Abstract
GB virus type C (GBV-C) is a single-stranded positive-sense RNA virus classified in the Flaviviridae family. Persistent coinfection with GBV-C is associated with lower human immunodeficiency virus type 1 (HIV-1) load, higher CD4+ T-cell count, and prolonged survival in HIV-1 coinfected patients. The GBV-C envelope glycoprotein E2 has been reported to interfere with HIV-1 entry. In this study, we showed that the expression of GBV-C E2 inhibited HIV-1 Gag assembly and release. Expression of glycosylated GBV-C E2 inhibited HIV-1 Gag precursor processing, resulting in lower production of CAp24 and MAp17, while the overall expression level of the Gag precursor Pr55 remained unchanged. Membrane floatation gradient and indirect immunofluorescence confocal microscopy analysis showed that glycosylated E2 disrupted HIV-1 Gag trafficking to the plasma membrane, resulting in Gag accumulation in subcellular compartments. This interference in HIV-1 Gag trafficking led to diminished HIV-1 particle production, which is a critical step for HIV-1 to infect new host cells. These findings shed light on a novel mechanism used by GBV-C E2 to inhibit HIV-1 replication and may provide insight into new approaches for suppressing HIV-1 replication.
Keywords: GBV-C, GBV-C E2, HIV-1, HIV-1 assembly, HIV-1 Gag, plasma membrane targeting
GB virus type C (GBV-C) is a nonpathogenic, positive-sense RNA virus classified in the Flaviviridae family [1]. GBV-C is primarily lymphotropic and does not replicate in hepatocytes [2, 3]. In 2001, Stapleton et al created the first complementary DNA (cDNA) clone of GBV-C and showed that RNA transcripts from this clone were able to effectively replicate in CD4+ T cells. Because of shared transmission routes, human immunodeficiency virus type 1 (HIV-1)/GBV-C coinfection is common. Clinical studies of HIV-1 positive individuals reported that up to 86% of patients had evidence of past or active GBV-Cinfection [4, 5]. More than 10 cross-sectional studies performed worldwide noted that GBV-C–coinfected patients had reduced HIV-1 loads, higher CD4+ T cell counts, a delay in AIDS prognosis. and a longer lifespan, compared with patients infected with HIV-1 only [4–12]. These studies suggest that GBV-C may have an adverse effect on HIV-1 replication.
Previous studies have reported that GBV-C affects the HIV-1 lifecycle by blocking HIV-1 attachment to target cells. The inhibitory effect is primarily mediated by modulating the expression of chemokines and chemokine receptors [13, 14]. The NS5A nonstructural phosphoprotein of GBV-C was shown to be responsible for this effect [15–17]. The E2 envelope glycoprotein of GBV-C was also shown to inhibit HIV-1 entry into target cells. Jung et al reported that preincubation of interleukin 2–stimulated peripheral blood mononuclear cells with secretable truncated E2 blocks HIV-1 entry [18]. In addition, synthetic peptides of E2 were shown to inhibit gp41-mediated liposome fusion or strain-specific inhibition of HIV-1 entry [19, 20]. Furthermore, Mohr et al showed that E2 also elicits antibodies that react with a cellular antigen on HIV-1 particles, resulting in neutralization of diverse HIV-1 isolates and inhibition of HIV-1 attachment to target cells [21]. Recently, GBV-C NS3 and E2 were also shown to play a role in inhibiting HIV-1 replication in a T-cell line through an unknown mechanism [22, 23]. Although GBV-C and HIV-1 target the same cells, the effect of GBV-C on the late stages of the HIV-1 lifecycle is yet to be defined.
HIV-1 Gag proteins play a major role in virus assembly and release [24, 25]. Gag is expressed as the Pr55Gag precursor polyprotein, which is processed into matrix (p17), capsid (p24), nucleocapsid (p7), and p6 proteins by the viral protease [24]. During HIV-1 replication, Pr55Gag is targeted to the plasma membrane via regions within the matrix domain [26–28]. Plasma membrane targeting of HIV-1 Gag is an important initiation step of Gag processing. It has been shown that a Gag matrix mutant that was not able to target to the plasma membrane failed to be processed [29, 30]. During viral budding from the plasma membrane, the viral protease within Gag-Pol complexes autocatalyzes its release from the Pol domain and subsequently processes Pol and Gag proteins into their mature constituent proteins. This process leads to reassembly of the immature virus particle into a mature infectious virion [31]. In this study, we analyzed the effect of GBV-C E2 expression on HIV-1 assembly and release. We found that the expression of GBV-C E2 protein inhibited HIV-1 assembly through interference with HIV-1 Gag intracellular trafficking. These observations may offer insights on novel approaches for the development of antiretroviral therapies.
METHODS
Plasmids, Antibodies, and Reagents
pAF121950 [3], the full-length cDNA of GBV-C (GenBank accession number AF121950), was obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (NIH-ARRRP). E2 gene was amplified from pAF121950, using the forward primer 5′-ATCTAGAGCCATGGCCCCCGCCTCCG-3′ and the reverse primer 5′-TGGATCCTTAAAGGTCTTCTTCTGAGATGAGTTTTTGTTCTGCCACGGCAGCG-3′, with the cMyc sequence incorporated into the C-terminus of the E2 open reading frame. The E2-cMyc gene was subcloned into the VR1012 expression vector [32] (generously provided by Vical San Diego, CA), using XbaI and BamHI restriction sites, to generate the E2 expression vector. The E2 gene was also amplified from pAF121950, using the forward primer 5′-AGGATCCGCCCCCGCCTCCG-3′ and the same reverse primer described above. The polymerase chain reaction (PCR) product was subcloned into the VR1020 vector (Vical, San Diego, CA) downstream of the tissue plasminogen activator (tPA) secretory signal sequence at the BamHI restriction site to generate the tPA-E2 expression vector. To establish the immunoglobulin G(IgG)–E2 construct, the signal sequence of IgG κ was fused to the N-terminus of E2-cMyc using PCR with 5'-AGTCGACATGGAGACAGACACACTCCTGCTATGGGTACTGCTGCTCTGGGTTCCAGGTTCCACTGGTGACGCCCCCGCCTCCG-3' as a forward primer and the same reverse primer. The PCR product was cloned into VR1012 using XbaI and BamHI restriction sites. The HIV-1 Gag-Pol expression construct pGPCINS was a generous gift from Xiao-Fang Yu (Johns Hopkins University, Baltimore, MD) [33]. The proviral DNA constructs HIV-1 pNL4-3 [34], simian immunodeficiency virus (SIV) pSIVagmTan-1 [35], and xenotropic murine leukemia virus–related virus (XMRV) VP62 [36] were from NIH-ARRRP. Deletions were introduced into IgG-E2 by PCR-mediated mutagenesis, using the following primer sets: forward primer 5′-AACCAGTGGCCCCTATC-3′ and reverse primer 5′-GCATGACTGCCACTTCAAC-3′ for E2DN1, forward primer 5′-TGGCCCGAAACCGG-3′ and reverse primer 5′-TTGTCCGTGGCTCCAAT-3′ for E2 DN2, forward primer 5′-CTAGGCACGGAAGTGTCTG-3′ and reverse primer 5′-GCAGTCCCGCACACAG-3′ for E2DC2, and forward primer 5′-AACCAGTTGGCGGTTCTA-3′ and reverse primer 5′-CATGCAGTTGTTAATGGGC-3′ for E2DMID. For this method, each primer was synthesized with a 5′-phosphate modification. The PCR products were self-ligated using the DNA Quick Ligation Kit (New England BioLabs) and were cloned to form the E2 deletion mutants. Anti-cMyc monoclonal antibody (4A6) was purchased from Millipore. The anti-p24 monoclonal antibody (183-H12-5C) [37], polyclonal anti-HIV-1 p17 (VU47) [38], and HIV immunoglobulin (HIV-Ig) were obtained from NIH-ARRRP. Alexa 546–conjugated goat anti-rabbit antibody and Alexa 488–conjugated goat anti-mouse antibody were purchased from Molecular Probes. Anti-MuLV antiserum was purchased from the American Type Culture Collection.
