Abstract
The triatomines vectors of Trypanosoma cruzi are principal factors in acquiring Chagas disease. For this reason, increased knowledge of domestic transmission of T. cruzi and control of its insect vectors is necessary. To contribute to genetic knowledge of North America Triatominae species, we studied genetic variations and conducted phylogenetic analysis of different triatomines species of epidemiologic importance. Our analysis showed high genetic variations between different geographic populations of Triatoma mexicana, Meccus longipennis, M. mazzottii, M. picturatus, and T. dimidiata species, suggested initial divergence, hybridation, or classifications problems. In contrast, T. gerstaeckeri, T. bolivari, and M. pallidipennis populations showed few genetics variations. Analysis using cytochrome B and internal transcribed spacer 2 gene sequences indicated that T. bolivari is closely related to the Rubrofasciata complex and not to T. dimidiata. Triatoma brailovskyi and T. gerstaeckeri showed a close relationship with Dimidiata and Phyllosoma complexes.
Introduction
The subfamily Triatominae includes species that are important vectors in the transmission of the protozoan parasite Trypanosoma cruzi, the causal agent of Chagas disease. Although more than 33 species of triatomines have been reported in Mexico,1,2 epidemiologic, biologic, and genetic data for these insects in the literature is scarce. These data are crucial for establishing an appropriate strategy of Chagas disease control. Currently, vector control is the only way to control this infection because of limited efficacy of treatment during the chronic infectious phase and the absence of a vaccine.3,4
Phylogenetic and genetic relationships between Triatoma bolivari, T. brailovskyi, T. mexicana, T. gerstaeckeri, T. recurva, and T. protracta have not been documented. Triatoma brailovskyi, T. bolivari, and T. mexicana are species that are endemic to Mexico and considered as part of the Phyllosoma complex species (Meccus bassolsae, M. longipennis, M. mazzottii, M. pallidipennis, M. picturatus, and M. phyllosomus).
Triatoma brailovskyi and T. bolivari have been found in the states of Colima, Nayarit, Oaxaca, and Jalisco.2,5–8 Their epidemiologic importance as potential vectors of T. cruzi is unknown. Triatoma mexicana has been found in the states of Guanajuato, Hidalgo, Queretaro, and San Luis Potosi, where the rate of T. cruzi infections is less than 4%.1,9,10 Triatoma gerstaeckeri has been found in the states of Chihuahua, Coahuila, Nuevo Leon, San Luis Potosi, Sonora, Hidalgo, Tamaulipas, and Veracruz2,10–13 and in the United States in Texas.14 In some of these states, this species was associated with human houses and T. cruzi infections rates of 0% to 63%.10,15–18 Triatoma recurva has been reported in the states of Baja California, Baja California Sur, Chihuahua, Guerrero, Nayarit, Sinaloa and Sonora, and a high level of T. cruzi infection greater than 90%,1,19 and has been reported in the United States in Arizona. Triatoma protracta has been reported in the states of Baja California Norte, Chihuahua, Coahuila, Durango, Nuevo Leon, Tamaulipas, Sinaloa, Sonora, San Luis Potosi, and Zacatecas and in several states in the southwestern United States (Arizona, California, Nevada, Texas, New Mexico, and Utah), with T. cruzi infections less than 19%.1,9,10,20,21
Triatoma brailovskyi has been proposed to be a member of a specific complex Dimidiata by using morphologic marker analysis; this complex includes T. dimidiata, T. hegneri, T. ryckmani and T. gomeznunezi.22 On the basis of internal transcribed spacer 2 (ITS-2) sequences, the species T. gerstaeckeri and T. mexicana have been included in the Phyllosoma complex, and T. bolivari has been included in the Rubrofasciata complex.23 Triatoma protracta is included in the Protracta complex, and T. recurva was proposed to be included in the Phyllosoma complex by using cytochrome B (cytB) sequences, although this last species reportedly shares intermediate characteristics with the genus Dipetalogaster.22,24
In North America, genetic population studies of triatomines by using nucleotide sequences are scarce. Most of this research focuses on T. dimidiata25–27 In some cases, large hybrid zones exist between sympatric sibling species.28 Triatoma recurva and T. rubida showed significant genetic differences when cytB and cytochrome oxidase I sequences were compared in populations from widely separated geographic localities.24 With respect to the Phyllosoma complex species, the principal studies have focused on taxonomic relationships. However, genetic population analyses are scarce. These types of studies are important because taxonomic status of these species would provide useful information.
