Abstract
Integrative genomics studies have greatly advanced our understanding of cardiovascular pathophysiology over the last decade. Here, we highlight the strengths and challenges of this cutting-edge approach and provide examples where novel insights have arisen through the integration of multi-level genomic information and cardiac physiology. Going forward, the integration of comprehensive next-generation sequencing data sets with quantitative phenotypes at the molecular, cellular, and whole-heart level using advanced modelling approaches provides an unprecedented opportunity for cardiovascular science.
Keywords: Integrative genomics
1. What is integrative genomics?
Genomics is the study of the sequence, structure, and biology of genome-level data sets (DNA and RNA). In the 1970 s two developments laid the foundations for the field as we recognize it today. Nucleotide sequencing technologies, and particularly the Sanger method,1 allowed for the sequencing of genomes of increasing size and complexity, starting with viral and mitochondrial genomes,1 and later progressing through bacteria (Haemophilus influenzae 19952) and model organisms (e.g. Caenorhabditis elegans, 19983) and finally to humans.4,5 Shortly afterwards Botstein et al.6 demonstrated how genetic markers (initially restriction fragment length polymorphisms) could be used to construct a map of the genome. By correlating phenotypes with these genome-wide markers, diseases could be genetically mapped to a genomic locus,7 and the underlying genes identified on the basis of their position in the genome without prior knowledge of their function (positional cloning or reverse genetics).8 In contrast to genetics, which is primarily focused on the study of inheritance, genomics seeks statistical associations between genomic data and phenotypes with relatively few assumptions. The term genomics became established in the late 1980s,9 captured by the publication of a new journal with that title in 1987, and during the 1980 and 1990s a number of diseases and traits were mapped to the genome, and many disease genes identified.
A limitation of genomic approaches to date is that a genotype–phenotype correlation typically identifies an association between a trait and a locus or list of genes, rather than a trait and a gene. The subsequent identification of THE causative gene remains a substantial challenge. Integrative Genomics describes approaches to this problem that use additional layers of data to inform the search space for candidate genes, for example overlaying data on the regulation of gene expression (coined genetical genomics).10,11 Integrative genomic methodologies are underpinned by the fundamental hypothesis that polymorphisms (non-disease causing genetic variation) and/or mutations in or near genes have an effect on the expression of the causative gene AND are also associated with the trait under study. It is possible for a gene to have a coding mutation affecting protein function but not directly influencing gene expression that could escape detection. However, in this instance homoeostatic effects acting to restore gene function often induce a transcriptional response, or the mutation affects RNA processing, and this can still be detected by studying gene expression.
In this review, we will outline some of the genome-level data sets that may be used in integrative genomics approaches and the methods on which they depend, review approaches to integrate these data sets (with an emphasis on DNA and RNA level data), and illustrate these with examples of their applications to cardiovascular biology.
2. Characterizing phenotypes
As with any statistical approach, integrative genomics demands an understanding of the type of trait under study: for instance, continuous (e.g. blood pressure) or categorical (e.g. hypertensive/normotensive). However, for complex disease traits the relationship between disease definition and physiology may not be straightforward. Some categorical traits (hypertension vs. normotension) represent binary classifications of an underlying continuous trait, in which case it is usually most powerful to apply genomic analyses to the underlying quantitative variable, if accurate. Indeed, categorization of continuous phenotypes may increase the risk of false association, and should generally be avoided. Conversely, a continuous variable may vary in response to quite distinct pathophysiological processes (e.g. left ventricular contractility may be affected by extracellular fibrosis and/or by myocyte contractility), in which case a more detailed multi-dimensional description of the phenotype is likely to be informative. Multivariate statistical methods are ideal in this instance as they preserve complexity (and information content) of the underlying biological processes. It is our opinion that the precision and reproducibility of all levels of phenotypic assessment is of fundamental importance to integrative genomic studies.
The identification of genetic determinants of a phenotype of interest is clearly predicated on that trait being under genetic control. Many traits are influenced by both genetic and environmental factors, in which case our ability to identify genetic determinants will depend on the relative contribution of these two groups of factors, and the effect size exerted by each contributing gene. It is therefore common to estimate the heritability of a trait (the proportion of the observed trait variation in a population due to genetic factors) in order to decide whether the genetic signal is likely to be sufficiently large to detect.
Genetic variation can have a range of effect sizes on the phenotype(s), and this influences strategies to detect such variation. When genome-wide association studies (GWAS) were first undertaken the field was dominated by the ‘common disease-common variant’ hypothesis, which predicted that association mapping would be powerful in dissecting the causes of complex phenotypes.12 In fact GWAS have revealed a complex genetic architecture underlying the regulation of disease, where the vast majority of identified gene variants exert a limited effect on the complex trait,13,14 and models have been revised.15 By way of illustration, the QT interval, a measure of cardiac repolarization, is a complex trait. Many common variants have been shown to influence the QT interval, though the effect size of each is typically small. Extreme QT intervals may also be inherited as Mendelian traits due to single variants, often in the same genes that harbour common non-coding variants of more modest effect. An awareness of the likely genetic architecture of our traits of interest is invaluable in considering genomic strategies to dissect them. A trait is most likely to be genetically tractable if it is highly heritable, and if the heritable component consists of a relatively small number of genes exerting a substantial effect, rather than many genes with small effects. These conditions are perhaps most likely to be met for molecular endo-phenotypes that are closely related to, or are direct consequences of, the activity of a small number of gene products. For instance, transcript expression levels (mRNA abundance) are tractable endo-phenotypes that are highly heritable and amenable for genetic mapping,16 with individual genetic variants having large effects on trait variation, as will be discussed later.