Cell Culture, Western Blot Analysis, HIV-1 Preparation, and Viral Infectivity (MAGI) Assay
The human embryonic kidney 293T, HeLa, and TZM-bl cell lines [39] (NIH-ARRRP) were grown in Dulbecco's modified essential medium supplemented with 10% fetal calf serum at 37°C in a 5% CO2 atmosphere. DNA transfections were performed with polyethylenimine for 293T cells, as described previously [40], or with Lipofectamine 2000 (Invitrogen) for HeLa cells, following the manufacturer's instructions. Western blot analysis, preparation of HIV-1, and the viral infectivity assay (ie, the MAGI assay) were performed as described previously [40, 41].
PNGase F Treatment
Cell lysates from 293T cells transfected with tPA-E2 or IgG-E2 were denatured with glycoprotein denaturing buffer (5% sodium dodecyl sulfate and 10% 2-mercaptoethanol) for 10 minutes at 100°C. The reaction was then activated by adding G7 reaction buffer (0.5 M sodium phosphate, pH 7.5), 10% NP-40, and PNGase F (New England BioLabs) at a 1:10 ratio and incubating the preparation at 37°C for 1 hour. Samples were then analyzed by Western blot analysis. All experiments in this study were performed at least 3 times independently.
HIV-1 p24 Enzyme-Linked Immunosorbent Assay (ELISA)
The HIV-1 p24 antigen capture assay kit was purchased from Biological Products Core, AIDS and Cancer Virus Program, SAIC-Frederick, National Cancer Institute (Frederick, MD). The lower limit of detection for this kit is around 100 pg. The ELISA was performed as detailed in the kit instructions. All analyses were performed at least 3 times for each experiment.
Membrane-Binding Analysis
Membrane floatation analysis of cell lysates was performed as detailed by Ono et al [42]. In brief, 2 days after transfection, 293T cells were washed with phosphate-buffered saline (PBS) and disrupted with homogenization buffer containing 10% sucrose, 10 mM Tris-HCl (pH 7.5), and 1 mM Tris–ethylenediaminetetraacetic acid (TE), with Complete Protease Inhibitor Cocktail (Roche). Lysis was performed by sonication (twice for 15 seconds each time). Nuclear fractions and unlysed cells were removed by adjusting the samples to 150 mM NaCl–1 mM MgCl2 prior to centrifugation at 1000 × g for 10 minutes at 4°C. Subsequently, 250-µL postnuclear supernatants were mixed with 1.25 mL of 85.5% sucrose in TE to adjust the sample to 73% sucrose. The sample was then placed at the bottom of a centrifuge tube and layered with 7 mL of 65% sucrose in TE and 3.25 mL of 10% sucrose in TE. The sucrose step gradient was centrifuged at 35 000 rpm for 18 hours at 4°C, using a Beckman SW41 rotor.
Immunofluorescence Microscopy
Confocal analysis was performed as described previously [43]. In brief, HeLa cells were plated on glass coverslips in a 6-well plate and grown overnight. Cells were then transfected using Lipofectamine 2000. Twenty-three hours after transfection, cells were fixed with 4% paraformaldehyde in PBS solution at room temperature for 10 minutes, permeabilized with 0.1% Triton X-100 for 10 minutes, and blocked with 5% bovine serum albumin for 1 hour. Immunofluorescent staining was performed using polyclonal anti–HIV-1 p17 (VU47) and anti-cMyc monoclonal antibody (4A6) as primary antibodies, which were detected via goat anti-rabbit Alexa 546–conjugated and goat anti-mouse Alexa 488–conjugated secondary antibodies, respectively. Images were captured using a Nikon A1R confocal microscope.