The present study used cytB and ITS-2 sequences and taxonomic analysis to examine genetic variation between the T. brailovskyi, T. bolivari, T. gerstaeckeri, T. mexicana, T. protracta, and T. recurva, and with species of their respective complexes (Phyllosoma and Protracta) and other sequences of triatomines of epidemiologic importance in North America.
Materials and Methods
Taxon sampling.
Adults of the species T. bolivari, T. brailovskyi, T. gerstaeckeri, T. mexicana, T. protracta, and T. recurva were collected in different states in Mexico (Table 1). Specimens were identified morphologically by using keys of Lent and Wygodzinsky,9 Martínez and others,5 and Carcavallo and others.6 Similar specimens are deposited in the collections of the Laboratorio de Tripanosomiasis of Instituto de Investigaciones Biomédicas, Universidad Nacional Autónoma de México and Centro Universitario del Sur, Universidad de Guadalajara.
Table 1.
GenBank accession numbers and genetic diversity of species of triatomines analyzed, Mexico*
| Species | Origin | cytB (no.) | ITS-2 (no.) | ||
|---|---|---|---|---|---|
| Meccus bassolsae (Triatoma bassolsae) | Puebla | AY85941029 (1) | AY860394,29 AY860400-230 AM28672423 (5) | π = 0.005, h = 3, θ = 0.7, S = 99%, T = −1.14 | |
| T. barberi | Oaxaca | AY83013729 (1) | |AJ29359030 (1) | ||
| T. bolivari | Jalisco | JQ282718-9† (2) | π = 0, h = 1, θ = 1, S = 100% | JQ282701-3† (3) | π = 0.005, h = 3, θ = 0.7, S = 99%, T = −1.14 |
| Oaxaca | AM28672523 (1) | ||||
| T. brailovskyi | Jalisco | JQ282720-2† (3) | π = 0.006, h = 2, θ = 0.67, S = 99% | JQ282704-6† (3) | π = 0, h = 1, θ = 0, S = 100% |
| T. gerstaeckeri | Chihuahua | JQ282723-5† (3) | π = 0.004, h = 3, θ = 1, S = 99.4% | JQ282707-9† (3) | π = 0.001, h = 2, θ = 0.5, S = 100% |
| San Luis Potosi | AM28673423 (1) | ||||
| T. lecticularia | Nuevo Leon | AY85941429 (1) | AY860405-729 (3) | π = 0.007, h = 3, θ = 1, S = 99% | |
| Meccus longipennis (Triatoma longipennis) | Nayarit | AY85941229(1) | π = 0.093, h = 2, θ = 1, S = 91% | AY860397-829 (2) | π = 0.004, h = 3, θ = 1, S = 99% |
| Zacatecas | DQ19881524 (1) | AJ28688345 (1) | |||
| T. mexicana | Guanajuato | JQ282726-7† (2) | π = 0.058, h = 3, θ = 1, S = 92% | JQ282710-2† (3) | π = 0.02, h = 3, θ = 0.8, S = 96% |
| Hidalgo | DQ11897629(1) | AM28672823 (1) | |||
| T. nitida | Guatemala | AF04572331 (1) | AM28673323 (1) | ||
| T. protracta | ? Sonora | AF04572731 (1) | π = 0.038, h = 3, θ = 1, S = 94% | JQ282713† (1) | π = 0.02, h = 2, θ = 0.7, S = 99% |
| Chihuahua | JQ282728-9† (2) | JQ282714-5† (2) | |||
| M. pallidipennis (Triatoma pallidipennis) | ? Morelos | AF04572431 (1), AY859419-20,29 DQ19881424 (3) | π = 0.015, h = 4, θ = 1, S = 97%, T = −0.15 | AJ286882,45 AM286729-30,23 AY860395,29 AY86040329 (5) | π = 0.003, h = 3, θ = 0.8, S = 99% |
