Abstract
The conventional reductionist approach to cardiovascular research investigates individual candidate factors or linear signalling pathways but ignores more complex interactions in biological systems. The advent of molecular profiling technologies that focus on a global characterization of whole complements allows an exploration of the interconnectivity of pathways during pathophysiologically relevant processes, but has brought about the issue of statistical analysis and data integration. Proteins identified by differential expression as well as those in protein–protein interaction networks identified through experiments and through computational modelling techniques can be used as an initial starting point for functional analyses. In combination with other ‘-omics’ technologies, such as transcriptomics and metabolomics, proteomics explores different aspects of disease, and the different pillars of observations facilitate the data integration in disease-specific networks. Ultimately, a systems biology approach may advance our understanding of cardiovascular disease processes at a ‘biological pathway’ instead of a ‘single molecule’ level and accelerate progress towards disease-modifying interventions.
Keywords: Proteins, Metabolites, Mass spectrometry, Systems biology, Bioinformatics
1. Introduction
Proteomics represents the large-scale analysis of proteins, particularly their structures and functions. The term ‘proteomics’ was coined to make an analogy with genomics, the study of the genome. Although the genome is just the ‘blueprint’ of the proteins, the proteins execute cellular function. Importantly, the transcriptome is not linearly proportional to proteome and many human diseases result from alterations in the proteome. In the first part of this review, we provide a short summary of proteomics techniques that have been extensively reviewed elsewhere.1–3 Knowing the major limitations and advantages of the different proteomic techniques is essential for their successful application. An overview of systems biology approaches and examples follows, along with some of the resources available. Computational methods for dealing with the unique challenges of proteomics data will be key to fulfilling the promise of systems biology.
2. Proteomics
Before summarizing different proteomic strategies (Figure 1), a few points should be emphasized4:
No proteomic technology can currently resolve the entire complexity of the mammalian proteome.
With any proteomic technique, there is bias towards more abundant proteins.
In general, there is a trade-off between how many proteins can be identified and how accurately they can be quantified.
-
Inevitably, information is lost by the propagation of quantitative peptide information to protein changes.
Table 1 gives a brief overview of the advantaged and disadvantages of the following proteomics techniques.
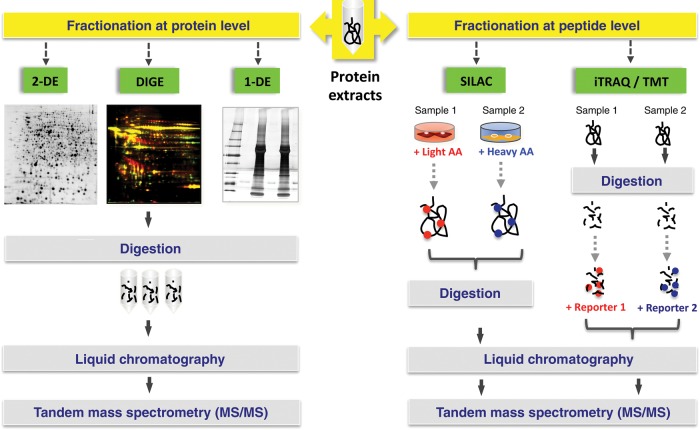
Figure 1.
Proteomic approaches. Protein extracts can either be fractionated at the protein level prior to digestion or after protein digestion at the peptide level. In DIGE, the protein extracts are labelled with different fluorescent dyes before they are separated by 2-DE. For SILAC, cells are metabolically labelled in culture by incorporation of heavy or light amino acids. Alternatively, labelling is performed at the peptide level, using iTRAQ or TMT isobaric tags. Peptides are then analysed by MS/MS. 2-DE, two-dimensional gel electrophoresis; DIGE, difference gel electrophoresis; 1-DE, one-dimensional gel electrophoresis; SILAC, stable isotope labelling with amino acids in cell culture; AA, amino acid; iTRAQ, isobaric tag for relative and absolute quantitation; TMT, tandem mass tag.
Table 1.
Comparison of proteomics methods
| Abbreviation | Full name and explanation | Advantages | Disadvantages |
|---|---|---|---|
| DIGE | Difference gel electrophoresis | Quantitation at the protein level | Low sensitivity |
| Visualization of posttranslational modifications and protein isoforms | Only the differentially expressed proteins tend to be identified by MS/MS | ||
| Good quantitative accuracy | Proteins with very high or low pI or Mw are not resolved on the gel | ||
| Gel-LC-MS/MS | Separation by SDS–PAGE before LC-MS/MS analysis | Ease of use | Prefractionation increases time requirements for MS/MS analysis |
| Prefractionation before LC-MS/MS analysis increases sensitivity | Poor quantitative accuracy in complex mixtures without peptide labelling | ||
| ‘Laddering’ as indication of proteolytic degradation | Proteins with very high or low Mw are not resolved on the gel | ||
| SILAC | Stable isotope labelling with amino acids in cell culture | Minimal experimental variation | Quantitation at the peptide level |
| Excellent quantitative accuracy | Not suitable for cells that do not proliferate in culture, i.e. cardiomyocytes | ||
| Ease of use for cells in culture that proliferate and tolerate filtered serum supplements | Metabolic labelling of animals is expensive | ||
| iTRAQ, TMT-tags | Isotopic labelling of peptides | Good quantitative accuracy | Quantitation at the peptide level |
| Can be used with tissues as well as cell cultures | Mixed MS/MS spectra will contain reporter ions from different peptides |
2.1. Two-dimensional gel electrophoresis
Two-dimensional gel electrophoresis (2-DE) allows separation of proteins based on their isoelectric point (pI) and molecular weight (Mw).5 The first dimension involves separating proteins according to their pI. A protein mixture is loaded onto a strip with an immobilized pH gradient. Once an electric field is applied, the proteins migrate to their pI, where they become zwitterionic, i.e. they lose their net charge and stop migrating (isoelectric focusing). After isoelectric focusing is complete, the immobilized pH gradient strips are transferred onto large-format gels for separation in the second dimension, where proteins are resolved according to their molecular mass by SDS–PAGE.
Unlike SDS–PAGE, 2-DE gels produce complex maps of proteomes that are visualized as discrete protein ‘spots’. Since pI and Mw are independent properties, 2-DE gels can resolve many more proteins than SDS–PAGE. Importantly, the same protein may be present in multiple spots on a gel. Shifts in pI or Mw indicate the presence of post-translational modifications, protein degradation, or protein isoforms.6,7 Protein features are visualized with Coomassie or silver staining, and differential expression between samples is determined using relative densitometric quantification. However, gel-to-gel variability can limit the quantitative accuracy and prohibit the detection of minor differences in expression.