RESULTS
Expression of Glycosylated GBV-C E2 Inhibits Proteolytic Processing of HIV-1 Gag and Virus-Like Particle (VLP) Release
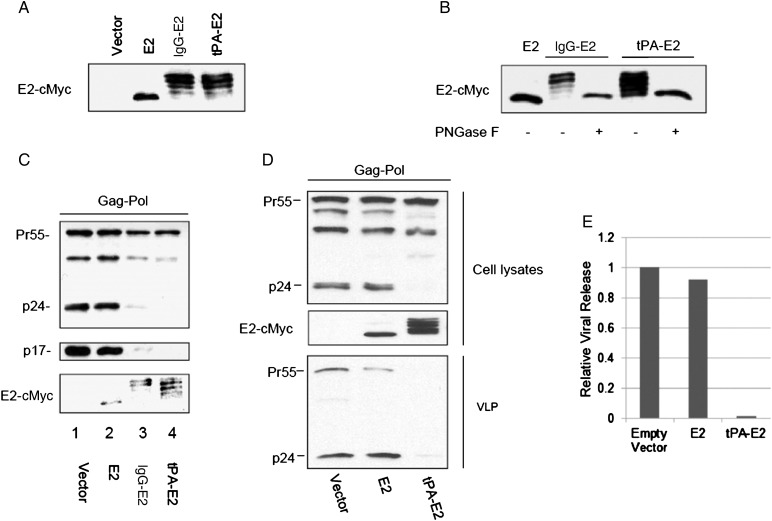
E2 is predicted to be expressed in a glycosylated form and targeted to the endoplasmic reticulum during GBV-C replication. To express E2 in its glycosylated state, the secretory signal peptide of tPA and IgG were fused to the N terminus of E2 to create tPA-E2 and IgG-E2, respectively. 293T cells were transfected with empty vector or E2, tPA-E2, or IgG-E2 expression constructs. Forty-eight hours after transfection, Western blot analysis of the cell lysates showed that the tPA and IgG leader sequence caused E2 to have an electrophoretic banding pattern commonly observed for glycosylated proteins, whereas the unglycosylated form of E2 migrated as a single band (Figure 1A). To confirm that sugar moieties were responsible for the banding pattern observed with tPA-E2 and IgG-E2, 293T cells lysates expressing tPA-E2 and IgG-E2 were digested using a protein deglycosylation mixture containing PNGase F, which deaminates asparagine to aspartic acid, to remove asparagine-linked oligosaccharide chains. Western blot analysis showed that the deglycosidase treatment caused tPA-E2 and IgG-E2 to mobilize as a 42-kD band that was indistinguishable from the band of the unglycosylated E2 control (Figure 1B). To determine whether E2 affects proteolytic processing of HIV-1 Gag, the HIV-1 Gag-Pol expression construct pGPCINS was cotransfected into 293T cells with empty vector, E2, IgG-E2, or tPA-E2. As shown by Western blot analysis in Figure 1C, when HIV-1 Gag-Pol was coexpressed with empty vector or E2, the Gag precursor was efficiently processed into p24 and p17 (Figure 1C). However, when Gag-Pol was coexpressed with tPA-E2 or IgG-E2, the p24 and p17 productions were dramatically inhibited, with no noticeable change in Gag precursor production (Figure 1C), suggesting that the expression of tPA-E2 or IgG-E2 inhibited HIV-1 Gag precursor processing. We also cotransfected the p24 expression construct with empty vector or with E2, tPA-E2, or IgG-E2 expression vectors. Western blot analysis showed that the expression of E2, tPA-E2, or IgG-E2 did not alter the p24 expression level (data not shown). These results ruled out the possibility that tPA-E2 or IgG-E2 affected p24 stability. They further demonstrated that the glycosylated E2 (designated “glycol-E2”) inhibited HIV-1 Gag processing. To investigate whether glycol-E2 affects Gag VLP release, the Gag-Pol expression construct was cotransfected with empty vector, E2, or the tPA-E2 expression construct. Forty-eight hours after transfection, the culture supernatants were harvested for collection of VLPs by ultracentrifugation, as described in Materials and Methods. Figure 1D shows that glycol-E2 dramatically inhibited Gag-Pol VLP release, as determined by densitometry analysis (Figure 1E). Cell viability and vitality analysis showed that E2 and IgG-E2 expression did not alter cell viability (Supplementary Figure S1).
Figure 1.
Expression of GB virus type C (GBV-C) glycosylated E2 inhibits proteolytic processing of HIV-1 Gag and virus-like particle (VLP) release. A, E2 or E2 featuring an N-terminal tissue plasminogen activator (tPA) or immunoglobulin G (IgG) leader sequence was expressed in 293T cells. Cells were lysed and analyzed by Western blotting with an anti-Myc antibody. B, 293T cells expressing IgG-E2 or tPA-E2 were treated with PNGase F (+) for 4 hours at 37°C or left untreated (−). E2 was expressed as a control. Samples were analyzed by Western blot with an anti-Myc antibody. C and D, The effect of GBV-C E2 on HIV-1 Gag processing was analyzed in 293T cells cotransfected with empty vector, E2, IgG-E2, or tPA-E2. Cells were harvested for Western blotting with c-Myc (E2, IgG-E2, and tPA-E2), HIV-1 anti-p24, or p17 antibodies to analyze protein expression in lysates. VLPs were also monitored by Western blot, using an anti-p24 antibody (D). E, The relative level of viral release was measured by densitometry analysis of the blot shown in panel D.
Glycol-E2 Inhibits HIV-1 Release but Not Maturation
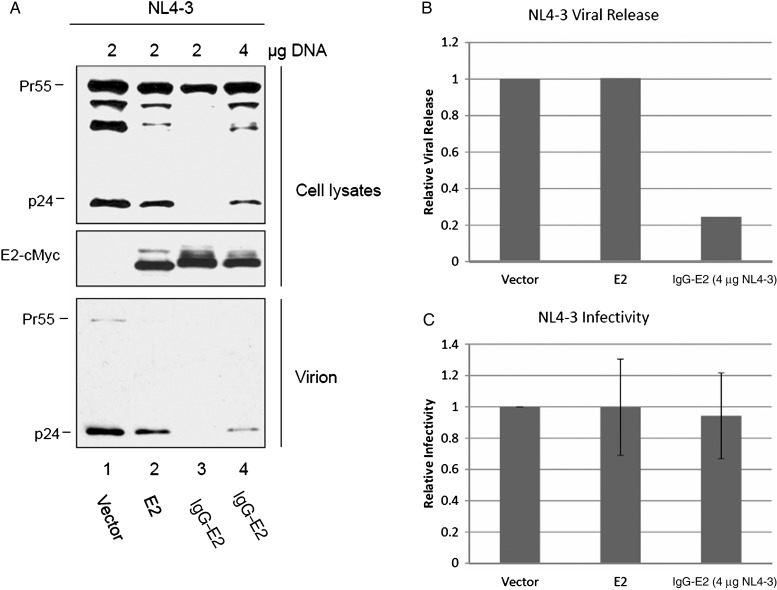
To determine whether glycol-E2 inhibits HIV-1 release, the HIV-1 proviral construct NL4-3 was cotransfected with empty vector or with E2 or IgG-E2 expression constructs. Cell lysates and viral pellets were analyzed by Western blotting. For an unknown reason, overall expression of NL4-3 Gag was reduced when the IgG-E2 and NL4-3 plasmids were cotransfected at a 1:1 ratio, using 2 μg of each plasmid (Figure 2A). To assess Gag processing, the expression of Pr55 has to be the same among all samples. Therefore, 4 µg of the NL4-3 plasmid were used to increase the overall Gag expression level (Figure 2A) to levels seen with 2 µg NL4-3 in cotransfections with vector or E2 (Figure 2A). As shown in Figure 2A, when similar levels of Gag precursors were expressed, glycol-E2 inhibited intracellular p24 production and NL4-3 viral release (Figure 2A). Densitometry analysis of the Western blot bands of Figure 2A showed that the NL4-3 viral release was reduced approximately 5-fold by the expression of glycol-E2 (Figure 2B). As seen in Supplementary Figure S2, similar results were obtained in more physiologically relevant Jurkat and SupT1 T cells. Viral supernatants were also collected for measuring infectivity by the MAGI assay. After the input viral amount was normalized by HIV-1 p24 ELISA, there was no significant change in viral infectivity among vector, E2, and IgG-E2 transfected samples (Figure 2C). These data suggest that the expression of glycol-E2 did not affect HIV-1 maturation.
