Abstract
Aims
Human genome-wide association studies (GWAS) of hypertension identified only few susceptibility loci with large effect that were replicated across populations. The vast majority of genes detected by GWAS has small effect and the regulatory mechanisms through which these genetic variants cause disease remain mostly unclear. Here, we used comparative genomics between human and an established rat model of hypertension to explore the transcriptional mechanisms mediating the effect of genes identified in 15 hypertension GWAS.
Methods and results
Time series analysis of radiotelemetric blood pressure (BP) was performed to assess 11 parameters of BP variation in recombinant inbred strains derived from the spontaneously hypertensive rat. BP data were integrated with ∼27 000 expression quantative trait loci (eQTLs) mapped across seven tissues, detecting >8000 significant associations between eQTL genes and BP variation in the rat. We then compiled a large catalogue of human genes from GWAS of hypertension and identified a subset of 2292 rat–human orthologous genes. Expression levels for 795 (34%) of these genes correlated with BP variation across rat tissues: 51 genes were cis-regulated, whereas 459 were trans-regulated and enriched for ‘calcium signalling pathway’ (P = 9.6 × 10−6) and ‘ion channel’ genes (P = 3.5 × 10−7), which are important determinants of hypertension. We identified 158 clusters of trans-eQTLs, annotated the underlying ‘master regulator’ genes and found significant over-representation in the human hypertension gene set (enrichment P = 5 × 10−4).
Conclusion
We showed extensive conservation of trans-regulated genes and their master regulators between rat and human hypertension. These findings reveal that small-effect genes associated with hypertension by human GWAS are likely to exert their action through coordinate regulation of pathogenic pathways.
Keywords: Integrative genomics, Expression QTLs, Time series analysis, Trans-acting regulation, Genome-wide association studies
1. Introduction
Hypertension, characterized by elevated blood pressure (BP), is one of the strongest risk factors for common cardiovascular diseases worldwide.1 Despite the high heritability of BP variation,2 human genome-wide association studies (GWAS) have been able to identify only few susceptibility loci with a large effect for hypertension that were replicated across populations3–7 and a large proportion of the underlying genetic determinants remains to be investigated. Moreover, the functional context and regulatory mechanisms through which the susceptibility variants identified by GWAS cause hypertension or elevated BP remain mostly unclear.
Common genetic variation underlying disease susceptibility might exert its function by altering gene expression in a specific tissue. Quantitative variation in gene expression that is due to underlying sequence variation can be mapped to the genome in segregating populations as expression quantitative trait loci (eQTLs).8 Genetic variants can affect the expression of genes that reside locally (i.e. cis-eQTLs) or remotely (i.e. trans-eQTLs), reflecting different regulatory mechanisms of transcription.8 In particular, trans-eQTLs are informative of downstream-regulated genes, which can be associated with disease susceptibility and hence might reveal pathophysiological pathways.9,10 GWAS focus on the identification of significant SNP(s)-disease associations and pinpoint local (cis-acting) genetic effects. Similarly, typical GWAS have limited power to detect secondary (trans-acting) regulatory effects of the associated genes on disease or complex phenotypes. Gene expression can be considered as an intermediate ‘endo-phenotype’ between genetic variation and whole body disease phenotypes, and can be used to investigate the regulatory mechanisms mediating genetic susceptibility to disease. For example, genome-wide expression levels can be directly correlated with variation in pathophysiological phenotypes by quantitative trait transcript (QTT) analysis to pinpoint eQTL genes and transcripts associated with disease.11 This approach was successfully exploited in the rat model to identify several susceptibility genes for cardiovascular traits including BP variation and hypertension.12,13
In this study, we investigate the regulatory mechanisms mediating the effect of the genetic associations with hypertension and BP variation identified in human GWAS, including both significant and replicated GWAS signals. To this aim we annotated a large catalogue of GWAS results and identified hypertension-associated genes in humans and carried out extensive comparative genomics analyses between rat and human genes (Figure 1). We used an established rat model of hypertension, the BXH/HXB panel of rat recombinant inbred (RI) lines, derived from the Brown Norway (BN) and spontaneously hypertensive rat (SHR)14,15 to explore the downstream effects of hypertension-associated genes on expression in multiple tissues. Here, we took advantage of high-resolution radiotelemetric BP measurements and employed time series analysis to capture different physiological aspects of BP variation and heart rate (HR). We analysed 11 BP parameters with global gene expression variation, and used these data to inform transcriptional regulatory mechanisms for genetic variants associated with hypertension in humans.
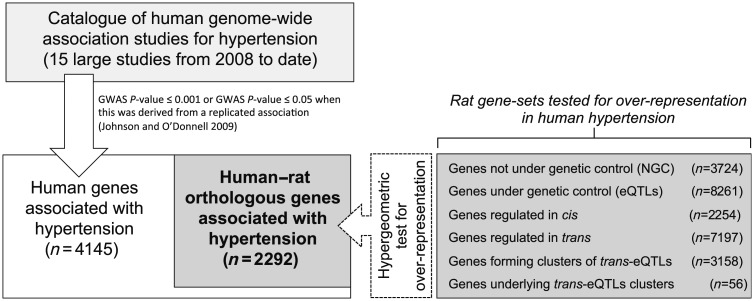
Figure 1.
Schematic representation of the comparative genomics analysis of rat and human hypertension genes. To assemble a comprehensive set of human hypertension genes we combined GWAS results from 15 large studies and adopted a relaxed threshold of significance (GWAS P-value) to ascertain associated genes (n = 4145), as previously described in Johnson and O'Donnell (see Section 2). Different gene sets in the rat where then tested for over-representation against the human-rat orthologues genes (n = 2292) associated with hypertension by GWAS.
2. Methods
2.1. Rat data sets
The BXH/HXB RI rat lines were derived by intercrossing BN and SHR strains as previously described.14 Genome-wide gene expression profiles are available in seven tissues using the Affymetrix 230 2.0 Array chip (aorta, liver, left ventricle, and skeletal muscle) and Affymetrix 230A Array chip (kidney, adrenal gland, and fat). Tissues were collected from four unfasted males (6 weeks old) of each RI strain between 9 a.m. and 10 a.m., and were frozen in liquid nitrogen and stored at –80°C. The gene expression levels were normalized using RMA16 and outliers within each RI strain removed using the Nalimov test, as previously described.15 Time series blood pressure measurements were collected from 28 BXH/HXB RI lines. We implanted indwelling aortic radiotelemetry transducers (Data Sciences International) and measured arterial pressure in conscious, unrestrained rats. The Dataquest IV radiotelemetry system (Data Sciences International, St Paul, MN, USA) was used for the measurement of systolic, diastolic, mean arterial pressure, and HR. Transmitters (TA11PA) were implanted to 12-week-old males after the calibration as recommended by the supplier. Briefly, the rats were anaesthetized with ketamine (100 mg/kg) and xylazine (5 mg/kg), a midline abdominal incision was made and abdominal aorta was exposed. The adequacy of anaesthesia was assessed by testing the loss of the pedal reflex. The catheter of the transmitter was inserted rostrally into the aorta through a small hole close to the bifurcation and covered by the piece of nitrocellulose sheet to protect the bleeding. The body of the transmitter was sutured to the inside of abdominal muscle wall, whereas muscle and skin incisions were suture closed. Rats were housed individually in standard cages, which were placed over the receivers that were connected to the personal computer for data acquisition. The animals were left to recover for 10 days after the surgery. After the end of experiment, the rats killed by overdose of the above mentioned anaesthetics. Five hundred measurements per second were collected over a period of 30 min in each animal (recorded between 9 a.m. and 12 p.m.), and there are between 2 and 11 rats per strain.12 All of the animal studies were performed in agreement with the Animal Protection Law of the Czech Republic (311/1997) and approved by the Ethics Committee of the Czech Academy of Sciences Institute of Physiology. This investigation conforms to the Directive 2010/63/EU of the European Parliament.