| M. picturatus (T. picturata) | Nayarit | AY85941329 (1) | π = 0.14, h = 2, θ = 1, S = 86% | AY860396,29 AY860399,30 AY86040429 (3) | π = 0.002, h = 3, θ = 0.8, S = 99% |
| Jalisco | DQ19881724 (1) | AJ28688446 (1) | |||
| M. phyllosomus (Triatoma phyllosoma) | Oaxaca | AY859411,29 DQ198818,24 HQ185139-6446 (28) | π = 0.016, h = 10, θ = 0.6, S = 94%, T = −1.7 | AJ286881,45 AY860403,29 HQ185165-9046(28) | π = 0.004, h = 11, θ = 0.7, S = 96%, T = −2.4 |
| T. mazzottii (Meccus mazzottii) | Guerrero | AY859421-2229 (2) | π = 0.09, h = 3, θ = 1, S = 87% | AY860392-329 (2) | π = 0.003, h = 2, θ = 0.7, S = 99.5% |
| Oaxaca | DQ19881624 (1) | AJ28688545 (1) | |||
| T. dimidiata | Several places | AY859417-8,29 AF301594,44 AY062149-64b, FJ197154-9,25 FN641804-1842 (40) | π = 0.08, h = 35, θ = 0.94, S = 87%, FSTA-2 = 0.89, FSTB-2 = 0.81, FSTA-B = 0.46, π1 = 0.04, π2 = 0.01, πA = 0.02, πB = 0.04. | AJ286875-80,45 AM286693-723,23 AY860408-17,26 DQ871354-56,43 EF383122-29,26 FJ197146-53,25 GQ214508-1342 (72) | π = 0.02, h = 25, θ = 0.9 S = 93%, FSTA-2 = 0.84, FSTB-2 = 0.85, FSTA–B = 0.47, π1 = 0.008, π2 = 0.002, πA = 0.004, πB = 0.005 |
| T. recurva | Sonora | DQ198812,24 JQ282730-1† | π = 0.009, h = 3, θ = 1, S = 99%, T = 0.04 | JQ282716-7† (2) | π = 0, h = 1, θ = 0, S = 100% |
| Arizona, USA | DQ19881324 (4) | ||||
| T. rubida | Nayarit | DQ198810,24 | π = 0.05, h = 5, θ = 1, S = 89%, T = 0.04 | AM28673523 (1) AY860389-9129 (3) | π = 0, h = 1, θ = 0 S = 100% |
| Sonora | AY859415-1630 (3) | ||||
| Baja California Sur | DQ19881124 (1) | ||||
| Arizona, USA | DQ198808-0924 (1) | ||||
Numbers in superscript indicate article that referred to the GenBank accession no. of the sequence analyzed in this study, cytB = cytochrome B; ITS-2 = internal transcribed spacer 2; ? = unknown location; π = nucleotide diversity; h = no. haplotypes; θ = haplotype diversity; S = similarity percentage; T = Tajima's D test; FST1–2 = coancestry coefficient (CC) between group 1 and 2; FSTA–B = CC between subgroup A and group 2; FSTB–2 = CC between subgroup B and group 2; π1 = nucleotide diversity group 1; π2 =ND group 2; πA = ND subgroup A; πB = ND subgroup B.
Sequences obtained in this study.
DNA extraction, amplification, cloning, and sequencing.
Genomic DNA was extracted from one leg of each insect by using the extraction method of Martínez and others.5 The oligonucleotides used for the amplification of cytB and ITS-2 sequences have been described.29,30 Fragments obtained by amplification were subcloned into the cloning vector pCR2.1 (Invitrogen, Carlsbad, CA) and sequenced by using universal vector primers (T7 promoter and M13 reverse). Sequencing for both chains was performed by using the 310 ABI prism sequencer (Applied Biosystems, Foster City, CA).
Population genetic analysis.