Figure 2.
Glycosylated E2 inhibits human immunodeficiency virus type 1 (HIV-1) release but not viral maturation. A, 293T cells were cotransfected with HIV-1 proviral construct pNL4-3 along with empty vector, E2, or immunoglobulin G (IgG)–E2 at a 1:1 or 1:2 ratio. Cell lysates and virions concentrated from cell culture supernatants were harvested 48 hours after transfection and analyzed by Western blotting with anti-p24 or anti-cMyc. B, The relative viral release was determined by densitometry analysis of panel A. C, After normalizing virus by p24 enzyme-linked immunosorbent assay (top), supernatants were used to infect TZM-β-gal indicator cells to measure viral infectivity, using the MAGI assay.
Expression of Glycol-E2 Inhibits HIV-1 Gag Membrane Targeting
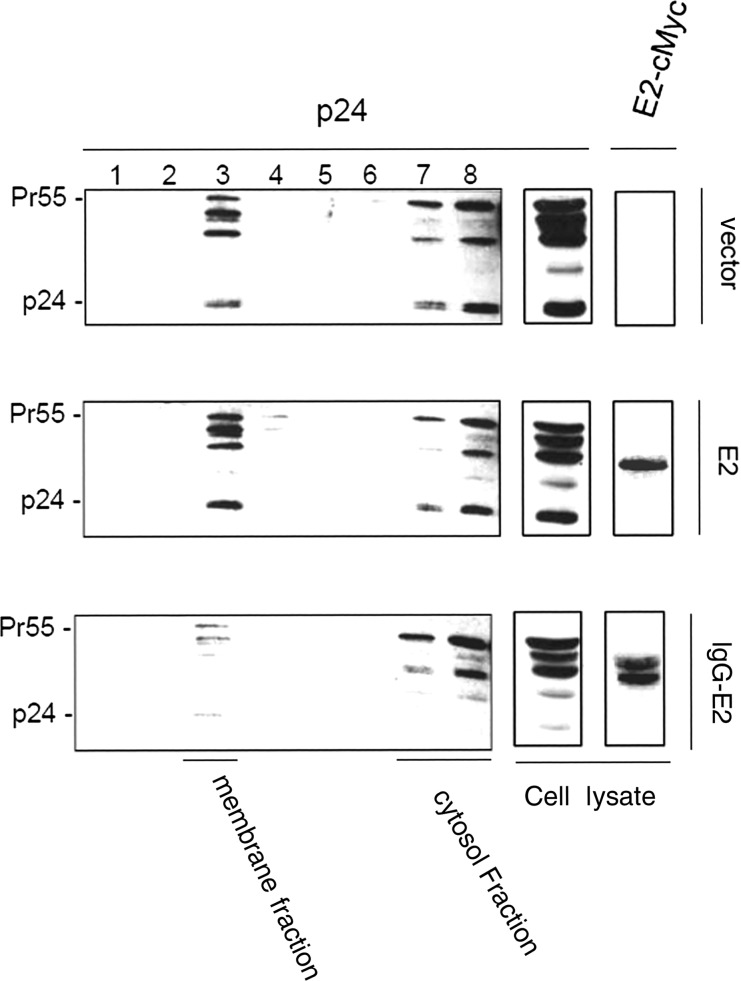
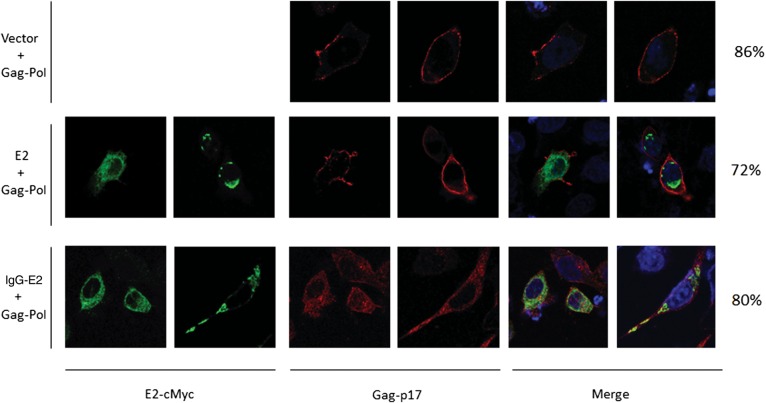
Gag association with cellular membranes is a critical step in HIV-1 assembly and release. To examine whether glyco-E2 expression alters Gag membrane targeting, the Gag-Pol expression construct was cotransfected into 293T cells with empty vector or with E2 or IgG-E2 expression vectors. Forty-eight hours after transfection, cell lysates were subjected to membrane floatation centrifugation and Western blot analysis, as described in Materials and Methods. In this assay, membrane-associated proteins migrate to the interface between 65% and 10% sucrose (Figure 3). When Gag-Pol was expressed alone with empty vector or with E2, significant amounts of Gag were localized in fraction 3, representing the membrane-associated protein (Figure 3A and 3B). By contrast, in cells coexpressing glycol-E2, most of the Gag was localized in fractions corresponding to cytosolic Gag. Only residual amounts of Gag were targeted to the membrane-associated fraction (Figure 3C). This result suggests that the expression of glycol-E2 inhibited HIV-1 Gag membrane targeting. We next sought to confirm the membrane targeting effect of glycol-E2 by immunofluorescence microscopy. The Gag-Pol expression construct was transfected into HeLa cells along with empty vector or with E2 or IgG-E2 expression vectors. Other experiments confirmed that the inhibitory effect of IgG-E2 on Gag assembly and release could be replicated in the HeLa cell line (data not shown). The cells were stained with antibodies directed against Gag-p17 for analysis of Gag subcellular localization. E2 and IgG-E2 were stained with a monoclonal anti-cMyc antibody. Gag is known to have 3 types of subcellular distribution patterns that can be observed through fluorescent microscopy: diffused, intracellular punctate, or plasma membrane localization. Twenty-three hours after transfection, the localization of Gag was assessed by confocal microscopy. As seen in Figure 4, in the absence of E2 or glycol-E2, Gag was shown to be punctate and primarily localized toward the periphery along the plasma membrane. A similar distribution pattern was also seen with cells containing a plasmid encoding unglycosylated E2. However, in the presence of glycol-E2, Gag was predominantly accumulated throughout the cytoplasm. These data indicate that the expression of glyco-E2 disrupted Gag trafficking to the plasma membrane, resulting in intracellular accumulation of Gag.