3. Sources of data for integrative approaches
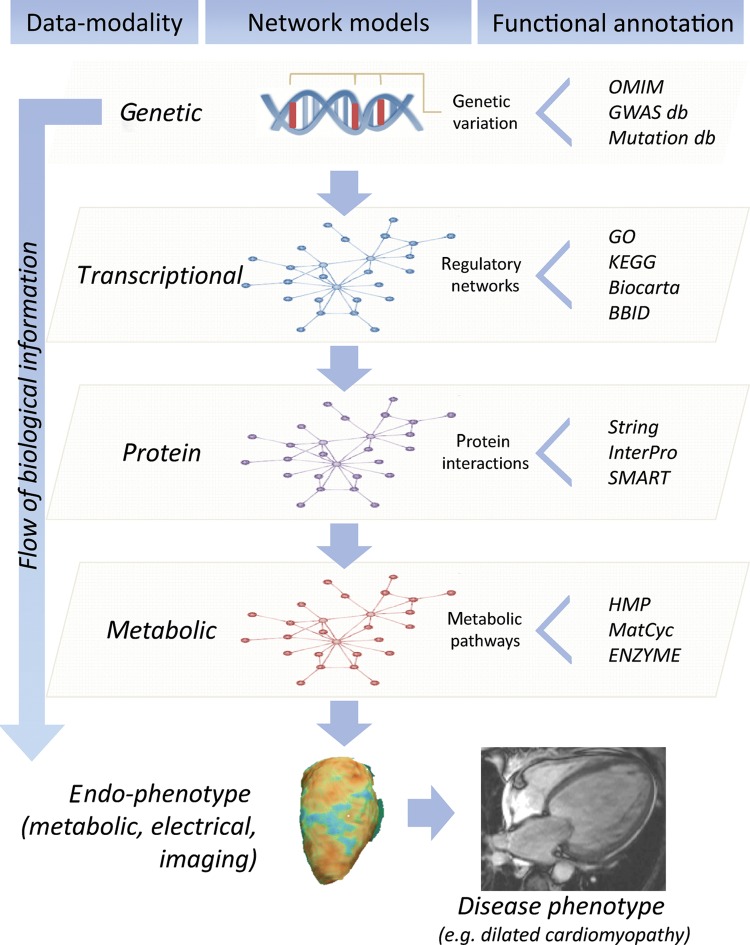
Francis Crick's central dogma17 captures the essence of the information flow from genome to protein, which then leads via a chain of intermediates, interacting with each other and the environment, to phenotype. Depending on our goals, information at any tier of this progression may be integrated to infer genome function. The foundational data set is the sequence of the genome, though epigenetic modifications (DNA methylation and histone modification) not addressed by the central dogma are also heritable and important. RNA transcripts form the next tier. In addition to simple transcript abundance, genome-wide assessment of splicing and RNA editing may also be informative, and the biological importance of non-coding RNAs is now recognized. Finally, proteomics, metabolomics/metabonomics, and other large-scale assays of biomarkers and intermediate phenotypes may be included (Figure 1).
Figure 1.
Integrative genomics across data modalities. The cartoon depicts the flow of biological information from the DNA to the disease level, and modelling of multi-modality data within different layers: genetic (DNA), transcriptional (RNA), protein, and metabolic. Interaction within each layer of biological data can be described using network-models and analysed in conjunction with endo-phenotypes at the cellular and organ level to understand human heart disease pathobiology. Network analyses of quantitative biochemical data sets provide information about complex gene–gene interactions and pathway annotation while increasing power to find individual ‘key players’ in disease, which is not possible in single gene studies. Each network model (genetic, transcriptional, protein, metabolic) can be annotated using extensive bioinformatics and database (db) resources. This allows inference of the ‘functional context’ in which individual genes or networks operate by combining experimental and -omics data. OMIM, Online Mendelian Inheritance in Man (http://www.ncbi.nlm.nih.gov/omim/); GWAS db (for instance: https://www.gwascentral.org/), Mutation db (http://reseq.biosciencedbc.jp/resequence/); GO, Gene Ontology (http://www.geneontology.org/); KEGG, Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/); Biocarta (http://www.biocarta.com/); BBID, Biological Biochemical Image Database (http://bbid.grc.nia.nih.gov/); String, Known and Predicted Protein-Protein Interactions (http://string-db.org/); InterPro, InterPro protein sequence analysis & classification (http://www.ebi.ac.uk/interpro/); SMART, Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de/); HMP, Human Metabolome Database (http://www.hmdb.ca/); MetaCyc, Metabolic Encyclopedia of enzymes and metabolic pathways (http://www.metacyc.org/); ENZIME, Enzyme nomenclature database (http://enzyme.expasy.org/).
4. Genomic data and the identification of genomic disease loci
A reference genome sequence is now available for humans and many model organisms used in cardiovascular research, and sites of common (and rare) DNA sequence variation have been variably catalogued for each species. Most genomic studies sample the genetic variation in an individual, genotyping a number of polymorphic genetic markers spaced across the genome and correlating genotypic variation with phenotype and/or disease. This may identify a locus associated with the trait, the size of the locus being dependent on the resolution of the marker genotyping and the genetic diversity of the population studied.