A more sophisticated 2-DE technique is difference gel electrophoresis (DIGE, Figure 2A).8 DIGE involves fluorescent labelling of protein mixtures with Cy-dyes in order to determine relative differences in protein expression. An internal standard comprising the pooled experimental samples is included, which is representative of all samples. The sensitivity of detection of DIGE is comparable with the sensitivity of silver staining9 and the dyes are matched for pI and Mw. The main advantage of DIGE over conventional 2-DE gels is that samples can be multiplexed on the same gel, thus reducing the number of gels needed and limiting experimental variation. DIGE employed with an internal standard reliably quantifies differences as low as 10% in protein expression.10 The gels are scanned using a fluorescence scanner, which specifically measures the emission wavelength of each Cy-dye. Commercial software packages match protein features and calculate differential expression from the scanned gel images. Normalization of protein levels across gels is performed by comparing the protein ratios with the internal standard that is co-detected on each gel.
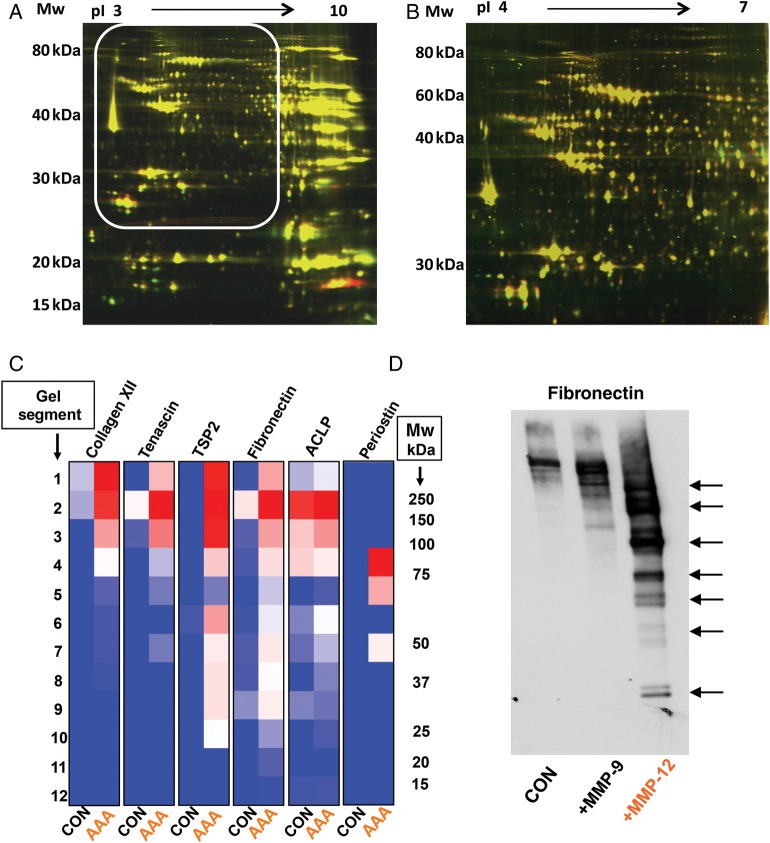
Figure 2.
Gel-based proteomics. Separation of the murine cardiac proteome by DIGE on different immobilized pH gradients: pH 3–10 NL (A) and pH 4–7 (B). The white box highlights the better resolution of the same area on the narrow pH gradient. (C) Mw distribution of six extracellular glycoproteins by SDS–PAGE. The characteristic ‘laddering’ in abdominal aortic aneurysms (AAA) compared with normal aortic tissue (CON) is indicative of proteolysis. Differences in spectral counts are color-coded (red high, blue low). (D) Incubation of healthy aortic tissues with matrix metalloproteinases-12 (MMP-12) induced a similar fragmentation pattern of fibronectin as observed in AAA. In comparison, degradation by matrix metalloproteinases-9 (MMP-9) was less pronounced (reproduced with permission from Didangelos et al.15).
Unlike other proteomic techniques, quantitation by 2-DE is performed at the protein level, not at the peptide level, and the quantitation is uncoupled from the identification by mass spectrometry (MS). Silver staining can be used to visualize protein features on a gel to facilitate excision of the relevant spots for MS. Alternatively, spots are directly picked from fluorescent gels using a robotic spot picker. Spots are then subjected to in-gel tryptic digestion before protein identification.
One of the main caveats of the 2-DE approach is that high-abundant proteins mask less-abundant proteins. This can be partially addressed by using gradients with a narrow pH range (Figure 2B). However, separation in the first dimension, in particular the transition from the first to the second dimension is not loss-free, and very large, small, and hydrophobic proteins remain difficult to resolve.
2.2. Liquid-chromatography tandem mass spectrometry
Liquid-chromatography tandem mass spectrometry (LC-MS/MS) is the current gold standard in proteomics. The basic principle of MS involves measuring the mass-to-charge ratio (m/z) of an ionized peptide and its fragmentation products. Proteins are initially digested by enzymes such as trypsin to produce peptide fragments that are easier to resolve by reverse-phase LC and ionize by electrospray MS.11 Depending on their hydrophobicity, the peptides elute at different time points from the reverse phase column (retention time). A typical workflow using LC-MS/MS involves a regular survey scan to record the masses and the intensities of the eluting peptides. The most abundant precursor ions eluting from the column are selected for fragmentation (MS/MS). The amino acid sequence information obtained from MS/MS data allows the identification of the protein. Peptide parameters, such as spectral counts, ion intensities, and chromatographic peak area, can provide a quantitative index for protein abundance (label-free quantitation).12 The versatility of mass spectrometric technology has spawned numerous different mass spectrometers, with MALDI-TOF-TOF, Q-TOF, and Orbitrap mass analysers13 being among the common ones currently in use for discovery proteomics.
2.3. Gel-LC-MS/MS analysis
Pre-fractionation by SDS–PAGE prior to MS has proved useful in the characterization of samples that are not amenable to separation by 2-DE. It also helps to overcome the single greatest cause of bias against low-abundant proteins—the stochastic under sampling of low-abundant peptides that arises because high-abundant peptides dominate the duty cycle of the mass spectrometer. For gel-LC-MS/MS analysis, proteins are separated by SDS–PAGE, the entire gel lane is divided into a series of bands, the bands are excised without leaving empty gel pieces behind, digested with trypsin, and LC-MS/MS analysis is performed on each of the bands.14,15 Since gel bands tend to be mixtures of proteins, LC separation is essential for protein identification and quantitation, i.e. by spectral counting.16 Spectral counting has become a popular strategy to quantitate relative protein abundance but is less reliable for complex mixtures. Generally, the more abundant a protein, the more likely it is detected by MS/MS. The spectral counts are derived from the number of MS/MS spectra corresponding to a particular protein.
In the gel-LC-MS/MS approach, information on the native Mw of a protein is preserved. If protein degradation has occurred prior to tryptic digestion, peptides are detected by MS in gel segments below the expected Mw of the native proteins (Figure 2C). Thus, verification regarding whether differentially expressed proteins are confined to the same gel bands is essential. Otherwise, a degraded protein may appear upregulated due to its characteristic ‘laddering’ on the SDS–PAGE (Figure 2D). Alternatively, protein fragments may be too small and escape detection because they migrated ahead of the buffer front. On the other hand, information on proteolytic degradation products is important and lost in conventional shotgun proteomics analysing tryptic peptides without prior separation at the protein level.
