015-6. [DOI] [PubMed] [Google Scholar]
- 2.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5(1):5–23. doi: 10.1016/j.molonc.2010.11.003. doi:S1574-7891(10)00127-4 [pii] 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. doi:bcr2635 [pii] 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prat A, Perou CM. Mammary development meets cancer genomics. Nat Med. 2009;15(8):842–844. doi: 10.1038/nm0809-842. doi:nm0809-842 [pii] 10.1038/nm0809-842. [DOI] [PubMed] [Google Scholar]
- 5.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, Feleppa F, Huschtscha LI, Thorne HJ, Fox SB, Yan M, French JD, Brown MA, Smyth GK, Visvader JE, Lindeman GJ. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–913. doi: 10.1038/nm.2000. doi:nm.2000 [pii] 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 6.Bockhorn J, Dalton R, Nwachukwu C, Huang S, Prat A, Yee K, Chang Y-F, Huo D, Wen Y, Swanson KE, Qiu T, Lu J, Park SY, Dolan ME, Perou CM, Olopade OI, Clarke MF, Greene GL, Liu H. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nature Communications. 2012 doi: 10.1038/ncomms2393. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14(6):818–829. doi: 10.1016/j.devcel.2008.05.009. doi:S1534-5807(08)00209-8 [pii] 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29(34):4741–4751. doi: 10.1038/onc.2010.215. doi:onc2010215 [pii] 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meacham CE, Ho EE, Dubrovsky E, Gertler FB, Hemann MT. In vivo RNAi screening identifies regulators of actin dynamics as key determinants of lymphoma progression. Nat Genet. 2009;41(10):1133–1137. doi: 10.1038/ng.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J, Wen S, Chang YF, Bachmann MH, Shimono Y, Dalerba P, Adorno M, Lobo N, Bueno J, Dirbas FM, Goswami S, Somlo G, Condeelis J, Contag CH, Gambhir SS, Clarke MF. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc Natl Acad Sci U S A. 2010;107(42):18115–18120. doi: 10.1073/pnas.1006732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2011. [Google Scholar]
- 12.Buffa FM, Camps C, Winchester L, Snell CE, Gee HE, Sheldon H, Taylor M, Harris AL, Ragoussis J. microRNA-Associated Progression Pathways and Potential Therapeutic Targets Identified by Integrated mRNA and microRNA Expression Profiling in Breast Cancer. Cancer research. 2011;71(17):5635–5645. doi: 10.1158/0008-5472.CAN-11-0489. [DOI] [PubMed] [Google Scholar]
- 13.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer research. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. doi:65/16/7065 [pii] 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 14.Liu H. MicroRNAs in breast cancer initiation and progression. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-1128-9. doi:10.1007/s00018-012-1128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antonov AV, Dietmann S, Wong P, Lutter D, Mewes HW. GeneSet2miRNA: finding the signature of cooperative miRNA activities in the gene lists. Nucleic Acids Res. 2009;37(Web Server issue):W323–328. doi: 10.1093/nar/gkp313. doi:gkp313 [pii] 10.1093/nar/gkp313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmgren S, Vartiainen M, Lappalainen P. Twinfilin, a molecular mailman for actin monomers. J Cell Sci. 2002;115(Pt 5):881–886. doi: 10.1242/jcs.115.5.881. [DOI] [PubMed] [Google Scholar]
- 17.Ojala PJ, Paavilainen VO, Vartiainen MK, Tuma R, Weeds AG, Lappalainen P. The two ADF-H domains of twinfilin play functionally distinct roles in interactions with actin monomers. Mol Biol Cell. 2002;13(11):3811–3821. doi: 10.1091/mbc.E02-03-0157. doi:10.1091/mbc.E02-03-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poukkula M, Kremneva E, Serlachius M, Lappalainen P. Actin-depolymerizing factor homology domain: a conserved fold performing diverse roles in cytoskeletal dynamics. Cytoskeleton (Hoboken) 2011;68(9):471–490. doi: 10.1002/cm.20530. doi:10.1002/cm.20530. [DOI] [PubMed] [Google Scholar]
- 19.Ernst M, Najdovska M, Grail D, Lundgren-May T, Buchert M, Tye H, Matthews VB, Armes J, Bhathal PS, Hughes NR, Marcusson EG, Karras JG, Na S, Sedgwick JD, Hertzog PJ, Jenkins BJ. STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. J Clin Invest. 2008;118(5):1727–1738. doi: 10.1172/JCI34944. doi:10.1172/JCI34944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuringa JJ, Wierenga AT, Kruijer W, Vellenga E. Constitutive Stat3, Tyr705, and Ser727 phosphorylation in acute myeloid leukemia cells caused by the autocrine secretion of interleukin-6. Blood. 2000;95(12):3765–3770. [PubMed] [Google Scholar]
- 21.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS biology. 2005;3(3):e85. doi: 10.1371/journal.pbio.0030085. doi:10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li N, Kaur S, Greshock J, Lassus H, Zhong X, Wang Y, Leminen A, Shao Z, Hu X, Liang S, Katsaros D, Huang Q, Butzow R, Weber BL, Coukos G, Zhang L. A combined array-based comparative genomic hybridization and functional library screening approach identifies mir-30d as an oncomir in cancer. Cancer research. 2012;72(1):154–164. doi: 10.1158/0008-5472.CAN-11-2484. doi:10.1158/0008-5472.CAN-11-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaziel-Sovran A, Segura MF, Di Micco R, Collins MK, Hanniford D, Vega-Saenz de Miera E, Rakus JF, Dankert JF, Shang S, Kerbel RS, Bhardwaj N, Shao Y, Darvishian F, Zavadil J, Erlebacher A, Mahal LK, Osman I, Hernando E. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer cell. 2011;20(1):104–118. doi: 10.1016/j.ccr.2011.05.027. doi:10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elson-Schwab I, Lorentzen A, Marshall CJ. MicroRNA-200 family members differentially regulate morphological plasticity and mode of melanoma cell invasion. PloS one. 2010;5(10) doi: 10.1371/journal.pone.0013176. doi:10.1371/journal.pone.0013176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng CW, Wang HW, Chang CW, Chu HW, Chen CY, Yu JC, Chao JI, Liu HF, Ding SL, Shen CY. MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast cancer research and treatment. 2012;134(3):1081–1093. doi: 10.1007/s10549-012-2034-4. doi:10.1007/s10549-012-2034-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.