Two principal statistical approaches are used for this correlation. Linkage seeks co-segregation of a genetic marker with a trait in a pedigree or a population of related individuals, while association usually looks for a correlation between allele frequency and a trait in unrelated individuals, though can be adapted to include families. Both techniques had been applied in a targeted way to individual loci before the description of genome-wide markers, but their genome-wide application opened the door to a dramatic acceleration in functional annotation of loci and genes across the genome.
Linkage studies in humans have been fruitful in identifying Mendelian disease genes. The first autosomal disease locus to be identified by linkage mapping with genome-wide markers was the Huntingdon's disease locus,7 and the first gene to be identified by positional-cloning was for chronic granulomatous disease.8 Cardiovascular disease genes followed swiftly, for example, the first genes underlying hypertrophic cardiomyopathy18,19 and long QT syndrome,20,21 two of the commoner inherited cardiac conditions, were identified by genome-wide linkage followed by positional cloning.
In contrast, linkage studies have been of limited benefit in dissecting continuous traits and common disease in humans. For example, coronary artery disease has been widely studied22 with less than overwhelming results. As linkage approaches require segregation, their application is limited by the identification and the recruitment of appropriate families. Large families are most powerful (e.g. Wang et al.23 identified a CAD locus in a single pedigree of 19 genotyped individuals, of which 13 were affected), but are rare and it may be difficult to retrieve DNA from all subjects. Alternatively large numbers of smaller families may be studied (e.g. Pajukanta et al.24 genotyped 364 individuals from 156 families), but often with small reproducibility of findings across populations, and reduced power to detect relatively common alleles of modest effect size that underlie such traits in the general population.
Association studies are more powerful in large human population samples, and in 2005 GWAS were made possible25,26 by a new high-resolution catalogue of common single nucleotide variation in the human genome (produced by the HapMap project), coupled with relatively cheap high-throughput genotyping technologies (DNA microarrays). The NIHR catalogue of GWAS (http://www.genome.gov/gwastudies; accessed 15 June 201227) now contains details of 1271 papers reporting >6000 disease associations with diverse traits, including ∼50 cardiovascular traits ranging from risk factors (blood pressure, diabetes, smoking behaviour), to traits and intermediate phenotypes (heart rate, electrocardiogram parameters), to diseases (myocardial infarction, atrial fibrillation, ventricular fibrillation), and drug responses (statins, clopidogrel, and warfarin).
It should be pointed out that linkage studies have been particularly informative for identifying complex disease loci when applied to genetically tractable model organisms. Experimental breeding strategies and careful environmental control combine to yield studies of much greater power and reproducibility than can be achieved in humans. For example, our groups have worked extensively with a powerful rodent genetic resource developed for linkage mapping of cardiovascular and other traits: a panel of recombinant inbred rat strains derived from the spontaneously hypertensive rat (displaying a number of extreme cardiovascular traits) and the Brown Norway (essentially normal CV physiology).28,29 By investigating the segregation of cardiovascular traits in experimental crosses derived from these strains, we and others have identified a large number of ‘quantitative trait loci’ underlying CV traits including blood pressure, insulin resistance, and left ventricular hypertrophy.28,30–3,4 The Rat Genome Database (http://rgd.mcw.edu/; accessed 3 July 2012) currently contains 351 QTL for blood pressure in the rat, 73 QTL for cardiac mass, and 21 for heart rate.
Whatever the organism or statistical approach, genome–phenotype correlations typically identify a disease or trait locus, rather than a gene. Moving from locus to underlying gene and causative genetic variation requires the integration of further information (for example, see Glazier et al.3,5), and remains the biggest challenge in this field of biology.
5. Integrating transcript expression data to identify cardiovascular disease genes
Transcription from DNA to RNA is the first step in the cascade from genotype to phenotype, and a number of techniques are available to determine which transcripts are present in a cell, tissue or sample of interest. Northern blotting, fluorescence in-situ hybridization and quantitative PCR are relatively low-throughput techniques that require prior knowledge of the sequence of the transcript to be quantified (or at least part of the sequence). DNA microarrays allow high-throughput genome-wide expression profiling of known transcripts at low cost. Sequencing-based approaches have an added advantage of detecting novel transcripts, but have been expensive to date, and initially only sequenced short tags, rather than whole transcripts. In the era of next generation sequencing (NGS), complete quantitative assessment of transcript expression, splicing, sequence variation, and RNA editing is now available through direct RNA sequencing.
How then can transcriptome data be used to find disease genes? At the simplest level, one can ask which genes are expressed in a tissue of interest. For example, having identified a locus for left ventricular mass, one may ask whether any genes at the locus are expressed in the heart. This requires careful consideration of the tissue of interest, which in turn relies upon refined phenotypic assessment. In the example above, left ventricular mass may of course be influenced by genes that are not expressed in the heart, for example, via effects on blood pressure. Nonetheless, a multivariate approach to the phenotype can discriminate tissue-specific effects that are independent of secondary causation. In our studies multivariate analyses have identified blood pressure-independent regulation of left ventricular mass with an increased myocyte size, in which context the gene of interest is expected to be expressed in the ventricle, and most likely the myocyte, rather than the kidney or adrenal.33,3,6 Another conceptually simple approach is to compare transcript levels in two physiological states, e.g. in cases and controls.3,7,3,8 The resulting list of genes that are up- and down-regulated may highlight pathways and networks underlying the trait under study, and by intersecting these genes with a genetic locus we may be able to identify a shortlist of candidates.