2.2. Time series analysis
2.2.1. Conventional phenotype measurements
Diastolic blood pressure (DBP) was obtained by averaging the minimum peak values within an individual time series and systolic blood pressure (SBP) was similarly obtained by averaging the maximum peak values within an individual time series. The mean blood pressure (MBP) was obtained by averaging all blood pressure measurements. The heart rate was obtained by dividing the number of maximum peaks in an individual time series by the length of the time series, in seconds, to obtain a measurement in beats per second. Peak-to-peak distance (PDIST) was calculated by averaging the distance between maximum peaks. Pulse pressure (PP) was calculated by averaging the difference between adjacent maximum and minimum peaks.
2.2.2. Spectral phenotype measurements
To better characterize the cyclical and periodic nature of the blood pressure time series, we performed fast Fourier transform (FFT) analysis. Since the time series must be (weakly) stationary to obtain an accurate estimate of the spectral power, the Kwiatkowski–Phillips–Schmidt–Shin (KPSS) test was used to assess the existence of a trend and the level stationarity of each time series.17 In all time series analysed, the null hypothesis was rejected and a differencing method of lag one (i.e. two consecutive measurements were subtracted) was used to correct for the non-stationarity. Each lagged time series was thinned by a factor of 10, so that the number of measurements was reduced from 500 to 50 to remove possible autocorrelation. All time series were then transformed from the time domain into the frequency domain via FFT. We defined SPEC as the height of the highest peak in the spectral power density of the original sequence, FREQ to be the frequency at which the peak occurs, and POWER is the average power calculated by integrating over the 95% confidence interval surrounding the peak. (for details, see Supplementary material online, Methods).
2.2.3. Wavelet phenotype measurements
Wavelet analysis allows to decompose a time series into time/frequency space simultaneously. First, the original time series with 500 measurements per second was filtered using a 1-D digital filter to reduce the trend in the time series. Secondly, a Morlet wavelet was applied and the local maximum of the wavelet coefficients was calculated within the HR range (4–8 Hz). Within this range, we defined WAVE as the local maximum peak and WFREQ as the frequency that corresponds to the local maximum (for details, see Supplementary material online, Methods). Finally, for each of the conventional, spectral and wavelet phenotype, a Grubbs test was applied to identify and remove outliers within each strain; strains means were then used in the subsequent analysis (for details on the phenotypes, see Supplementary material online, Table S1). Conventional, spectral and wavelet phenotypes were clustered using partial correlation analysis by means of graphical Gaussian models, as implemented in GeneNet R package.18
2.3. Expression QTL mapping
We employed a sparse Bayesian regression model19 to map eQTLs using genome-wide gene expression levels measured in the adrenal gland, aorta, fat, kidney, left ventricle, liver, and skeletal muscle tissues. A genome-wide SNP panel consisting of 1304 non-redundant SNPs20 was used and missing genotypes were imputed using FastPhase.21 Microarray probesets that map to multiple locations in the genome and non-expressed transcripts were removed prior to the analysis. This defined a set of 11 671 genes in the aorta, liver, left ventricle, skeletal muscle and 8444 genes in the adrenal, fat, kidney, respectively. Significance of the strength of transcript–SNP association [i.e. posterior probability of association (PPA)] was assessed using the method proposed in Chen et al.22 In detail, SNPs are ranked in a decreasing order according to their local false discovery rate (FDR) (i.e. 1-PPA) and the top-ranked SNPs with an average FDR <10% are declared significant. For each microarray probeset whose expression mapped to a SNP (hence forming an eQTL), we defined a cis-eQTL when the location of the probeset is within 10 Mb of the physical location of the SNP. Trans-eQTLs were identified when the probeset was located >10 Mb away from the SNP or on another chromosome.15 Trans-eQTL clusters were defined to be groups of 10 or more genes, which map in trans to the same location of the genome (i.e. the same SNP). Genes located within a 2 kb region around the SNP defining the trans-eQTL cluster were called putative ‘master regulators’ and represent candidate regulators of the trans-eQTLs. When multiple genes were found within the 2 kb region the gene nearest to the SNP defining the trans-cluster was prioritized.
2.4. Quantitative trait transcript analysis
To determine the association between blood pressure phenotypes and gene expression, we correlated each phenotype with genome-wide expression levels across seven tissues using pairwise Pearson correlation, as previously described.12 Using the Westfall-Young procedure, we corrected P-values of each pairwise correlation using 1000 permutations.23 Associations were deemed significant with an empirical P-value <0.05.
2.5. Functional annotation analysis
The Kyoto Encyclopedia of Genes and Genomes pathway and gene ontology enrichment analysis was performed using DAVID.24 The significance of the enrichment was assessed by the Fisher's exact test and correction for multiple testing was performed using the Benjamini FDR.
2.6. Human genome-wide association studies and comparative genomics analysis
We used and expand the database of human GWAS results published by Johnson and O'Donnell,25 to obtain a list of rat genes with human orthologues associated with hypertension. Specifically, the database contained all SNPs associated with many human diseases/traits from 118 GWAS published until 2008, and included SNPs that were significant based upon four conditions:
The SNP had an identifiable ID or verifiable genomic position.
A statistical P-value for association was reported.
The P-value was ≤0.001 (allowing for rounding) if the association was from a raw, unadjusted genome-wide association scan.
The P-value was ≤0.05 if the association was derived from replication, fine mapping, or re-sequencing efforts, or if it was identified as belonging to a locus or region that was specifically identified as an a priori candidate by the authors.
We filtered the list of GWAS data to look only at hypertension related phenotypes (hypertension case–control, blood pressure, SBP, and DBP) resulting in five published studies. We expanded upon this list using the same criteria from GWAS published since 2008, which included an additional 10 research articles.6,7,26–33 The genes closest to, or encompassing, the human SNPs and the rat orthologues of those human genes were identified using Ensembl's Biomart 67 (www.ensembl.org/biomart/martview). We looked for an over-representation in the human GWAS genes for the following rat gene sets: (i) cis and trans-eQTLs mapped in seven tissues; (ii) non-eQTL genes; (iii) trans-eQTL clusters and genes underlying trans-eQTL clusters; (iv) the subset of each of the above (i–iii) that significantly correlated with any blood pressure trait by QTT analysis (FDR <5%). Over-representation of rat genes in the set of human GWAS data was assessed using the hypergeometric test, and empirical P-values of significance were assessed using 10 000 permutations, with the size of the sample drawn equal to that of the set analysed.34
3. Results
3.1. Spectral and wavelet analysis of blood pressure variation in the rat
Radiotelemetric BP data were collected in the BXH/HXB RI strains panel14 at a frequency of 500 Hz over the course of 30 min.12 In addition to conventional approaches to measure standard BP indices (SBP, DBP, MBP, PP, PDIST, and HR), we used spectral and wavelet analysis to assess different physiological parameters of BP and accurately quantify variability of BP and HR signals. Spectral analysis transforms the BP time series from the time domain to the frequency domain, allowing the decomposition of the signal into periodic components that can be informative of variation of cardiovascular functions and circadian BP rhythm.35 Similarly, wavelet analysis transforms the time series from the time domain to the time and frequency domain, retaining information about the periodic and variable components of BP and HR.36 We derived 11 phenotypes and showed that the new BP parameters (FREQ, POWER, SPEC, WAVE, and WFREQ) have heritability estimates that are comparable with the standard phenotypes (heritability range: 13–32%), suggesting a genetic component underlying the regulation of these traits (Table 1 and Supplementary material online, Table S1).
Table 1.