2.4. Shotgun proteomics
Apart from gel-based approaches, there are gel-free methods to quantify differences in protein expression based on peptide abundance. Although these shot-gun proteomic methods can mine deeper into the proteome, problems arise with quantitation if samples are too complex. MS is not inherently quantitative because of differences in the ionization efficiency. The most abundant ions will attract the most charges during electrospray ionization, making it less likely for low-level peptides to get ionized. To avoid false-positive protein changes due to co-eluting high-abundant peptides, labelling techniques should be used for reliable quantitation. Popular labelling methods include isobaric tagging for relative and absolute quantification (iTRAQ), tandem mass tags (TMT), and stable isotope labelling by amino acids in cell culture (SILAC).17 iTRAQ is currently available as four-plex and eight-plex, allowing the relative quantification of up to eight samples, whereas labelling of TMT and SILAC can been used with six and three samples, respectively.18 However, peptides are just a surrogate measure and not always reliable for protein quantitation, i.e. if they are subject to post-translational modifications or proteolysis.
2.4.1. Stable isotope labelling by amino acids in cell culture
SILAC makes use of non-radioactive isotope labels to label proteins with light (e.g. 12C) and heavy isotopes (e.g. 13C).18 Samples can be multiplexed and analysed during the same MS run, thereby minimizing experimental error.19 The SILAC pairs co-elute during chromatography but the corresponding peptides of the heavy and light isoform appear with a characteristic mass shift. The relative quantity of each protein can be calculated by the differences in the peak intensities of SILAC-labelled peptides. The use of SILAC to quantify differential levels of proteins goes beyond using cells in culture. SILAC-labelled mice have been described with near-complete labelling of all proteins, although the SILAC diet is expensive.20 Metabolic labelling also introduces information on amino acid synthesis and sourcing, protein assembly, and turnover kinetics.
2.4.2. Isobaric tagging for relative and absolute quantification/tandem mass tags
In instances where human tissue is used, iTRAQ or TMT is an option for multiplexing clinical samples for differential expression studies by LC-MS/MS,21 but these techniques are not without caveats.22 (i) One disadvantage of the iTRAQ and the TMT system over SILAC is the fact that labelling is performed at the peptide level and occurs late in the experimental process. Before labelling, proteins are first extracted from cells or tissues and digested to peptides. This is a potential source of variation. (ii) Unlike SILAC, quantitation is performed at the MS/MS level, not at the MS level. The peptides from different samples maintain their identical m/z ratios after labelling (MS). Only upon fragmentation (MS/MS), the isobaric mass tags release their different reporter ions with a single isotopic substitution per tag and provide quantitative information for each individual sample. A commonly observed problem in iTRAQ experiments is that a complex background can lead to underestimation of protein fold changes. During precursor ion selection, more than one peptide may be within the mass window selected for fragmentation. In such mixed MS/MS spectra, reporter ions originating from peptides of different proteins are erroneously combined for quantification.
2.5. Protein identification
Although accurate and accessible databases are needed for each of the ‘-omics’ fields, proteomics is perhaps the most dependent on these resources. The technologies for identifying and quantifying proteins are reliant on comprehensive databases for protein identification and peptide quantification. These databases are not directly under the scope of systems biology, but they provide a foundation for the latter analyses, as the curation and maintenance of these databases are vital for the correct identification and quantification of the examined proteins.
For functional and sequence-based databases, UniProt is one of the most comprehensive. UniProt consists of several classifications: Swiss-Prot and TrEMBL contain sequence and functional information about proteins, UniRef and UniParc contain sequence and archived sequence records and, when available, supporting data such as literature references and cross-referenced databases.23 Programs such as Mascot, SEQUEST, or X!Tandem search FASTA protein sequences obtained from public databases such as UniProt. After performing an ‘in silico fragmentation’ with known enzyme specificity, the peak mass lists with intensities (the experimental data) are searched against the in silico-fragmented database. Parent ion masses are scanned against the masses derived from the database sequences. If there is a match within a certain mass tolerance, the observed MS/MS spectra are then compared with the theoretical sequence-derived ion series. Although not explicitly covered here, the review and commentary by Noble and MacCoss24 provide insight into these methodologies and techniques. The scoring algorithms can produce different results and the reliance on single-peptide identifications in large-scale data sets is a potential cause of false identifications. Most proteomic studies only report identifications with a minimum of two unique peptides or include the MS/MS spectra for single-peptide identifications.
3. Systems biology approaches
3.1. Cellular and subcellular proteome identification
Technological advances in the past 5–10 years have made large ‘-omics’ experiments feasible, where biological changes can be assessed at the systems level (Figure 3). One can now identify and quantify the proteins present in a specific cell type or subcellular fraction. As proteins can be present at varying levels in different cellular systems, it is imperative to know the baseline measurements for cells and systems related to cardiovascular disease. In that respect, the proteomes of several cardiovascular-specific cell types have been characterized in the past few years, including human arterial smooth muscle cells (SMC),25 human early pro-angiogenic cells,26 rat cardiac stem cells and neonatal cardiomyocytes,27 and human left ventricle.28 Recently, Burkhart et al.29 characterized the proteome of human platelets within and between healthy subjects. They identified approximately 4000 unique proteins and showed that 85% of the platelet proteome did not vary across subjects. Subcellular fractions can also be informative for cardiovascular disease. Several of these fractions have been analysed, including the extracellular matrix (ECM) in human aorta,14 the mitochondrial proteome in mouse,30 and the rodent cardiac myofilament.31
Figure 3.
Computational approaches in proteomics. Bioinformatics has become an essential part of the proteomic workflow to comprehensively analyse and visualize global changes in proteins as biological networks.
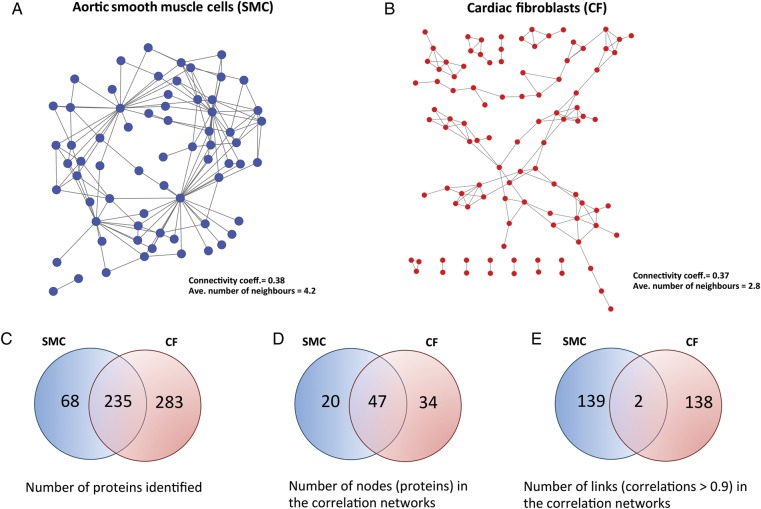
As an example, Figure 4 illustrates the proteomic network structure of ECM proteins in two different murine cells types: primary aortic SMC (Figure 4A) and cardiac fibroblasts (CF) (Figure 4B). Both are correlation networks where the links between nodes (proteins) represent correlation values >0.9. The Venn diagrams show the total number of proteins identified in the two cell types and those in common between the two (Figure 4C); the number of nodes (proteins) in the networks as well as the number of shared nodes between the two (Figure 4D); finally, the number of links (correlations >0.9) for SMC and CF as well as the ones in common between the two (Figure 4E). The two networks and the corresponding nodes and links highlight the differences in the relationship between the ECM proteins in two different cell types.