A powerful approach focuses instead on the genetic control of gene expression (genetical genomics10). mRNA transcript levels are quantitative traits, under the combined influence of genetic and environmental factors. Having identified a new genetic locus that determines a trait, we hypothesise that the specific causative variant responsible exerts its effect by altering transcription of a gene—either by altering the protein product itself (e.g. truncating variants and single amino acid substitutions underlying Mendelian disease), or by altering expression levels of gene transcript (particularly for quantitative traits).
If the expression level of a gene transcript can be mapped to the genome, it is termed an expression quantitative trait locus (eQTL). When the eQTL coincides with the location of the gene itself it is termed a cis-eQTL (for example, due to a polymorphism in the gene promoter), and where the eQTL is distant it is termed a trans-eQTL (for example, genetic variation in a transcription factor that regulates in trans the expression of its target genes32). Usually, disease-genes can be identified where cis-eQTLs are found in (or co-localize with) loci that are correlated with disease (i.e. QTL for physiological traits).
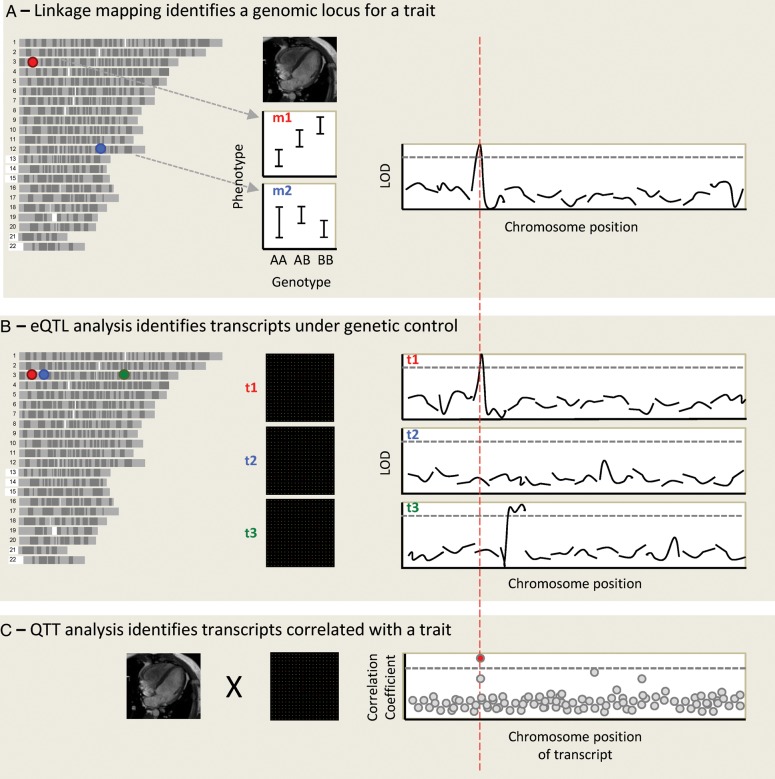
In addition, gene expression data can also be used for quantitative trait transcript analysis (QTT), in which transcript levels are correlated with a phenotype to directly prioritize candidate genes for the trait.39 Finally, by overlaying data on genome sequence variation, transcript expression, and phenotype variation, we can powerfully move from a list of 100 s of genes at a genetic locus, to a few or even a single gene that is dysregulated in a given cell-type or tissue, found at the locus of interest, and whose expression correlated with the trait (Figure 2). This rationale to select candidate genes assumes that genetically controlled transcriptional regulation of a given gene is the mechanism of action that determines susceptibility to disease or complex trait variation.
Figure 2.
An integrative genomic approach to identify cardiovascular disease genes. Genotypes and phenotypes are measured in a population of related individuals, and each genomic marker position is assessed for linkage with the phenotypes. In this case, left ventricular mass (LVM) is studied in a rodent population. The allelic effect is shown for two genomic markers, marker 1 (m1) on chromosome 3 (chr3) and m2 on chr12. A linkage plot is shown for the first 12 chromosomes, showing linkage of LVM to a locus on chr3, at the position of marker m1. The y-axis is the LOD score (logarithm10 of odds), and the dotted line represents genome-wide statistical significance. (A) Microarrays are used to obtain a genome-wide transcript expression profile for RNA expression in the left ventricle in the same population, and the expression of each transcript is mapped as a quantitative trait. The expression of transcript 1 (t1) maps to chr3, where this transcript is encoded: it is hence termed a cis-eQTL. t2, encoded at the same genomic locus, does not appear to be genetically regulated. Although both genes lie within the original LVM locus, t1 is prioritized as the best candidate after eQTL analysis. Expression of t3, also encoded on chr3, maps to chr4: it is a trans-eQTL. (B) Quantitative trait transcript analysis involves the direct correlation of phenotype and expression data. After correction for multiple testing a single transcript emerges as most highly correlated with LVM. If this is t1 this adds further weight to its candidacy.