Blood pressure and heart rate phenotypes derived from time series, spectral, and wavelet analyses of readiotelemetric blood pressure measurements in the rat RI strains
| Trait name | Abbreviation | Type | Groupinga | Median | Range | Heritabilityb (h2) (%) |
|---|---|---|---|---|---|---|
| Diastolic blood pressure | DBP | Conventional | BP | 80.55 | 59.03–94.63 | 21 |
| Heart rate | HR | Conventional | Rate | 298.5 | 276.5–352.4 | 23 |
| Mean blood pressure | MBP | Conventional | BP | 91.44 | 68.44–108.61 | 24 |
| Peak-to-peak distance | pDist | Conventional | Rate | 0.20 | 0.16–0.22 | 21 |
| Pulse pressure | PP | Conventional | Pressure | 32.82 | 22.16–40.49 | 21 |
| Systolic blood pressure | SBP | Conventional | BP | 112.92 | 87.26–142.93 | 31 |
| Maximum frequency | Freq | Spectral | Rate | 4.79 | 4.41–5.50 | 21 |
| Total power | Power | Spectral | Pressure | 67.03 | 28.66–133.18 | 25 |
| Maximum spectra | Spec | Spectral | Pressure | 0.17 | 0.10–0.44 | 18 |
| Wave amplitude | Wave | Wavelet | Wave | 0.25 | 0.08–0.49 | 13 |
| Wave frequency | Wfreq | Wavelet | Rate | 4.88 | 4.34–5.84 | 19 |
aGrouping was determined by clustering the blood pressure traits using partial correlations (FDR <5%), which were estimated by graphical Gaussian models (see Section 2).
bFor each blood pressure trait the narrow sense heritability (h2) in the RI strains was calculated, as previously described.37
3.2. Extensive trans-acting regulation of gene expression across seven rat tissues
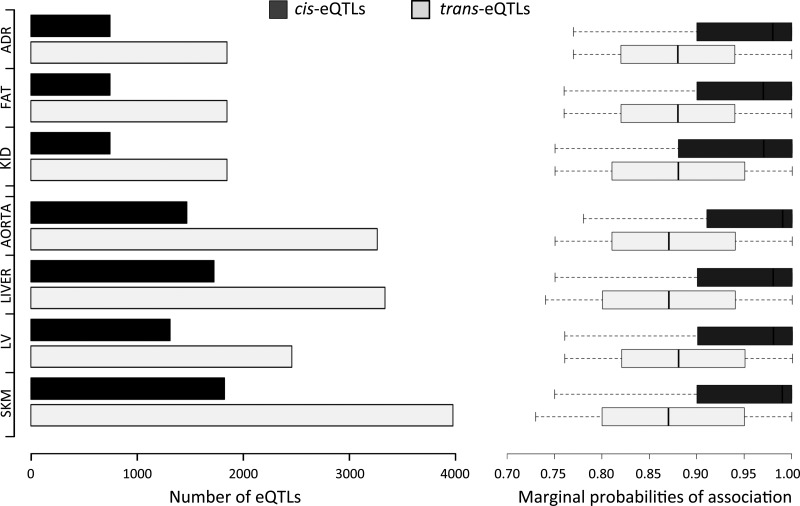
To identify primary and secondary regulators of BP variation in the rat, we integrated all BP parameters with genome-wide gene expression and genetic variation in the BXH/HXB RI strains. We performed genome-wide eQTL mapping in seven rat tissues (adrenal, aorta, fat, kidney, liver, left ventricle, and skeletal muscle) using sparse Bayesian regression methods,19 and identified 26 990 eQTLs (FDR <10%) across the seven tissues, both cis- and trans-acting (Figure 2 and Supplementary material online, Table S2). Overall, 4.0–7.0% of transcripts were found to be cis-regulated and 9.8–15.1% were found to be trans-regulated across tissues. In keeping with previous data,19,37 the stronger associations were observed for the cis-eQTLs (average marginal PPA = 0.94) when compared with the trans-eQTLs (average marginal PPA = 0.88), and on average we found 2.3-fold more trans- than cis-eQTLs (Figure 2). We looked at previously mapped blood pressure QTLs in SHR and SHR/BN strains (Rat Genome Database, http://rgd.mcw.edu/) and found that 3713 eQTLs (∼15% of all eQTLs identified) were located within these QTL regions (Supplementary material online, Table S3). In particular, 2439 (66%) of these eQTLs were trans-regulated, suggesting a role for trans-eQTLs in the regulation of known BP loci in the rat.
Figure 2.
Cis- and trans-eQTLs detected in seven rat tissues. Left, number of cis- and trans-eQTL detected at a 10% FDR level across tissues. The Affymetrix 230 a chip with ∼15 000 microarray probesets was used for expression profiling in adrenal (ADR), fat (FAT), and kidney (KID) tissues, whereas the Affymetrix 230.2 with ∼30 000 microarray probesets was used for expression profiling in the aorta (AORTA), liver (LIVER), left ventricle (LV) and skeletal muscle (SKM) tissues, resulting in more eQTLs detected in the latter four tissues. Right, box-plots for the marginal probability of association of cis- and trans-eQTLs detected across tissues (see Section 2). Each box-plot shows the distribution of marginal probability of association from 25–75th percentile, and the thick line indicates the median for each distribution. Whiskers denote the intervals between the 5 and 95th percentiles.
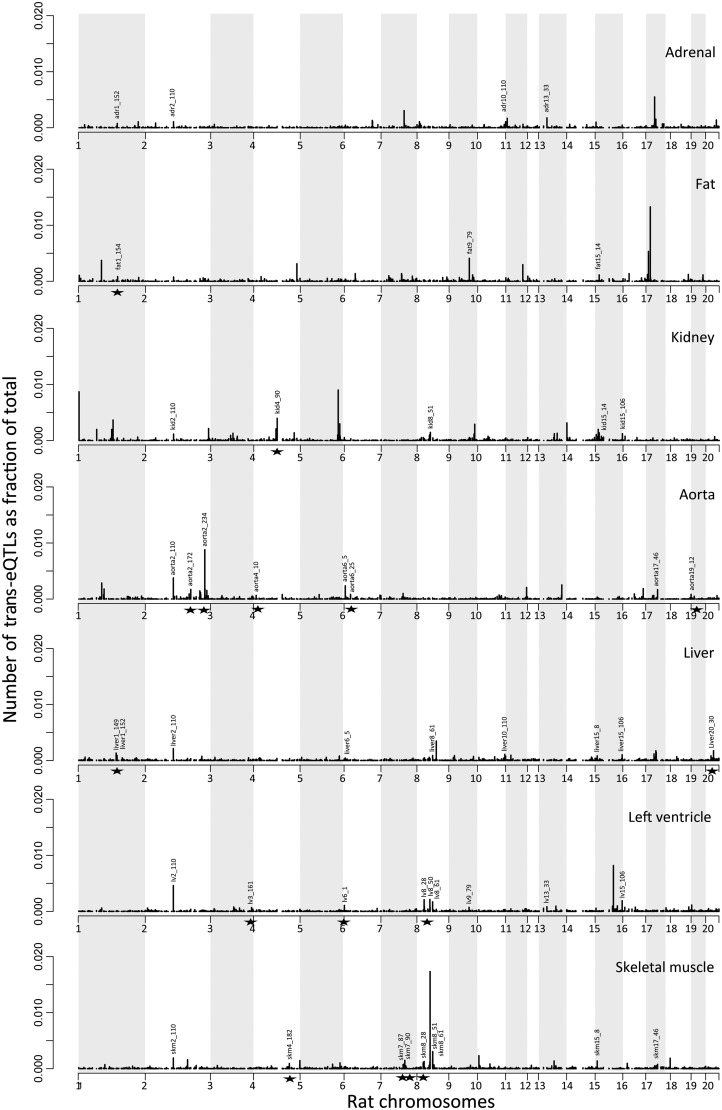
Multiple trans-regulated genes can map to the same locus forming clusters of eQTLs (or hot spots),38 which can provide direct insights into the co-ordinated regulatory mechanisms underlying disease.9,10 At FDR <10% we identified 158 trans-eQTL clusters across the seven tissues (Figure 3 and Supplementary material online, Table S4), where we defined a trans-eQTL cluster to be a group of 10 or more genes whose mRNA expression map in trans to the same genetic locus. Fourteen trans-eQTL clusters were detected in multiple tissues (Figure 3), suggesting common regulatory mechanisms that are conserved between multiple organs and systems. Since 14 clusters were detected in multiple tissues and pinpointed to the same QTL regions (Figure 3), only 138 trans-eQTL cluster loci mapped uniquely to the genome. One of these ‘replicated’ trans-eQTL clusters, detected in the kidney (kid15_106), liver (liver15_106), and left ventricle (lv15_106) tissues (Figure 3), was identified in previous studies and harboured a ‘master regulator’ gene at the locus, which was conserved between rats and humans.9 Here, we have systematically annotated all genes located at (or nearby) the SNP that define the trans-eQTL cluster loci identified across tissues (Figure 3). For each unique trans-eQTL cluster, we conservatively identified the closest gene to SNP defining the trans-eQTL cluster using a 2 kb region centred on the SNP marker. These genes were called putative ‘master regulators’ for the trans-eQTL clusters. Fifty-six of the 138 unique trans-eQTL clusters loci had an annotated gene within the 2 kb region and 12 of these genes were cis-regulated (Supplementary material online, Table S5).