Figure 4.
Protein identification. The visualization of ECM proteins, and the corresponding correlation networks, identified by proteomics in the secretome of two murine cell types: (A) aortic SMC and (B) CF. Correlation networks have been thresholded at a correlation coefficient of >0.90. (C) Number of ECM proteins identified. Number of nodes (proteins) (D) and links (correlations) (E) found in each cell type, as well as those in common.
For a systems biology approach of cardiovascular disease, it is important to identify and quantify proteomes in different species, tissue, cellular or subcellular compartments, as the differences, shown here in two ECM-producing cell types, may be specific to the defined system. Including these proteomes in public repositories will aid further systems biology studies as the proteomics data will be available to other researchers. One of the biggest public repository of proteomics data is the PRoteomics IDEntifications (PRIDE) database supported by the European Bioinformatics Institute (EBI).32 As of the data of submission for this review, PRIDE contained over 26 000 proteomics experiments with the associated studies.
3.2. Differential protein expression analysis
With a defined system, one can study changes that result after a systematic perturbation. These perturbations can come in the form of inhibition, over-expression, incubation, or a number of other cellular manipulation techniques, but also as a comparison between normal and disease samples. The system perturbation approach is not unique to proteomics; the other ‘-omics’ fields use similar approaches. A large number of studies have been performed on transcriptomics and differential gene expression analysis and several methods have been developed. Proteomics faces similar statistical and computational consideration genomics, but there are also challenges specific to proteomics, especially label-free techniques. Several studies, including our own,14,15,33,34 have applied standard statistical methods to cardiovascular proteomics data.35–37 Although these methods may be appropriate, i.e. for the analysis of DIGE data, they are not optimal for label-free, spectral count protein expression data. Although many proteins are identified, less-abundant proteins often contain one or more missing values across samples.38 This presence/absence dichotomy is not suitable for basic imputation methods. Missing values will skew statistical tests that assume normality and standard statistical methods (Student's t-test, analysis of variance—ANOVA, linear regression) may not accurately determine differential expression.39–41 On the other hand, excluding proteins with missing spectral counts will inherently create a bias towards high-abundant proteins.39 Low-abundant proteins, however, are often informative, especially when comparing disease states, as the presence in one state and the absence in another can suggest a functional role for that protein.
The small number of replicates within an experiment reduces the robustness and increases the noise-to-signal ratio.42 Non-parametric tests, like the Wilcoxon rank-sum test, have limited power when used with the small sample sizes often found in proteomic studies.41 Applying multiple testing corrections becomes problematic, as permutation or bootstrapping techniques are not feasible with small sample sizes. With this in mind, several methods have been developed specifically for evaluating protein differential expression, which take into consideration these limitations. Some methods address the non-normal distribution properties of the data, where the data are normalized and transformed to better fit the standard statistical tests. Three of the commonly used methodologies for this approach are the Normalized Spectral Abundance Factor (NSAF),43 the Power Law Global Error Model (PLGEM),44 and the Normalized Spectral Index (SIN)45 (Table 2). These methods, however, do not take into consideration the small sample sizes that are common in proteomics experiments, nor do they directly correct for multiple testing.
Table 2.
Differential expression methods
Other methods incorporate techniques that address both the non-normal distributions and the limited number of replications. The Spectral Index (SpI),46 Qspec,47 and the hybrid approach proposed by Wang et al.39 are three examples of methods that account for small sample sizes and do not require the data to be normally distributed (Table 2). These methods also directly incorporate multiple testing corrections. Unlike gene expression microarray analyses, there is no standard method for normalization and differential expression analyses in proteomics. Owing to the variability between experiments and methodologies, statisticians and computational biologists should guide proteomic analyses.
3.3. Incorporating functional and pathway information
Functional information such as Gene Ontology (GO) and pathway resources can inform on the biological function of proteins and their interactions and on the relevance of the proteins to the disease. GO48 and Kyoto Encyclopedia of Genes and Genomes (KEGG)49 are the two resources widely used in the literature but there are other pathway and functional databases available (Table 3). The KEGG database contains manually curated pathways within five areas: metabolism, genetic information processing, environmental information processing, cellular processes, and human disease. Unlike GO, these pathways are species dependent. The GO database contains species-independent terms relating to genes and their products. There are three main classifications of ontologies, cellular component, biological process, and molecular function with several sub-classifications under each of the three. In addition, there is a GO consortium specifically focused on annotating genes relevant to cardiovascular disease (http://www.geneontology.org/GO.cardio.shtml), and, to date, has identified over 4000 genes with a cardiovascular disease association. The GO Cardiovascular Consortium also annotates gene products, including proteins and microRNAs. Instead of a one-way exchange, the relationship between the cardiovascular proteomics community, including our group, and the consortium is circular. Researchers not only use GO annotation to inform their research, but can also submit data from their experiments to validate annotations and suggest novel cardiovascular GO terms.
Table 3.
Functional annotation databases
| Abbreviation | Name | Website |
|---|---|---|
| BBID | Biological Biochemical Image Database | http://bbid.grc.nia.nih.gov/ |
| BioCarta | BioCarta Pathways | http://www.biocarta.com/genes/index.asp |
| GO | Gene Ontology | http://www.geneontology.org/ |
| KEGG | Kyoto Encyclopedia of Genes and Genomes | http://www.genome.jp/kegg/ |
| NCBI BioSystems | NCBI BioSystems Database | http://www.ncbi.nlm.nih.gov/biosystems |
| OMIM | Online Mendelian Inheritance in Man | http://omim.org/ |
| PANTHER | Protein ANalysis THrough Evolutionary Relationships | http://www.pantherdb.org/ |
| PID | NCI-Pathway Interaction Database | http://pid.nci.nih.gov/ |
| Reactome | Reactome | http://www.reactome.org/ |
| WikiPathways | WikiPathways | http://www.wikipathways.org/index.php/WikiPathways |
As an example, Isserlin et al.36 incorporated a differential expression analysis with a Gene Set Enrichment Analysis (GSEA) to identify sets of differentially expressed proteins that were enriched for functional terms relating to dilated cardiomyopathy. They utilized GO as well as several other sources of publicly available functional data to perform the GSEA and derived functional networks, which show novel processes in the progression from pre-symptomatic to dilated cardiomyopathy.