The first genome-wide eQTL studies were performed in yeast,40 and subsequently most studies have been carried out in model organisms or cell lines, due to the ready availability of tissue for RNA work in these systems. Studies in humans have typically used eQTLs identified in blood, which may overlap with eQTLs in other tissues, but are likely to miss many tissue-specific eQTLs.4,1,4,2 Nonetheless, the approach has been applied to integrate GWAS hits with cis-eQTLs in humans. For example, in the cardiovascular domain LIPA (lysosomal acid lipase A) has been identified as a candidate gene for coronary artery disease through GWAS,4,3 supported by evidence that LIPA is a cis-eQTL associated with the same locus. QTT analysis also demonstrated a correlation between LIPA and endothelial function. This demonstrates the added power of an integrated approach over GWAS alone.4,4
Further examples are mostly derived from model systems. For instance, in the rat >200 traits have been quantified in the BXH/HXB panel previously described, and genetic loci for many of these have been refined using integrative genomic approaches.3,4 In our group, we have used this model to identify genes for cardiac hypertrophy. Osteoglycin (Ogn) is a protein that regulates left ventricular mass in rats, mice, and humans, possibly through the TGF-β signalling pathway,3,6 and Endonuclease G (Endog) regulates mitochondrial biogenesis and cardiac hypertrophy.33 In each case, we were able to translate our findings to humans using cardiac transcript level data. In the Ogn study, we showed that left ventricular mass in patients with aortic stenosis was only weakly correlated with the severity of stenosis, but strongly correlated with OGN levels in QTT analysis, whereas in the EndoG study we carried out co-expression network analysis using a large human transcriptome data set, and identified EndoG in a network highly enriched for mitochondrial genes and oxidative metabolism processes that supported a novel role for EndoG in mitochondrial biogenesis.
6. Higher-order integration of multiple data sets
The simple overlay of genetic, expression, and phenotypic variation described above can be extended to various data multi-modalities, including for instance microRNAs, proteins, metabolites, and high-resolution imaging data (e.g. cardiac MRI). The large dimensionality of individual data sets and their integration does require, and advocate, the use of sophisticated multivariate modelling able to capture complex linear and non-linear interactions predictive of physiological states. In the first instance, these multi-modalities can be considered (and treated) as distinct layers of data where biological information is sought to flow from the DNA level (genetically encoded) to the phenotypic level (Figure 1). Each of these layers can be analysed to model different kinds of interaction (gene–gene, mRNA–mRNA, protein–protein, metabolites, structural, etc.), and prior biological information can be taken into account in the modelling procedure (i.e. Bayesian modelling4,5–4,7).
Particular attention is paid to predictive networks from this type of modelling, and how networks can help link DNA-level variation to physiological states or disease.4,4 Several commonly used algorithms to infer gene co-expression networks from microarray data,4,8–5,1 enabled analysis of transcriptional networks across multiple tissues, and cell-types and their relevance to disease and complex whole-body phenotypes.
A distinct advantage provided by the network description is the ability to provide a framework for exploring the functional and molecular context within which single genes operate. For instance, as mentioned above, using linkage mapping approaches we identified EndoG as a major determinant of cardiac hypertrophy, but only by looking at the network where EndoG was ‘operating’ in the human heart, were we able to re-annotate EndoG function and reveal a novel role for the gene in mitochondrial biogenesis.33
Beyond standard eQTL identification approaches, the integration of eQTLs with network modelling can identify ‘master regulator’ genes that control in a coordinated fashion the behaviour of multiple genes (down-stream). For instance, our group identified Ebi2, a G-protein couple receptor, as the master regulator gene of an inflammatory co-expression gene network that was conserved between rats and humans, and that was associated with increased risk of Type 1 diabetes.32 The latter example also showed how network models could be extended across multiple species,5,2 revealing conserved pathogenic pathways and opening the door to functional validation experiments that are not possible in humans.
7. Future directions using integrative approaches
We have seen how the integration of multiple genome-wide data sets has led to the identification of many human disease genes and pathways, increased our understanding of disease pathogenesis, and suggested new targets for therapy. How do we see this developing in the future?
7.1. Increasing dimensionality and publically available data
We anticipate that the breadth of data incorporated in integrative approaches will continue to increase, with particular emphases on detailed, multi-varied, high-resolution phenotyping, the integration of electronic patient records,5,3 and an increasing awareness of the importance of making high-quality data sets publically available, resonating with the recent discussions on open-access publishing.5,4
7.2. Next-generation sequencing
NGS has increased the availability of sequencing by orders of magnitude, bringing genomic studies that would have required huge international collaborations a decade ago within reach of individual labs, and has contributed hugely to the growth of data sets available for integrative approaches.
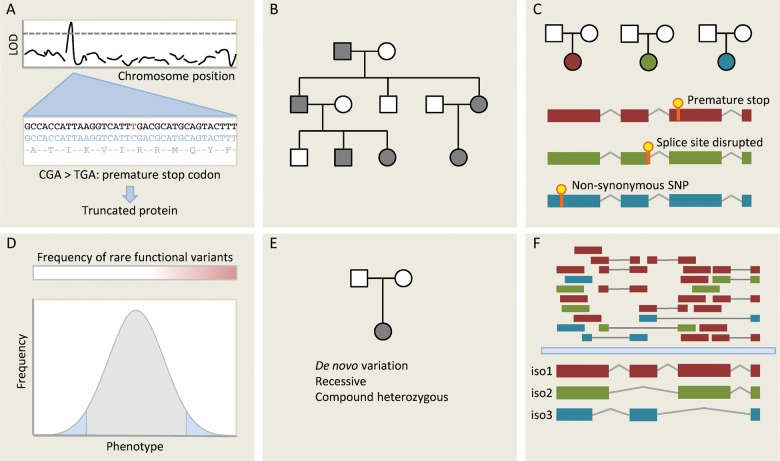
In our discussion of genomic data, we focused on approaches that sample genetic variation in an individual (e.g. DNA microarray genotyping), as these have formed the basis of most high-throughput studies to date. NGS now allows us to probe the full spectrum of genetic variation within an individual. Whole genome sequencing is becoming increasingly available as costs fall, and whole exome sequencing has been widely adopted as an intermediate genome-wide technique capable of detecting much disease-causing variation.5,5,5,6 A full discussion of NGS is beyond the scope of this review, but Figure 3 gives an overview of some of the applications of NGS for disease gene discovery.