Figure 3.
Trans-eQTL clusters detected across rat tissues. For each tissue the number of trans-eQTLs is reported as a fraction of the total number of eQTLs across the rat genome (chromosomes). A trans-eQTL cluster comprises at least 10 unique genes regulated in trans by a single SNP. The clusters that replicate across tissues are indicated by their names and are detailed in Supplementary material online, Table S3. Star symbols indicate the sixteen trans-eQTL cluster loci that harboured putative ‘master regulator’ genes (Table 3), which in humans were associated with hypertension by GWAS.
3.3. Identification of rat genes associated with blood pressure variation
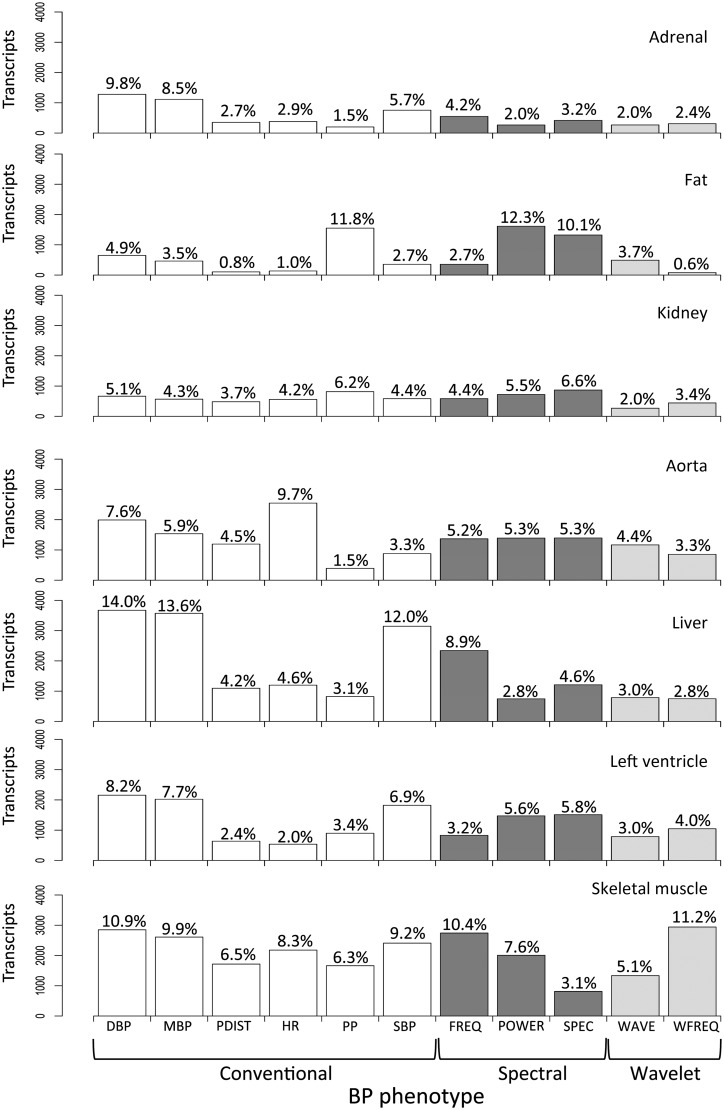
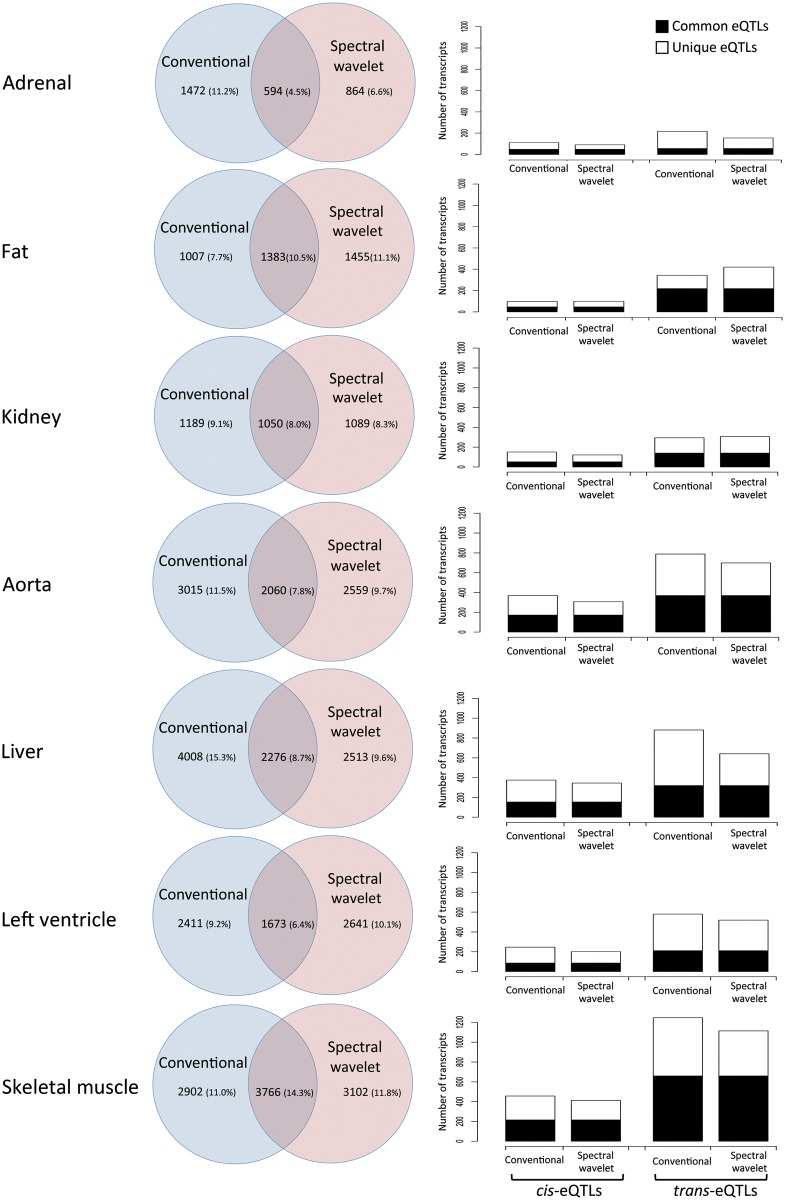
To prioritize genes associated with the regulation of blood pressure in the rat, we carried out a genome-wide correlation analysis between eleven BP parameters and gene transcript abundances in seven tissues (QTT analysis).11 Figure 4 gives the number and distribution of genes [cis-regulated, trans-regulated and not under genetic control (NGC transcripts)] that were most strongly associated with any BP phenotype across tissues (FDR <5%). Overall, we found more than four-fold more NGC transcripts associated with any BP phenotype (ranging from 18.8–28.2% of transcripts across tissues) than either cis- (1.1–2.5%) or trans-eQTLs (2.2–6.5%). This suggests that variation in gene expression that is not under genetic control in the rat may either contribute to regulation of BP traits or might reflect changes secondary to BP (Supplementary material online, Table S6). In particular, using spectral and wavelet analysis we derived new BP parameters (Table 1), which significantly correlated with variation in gene expression across tissues similarly to that observed for standard BP indices (Figure 5). A large proportion of genes (37–59%, across tissues) were uniquely associated with the newly derived BP parameters (i.e. spectral and wavelet phenotypes), suggesting that these traits reflect different aspects of BP variation that was not captured by standard BP indices. Although we found more trans-eQTL genes associated with BP phenotypes overall, no differences were observed between the relative proportions of cis- and trans-eQTLs associated with standard and spectral/wavelet-derived BP traits (Figure 5).