3.4. Network biology
The identification of differentially expressed proteins is only one part of a systems biology approach to proteomics. Further analyses are often performed on the set of differentially expressed proteins to elucidate their functional role in the disease pathology. The post-genomic shift in paradigm acknowledges the fact that many biological systems can be represented using concepts of network biology. Different pathways cross-talk with each other at points that can be graphically represented as well-connected nodes or nexuses within a map of signalling networks.50 In addition to high-throughput data acquisition, the last decade introduced a number of sophisticated methodologies that intend to interrogate cellular interactions.51–55 Preliminary analyses of these interactomes revealed the complexity of molecular signalling, which presents a challenge for accurate interpretation and application. It is now believed that the human interactome comprises approximately 20 000 protein-coding genes, approximately 1000 metabolites, and an undefined number of distinct proteins, whereas the number of functional links between these components is expected to be approximately 130 000.51 An emerging computational discipline, network biology, has been proposed as a tool that may supplement traditional quantitative analysis and uncover relational properties that control the behaviour of a cell through data integration and computational modelling.56 Network biology was successfully used to define gene regulatory patterns in physiological cardiac hypertrophy57 and highlight network topology of heart development and failure.58 Increasing evidence suggests that combination of network concepts, such as centrality, with gene, protein, or microRNA expression information, may contribute to better prioritization of relevant biological targets.59–61
Despite their usefulness, networks analyses should be used with some degree of caution. It is currently not feasible to access and characterize the entire human proteome and so each proteomic network will consist of a subset of all possible proteins. Network studies, especially those focusing on protein–protein interactions, have shown that network properties from a sample or subset of a global network differ from the properties of the global network.62–64 For example, biological networks have been described as ‘scale-free’ networks where there are several nodes with a high degree of connectivity and many nodes with a low degree of connectivity.65 Although this may be an appropriate assumption for a large-scale biological network, no statistical tests were applied to prove scale-freeness in biological networks, and the smaller sub-networks, including most protein–protein interaction networks, do not follow the same assumptions.
4. Is a systems-level integration of ‘-omics’ data the way forward?
Within a biological system, proteins do not act on their own, but rather, through complex interactions with metabolites, RNA, and other proteins. As we learn more about the pathophysiology of cardiovascular diseases, the underlying complexity becomes apparent, and the integration of ‘-omics’ fields provides an unbiased way to elucidate the mechanisms. The advancements in the technologies and the data availability of each of the ‘-omics’ give rise to a finer assessment but also provide a greater opportunity to study the interactions between genes, gene expression, proteins, and metabolites.
4.1. Integrating transcriptomics and proteomics
Initial investigations into the correlation between mRNA levels and protein levels have shown poor-to-moderate associations between the two. These low correlations can be attributed to epigenetic factors, translation rates, and protein degradation rates, but they can also be due to the levels being assessed for different samples, across different time points.66 To overcome some of these issues, Schwanhausser et al.67 used mouse fibroblasts to quantify and analyse global mRNA and protein levels along with their associated half-lives, transcription, and translation rates. To get an accurate picture of the strength of association between mRNA and protein levels, the authors used metabolic pulse labelling in an experimentally growing population of embryonic fibroblasts to record mRNA and proteins levels occurring at the same time point. They found that 40% of the variation in proteins levels can be attributed to variation in mRNA levels but translation efficiency was the best predictor. Certain combinations of half-lives and mRNA levels correspond to shared functional role, indicating shared selective pressures. In another transcriptomic and proteomic analysis, Zhao et al.68 reconstructed a heart-specific metabolic network using transcriptome and proteome data with a model-building algorithm. Using generic genome-wide metabolic networks, they constructed heart-specific models by mapping transcriptomics and proteomics data from the heart onto the genetic networks. The resulting model contained 2803 reactions with 1721 active enzymes in the heart. With this metabolic network, they were able to estimate the lethality, in silico, of house-keeping and heart-specific genes and identify potential CVD biomarkers.
4.2. Integrating proteomics and metabolomics
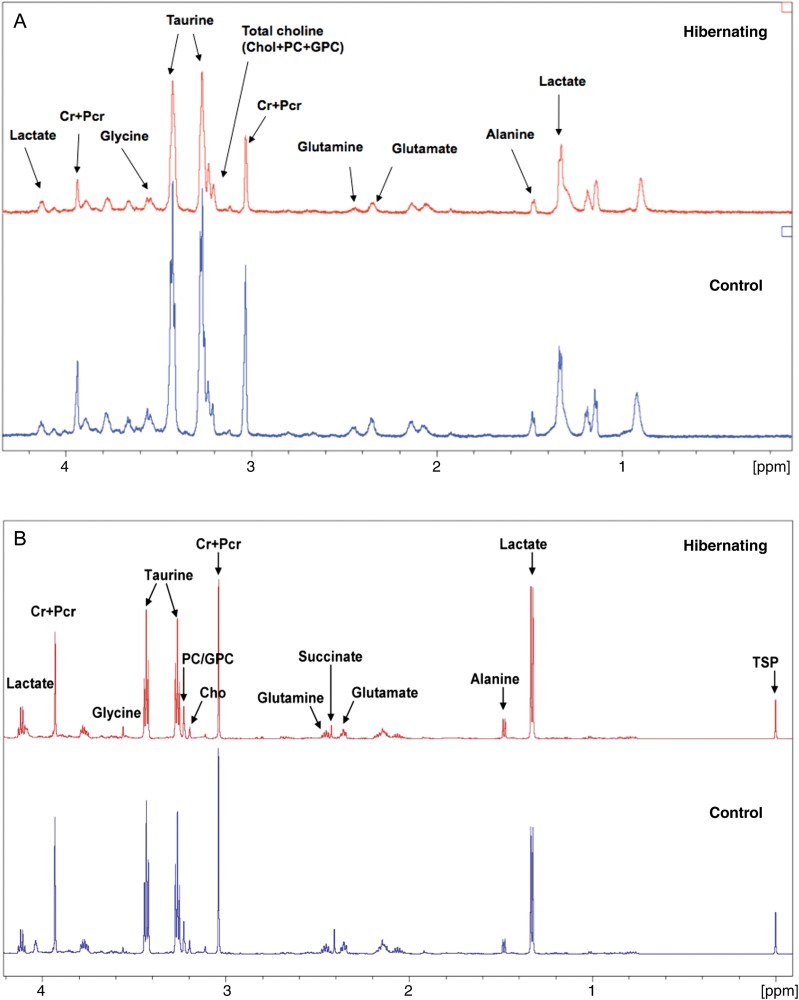
Currently, proteomics and metabolomics are rarely used in tandem, but this technological platform offers advantages. It has the potential to identify the emergent behaviour that cannot be found by studying proteins or metabolites in isolation. Besides, proteomic and metabolomics findings can effectively reinforce or cross-validate each other. We utilized a combined proteomics and metabolomics approach to investigate cardiovascular diseases.69 Our aim was to contribute to a better understanding of enzymatic and metabolite changes associated with cardiovascular diseases, including atherosclerosis,6,70 ischaemic preconditioning,71 cardioprotective signalling,72,73 and atrial fibrillation.74 In a recent study on myocardial hibernation, murine hearts were analysed by a combined transcriptomic, proteomic, and metabolomic approach (Figure 5).75,76 Unguided network analysis correctly identified hypoxia-inducible factor 1 alpha (HIF1α) activation as the top signalling pathway, and provided independent confirmation that anaerobic glycolysis is affected. A direct link to cardiac remodelling was also provided by the activation of collagen hydroxylases, which produce hydroxyproline. By combining the ‘-omics’ data, the P-value of the HIF1α signalling pathway decreased by two orders of magnitude, and became the top-ranking pathway even though it was not the top-ranked pathway based on either dataset individually. The proteomics and transcriptomics focused on, and contributed different molecules to, the protein network, which enabled the HIF1α signalling pathway to rise to the top.