Figure 3.
Expression of glycosylated E2 inhibits HIV-1 Gag membrane targeting. The Gag-Pol expression vector was cotransfected with empty vector or with E2 or immunoglobulin G (IgG)–E2 expression vector. Two days after transfection, 293T cells were sonicated. Nuclear fractions and unlysed cells were removed by centrifugation. The clear postnuclear supernatants were mixed with 85.5% sucrose to adjust the sample to 2 mL with 73% sucrose. The sample was then placed at the bottom of a centrifuge tube and layered with 7 mL of 65% sucrose in Tris–ethylenediaminetetraacetic acid (TE) and 3.25 mL of 10% sucrose in TE. The sucrose step gradient was centrifuged at 35 000 rpm for 18 hours at 4°C in a Beckman SW41 rotor. Eight fractions were collected from each sample after ultracentrifugation. The fraction samples and total cell lysates of Gag-Pol cotransfected vector (Figure 3A), E2 (Figure 3B), or IgG-E2 (Figure 3C) were analyzed by Western blot.
Figure 4.
Glycosylated E2 inhibits Gag targeting to the plasma membrane. HeLa cells were transfected with Gag-Pol along with empty vector, E2, or immunoglobulin G (IgG)–E2. Twenty-three hours after transfection, cells were fixed, permeabilized, and immunostained, as described in Materials and Methods. Gag and E2 proteins were detected with anti-p17 polyclonal antisera and anti-cMyc, respectively. Gag is shown in red (center), and E2 proteins are shown in green (left). Fifty cells were counted in each group. The percentage of cells showing the presented pattern was calculated and is shown in the far right of the figure.
Membrane Interaction Domain (MID) Is Essential for the Inhibitory Effect of Glycol-E2 on HIV-1 Gag Processing and Release
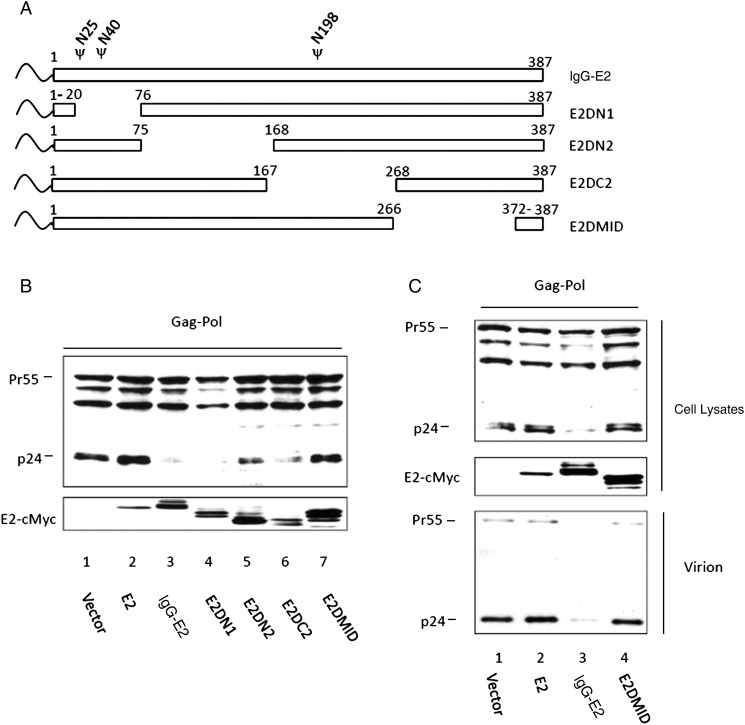
To determine the essential domain for the inhibitory effect of glycol-E2, a series of internal deletion mutants of IgG-E2 were generated, as shown in Figure 5A. The Gag-Pol expression vector was cotransfected with empty vector, E2, IgG-E2, and the IgG-E2 internal deletion mutants. Forty-eight hours after transfection, cell lysates were analyzed by Western blot analysis. As shown in Figure 5B, among the 4 internal deletion mutants, E2DMID lost the inhibitory effect on Gag processing (Figure 5B). Moreover, the inhibitory effect of E2DN2 was partially lost (Figure 5B). However, when increasing amounts of E2 plasmids were cotransfected with Gag-Pol, E2DMID continued to have no effect on Gag precursor processing, whereas E2DN2 behaved more like wild-type IgG-E2 (data not shown). This result indicates that the MID of E2 was essential for its inhibitory effect. The Gag-Pol expression vector was also cotransfected with empty vector and with E2, IgG-E2, and E2DMID expression vectors. Forty-eight hours after transfection, cell and viral lysates were analyzed by Western blot. E2DMID showed no inhibition in Gag processing and viral release (Figure 5C). The results confirmed that the MID was essential for the inhibitory effect. The MID comprises 2 membrane interaction domains (aa 267–298 and aa 347–364) [44–46] and the putative transmembrane domain (aa 336–387) [47].
Figure 5.
The membrane interaction domains (MIDs) of GB virus type C E2 are essential for its inhibitory effect on human immunodeficiency virus type 1 (HIV-1) Gag processing and release. A, Schematic of immunoglobulin G (IgG)–E2 and mutant constructs. B, Gag-Pol was expressed in the presence of E2, IgG-E2, or truncated IgG-E2 constructs (E2DN1, E2DN2, E2DC2, and E2DMID) by transfection of 293T cells. Cells were harvested and lysed 2 days after transfection to analyze processing of the Gag-Pol precursor by Western blot analysis. C, E2, IgG-E2, or the E2DMID mutant was cotransfected with Gag-Pol into 293T cells. Two days after transfection, Western blotting was used to assess processing of HIV-1 Gag and virus-like particle release.