Figure 4.
Rat genes associated with blood pressure phenotypes. For each tissue the number (and %) of transcripts that were significantly associated with each BP phenotype by QTT analysis is reported (FDR <5%). Eleven BP phenotypes were grouped as ‘conventional’ (diastolic blood pressure, DBP; mean blood pressure, MBP, peak-to-peak distance, PDIST; heart rate, HR; pulse pressure, PP; and systolic blood pressure, SBP), ‘spectral’ (maximum frequency, FREQ; total power, POWER, and maximum spectra, SPEC), and ‘wavelet’ (wave amplitude, WAVE and wave frequency, WFREQ).
Figure 5.
Rat genes that associated with conventional and newly derived (spectral and wavelet) blood pressure traits. Left, for each tissue the Venn diagrams give the number of transcripts that significantly correlated with the ‘conventional’ phenotypes (DBP, HR, MBP, PDIST, PP, and SBP) when compared with the ‘spectral’ and ‘wavelet’ phenotypes (FREQ, POWER, SPEC, WAVE, and WFREQ). The percentages are given with respect to the total number of transcripts analysed and are corrected for the number for probesets present in the Affymetrix microarrays. Right, number of cis- and trans-eQTLs that across tissues were associated with ‘conventional’ and ‘spectral/wavelet’ BP phenotypes, respectively. ‘Common eQTLs’ indicate eQTLs that were associated with both ‘standard’ and ‘spectral/wavelet’ BP-phenotypes, whereas ‘unique eQTLs’ indicate eQTLs that were specifically associated with either ‘conventional’ or ‘spectral/wavelet’ BP phenotypes.
3.4. Widespread conservation of eQTL genes between the rat and human hypertension
Genes that associated with cardiovascular traits in the rat, when conserved across species, can have similar functional consequences on related human phenotypes and disease.12,13 Here, have performed a systematic investigation of all eQTL genes and transcripts analysed in seven tissues in the context of BP variation in the rat. We then carried out comparative genomics analysis between rat and human genes, focusing on human genes that were associated with hypertension in human GWAS. To this aim we compiled and annotated all genetic associations from 15 publicly available human GWAS and meta-analyses of hypertension and BP (see Section 2 and Supplementary material online, Table S7). We first explored whether rat eQTL genes whose expression correlated with BP variation were conserved between the rat and human genes associated with hypertension by GWAS (Supplementary material online, Table S8). Out of 2292 rat genes that were orthologue to human genes identified by GWAS, 795 (35%) were associated with BP phenotypes in the rat (FDR <5%, Supplementary material online, Table S9). Five hundred and ten (64%) of these genes formed eQTLs in the rat and were regulated in cis (n = 51), trans (n = 342), or showed both cis- and trans-regulation across tissues (n = 117). Overall, we found about seven-fold more trans- than cis-eQTLs that correlated with BP variation in the rat and that in human were associated (marginally or significantly) with hypertension in GWAS. Functional annotation analysis revealed that BP-associated trans-eQTLs were most highly enriched for ‘calcium signalling pathway’ (P = 9.6 × 10−6) and ‘ion channel’ related functions (P = 3.5 × 10−7), possibly reflecting the downstream effect of primary hypertension genes on pathophysiological pathways.
We then investigated to what extent rat gene sets (cis-, trans-regulated, and NGC-transcripts) were significantly enriched among the rat–human orthologous genes associated with hypertension by GWAS (Figure 1). In contrast with NGC transcripts, eQTLs were significantly over-represented in the rat–human orthologous genes (enrichment P < 0.017), irrespective of whether they associated with BP traits in the rat or the tissue where they were expressed (Table 2). Specifically, both cis-eQTLs (14.4%, enrichment P = 0.018) and trans-eQTLs (13.6%, enrichment P = 0.021) were similarly over-represented in the human GWAS results more than what expected by chance. We also looked at eQTLs that overlapped within known BP-QTLs in the rat (n = 3713 eQTLs; Supplementary material online, Table S3) and found a significant over-representation for these genes in the human GWAS data (enrichment P = 2 × 10−3).
Table 2.
Enrichment of rat genes in the rat–human orthologous genes that were genetically associated with hypertension in human GWAS
| Number of rat genes found in the rat–human orthologous gene seta (%, enrichment P-valueb) |
||
|---|---|---|
| Rat gene set | Genes profiled by microarrayc | Genes associated with any rat BP traitd |
| Genes not under genetic control in any tissue | 405 (10.8%, P = 1) | 289 (10.7%, P = 1) |
| Genes under genetic control (eQTLs)e | 1314 (13.4%, P = 0.016) | 506 (14.2%, P = 0.017) |
| Genes regulated in cis | 354 (14.4%, P = 0.018) | 129 (14.4%, P = 0.12) |
| Genes regulated in trans | 1128 (13.6%, P = 0.021) | 406 (14.2%, P = 0.017) |
| Clusters of genes (n > 10) regulated in transf | 419 (13.2%, P = 0.37) | 176 (13.4%, P = 0.36) |
| Genes underlying trans-eQTL clustersg | 16 (28.6%, P = 5 × 10−4) | — |
aRat–human orthologous genes (n = 2292) that were genetically associated with hypertension in human GWAS using a relaxed ‘discovery scan’ P-value threshold of P < 0.001 (see Section 2 and Supplementary material online, Table S9).
bEmpirical P-values of significance for the observed enrichment were assessed using 10 000 permutations.
cRat genes that were profiled by microarray analysis and were expressed across seven tissues (see Section 2).
dRat genes that were significantly associated (FDR <5%) with any BP phenotype by QTT analysis (reported in Figure 4).
eRat genes that mapped as eQTLs in at least one tissue (reported in Figure 2).
fClusters of trans-eQTLs (n = 156, comprising a total 3158 unique genes) were defined as a group of 10 or more genes mapping to the same locus (reported in Figure 3 and Supplementary material online, Table S3).
gGene closest to each SNP defining the trans-eQTL cluster locus, which was defined here as a 2 kb region centred around the SNP. These genes represent the putative ‘master regulators’ for the trans-clusters and are detailed in Table 3. Similar significant enrichment was found for putative ‘master regulator’ genes located within 20 kb and 2 Mb regions centred around the trans-eQTL cluster loci (data not shown).
Although clusters of trans-eQTLs as a whole were not significantly over-represented in human GWAS, 22 individual clusters showed significant over-representation (enrichment P < 0.05), two of these overlapped with known BP-QTLs in the rat and seven clusters contained at least 50% of genes significantly associated with BP traits in the rat (FDR<5%). The most important over-representation of rat genes in human GWAS results was observed for the putative ‘master regulators’ that underlie trans-eQTL cluster loci: 16 out of 56 ‘master regulator’ genes were found to be associated to hypertension in human GWAS, representing a significant enrichment over what expected by chance (28.6%; enrichment P = 5 × 10−4, Table 2). Three of these genes (Dpysl5, Bcat1, and D3ZCG8_RAT) were located at trans-eQTL clusters (aorta6_25, skm4_182, skm7_90, Figure 3) comprising genes that were over-represented among the human GWAS gene set (enrichment P < 0.05, Table 3). These analyses identified trans-eQTL clusters and their putative ‘master regulators’ that are conserved between rats and humans, and which are likely to play a role in human hypertension as primary and secondary determinants, respectively.
Table 3.