Figure 3.
Different approaches for gene discovery in humans using next-generation sequencing (NGS). (A) One of the simplest applications of NGS is deep sequencing genes at a known disease locus to identify functional variants that may be responsible for the observed effect. Here, a C/T substitution generates a novel stop codon, which truncates a gene product that may be functionally important. (B) Mendelian disease within a single family is typically genetically homogenous: in the absence of phenocopies affected individuals will all share the same causative variant. Whole-genome or whole-exome sequencing can be used to identify functional variants segregating with disease in a family. Many variants will be shared through simple relatedness, so large families are needed for this approach. It also makes assumptions about which classes of variants are likely to underlie Mendelian diseases—typically truncating variants and very rare non-synonymous SNPs. If the causative variant is synonymous or non-coding then it is unlikely to be detected by targeted NGS approaches. (C) An alternative methodology is to sequence unrelated individuals with the same phenotype or endophenotype. Here, we do not expect that affected individuals will carry the same variant, but we hypothesize that they may carry distinct variants in the same gene. This is a powerful approach for genetically homogenous conditions, but inherited cardiac conditions are typically heterogenous (variation in many different genes yields the same phenotype) limiting the applicability of this approach. (D) Strategy C can be extended to continuous traits. If rare variants of moderately large effect contribute to the phenotype then we may be able to detect these by focusing sequencing efforts on the extremes of a very large population, seeking genes that are enriched for rare functional variants using burden testing. (E) Where disease is caused by de novo variation, this may be detected by sequencing trios (proband and both parents). This approach may also be applied to recessive phenotypes. (F) RNA sequencing not only measures total transcript abundance, but can also quantify different isoforms, detect novel transcripts and novel splicing events, and identify sequence variants. Here, isoform 1 is the predominant transcript, but RNAseq provides evidence for two other isoforms. This can be used in integrative genomic studies.
For integrative genomic studies, high-throughput DNA sequencing is a double-edged sword. On the one hand, we can identify variants correlated with phenotypes directly, rather than identifying intermediate loci. On the other hand, NGS studies using association or segregation often identify many variants that correlate with a phenotype, and the integration of additional informative data sets remains important in refining candidate gene/variant lists.
We have already alluded to the role that NGS has played in transcriptomics—in particular the development of RNA sequencing that permits the quantification of both known and novel transcripts with a wide dynamic range, as well as identifying RNA splicing and editing events. NGS can also be applied to identify sites of epigenetic modification and transcriptional control—for example, using ChIP-Seq (Chromatin Immunoprecipitation sequencing) to identify transcription factor-binding sites or histone modifications, or bisulphite sequencing to detect DNA methylation.
7.3. The role of model organisms
We see a changing relationship between model organisms and humans for gene discovery. Rather than identifying candidate disease genes in model systems, and validating findings in humans (for example, see Petretto et al.3,6), we see a shift towards primary discovery through reverse genetics in humans, largely enabled by increasing availability of sequencing technology coupled with efforts to collate and curate large-scale human phenotypic data. We believe that primary discovery in model organisms will increasingly take the form of ambitious (and costly) high-throughput forward genetic screens in lower organisms, as have been applied in yeast,5,7 flies,5,8 and fish.59
Whatever the organism used for the primary genetic discovery, subsequent validation remains essential to prove gene effects. Here, we are seeing more ‘disease in a dish’ models, with patient-derived-induced pluripotent stem (iPS) cells a particularly exciting modality for functional genetic studies. Finally, an increasingly sophisticated range of tools for genome manipulation is available for translation to animals, such as zinc finger nucleases and TALENs. Rats offer several advantages over mice for cardiovascular research, and effective knock-out and transgenic technologies applicable to species other than the mouse are extremely welcome.
Conflict of interest: none declared.
Funding
We acknowledge funding from the Wellcome Trust, the British Heart Foundation, the Medical Research Council (UK), and the NIHR Royal Brompton Cardiovascular Biomedical Research Unit.