Figure 5.
Metabolomics. A comparison of control and hibernating murine hearts by high-resolution magic-angle-spinning 1H-magnetic resonance spectroscopy (HRMAS 1H-MRS) analysis from solid hearts (A) and 1H-nuclear magnetic resonance spectroscopy (1H-NMR) of cardiac tissue extracts (B, reproduced with permission from Mayr et al.75). Both techniques showed consistent changes in metabolites, i.e. the ratio of glutamate, lactate, and taurine in hibernating compared with control hearts as determined by HRMAS 1H-MRS was 0.81, 1.09, and 0.79, respectively, which is in good agreement with the measurements of 0.68, 1.13, and 0.72 for the same metabolites by 1H-NMR. HRMAS 1H-MRS provides a means for measuring metabolites in intact hearts ex vivo. 1H-NMR of tissue extracts offers better resolution and allows the identification of more metabolites than HRMAS 1H-MRS spectra obtained from solid tissue.
4.3. Personal ‘-omics’ profiles
In a proof of concept study, Chen et al.77 presented an integrative personal ‘-omics’ profile analysis, where the genetic, transcripomic, proteomic, metabolomic, and autoantibody profiles were measured and integrated for one healthy individual over the course of 14 months. The measurements were assessed in blood components (plasma, serum, and peripheral blood mononuclear cells) at several time points during the course of the study. The ‘-omics’ responses were studied in greater detail during two viral infections, showing the dynamic response of the immune system. Interestingly, the authors identified a genetic predisposition to type II diabetes at the start of the study and noticed a pronounced change in insulin-related responses after the second infection. Although the causal relationship between the infection and the onset of diabetes cannot be determined from one individual, these tightly linked events, and the indication that they are related, were detected only through the combination of ‘-omics’ profiles. As the technology to measure ‘-omics’ profiles becomes feasible, the greatest challenge will not be the generation of data, but their analysis.
5. Conclusions
A discrete biological function is very rarely attributed to one single molecule; more often it is the combined input of many proteins. The studies mentioned above, which integrate protein data with other ‘-omics’ data including transcriptomics67,68,77 and metabolomics,75–77 illustrate the utility of an integrative ‘-omics’ approach to cardiovascular diseases. However, variants and changes from the genetic to the phenotypic level are not linearly associated and often variations seen at one level are absent at another. Although the integration of data from different ‘-omics’ techniques is still a challenge, the incorporation of proteomics with systems biology, and the application to study metabolism, is a promising area for future applications in cardiovascular diseases.78,79 Combining proteomics with stringent statistics, bioinformatics, and other ‘-omics’ technologies, such as metabolomics, can aid in identifying targets that have clinical relevance for working towards new therapies for cardiovascular disease.80 Improvements in protein identification and quantification technologies as well as the availability of more proteomics data sets in public data repositories such as PRIDE,32 combined with focused GO curation,81,82 will facilitate the application of systems biology to cardiovascular research.
Funding
This work was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at Guy's and St Thomas' NHS Foundation Trust and King's College London. M.M. is a Senior Fellow of the British Heart Foundation.
Acknowledgements
We are grateful to Drs Basetti Madhu, Melanie Abbonenc, and Athanasios Didangelos for providing figures.
Conflict of interest: none declared.
References
- 1.Prokopi M, Mayr M. Proteomics: a reality-check for putative stem cells. Circ Res. 2011;108:499–511. doi: 10.1161/CIRCRESAHA.110.226902. [DOI] [PubMed] [Google Scholar]
- 2.McGregor E, Dunn MJ. Proteomics of heart disease. Hum Mol Genet. 2003;12:R135–R144. doi: 10.1093/hmg/ddg278. [DOI] [PubMed] [Google Scholar]
- 3.Arrell DK, Neverova I, Van Eyk JE. Cardiovascular proteomics: evolution and potential. Circ Res. 2001;88:763–773. doi: 10.1161/hh0801.090193. [DOI] [PubMed] [Google Scholar]
- 4.Mayr M, Zhang J, Greene AS, Gutterman D, Perloff J, Ping P. Proteomics-based development of biomarkers in cardiovascular disease: mechanistic, clinical, and therapeutic insights. Mol Cell Proteomics. 2006;5:1853–1864. doi: 10.1074/mcp.R600007-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Gorg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 2004;4:3665–3685. doi: 10.1002/pmic.200401031. [DOI] [PubMed] [Google Scholar]
- 6.Mayr M, Chung YL, Mayr U, Yin X, Ly L, Troy H, et al. Proteomic and metabolomic analyses of atherosclerotic vessels from apolipoprotein E-deficient mice reveal alterations in inflammation, oxidative stress, and energy metabolism. Arterioscler Thromb Vasc Biol. 2005;25:2135–2142. doi: 10.1161/01.ATV.0000183928.25844.f6. [DOI] [PubMed] [Google Scholar]
- 7.McManus CA, Donoghue PM, Dunn MJ. A fluorescent codetection system for immunoblotting and proteomics through ECL-Plex and CyDye labeling. Methods Mol Biol. 2009;536:515–526. doi: 10.1007/978-1-59745-542-8_52. [DOI] [PubMed] [Google Scholar]
- 8.Unlu M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 9.Westermeier R, Scheibe B. Difference gel electrophoresis based on lys/cys tagging. Methods Mol Biol. 2008;424:73–85. doi: 10.1007/978-1-60327-064-9_7. [DOI] [PubMed] [Google Scholar]
- 10.McGregor E, Dunn MJ. Proteomics of the heart: unraveling disease. Circ Res. 2006;98:309–321. doi: 10.1161/01.RES.0000201280.20709.26. [DOI] [PubMed] [Google Scholar]
- 11.Wilm M. Principles of electrospray ionization. Mol Cell Proteomics. 2011;10:M111.009407. doi: 10.1074/mcp.M111.009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Sadygov RG, Yates JR., III A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 13.Scigelova M, Hornshaw M, Giannakopulos A, Makarov A. Fourier transform mass spectrometry. Mol Cell Proteomics. 2011;10:M111.009431. doi: 10.1074/mcp.M111.009431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Didangelos A, Yin X, Mandal K, Baumert M, Jahangiri M, Mayr M. Proteomics characterization of extracellular space components in the human aorta. Mol Cell Proteomics. 2010;9:2048–2062. doi: 10.1074/mcp.M110.001693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Didangelos A, Yin X, Mandal K, Saje A, Smith A, Xu Q, et al. Extracellular matrix composition and remodeling in human abdominal aortic aneurysms: a proteomics approach. Mol Cell Proteomics. 2011;10:M111.008128. doi: 10.1074/mcp.M111.008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnott D, Kishiyama A, Luis EA, Ludlum SG, Marsters JC, Jr, Stults JT. Selective detection of membrane proteins without antibodies: a mass spectrometric version of the western blot. Mol Cell Proteomics. 2002;1:148–156. doi: 10.1074/mcp.m100027-mcp200. [DOI] [PubMed] [Google Scholar]