Rat genes underlying trans-eQTLs clusters and that have a human orthologous gene associated with hypertension by GWAS
| Trans-eQTLs cluster (number of genes)a | GO enrichment for the trans-eQTLs cluster genes (P-value)b | P-value of enrichment of the trans-eQTL cluster genes in human GWAS datac | Rat gene underlying trans-eQTL cluster (within 2 kb window) | Significantly correlated blood pressure phenotype in the rat (FDR <5%) | Human orthologue gene symbol | Human orthologue gene name | GWAS SNPd | Gene Section | GWAS P-valuee | PMID for the GWAS |
|---|---|---|---|---|---|---|---|---|---|---|
| kid4_90 (n = 12) | — | 0.07 | Fam190a | — | FAM190A | Family with sequence similarity 190, member A | rs17018584 | 3′UTR | 2.45 × 10−11 | 17554300 |
| skm7_90 (n = 10) | — | 0.04 | D3ZCG8_RAT | — | SAMD12 | Sterile alpha motif domain containing 12 | rs17829808 | 20kb from TSS | 4.60 × 10−7 | 17554300;17903302 |
| skm4_182 (n = 25) | — | 0.02 | Bcat1 | MBP, SBP | BCAT1 | Branched chain amino-acid transaminase 1, cytosolic | rs7961152 | Intron | 7.39 × 10−6 | 17554300 |
| aorta19_12 (n = 10) | GO:0001890∼placenta development (P = 7.5 × 10−4) | 0.19 | Large | — | LARGE | Lke-glycosyltransferase | rs2277840 | Intron | 1.42 × 10−5 | 17554300;21909115 |
| skm7_87 (n = 14) | — | 0.10 | Trps1 | MBD, DBP | TRPS1 | Trichorhinophalangeal syndrome I | rs16887447 | Intron | 5.79 × 10−5 | 17554300 |
| fat1_154 (n = 10) | — | 0.20 | Odz4 | — | ODZ4 | odz, odd Oz/ten-m homolog 4 (Drosophila) | rs648443 | Intron | 3.50 × 10−4 | 17554300;21909115 |
| lv6_1 (n = 10) | — | 0.20 | Vit | — | VIT | Vitrin | rs1468811 | Intron | 3.64 × 10−4 | 17554300 |
| aorta2_172 (n = 12) | — | 0.27 | Pdgfc | SPEC | PDGFC | Platelet-derived growth factor C | rs17230544 | ncRNA | 3.83 × 10−4 | 17463246 |
| aorta2_234 (n = 153) | GO:0030029∼actin filament-based process (P = 7.7 × 10−6) | 0.38 | Ppp3ca | — | PPP3CA | Protein phosphatase 3, catalytic subunit, alpha isozyme | rs2850341 | Intron | 3.96 × 10−4 | 17554300 |
| skm17_92 (n = 35) | — | 0.08 | Myo3a | — | MYO3A | Myosin IIIA | rs982082 | Intron | 4.91 × 10−4 | 21909115 |
| liver1_149 (n = 30) | GO:0005739∼mitochondrion (P = 3.8 × 10−5) | 0.24 | Dlg2 | — | DLG2 | Discs, large homolog 2 (Drosophila) | rs17146788 | Intron | 5.36 × 10−4 | 17463246;17554300;21909115 |
| lv8_28 (n = 31) | GO:0009057∼macromolecule catabolic process (P = 2.6 × 10−3) | 0.93 | Opcml | — | OPCML | Opioid binding protein/cell adhesion molecule-like | rs11223077 | Intron | 5.50 × 10−4 | 17554300;21909115 |
| skm8_28 (n = 19) | — | 0.20 | Opcml | — | OPCML | Opioid-binding protein/cell adhesion molecule-like | rs11223077 | Intron | 5.50 × 10−4 | 17554300;21909115 |
| aorta6_25 (n = 15) | — | 0.03 | Dpysl5 | — | DPYSL5 | Dihydropyrimidinase-like 5 | rs827884 | Intron | 6.67 × 10−4 | 17554300 |
| lv3_161 (n = 10) | — | 0.57 | Cyp24a1 | — | CYP24A1 | Cytochrome P450, family 24, subfamily A, polypeptide 1 | rs6022994 | Intron | 6.91 × 10−4 | 17554300 |
| aorta4_10 (n = 12) | GO:0006955∼immune response (P = 4.4 × 10−5) | 0.64 | Magi2 | — | MAGI2 | Membrane-associated guanylate kinase, WW and PDZ domain containing 2 | rs17150624 | Intron | 7.12 × 10−4 | 17554300;21909115 |
aClusters of trans-eQTLs were defined as a group of 10 or more genes mapping to the same locus (reported in Figure 3 and Supplementary material online, Table S3). Clusters detected in multiple tissues are highlighted with bold fonts.
bSignificance for gene ontology (GO) enrichment reported with FDR<5%.
cEnrichment of rat trans-eQTLs cluster genes in the rat–human orthologous genes that were genetically associated with hypertension by GWAS (P < 0.001).
dSNP from the GWAS which were reported to associated with hypertension.
eSignificance of the genetic association with hypertension reported in the GWAS.
4. Discussion
In this study we have shown that genes associated with hypertension in human GWAS, when conserved with the rat, are likely to form both cis- and trans-acting eQTLs in multiple tissues. Our analysis of genetic regulation of gene expression and BP in the rat suggested that BP-associated genes are more likely to exert downstream effects on gene expression in trans. This emphasizes the role of secondary regulatory mechanisms in disease susceptibility, which are usually undetectable and unappreciated in human GWAS.
We used the HXB/BXH rat RI strains, a well-characterized model of hypertension, to assess the genetic regulation of gene expression in tissues important for the primary regulation of blood pressure or that possess an intrinsic capability to regulate the local blood flow (adrenal, kidney, and aorta). Likewise, we investigated genome-wide expression in other four tissues (liver, skeletal muscle, heart, and fat) that may respond to differences in blood pressure. Using a large data set of ∼27 000 eQTLs we observed that, in addition to cis-regulated genes correlating with BP traits, trans-eQTLs can be associated with BP variation. Clustering of trans-acting eQTLs forming ‘hot spots’38 has been reported in several model organisms including yeast,39 drosophila,40 mice,41 rat,15,37 and suggested to be important in humans as well.42,43 Although clusters of trans-regulated genes can harbour genes directly associated with several traits, including metabolic44 and autoimmune phenotypes,9 these have commonly smaller effect sizes than cis-regulated genes,45 hence are more difficult to detect in model organisms and human GWAS. Here, taking advantage of powerful Bayesian eQTL mapping methods,19 we identified a larger set of trans-eQTLs and trans-eQTL clusters, where many clusters were replicated across tissues (Figure 3). Thirteen clusters (8%) were found at loci that overlap with known BP QTLs identified in SHR or SHR/BN strain combinations (Supplementary material online, Table S3), suggesting a role for these genes in blood pressure regulation in the rat.
To explore on a broader scale the contribution of eQTL genes to hypertension and identify candidate genes, we used radiotelemetric BP measurements and derived 11 parameters of BP variation, which were assessed by time series, spectral, and wavelet analyses (Table 1). Within in each tissue, ∼30% of analysed genes were associated with any BP parameter (Figure 4) and >8000 eQTLs, cis- and trans-acting, were associated with BP variation, identifying primary or secondary candidates for the regulation of blood pressure in the rat. Using wavelet and spectral analysis, we generated new, heritable traits for BP variation, which captured variation of cardiovascular functions and circadian BP that was not recapitulated by the ‘conventional’ BP parameters. This allowed for the identification of 2042 rat genes whose expression associated only with the new BP phenotypes (FREQ, POWER, SPEC, WAVE, and WFREQ) across tissues (Supplementary material online, Table S6).