References
- 1.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. doi:10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. doi:10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 3.Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. doi:10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 4.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. doi:10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 5.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. doi:10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314. [PMC free article] [PubMed] [Google Scholar]
- 7.Gusella JF, Wexler NS, Conneally PM, Naylor SL, Anderson MA, Tanzi RE, et al. A polymorphic DNA marker genetically linked to Huntington's disease. Nature. 1983;306:234–238. doi: 10.1038/306234a0. doi:10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- 8.Royer-Pokora B, Kunkel LM, Monaco AP, Goff SC, Newburger PE, Baehner RL, et al. Cloning the gene for an inherited human disorder—chronic granulomatous disease—on the basis of its chromosomal location. Nature. 1986;322:32–38. doi: 10.1038/322032a0. doi:10.1038/322032a0. [DOI] [PubMed] [Google Scholar]
- 9.McKusick VA, Ruddle FH. A new discipline, a new name, a new journal. Genomics. 1987;1:1–2. doi:10.1016/0888-7543(87)90098-X. [Google Scholar]
- 10.Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/s0168-9525(01)02310-1. doi:10.1016/S0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- 11.Jorgensen P, Breitkreutz BJ, Breitkreutz K, Stark C, Liu G, Cook M, et al. Harvesting the genome's bounty: integrative genomics. Cold Spring Harb Symp Quant Biol. 2003;68:431–443. doi: 10.1101/sqb.2003.68.431. doi:10.1101/sqb.2003.68.431. [DOI] [PubMed] [Google Scholar]
- 12.Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease—common variant… or not? Hum Mol Genet. 2002;11:2417–2423. doi: 10.1093/hmg/11.20.2417. doi:10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 13.Stranger BE, Stahl EA, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187:367–383. doi: 10.1534/genetics.110.120907. doi:10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. doi:10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2011;13:135–145. doi: 10.1038/nrg3118. doi:10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petretto E, Mangion J, Dickens NJ, Cook SA, Kumaran MK, Lu H, et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet. 2006;2:e172. doi: 10.1371/journal.pgen.0020172. doi:10.1371/journal.pgen.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. doi:10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 18.Jarcho JA, McKenna W, Pare JA, Solomon SD, Holcombe RF, Dickie S, et al. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N Engl J Med. 1989;321:1372–1378. doi: 10.1056/NEJM198911163212005. doi:10.1056/NEJM198911163212005. [DOI] [PubMed] [Google Scholar]
- 19.Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg HP, McKenna W, Seidman CE, et al. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. doi:10.1016/0092-8674(90)90274-I. [DOI] [PubMed] [Google Scholar]
- 20.Keating M, Atkinson D, Dunn C, Timothy K, Vincent GM, Leppert M. Linkage of a cardiac arrhythmia, the long QT syndrome, and the Harvey ras-1 gene. Science. 1991;252:704–706. doi: 10.1126/science.1673802. doi:10.1126/science.1673802. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q, Curran ME, Splawski I, Burn TC, Millholland JM, VanRaay TJ, et al. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet. 1996;12:17–23. doi: 10.1038/ng0196-17. doi:10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- 22.Lee WK, Padmanabhan S, Dominiczak AF. Molecular genetics of cardiovascular disease. eLS. 2010 Chichester, UK: John Wiley & Sons, Ltd; [Google Scholar]
- 23.Wang L, Fan C, Topol SE, Topol EJ, Wang Q. Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science. 2003;302:1578–1581. doi: 10.1126/science.1088477. doi:10.1126/science.1088477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pajukanta P, Cargill M, Viitanen L, Nuotio I, Kareinen A, Perola M, et al. Two loci on chromosomes 2 and X for premature coronary heart disease identified in early- and late-settlement populations of Finland. Am J Hum Genet. 2000;67:1481–1493. doi: 10.1086/316902. doi:10.1086/316902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. doi:10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 26.Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Sackler RS, Haynes C, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. doi:10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hindorff L, MacArthur J, Wise A, Junkins H, Hall P, Klemm A, et al. A Catalog of Published Genome-Wide Association Studies. Available at http://www.genome.gov/gwastudies. Accessed 15th June 2012) [Google Scholar]
- 28.Pravenec M, Klír P, Kren V, Zicha J, Kunes J. An analysis of spontaneous hypertension in spontaneously hypertensive rats by means of new recombinant inbred strains. J Hypertens. 1989;7:217–221. [PubMed] [Google Scholar]
- 29.Printz MP, Jirout M, Jaworski R, Alemayehu A, Kren V. HXB/BXH rat recombinant inbred strain platform: a newly enhanced tool for cardiovascular, behavioral, and developmental genetics and genomics. J Appl Physiol. 2003;94:2510–2522. doi: 10.1152/japplphysiol.00064.2003. doi:10.1063/1.1590051. [DOI] [PubMed] [Google Scholar]
- 30.Pravenec M, Gauguier D, Schott JJ, Buard J, Kren V, Bila V, et al. Mapping of quantitative trait loci for blood pressure and cardiac mass in the rat by genome scanning of recombinant inbred strains. J Clin Invest. 1995;96:1973–1978. doi: 10.1172/JCI118244. doi:10.1172/JCI118244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pravenec M, Kurtz TW. Recent advances in genetics of the spontaneously hypertensive rat. Curr Hypertens Rep. 2010;12:5–9. doi: 10.1007/s11906-009-0083-9. doi:10.1007/s11906-009-0083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinig M, Petretto E, Wallace C, Bottolo L, Rotival M, Lu H, et al. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature. 2010;467:460–464. doi: 10.1038/nature09386. doi:10.1038/nature09386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDermott-Roe C, Ye J, Ahmed R, Sun X-M, Serafín A, Ware J, et al. Endonuclease G is a novel determinant of cardiac hypertrophy and mitochondrial function. Nature. 2011;478:114–118. doi: 10.1038/nature10490. doi:10.1038/nature10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrissey C, Grieve IC, Heinig M, Atanur S, Petretto E, Pravenec M, et al. Integrated genomic approaches to identification of candidate genes underlying metabolic and cardiovascular phenotypes in the spontaneously hypertensive rat. Physiol Genomics. 2011;43:1207–1218. doi: 10.1152/physiolgenomics.00210.2010. doi:10.1152/physiolgenomics.00210.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glazier AM, Nadeau JH, Aitman TJ. Finding genes that underlie complex traits. Science. 2002;298:2345–2349. doi: 10.1126/science.1076641. doi:10.1126/science.1076641. [DOI] [PubMed] [Google Scholar]