Expression of Glycol-E2 Also Inhibits SIV and XMRV Gag Processing
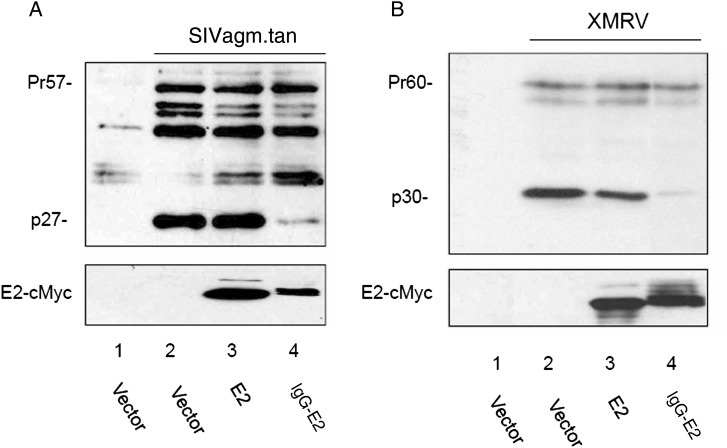
To test whether glycol-E2 also has an inhibitory effect on the processing of other retroviral Gag proteins, the SIVagm.tan proviral construct and XMRV proviral construct XMRV VP62 were cotransfected with vector and E2 and IgG-E2 expression vectors. Forty-eight hours after transfection, cell lysates were analyzed by Western blot analysis. IgG-E2 efficiently inhibited both SIV and XMRV Gag processing (Figure 6A and 6B). These data suggest that GBV-C glycol-E2 inhibits not only HIV-1 Gag processing and release, but also Gag expressed from other retroviruses, such as SIV and XMRV.
Figure 6.
Expression of glycol-E2 also inhibits simian immunodeficiency virus (SIV) and xenotropic murine leukemia virus–related virus (XMRV) Gag processing. Empty vector, E2, or immunoglobulin G (IgG)–E2 were cotransfected with the SIV (A) or XMRV (B) proviral construct into 293T cells. Cells were lysed 2 days after transfection for Western blot analysis. Human immunodeficiency virus type 1 immunoglobulin (HIV-IG), anti-MuLV p30 antisera, and anti-Myc antibody (4A6) were used to stain SIV, XMRV, and E2 proteins, respectively.
DISCUSSION
GBV-C and HIV-1 coinfection has been associated with prolonged survival among HIV-1–infected patients in clinical studies [48]. It has been shown that E2, NS5A, and NS3 have inhibitory effects on HIV-1 replication. In this study, we showed that the expression of E2 was able to inhibit HIV-1 Gag processing and viral release. The inhibitory effects were glycosylation dependent because they were eliminated when E2 was expressed in the unglycosylated form (Figure 1C and 1D). Plasma membrane targeting of HIV-1 Gag is an important initiation step of Gag processing and release. A Gag matrix mutant has been shown to be unable to target to the plasma membrane, failing to be processed and resulting in a lower level of particle release [29, 30]. In this study, the membrane floatation and confocal microscopy analysis showed that the expression of glycol-E2 disrupted targeting of HIV-1 Gag to the plasma membrane (Figures 3 and 4), eventually inhibiting Gag processing and release. Furthermore, this inhibitory effect did not appear to be limited to HIV-1, as glycol-E2 was also able to disrupt processing of SIVagm.tan and XMRV Gag proteins. The inhibitory effect was not observed with E2 lacking a signal sequence, indicating the importance of glycosylation and/or trafficking for E2 to exert an inhibitory effect. An recent study by Xiang et al showed that expression of the glycosylated form of an E2 fragment in a CD4+ T-cell line inhibited HIV-1 replication [22]. More interestingly, when E2 expressing Jurkat cells and naive MT-2 cells were cocultured, E2 significantly inhibited HIV-1 replication in MT-2 cells. However, the inhibitory effect was not observed when Jurkat/E2 and MT-2 cells were separated using a transwell system. This suggests that the inhibitory effect of E2 is dependent on cell-to-cell contact. It would be interesting to see whether the inhibitory effect is due to the defect of Gag membrane targeting.
The observed defect in Gag plasma membrane targeting in the presence of glycol-E2 expression may be attributed to several factors. One possibility is that glycol-E2 may redirect Gag trafficking to intracellular compartments. As illustrated in Figure 4, HIV-1 Gag was found to primarily localize on the plasma membrane or intracellularly, in a punctate pattern within the perinuclear region. In contrast, coexpression with glycol-E2 markedly changed Gag distribution to a pattern within the cytoplasm. This observation suggests that glycol-E2 mislocalizes Gag trafficking to other compartments, thereby preventing it from trafficking to the plasma membrane. The observation that E2 lacking a signal sequence did not alter Gag trafficking reveals that E2 trafficking through the endoplasmic reticulum and Golgi apparatus is important for its inhibitory effect on Gag trafficking. It is well known that cellular factors, such as phospholipid phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2], clathrin adaptor protein 3 (AP-3), Golgi-localized γ-ear containing Arf-binding (GGA), and ADP ribosylation factor (Arf), play important roles in HIV-1 Gag plasma membrane targeting [43, 49, 50]. The expression of glycol-E2 may render the cellular factor(s) unavailable to assist in Gag targeting to the plasma membrane. This warrants further investigation.
Because of the ability of HIV-1 to develop drug-resistant mutations, there is a constant need for the development of new therapeutic options for HIV-1–infected patients. In this study, we showed that E2 was able to inhibit HIV-1 replication by interfering with Gag trafficking to the plasma membrane. It suggests that when glycol-E2 is introduced into HIV-1–susceptible cells, these cells will be converted to nonsusceptible cells by blocking viral release. Importantly, glycol-E2 did not dramatically alter HIV-1 Pr55 precursor expression (Figures 1 and 2) but, instead, caused it to be retained intracellularly. This observation suggests that the accumulated viral proteins may eventually be presented to the host immune system to boost host immune responses against HIV-1 infection.