We went on and investigated whether cis-, trans-eQTLs, trans-eQTL clusters and their putative ‘master regulators’ are conserved between rats and humans and associated with hypertension. Using human GWAS data, we tested whether rat gene sets were more likely to be over-represented in human genes associated with hypertension and blood pressure than by chance (Figure 1). Following Johnson and O'Donnell25 we adopted a relaxed ‘discovery scan’ P-value threshold of P < 0.001 to create a sizeable set of human association results from 15 large GWAS studies, permitting rat gene sets to be formally tested for over-representation (see Section 2). This strategy allowed us to compile a large database of human hypertension genes (n = 4145; Supplementary material online, Table S7), and the choice of a relaxed ‘discovery scan’ P-value threshold enabled an even representation of association results from all studies, including those that released complete GWAS data (i.e. results not filtered for genome-wide significance, P = 10−8).25 In addition to highly significant (and replicated) GWAS hits, we explicitly aimed to use the genetic association signal of small effect (i.e. detected with low significance, P < 0.001) by GWAS, which can shed light on trans-regulatory mechanisms underlying disease, as previously demonstrated.9
Although we detected substantially more NGC transcripts (more than four-fold) associated with any BP phenotype in the rat than either cis- or trans-eQTLs, we found a significant over-representation of genes in the human GWAS results only for genes that in the rat formed eQTLs (Table 2). This observation is in keeping with recent data on annotation of human eQTL from lymphoblastoid cell lines that are enriched for trait-associated SNPs in GWAS studies.46 Intriguingly, the group of cis-regulated genes that associated with BP parameters in the rat (when considered altogether) was less over-represented within human GWAS hits than BP-associated trans-eQTLs. Similar over-representation of trans-eQTLs has been reported for other complex traits, as shown in recent studies of type 2 diabetes in humans, which revealed marked excess of trans-acting eQTLs within the top association signals.47 The 2859 trans-eQTLs that associated with BP in the rat might be representative of ‘pathogenic pathways’ that can also play a role in human hypertension. Functional annotation of the BP-associated trans-eQTLs that were conserved across species showed the most significant enrichment for ‘calcium signalling pathway’ genes (P = 9.6 × 10−6), reflecting an over-representation for ion transporters, sodium/potassium voltage channels, and glutamate receptors in this gene set. This observation is in keeping with several genetic, pathophysiological, and pharmacological studies providing evidence that ion channel functions are important in regulating BP levels,48,49 and suggest a role for conserved trans-regulated rat genes as secondary determinants of BP and hypertension.
Although strong enrichment was observed for the set of eQTLs that overlapped within known BP-QTLs in the rat (enrichment P = 2 × 10–3), the most suggestive result was found for the set of putative ‘master regulators’. Sixteen genes (out of 56) that were located at the trans-cluster loci were conserved to humans and detected in GWAS, representing a significant enrichment of what expected by chance (enrichment P = 5 × 10−4, Table 3). Since the putative ‘master regulator’ genes might regulate (in trans) clusters or networks of genes in the rat, these are likely to be primary (up-stream) regulators of pathways for blood pressure and hypertension. These ‘master regulator’ genes in humans were associated with hypertension with different levels of significance (GWAS P-value range from 10−4 to 10−11), with the strongest association signal for the FAM190A gene (GWAS P-value = 2.4 × 10−4).2 In the rat, Fam190a underlies a trans-eQTL cluster in the kidney tissue, which is moderately but not significantly enriched for human GWAS signals (enrichment P = 0.07, Table 3). Among the trans-regulated genes of this cluster, potassium voltage-gated channel, shaker-related subfamily, member 5 (KCNA5) has been implicated with pulmonary hypertension,50,51 and atrial fibrillation.52 The second strongest GWAS signal within the set of putative ‘master regulators’ in humans was observed for sterile alpha motif domain containing 12 (SAMD12) (GWAS P-value = 4.6 × 10−7),53 which in the rat lies at a trans-eQTL cluster that is enriched for genetic associations in human hypertension (enrichment P = 0.04, Table 3). Another putative ‘master regulator’ rat gene that exerts a function in human cardiovascular diseases is branched chain amino-acid transaminase 1, cytosolic or BCAT1. BCAT1 was previously reported to be associated with hypertension (GWAS P-value = 7.4 × 10−6),2 as well as other cardiovascular traits, including salt sensitivity54 and resting heart rate.55 Rat Bcat1 resides at the locus regulating in trans 25 genes in skeletal muscle tissue, where 5 of these (Fstl1, Mlf1, A2bp1, Tyr, and Anxa5) were associated with hypertension in human GWAS, and Bcat1 skeletal muscle expression significantly correlated with MBP and SBP in the rat.
Genetically regulated changes in gene expression can determine phenotypic variation and susceptibility to disease. Cis-acting genetic control of gene expression has been widely investigated since it can be detected with relatively higher confidence than trans-acting control, and cis-acting eQTL genes can have large effects on whole body phenotypes and disease.39 In contrast, the specific role of trans-regulated genes in the context of disease susceptibility has been explored to a less extent. Our data support the hypothesis that secondary regulatory mechanisms of gene expression (trans-eQTLs) are largely conserved between rats and human disease. Extensive comparative genomics analysis of GWAS data showed that human hypertension genes, when conserved with the rat, are enriched for trans-eQTLs that associated with BP variation across multiple tissues. This most likely reflects the downstream effect of primary disease-associated genes on secondary regulatory mechanisms important for the pathogenesis of hypertension. These findings can help to elucidate the functional context and pathways mediating the effect of primary susceptibility genes identified in human GWAS of hypertension, highlighting the important contribution of trans-regulatory mechanisms for disease pathogenesis.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
We acknowledge funding from European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement no. HEALTH-F4-2010-241504 (EURATRANS) (E.P., T.J.A., S.A.C); the Medical Research Council [(E.P., T.J.A., S.A.C., L.B. (G1002319)]; the Wellcome Trust (S.R.L., L.B.); the Imperial BHF Centre of Excellence (T.J.A.); the Leduq Transatlantic Network of Excellence (T.J.A., S.A.C.). M.P. was supported by grants 7E10067, LH11049 and grant LL1204 (within the ERC CZ program) from the Ministry of Education, Youth, and Sports of the Czech Republic.
Supplementary Material
Acknowledgements
We thank the anonymous Reviewers for their critical and constructive comments on the manuscript.
Conflict of interest: none declared.