- 36.Petretto E, Sarwar R, Grieve I, Lu H, Kumaran MK, Muckett PJ, et al. Integrated genomic approaches implicate osteoglycin (Ogn) in the regulation of left ventricular mass. Nat Genet. 2008;40:546–552. doi: 10.1038/ng.134. doi:10.1038/ng.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanton LW, Garrard LJ, Damm D, Garrick BL, Lam A, Kapoun AM, et al. Altered patterns of gene expression in response to myocardial infarction. Circ Res. 2000;86:939–945. doi: 10.1161/01.res.86.9.939. doi:10.1161/01.RES.86.9.939. [DOI] [PubMed] [Google Scholar]
- 38.Yang J, Moravec CS, Sussman MA, DiPaola NR, Fu D, Hawthorn L, et al. Decreased SLIM1 expression and increased gelsolin expression in failing human hearts measured by high-density oligonucleotide arrays. Circulation. 2000;102:3046–3052. doi: 10.1161/01.cir.102.25.3046. doi:10.1161/01.CIR.102.25.3046. [DOI] [PubMed] [Google Scholar]
- 39.Passador-Gurgel G, Hsieh W-P, Hunt P, Deighton N, Gibson G. Quantitative trait transcripts for nicotine resistance in Drosophila melanogaster. Nat Genet. 2007;39:264–268. doi: 10.1038/ng1944. doi:10.1038/ng1944. [DOI] [PubMed] [Google Scholar]
- 40.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. doi:10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 41.Powell JE, Henders AK, McRae AF, Wright MJ, Martin NG, Dermitzakis ET, et al. Genetic control of gene expression in whole blood and lymphoblastoid cell lines is largely independent. Genome Res. 2012;22:456–466. doi: 10.1101/gr.126540.111. doi:10.1101/gr.126540.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hernandez DG, Nalls MA, Moore M, Chong S, Dillman A, Trabzuni D, et al. Integration of GWAS SNPs and tissue specific expression profiling reveal discrete eQTLs for human traits in blood and brain. Neurobiol Dis. 2012;47:20–28. doi: 10.1016/j.nbd.2012.03.020. doi:10.1016/j.nbd.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wild PS, Zeller T, Schillert A, Szymczak S, Sinning CR, Deiseroth A, et al. A genome-wide association study identifies LIPA as a susceptibility gene for coronary artery disease. Circ Cardiovasc Genet. 2011;4:403–412. doi: 10.1161/CIRCGENETICS.110.958728. doi:10.1161/CIRCGENETICS.110.958728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schadt EE. Molecular networks as sensors and drivers of common human diseases. Nature. 2009;461:218–223. doi: 10.1038/nature08454. doi:10.1038/nature08454. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Jiang B, Zhu J, Liu JS. Bayesian models for detecting epistatic interactions from genetic data. Ann Hum Genet. 2011;75:183–193. doi: 10.1111/j.1469-1809.2010.00621.x. doi:10.1111/j.1469-1809.2010.00621.x. [DOI] [PubMed] [Google Scholar]
- 46.Stephens M, Balding DJ. Bayesian statistical methods for genetic association studies. Nat Rev Genet. 2009;10:681–690. doi: 10.1038/nrg2615. doi:10.1038/nrg2615. [DOI] [PubMed] [Google Scholar]
- 47.Bumgarner RE, Yeung KY. Methods for the inference of biological pathways and networks. Methods Mol Biol. 2009;541:225–245. doi: 10.1007/978-1-59745-243-4_11. doi:10.1007/978-1-59745-243-4_11. [DOI] [PubMed] [Google Scholar]
- 48.Kramer N, Schafer J, Boulesteix A-L. Regularized estimation of large-scale gene association networks using graphical Gaussian models. BMC Bioinformatics. 2009;10:384. doi: 10.1186/1471-2105-10-384. doi:10.1186/1471-2105-10-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. doi:10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schafer J, Strimmer K. An empirical Bayes approach to inferring large-scale gene association networks. Bioinformatics. 2005;21:754–764. doi: 10.1093/bioinformatics/bti062. doi:10.1093/bioinformatics/bti062. [DOI] [PubMed] [Google Scholar]
- 51.Zhao W, Langfelder P, Fuller T, Dong J, Li A, Hovarth S. Weighted gene coexpression network analysis: state of the art. J Biopharm Stat. 2010;20:281–300. doi: 10.1080/10543400903572753. doi:10.1080/10543400903572753. [DOI] [PubMed] [Google Scholar]
- 52.Wiles AM, Doderer M, Ruan J, Gu T-T, Ravi D, Blackman B, et al. Building and analyzing protein interactome networks by cross-species comparisons. BMC Syst Biol. 2010;4:36. doi: 10.1186/1752-0509-4-36. doi:10.1186/1752-0509-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012;13:395–405. doi: 10.1038/nrg3208. doi:10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 54.Openness costs. Nature. 2012;486:439–439. doi: 10.1038/486439a. [DOI] [PubMed] [Google Scholar]
- 55.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, et al. Exome sequencing identifies the cause of a Mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. doi:10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. doi:10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 57.Steinmetz LM, Scharfe C, Deutschbauer AM, Mokranjac D, Herman ZS, Jones T, et al. Systematic screen for human disease genes in yeast. Nat Genet. 2002;31:400–404. doi: 10.1038/ng929. [DOI] [PubMed] [Google Scholar]
- 58.Neely GG, Kuba K, Cammarato A, Isobe K, Amann S, Zhang L, et al. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell. 2010;141:142–153. doi: 10.1016/j.cell.2010.02.023. doi:10.1016/j.cell.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deo RC, MacRae CA. The zebrafish: scalable in vivo modeling for systems biology. Wiley Interdiscip Rev Syst Biol Med. 2011;3:335–346. doi: 10.1002/wsbm.117. doi:10.1002/wsbm.117. [DOI] [PMC free article] [PubMed] [Google Scholar]