- 17.Bouyssie D, Gonzalez de Peredo A, Mouton E, Albigot R, Roussel L, Ortega N, et al. Mascot file parsing and quantification (MFPaQ), a new software to parse, validate, and quantify proteomics data generated by ICAT and SILAC mass spectrometric analyses: application to the proteomics study of membrane proteins from primary human endothelial cells. Mol Cell Proteomics. 2007;6:1621–1637. doi: 10.1074/mcp.T600069-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 19.Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 20.Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, et al. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 21.Dayon L, Sanchez JC. Relative protein quantification by MS/MS using the tandem mass tag technology. Methods Mol Biol. 2012;893:115–127. doi: 10.1007/978-1-61779-885-6_9. [DOI] [PubMed] [Google Scholar]
- 22.Karp NA, Huber W, Sadowski PG, Charles PD, Hester SV, Lilley KS. Addressing accuracy and precision issues in iTRAQ quantitation. Mol Cell Proteomics. 2010;9:1885–1897. doi: 10.1074/mcp.M900628-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noble WS, MacCoss MJ. Computational and statistical analysis of protein mass spectrometry data. PLoS Comput Biol. 2012;8:e1002296. doi: 10.1371/journal.pcbi.1002296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dupont A, Corseaux D, Dekeyzer O, Drobecq H, Guihot A-L, Susen S, et al. The proteome and secretome of human arterial smooth muscle cells. Proteomics. 2005;5:585–596. doi: 10.1002/pmic.200400965. [DOI] [PubMed] [Google Scholar]
- 26.Urbich C, De Souza AI, Rossig L, Yin X, Xing Q, Prokopi M, et al. Proteomic characterization of human early pro-angiogenic cells. J Mol Cell Cardiol. 2011;50:333–336. doi: 10.1016/j.yjmcc.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 27.Stastna M, Chimenti I, Marban E, Van Eyk JE. Identification and functionality of proteomes secreted by rat cardiac stem cells and neonatal cardiomyocytes. Proteomics. 2010;10:245–253. doi: 10.1002/pmic.200900515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aye TT, Scholten A, Taouatas N, Varro A, Van Veen TAB, Vos MA, et al. Proteome-wide protein concentrations in the human heart. Mol BioSyst. 2010;6:1917–1927. doi: 10.1039/c004495d. [DOI] [PubMed] [Google Scholar]
- 29.Burkhart JM, Vaudel M, Gambaryan S, Radau S, Walter U, Martens L, et al. The first comprehensive and quantitative analysis of human platelet protein composition allows the comparative analysis of structural and functional pathways. Blood. 2012;120:e73–e82. doi: 10.1182/blood-2012-04-416594. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Li X, Mueller M, Wang Y, Zong C, Deng N, et al. Systematic characterization of the murine mitochondrial proteome using functionally validated cardiac mitochondria. Proteomics. 2008;8:1564–1575. doi: 10.1002/pmic.200700851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin X, Cuello F, Mayr U, Hao Z, Hornshaw M, Ehler E, et al. Proteomics analysis of the cardiac myofilament subproteome reveals dynamic alterations in phosphatase subunit distribution. Mol Cell Proteomics. 2010;9:497–509. doi: 10.1074/mcp.M900275-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vizcaíno JA, Côté R, Reisinger F, Barsnes H, Foster JM, Rameseder J, et al. The Proteomics Identifications database: 2010 update. Nucleic Acids Res. 2010;38(Database issue):D736–D742. doi: 10.1093/nar/gkp964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayr M, Siow R, Chung YL, Mayr U, Griffiths JR, Xu Q. Proteomic and metabolomic analysis of vascular smooth muscle cells: role of PKCdelta. Circ Res. 2004;94:e87–e96. doi: 10.1161/01.RES.0000131496.49135.1d. [DOI] [PubMed] [Google Scholar]
- 34.Barallobre-Barreiro J, Didangelos A, Schoendube FA, Drozdov I, Yin X, Fernandez-Caggiano M, et al. Proteomics analysis of cardiac extracellular matrix remodeling in a porcine model of ischemia/reperfusion injury. Circulation. 2012;125:789–802. doi: 10.1161/CIRCULATIONAHA.111.056952. [DOI] [PubMed] [Google Scholar]
- 35.Dai D-F, Hsieh EJ, Liu Y, Chen T, Beyer RP, Chin MT, et al. Mitochondrial proteome remodelling in pressure overload-induced heart failure: the role of mitochondrial oxidative stress. Cardiovasc Res. 2012;93:79–88. doi: 10.1093/cvr/cvr274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isserlin R, Merico D, Alikhani-Koupaei R, Gramolini A, Bader GD, Emili A. Pathway analysis of dilated cardiomyopathy using global proteomic profiling and enrichment maps. Proteomics. 2010;10:1316–1327. doi: 10.1002/pmic.200900412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammer E, Goritzka M, Ameling S, Darm K, Steil L, Klingel K, et al. Characterization of the human myocardial proteome in inflammatory dilated cardiomyopathy by label-free quantitative shotgun proteomics of heart biopsies. J Proteome Res. 2011;10:2161–2171. doi: 10.1021/pr1008042. [DOI] [PubMed] [Google Scholar]
- 38.Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, et al. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal Chem. 2003;75:4818–4826. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Anderson G, Smith RD, Dabney AR. A hybrid approach to protein differential expression in mass spectrometry-based proteomics. Bioinformatics. 2012;28:1586–1591. doi: 10.1093/bioinformatics/bts193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karp NA, Lilley KS. Design and analysis issues in quantitative proteomics studies. Proteomics. 2007;7(Suppl. 1):42–50. doi: 10.1002/pmic.200700683. [DOI] [PubMed] [Google Scholar]
- 41.Cairns DA. Statistical issues in quality control of proteomic analyses: good experimental design and planning. Proteomics. 2011;11:1037–1048. doi: 10.1002/pmic.201000579. [DOI] [PubMed] [Google Scholar]
- 42.Käll L, Vitek O. Computational mass spectrometry-based proteomics. PLoS Comp Biol. 2011;7:e1002277. doi: 10.1371/journal.pcbi.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 44.Pavelka N, Fournier ML, Swanson SK, Pelizzola M, Ricciardi-Castagnoli P, Florens L, et al. Statistical similarities between transcriptomics and quantitative shotgun proteomics data. Mol Cell Proteomics. 2008;7:631–644. doi: 10.1074/mcp.M700240-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Griffin NM, Yu J, Long F, Oh P, Shore S, Li Y, et al. Label-free, normalized quantification of complex mass spectrometry data for proteomic analysis. Nat Biotech. 2010;28:83–89. doi: 10.1038/nbt.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu X, Gharib SA, Green PS, Aitken ML, Frazer DA, Park DR, et al. Spectral index for assessment of differential protein expression in shotgun proteomics. J Proteome Res. 2008;7:845–854. doi: 10.1021/pr070271+. [DOI] [PubMed] [Google Scholar]