References
- 1.Staessen J, Wang J, Bianchi G. Essential hypertension. Lancet. 2003;361:1629–1641. doi: 10.1016/S0140-6736(03)13302-8. [DOI] [PubMed] [Google Scholar]
- 2.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rafiq S, Anand S, Roberts R. Genome-wide association studies of hypertension: have they been fruitful? J Cardiovasc Transl Res. 2010;3:189–196. doi: 10.1007/s12265-010-9183-9. [DOI] [PubMed] [Google Scholar]
- 4.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Gen. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, et al. Genome-wide association study of blood pressure and hypertension. Nat Gen. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.International Consortium for Blood Pressure Genome-Wide Association Studies. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wain LV, Verwoert GC, O'Reilly PF, Shi G, Johnson T, Johnson AD, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Gen. 2011;43:1005–1011. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jansen R. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- 9.Heinig M, Petretto E, Wallace C, Bottolo L, Rotival M, Lu H, et al. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature. 2010;467:460–464. doi: 10.1038/nature09386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fehrmann RSN, Jansen RC, Veldink JH, Westra H-J, Arends D, Bonder MJ, et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet. 2011;7:e1002197. doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Passador-Gurgel G, Hsieh W-P, Hunt P, Deighton N, Gibson G. Quantitative trait transcripts for nicotine resistance in Drosophila melanogaster. Nat Gen. 2007;39:264–268. doi: 10.1038/ng1944. [DOI] [PubMed] [Google Scholar]
- 12.Petretto E, Sarwar R, Grieve I, Lu H, Kumaran MK, Muckett PJ, et al. Integrated genomic approaches implicate osteoglycin (Ogn) in the regulation of left ventricular mass. Nat Gen. 2008;40:546–552. doi: 10.1038/ng.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pravenec M, Churchill PC, Churchill MC, Viklicky O, Kazdova L, Aitman TJ, et al. Identification of renal Cd36 as a determinant of blood pressure and risk for hypertension. Nat Gen. 2008;40:952–954. doi: 10.1038/ng.164. [DOI] [PubMed] [Google Scholar]
- 14.Pravenec M, Klír P, Kren V, Zicha J, Kunes J. An analysis of spontaneous hypertension in spontaneously hypertensive rats by means of new recombinant inbred strains. J Hyperten. 1989;7:217–221. [PubMed] [Google Scholar]
- 15.Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, Maciver F, et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Gen. 2005;37:243–253. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- 16.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 17.Kwiatkowski D, Phillips PCB, Schmidt P, Shin Y. Testing the null hypothesis of stationarity against the alternative of a unit root: how sure are we that economic time series have a unit root? J Econometrics. 1992;54:159–178. [Google Scholar]
- 18.Opgen-Rhein R, Strimmer K. From correlation to causation networks: a simple approximate learning algorithm and its application to high-dimensional plant gene expression data. BMC Sys Biol. 2007;1:37. doi: 10.1186/1752-0509-1-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petretto E, Bottolo L, Langley SR, Heinig M, McDermott-Roe C, Sarwar R, et al. New insights into the genetic control of gene expression using a Bayesian multi-tissue approach. PLoS Comp Biol. 2010;6:e1000737. doi: 10.1371/journal.pcbi.1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.STAR Consortium. SNP and haplotype mapping for genetic analysis in the rat. Nat Gen. 2008;40:560–566. doi: 10.1038/ng.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Gen. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen W, Ghosh D, Raghunathan TE, Sargent DJ. A false-discovery-rate-based loss framework for selection of interactions. Stat Med. 2008;27:2004–2021. doi: 10.1002/sim.3118. [DOI] [PubMed] [Google Scholar]
- 23.Westfall P. Resampling-based Multiple Testing: Examples and Methods for P-value Adjustment. New York: Wiley; 1993. [Google Scholar]
- 24.Dennis G, Jr, Sherman B, Hosack D. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:R60.1–R60.11. [PubMed] [Google Scholar]
- 25.Johnson AD, O'Donnell CJ. An open access database of genome-wide association results. BMC Med Genet. 2009;10:6. doi: 10.1186/1471-2350-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, et al. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 2009;5:e1000564. doi: 10.1371/journal.pgen.1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fox ER, Young JH, Li Y, Dreisbach AW, Keating BJ, Musani SK, et al. Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Hum Mol Genet. 2011;20:2273–2284. doi: 10.1093/hmg/ddr092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Org E, Eyheramendy S, Juhanson P, Gieger C, Lichtner P, Klopp N, et al. Genome-wide scan identifies CDH13 as a novel susceptibility locus contributing to blood pressure determination in two European populations. Hum Mol Genet. 2009;18:2288–2296. doi: 10.1093/hmg/ddp135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban H-J, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Gen. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 30.Lettre G, Palmer CD, Young T, Ejebe KG, Allayee H, Benjamin EJ, et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Padmanabhan S, Melander O, Johnson T, Di Blasio AM, Lee WK, Gentilini D. Genome-Wide Association Study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet. 2010;6:e1001177. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabatti C, Service SK, Hartikainen A-L, Pouta A, Ripatti S, Brodsky J, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Gen. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong K-W, Jin H-S, Lim J-E, Kim S, Go MJ, Oh B. Recapitulation of two genomewide association studies on blood pressure and essential hypertension in the Korean population. J Hum Genet. 2010;55:336–341. doi: 10.1038/jhg.2010.31. [DOI] [PubMed] [Google Scholar]
- 34.Good PI. Permutation, Parametric and Bootstrap Tests of Hypotheses. 3rd edn. New York: Springer; 2005. [Google Scholar]
- 35.Basset A, Laude D, Laurent S, Elghozi J-L. Contrasting circadian rhythms of blood pressure among inbred rat strains: recognition of dipper and non-dipper patterns. J Hypertens. 2004;22:727–737. doi: 10.1097/00004872-200404000-00015. [DOI] [PubMed] [Google Scholar]
- 36.Melek WW, Lu Z, Kapps A, Fraser WD. Comparison of trend detection algorithms in the analysis of physiological time-series data. IEEE Trans Biomed Eng. 2005;52:639–651. doi: 10.1109/TBME.2005.844029. [DOI] [PubMed] [Google Scholar]
- 37.Petretto E, Mangion J, Dickens NJ, Cook SA, Kumaran MK, Lu H, et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genetics. 2006;2:e172. doi: 10.1371/journal.pgen.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Breitling R, Li Y, Tesson BM, Fu J, Wu C, Wiltshire T, et al. Genetical genomics: spotlight on QTL hotspots. PLoS Genet. 2008;4:e1000232. doi: 10.1371/journal.pgen.1000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, et al. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Gen. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 40.Ruden DM, Chen L, Possidente D, Possidente B, Rasouli P, Wang L, et al. Genetical toxicogenomics in Drosophila identifies master-modulatory loci that are regulated by developmental exposure to lead. Neurotoxicology. 2009;30:898–914. doi: 10.1016/j.neuro.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu C, Delano DL, Mitro N, Su SV, Janes J, McClurg P, et al. Gene set enrichment in eQTL data identifies novel annotations and pathway regulators. PLoS Genet. 2008;4:e1000070. doi: 10.1371/journal.pgen.1000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KCC, et al. A genome-wide association study of global gene expression. Nat Gen. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 44.Small KS, Hedman AK, Grundberg E, Nica AC, Thorleifsson G, Kong A, et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Gen. 2011;43:561–564. doi: 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilad Y, Rifkin SA, Pritchard JK. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24:408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Below JE, Gamazon ER, Morrison JV, Konkashbaev A, Pluzhnikov A, McKeigue PM, et al. Genome-wide association and meta-analysis in populations from Starr County, Texas, and Mexico City identify type 2 diabetes susceptibility loci and enrichment for expression quantitative trait loci in top signals. Diabetologia. 2011;54:2047–2055. doi: 10.1007/s00125-011-2188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cowley AW. The genetic dissection of essential hypertension. Nat Rev Gen. 2006;7:829–840. doi: 10.1038/nrg1967. [DOI] [PubMed] [Google Scholar]
- 49.Martens JR, Gelband CH. Ion channels in vascular smooth muscle: alterations in essential hypertension. Proc Soc Exp Biol Med. 1998;218:192–203. doi: 10.3181/00379727-218-44286. [DOI] [PubMed] [Google Scholar]
- 50.Remillard CV, Tigno DD, Platoshyn O, Burg ED, Brevnova EE, Conger D, et al. Function of Kv1.5 channels and genetic variations of KCNA5 in patients with idiopathic pulmonary arterial hypertension. Am J Physiol. 2007;292:C1837–53. doi: 10.1152/ajpcell.00405.2006. [DOI] [PubMed] [Google Scholar]
- 51.Pozeg ZI, Michelakis ED, McMurtry MS, Thébaud B, Wu X-C, Dyck JRB, et al. In vivo gene transfer of the O2-sensitive potassium channel Kv1.5 reduces pulmonary hypertension and restores hypoxic pulmonary vasoconstriction in chronically hypoxic rats. Circulation. 2003;107:2037–2044. doi: 10.1161/01.CIR.0000062688.76508.B3. [DOI] [PubMed] [Google Scholar]
- 52.Yang T, Yang P, Roden DM, Darbar D. Novel KCNA5 mutation implicates tyrosine kinase signaling in human atrial fibrillation. Heart Rhythm. 2010;7:1246–1252. doi: 10.1016/j.hrthm.2010.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simino J, Shi G, Arnett D, Broeckel U, Hunt SC, Rao DC. Variants on Chromosome 6p22.3 associated with blood pressure in the HyperGEN Study: follow-Up of FBPP quantitative trait loci. Am J Hypertens. 2011;24:1227–1233. doi: 10.1038/ajh.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rhee M-Y, Yang SJ, Oh SW, Park Y, Kim C-I, Park H-K, et al. Novel genetic variations associated with salt sensitivity in the Korean population. Hypertens Res. 2011;34:606–611. doi: 10.1038/hr.2010.278. [DOI] [PubMed] [Google Scholar]
- 55.Eijgelsheim M, Newton-Cheh C, Sotoodehnia N, deBakker PIW, Müller M, Morrison AC, et al. Genome-wide association analysis identifies multiple loci related to resting heart rate. Hum Mol Genet. 2010;19:3885–3894. doi: 10.1093/hmg/ddq303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.