GBV-C E2 has been documented as an HIV-1–entry inhibitor in a number of studies. Our study suggests that E2 blocks Gag targeting to the plasma membrane, resulting in the disruption of virus release. These results and those of previous studies support the hypothesis that E2 can inhibit multiple stages of HIV-1 replication. Taken together, along with the inhibitory effects of GBV-C NS5A, NS3, and other advantages of GBV-C, such as its nonpathogenicity, its ability to replicate in CD4+ T cells, and its shared transmission pathway with HIV-1, the inhibitory effects of E2 render GBV-C an attractive source for the development of novel anti-HIV/AIDS therapies.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Xiao-Fang Yu (Johns Hopkins University) and Vical (San Diego, CA), for reagents, and Phuong Thi Sarkis, for assistance in editing the manuscript. The following reagents were obtained through the NIH-ARRRP, Division of AIDS, NIAID, NIH: pAF121950, from Jinhua Xiang and Jack Stapleton; pNL4-3, from Malcolm Martin; pSIVagmTan-1, from Marcelo Soares and Beatrice Hahn; XMRV VP62 cDNA, from Robert H. Silverman and Beihua Dong; HIV-1 p24 hybridoma (183-H12-5C), from Bruce Chesebro; antiserum to HIV-1 p17, from Paul Spearman; HIV-IG, from NABI and NHLBI; and TZM-bl, from John C. Kappes, Xiaoyun Wu, and Tranzyme.
Financial support. This work was supported by the National Institutes of Health (grants U54MD007593/U54RR026140, SC1GM089269, G12MD007586, and P30AI054999 to B. L.; and training grants 5T32HL007737, 5T32AI007281, and 2R25GM059994 to C. L. T).
Potential conflict of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed
References
- 1.Simons JN, Desai SM, Schultz DE, Lemon SM, Mushahwar IK. Translation initiation in GB viruses A and C: evidence for internal ribosome entry and implications for genome organization. J Virol. 1996;70:6126–35. doi: 10.1128/jvi.70.9.6126-6135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fogeda M, Navas S, Martin J, et al. In vitro infection of human peripheral blood mononuclear cells by GB virus C/hepatitis G virus. J Virol. 1999;73:4052–61. doi: 10.1128/jvi.73.5.4052-4061.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiang J, Wunschmann S, Schmidt W, Shao J, Stapleton JT. Full-length GB virus C (hepatitis G virus) RNA transcripts are infectious in primary CD4-positive T cells. J Virol. 2000;74:9125–33. doi: 10.1128/jvi.74.19.9125-9133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiang J, Wunschmann S, Diekema DJ, et al. Effect of coinfection with GB virus C on survival among patients with HIV infection. N Engl J Med. 2001;345:707–14. doi: 10.1056/NEJMoa003364. [DOI] [PubMed] [Google Scholar]
- 5.Stapleton JT. GB virus type C/Hepatitis G virus. Semin Liver Dis. 2003;23:137–48. doi: 10.1055/s-2003-39943. [DOI] [PubMed] [Google Scholar]
- 6.Heringlake S, Ockenga J, Tillmann HL, et al. GB virus C/hepatitis G virus infection: a favorable prognostic factor in human immunodeficiency virus-infected patients? J Infect Dis. 1998;177:1723–6. doi: 10.1086/517431. [DOI] [PubMed] [Google Scholar]
- 7.Toyoda H, Fukuda Y, Hayakawa T, Takamatsu J, Saito H. Effect of GB virus C/hepatitis G virus coinfection on the course of HIV infection in hemophilia patients in Japan. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;17:209–13. doi: 10.1097/00042560-199803010-00004. [DOI] [PubMed] [Google Scholar]
- 8.Lefrere JJ, Roudot-Thoraval F, Morand-Joubert L, et al. Carriage of GB virus C/hepatitis G virus RNA is associated with a slower immunologic, virologic, and clinical progression of human immunodeficiency virus disease in coinfected persons. J Infect Dis. 1999;179:783–9. doi: 10.1086/314671. [DOI] [PubMed] [Google Scholar]
- 9.Yeo AE, Matsumoto A, Hisada M, Shih JW, Alter HJ, Goedert JJ. Effect of hepatitis G virus infection on progression of HIV infection in patients with hemophilia. Multicenter Hemophilia Cohort Study. Ann Intern Med. 2000;132:959–63. doi: 10.7326/0003-4819-132-12-200006200-00006. [DOI] [PubMed] [Google Scholar]
- 10.Tillmann HL, Heiken H, Knapik-Botor A, et al. Infection with GB virus C and reduced mortality among HIV-infected patients. N Engl J Med. 2001;345:715–24. doi: 10.1056/NEJMoa010398. [DOI] [PubMed] [Google Scholar]
- 11.Takamatsu J, Toyoda H, Fukuda Y. GB virus C and mortality from HIV infection. N Engl J Med. 2002;346:377–9. [PubMed] [Google Scholar]
- 12.Nunnari G, Nigro L, Palermo F, et al. Slower progression of HIV-1 infection in persons with GB virus C co-infection correlates with an intact T-helper 1 cytokine profile. Ann Intern Med. 2003;139:26–30. doi: 10.7326/0003-4819-139-1-200307010-00009. [DOI] [PubMed] [Google Scholar]
- 13.Xiang J, George SL, Wunschmann S, Chang Q, Klinzman D, Stapleton JT. Inhibition of HIV-1 replication by GB virus C infection through increases in RANTES, MIP-1alpha, MIP-1beta, and SDF-1. Lancet. 2004;363:2040–6. doi: 10.1016/S0140-6736(04)16453-2. [DOI] [PubMed] [Google Scholar]
- 14.Jung Sa, Knauer Oa, Donhauser Na, et al. Inhibition of HIV strains by GB virus C in cell culture can be mediated by CD4 and CD8 T-lymphocyte derived soluble factors. AIDS. 2005;19:1267–72. doi: 10.1097/01.aids.0000180097.50393.df. [DOI] [PubMed] [Google Scholar]
- 15.Xiang J, McLinden JH, Chang Q, Kaufman TM, Stapleton JT. An 85-aa segment of the GB virus type C NS5A phosphoprotein inhibits HIV-1 replication in CD4+ Jurkat T cells. Proc Natl Acad Sci U S A. 2006;103:15570–5. doi: 10.1073/pnas.0604728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang Q, McLinden JH, Stapleton JT, Sathar MA, Xiang J. Expression of GB virus C NS5A protein from genotypes 1, 2, 3 and 5 and a 30 aa NS5A fragment inhibit human immunodeficiency virus type 1 replication in a CD4+ T-lymphocyte cell line. J Gen Virol. 2007;88:3341–6. doi: 10.1099/vir.0.83198-0. [DOI] [PubMed] [Google Scholar]
- 17.Xiang J, McLinden JH, Chang Q, Jordan EL, Stapleton JT. Characterization of a peptide domain within the GB virus C NS5A phosphoprotein that inhibits HIV replication. PLoS One. 2008;3:e2580. doi: 10.1371/journal.pone.0002580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung S, Eichenmuller M, Donhauser N, et al. HIV entry inhibition by the envelope 2 glycoprotein of GB virus C. AIDS. 2007;21:645–7. doi: 10.1097/QAD.0b013e32803277c7. [DOI] [PubMed] [Google Scholar]