- 47.Choi H, Fermin D, Nesvizhskii AI. Significance analysis of spectral count data in label-free shotgun proteomics. Mol Cell Proteomics. 2008;7:2373–2385. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2011;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.King JY, Ferrara R, Tabibiazar R, Spin JM, Chen MM, Kuchinsky A, et al. Pathway analysis of coronary atherosclerosis. Physiol Genomics. 2005;23:103–118. doi: 10.1152/physiolgenomics.00101.2005. [DOI] [PubMed] [Google Scholar]
- 51.Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 53.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 54.Wu G, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 2010;11:R53. doi: 10.1186/gb-2010-11-5-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drozdov I, Tsoka S, Ouzounis CA, Shah AM. Genome-wide expression patterns in physiological cardiac hypertrophy. BMC Genomics. 2010;11:557. doi: 10.1186/1471-2164-11-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dewey FE, Perez MV, Wheeler MT, Watt C, Spin J, Langfelder P, et al. Gene coexpression network topology of cardiac development, hypertrophy, and failure. Circ Cardiovasc Genet. 2011;4:26–35. doi: 10.1161/CIRCGENETICS.110.941757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barrenas F, Chavali S, Holme P, Mobini R, Benson M. Network properties of complex human disease genes identified through genome-wide association studies. PLoS One. 2009;4:e8090. doi: 10.1371/journal.pone.0008090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, et al. Plasma microRNA profiling reveals loss of endothelial mir-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 61.Zampetaki A, Willeit P, Tilling L, Drozdov I, Prokopi M, Renard JM, et al. Prospective study on circulating microRNAs and risk of myocardial infarction. J Am Coll Cardiol. 2012;60:290–299. doi: 10.1016/j.jacc.2012.03.056. [DOI] [PubMed] [Google Scholar]
- 62.Stumpf MP, Wiuf C, May RM. Subnets of scale-free networks are not scale-free: sampling properties of networks. Proc Natl Acad Sci USA. 2005;102:4221–4224. doi: 10.1073/pnas.0501179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fernandes LP, Annibale A, Kleinjung J, Coolen ACC, Fraternali F. Protein networks reveal detection bias and species consistency when analysed by information-theoretic methods. PloS One. 2010;5:e12083. doi: 10.1371/journal.pone.0012083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Annibale A, Coolen ACC. What you see is not what you get: how sampling affects macroscopic features of biological networks. Interface Focus. 2011;1:836–856. doi: 10.1098/rsfs.2011.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stumpf MPH. From the cover: subnets of scale-free networks are not scale-free: sampling properties of networks. Proc Natl Acad Sci USA. 2005;102:4221–4224. doi: 10.1073/pnas.0501179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Sousa Abreu R, Pentalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 68.Zhao Y, Huang J. Reconstruction and analysis of human heart-specific metabolic network based on transcriptome and proteome data. Biochem Biophys Res Commun. 2011;415:450–454. doi: 10.1016/j.bbrc.2011.10.090. [DOI] [PubMed] [Google Scholar]
- 69.Mayr M. Metabolomics: ready for the prime time? Circ Cardiovasc Genet. 2008;1:58–65. doi: 10.1161/CIRCGENETICS.108.808329. [DOI] [PubMed] [Google Scholar]
- 70.Mayr M, Grainger D, Mayr U, Leroyer AS, Leseche G, Sidibe A, et al. Proteomics, metabolomics, and immunomics on microparticles derived from human atherosclerotic plaques. Circ Cardiovasc Genet. 2009;2:379–388. doi: 10.1161/CIRCGENETICS.108.842849. [DOI] [PubMed] [Google Scholar]
- 71.Mayr M, Metzler B, Chung YL, McGregor E, Mayr U, Troy H, et al. Ischemic preconditioning exaggerates cardiac damage in PKC-delta null mice. Am J Physiol Heart Circ Physiol. 2004;287:H946–H956. doi: 10.1152/ajpheart.00878.2003. [DOI] [PubMed] [Google Scholar]
- 72.Mayr M, Chung YL, Mayr U, McGregor E, Troy H, Baier G, et al. Loss of PKC-delta alters cardiac metabolism. Am J Physiol Heart Circ Physiol. 2004;287:H937–H945. doi: 10.1152/ajpheart.00877.2003. [DOI] [PubMed] [Google Scholar]
- 73.Mayr M, Liem D, Zhang J, Li X, Avliyakulov NK, Yang JI, et al. Proteomic and metabolomic analysis of cardioprotection: interplay between protein kinase c epsilon and delta in regulating glucose metabolism of murine hearts. J Mol Cell Cardiol. 2009;46:268–277. doi: 10.1016/j.yjmcc.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mayr M, Yusuf S, Weir G, Chung YL, Mayr U, Yin X, et al. Combined metabolomic and proteomic analysis of human atrial fibrillation. J Am Coll Cardiol. 2008;51:585–594. doi: 10.1016/j.jacc.2007.09.055. [DOI] [PubMed] [Google Scholar]
- 75.Mayr M, May D, Gordon O, Madhu B, Gilon D, Yin X, et al. Metabolic homeostasis is maintained in myocardial hibernation by adaptive changes in the transcriptome and proteome. J Mol Cell Cardiol. 2011;50:982–990. doi: 10.1016/j.yjmcc.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.May D, Gilon D, Djonov V, Itin A, Lazarus A, Gordon O, et al. Transgenic system for conditional induction and rescue of chronic myocardial hibernation provides insights into genomic programs of hibernation. Proc Natl Acad Sci USA. 2008;105:282–287. doi: 10.1073/pnas.0707778105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HYK, Chen R, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mayr M, Mayr U, Chung YL, Yin X, Griffiths JR, Xu Q. Vascular proteomics: linking proteomic and metabolomic changes. Proteomics. 2004;4:3751–3761. doi: 10.1002/pmic.200400947. [DOI] [PubMed] [Google Scholar]
- 79.Mayr M, Madhu B, Xu Q. Proteomics and metabolomics combined in cardiovascular research. Trends Cardiovasc Med. 2007;17:43–48. doi: 10.1016/j.tcm.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 80.Arrell DK, Terzic A. Network systems biology for drug discovery. Clin Pharmacol Ther. 2010;88:120–125. doi: 10.1038/clpt.2010.91. [DOI] [PubMed] [Google Scholar]
- 81.Alam-Faruque Y, Huntley RP, Khodiyar VK, Camon EB, Dimmer EC, Sawford T, et al. The impact of focused Gene Ontology curation of specific mammalian systems. PLoS ONE. 2011;6:e27541. doi: 10.1371/journal.pone.0027541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khodiyar VK, Hill DP, Howe D, Berardini TZ, Tweedie S, Talmud PJ, et al. The representation of heart development in the gene ontology. Dev Biol. 2011;354:9–17. doi: 10.1016/j.ydbio.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]