- 19.Herrera E, Gomara MJ, Mazzini S, Ragg E, Haro I. Synthetic peptides of hepatitis G virus (GBV-C/HGV) in the selection of putative peptide inhibitors of the HIV-1 fusion peptide. J Phys Chem B. 2009;113:7383–91. doi: 10.1021/jp900707t. [DOI] [PubMed] [Google Scholar]
- 20.Koedel Y, Eissmann K, Wend H, Fleckenstein B, Reil H. Peptides derived from a distinct region of GB virus C glycoprotein E2 mediate strain-specific HIV-1 entry inhibition. J Virol. 2011;85:7037–47. doi: 10.1128/JVI.02366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohr EL, Xiang J, McLinden JH, et al. GB virus type C envelope protein E2 elicits antibodies that react with a cellular antigen on HIV-1 particles and neutralize diverse HIV-1 isolates. J Immunol. 2010;185:4496–505. doi: 10.4049/jimmunol.1001980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiang J, McLinden JH, Kaufman TM, et al. Characterization of a peptide domain within the GB virus C envelope glycoprotein (E2) that inhibits HIV replication. Virology. 2012;430:53–62. doi: 10.1016/j.virol.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.George SL, Varmaz D, Tavis JE, Chowdhury A. The GB virus C (GBV-C) NS3 serine protease inhibits HIV-1 replication in a CD4+ T lymphocyte cell line without decreasing HIV receptor expression. PLoS One. 2012;7:e30653. doi: 10.1371/journal.pone.0030653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freed EO. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 25.Scarlata S, Carter C. Role of HIV-1 Gag domains in viral assembly. Biochim Biophys Acta. 2003;1614:62–72. doi: 10.1016/s0005-2736(03)00163-9. [DOI] [PubMed] [Google Scholar]
- 26.Ono A. HIV-1 assembly at the plasma membrane: gag trafficking and localization. Future Virol. 2009;4:241–57. doi: 10.2217/fvl.09.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan X, Yu X, Lee TH, Essex M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol. 1993;67:6387–94. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou W, Parent LJ, Wills JW, Resh MD. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–69. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci U S A. 1990;87:523–7. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spearman P, Wang JJ, Vander Heyden N, Ratner L. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J Virol. 1994;68:3232–42. doi: 10.1128/jvi.68.5.3232-3242.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunn BM, Goodenow MM, Gustchina A, Wlodawer A. Retroviral proteases. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-4-reviews3006. REVIEWS3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wheeler CJ, Felgner PL, Tsai YJ, et al. A novel cationic lipid greatly enhances plasmid DNA delivery and expression in mouse lung. Proc Natl Acad Sci U S A. 1996;93:11454–9. doi: 10.1073/pnas.93.21.11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo K, Liu B, Xiao Z, et al. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J Virol. 2004;78:11841–52. doi: 10.1128/JVI.78.21.11841-11852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adachi A, Gendelman HE, Koenig S, et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–91. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soares MA, Robertson DL, Hui H, Allan JS, Shaw GM, Hahn BH. A full-length and replication-competent proviral clone of SIVAGM from tantalus monkeys. Virology. 1997;228:394–9. doi: 10.1006/viro.1996.8387. [DOI] [PubMed] [Google Scholar]
- 36.Urisman A, Molinaro RJ, Fischer N, et al. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–54. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varthakavi V, Browning PJ, Spearman P. Human immunodeficiency virus replication in a primary effusion lymphoma cell line stimulates lytic-phase replication of Kaposi's sarcoma-associated herpesvirus. J Virol. 1999;73:10329–38. doi: 10.1128/jvi.73.12.10329-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–64. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Shao Q, Yu X, Kong W, Hildreth JE, Liu B. N-terminal hemagglutinin tag renders lysine-deficient APOBEC3G resistant to HIV-1 Vif-induced degradation by reduced polyubiquitination. J Virol. 2011;85:4510–9. doi: 10.1128/JVI.01925-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao Q, Wang Y, Hildreth JE, Liu B. Polyubiquitination of APOBEC3G is essential for its degradation by HIV-1 Vif. J Virol. 2010;84:4840–4. doi: 10.1128/JVI.01911-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ono A, Freed EO. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J Virol. 1999;73:4136–44. doi: 10.1128/jvi.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong X, Li H, Derdowski A, et al. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell. 2005;120:663–74. doi: 10.1016/j.cell.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 44.Casas J, Espina M, Haro M, et al. Interfacial properties of a synthetic peptide derived from hepatitis G virus E2 protein: interaction with lipid monolayers. Langmuir. 2006;22:246–54. doi: 10.1021/la051812h. [DOI] [PubMed] [Google Scholar]
- 45.Larios C, Casas J, Alsina MA, Mestres C, Gomara MJ, Haro I. Characterization of a putative fusogenic sequence in the E2 hepatitis G virus protein. Arch Biochem Biophys. 2005;442:149–59. doi: 10.1016/j.abb.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 46.Perez-Lopez S, Nieto-Suarez M, Mestres C, Alsina MA, Haro I, Vila-Romeu N. Behaviour of a peptide sequence from the GB virus C/hepatitis G virus E2 protein in Langmuir monolayers: its interaction with phospholipid membrane models. Biophys Chem. 2009;141:153–61. doi: 10.1016/j.bpc.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 47.Pilot-Matias TJ, Carrick RJ, Coleman PF, et al. Expression of the GB virus C E2 glycoprotein using the Semliki Forest virus vector system and its utility as a serologic marker. Virology. 1996;225:282–92. doi: 10.1006/viro.1996.0602. [DOI] [PubMed] [Google Scholar]
- 48.Bhattarai N, Stapleton JT. GB virus C: the good boy virus? Trends Microbiol. 2012;20(3):124–30. doi: 10.1016/j.tim.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joshi A, Garg H, Nagashima K, Bonifacino JS, Freed EO. GGA and Arf proteins modulate retrovirus assembly and release. Mol Cell. 2008;30:227–38. doi: 10.1016/j.molcel.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci U S A. 2004;101:14889–94. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.