The percentage of polymorphic sites, nucleotide diversity (π), and haplotype polymorphism (θ) were calculated within populations of the same species. When the samples size was sufficient, the coancestry coefficient statistics (FST) and Tajima's D test were evaluated between populations. All tests were estimated using DnaSP version 4 software (www.ub.edu/dnasp/).
Sequence alignment and phylogenetic analysis.
Multiple alignments were made by using MEGA version 5 software (www.megasoftware.net/). Modeltest version 3.7 software www.molecularevolution.org/software/phylogenetics/modeltest) was used to determine the appropriate model of molecular evolution. For cytB sequences, the best model was the Hasegawa Kishino Yano model with gamma distribution and invariable sites. For ITS-2 sequences and both markers (cytB and ITS-2), the best model was the general time reversible model with gamma distribution. The phylogenetic reconstruction using Bayesian inference was performed with the Mr. Bayes 3.1.2 program (http://mrbayes.sourceforge.net/). The analysis ran for 10 million generations, sampling trees every 100 generations. Trees with scores lower than those at stationery (burn-in) were discarded from the analysis. The sampled trees that reached the stationary phase were collected and used to build majority consensus trees. Other species used in this study were Dipetalogaster maximus AF045728; T. infestans AF045721, DQ118975; T. sanguisuga AF045725; Rhodnius prolixus AF045718, DQ118977 with cytB and D. maximus AJ286887; T. infestans AY860387-8; and R. prolixus DQ118978 with ITS-2 sequences.29–31
Phylogenetic inferences based on RNA secondary structure.
RNA secondary structure of ITS-2 sequences from M. bassolsae, T. bolivari, T. brailovskyi, T. dimidiata, T. gerstaeckeri, M. mazzottii, T. mexicana, M. pallidipennis, M. phyllosomus, and T. recurva were predicted by using consensus alignment and singles sequences in three different web software: RNAalifold WebServer (http://rna.tbi.univie.ac.at/cgi-bin/alifold.cgi), RNA Folding form (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form), and RNA secondary structure prediction (www.genebee.msu.su/services/rna2_reduced.html). Structural domains in ITS-2 initially were predicted according to the frequency of bases and mutation information in the multiple alignments and then identified based on the structure logo program RNA Structure Logo (www.cbs.dtu.dk/∼gorodkin/appl/slogo.html).32,33
Results
Sequence analysis.
Thirty-one sequences with both markers were obtained and deposited in GenBank (Table 1). The size of the amplified cytB gene fragment was 313 basepairs; for ITS-2, it ranged from 454 to 584 basepairs. The cytB sequences showed a larger number of variables (49.8%) and informative sites (43.5%) than ITS-2 (39.9% and 34.8%, respectively). As expected for a cytB protein–coding gene, the third codon positions had the most variation, (75%), followed by the first (19.6%) and second (5.4%) positions.
Intraspecific variation in cytB of the species in the Phyllosoma complex (T. mexicana, M. mazzottii, M. picturatus, M. longipennis, and M. pallidipennis) from different geographic areas was analyzed. Sequences of T. mexicana from Hidalgo and Guanajuato showed 92% similarity and π = 0.058 between them; M. mazzottii from Guerrero and Oaxaca showed 87% similarity and π = 0.09; M. picturatus from Nayarit and Jalisco showed 86% similarity (43 of 313) and π = 0.14; M. longipennis from Nayarit and Zacatecas showed 91% similarity (29 of 313) and π = 0.093; and M. pallidipennis from Jalisco and other regions in Morelos showed 97% similarity (9 of 313) and π = 0.015. Triatoma dimidiata had two groups (groups 1 and 2), and group 1 had two subgroups (subgroups A and B) (Figure 1). The percentage of nucleotides variability between subgroups was 85% (47 of 313) with FSTA–B = 0.46, and the variation between subgroups A and B compared with group 2 was 79% (67 of 313) and FSTA-2 = 0.89 and 80% (64 of 313) and FST2B–2 = 0.81, respectively. For T. protracta from Chihuahua and unknown regions of Mexico, we observed 94% similarity (18 of 313) and π = 0.038.
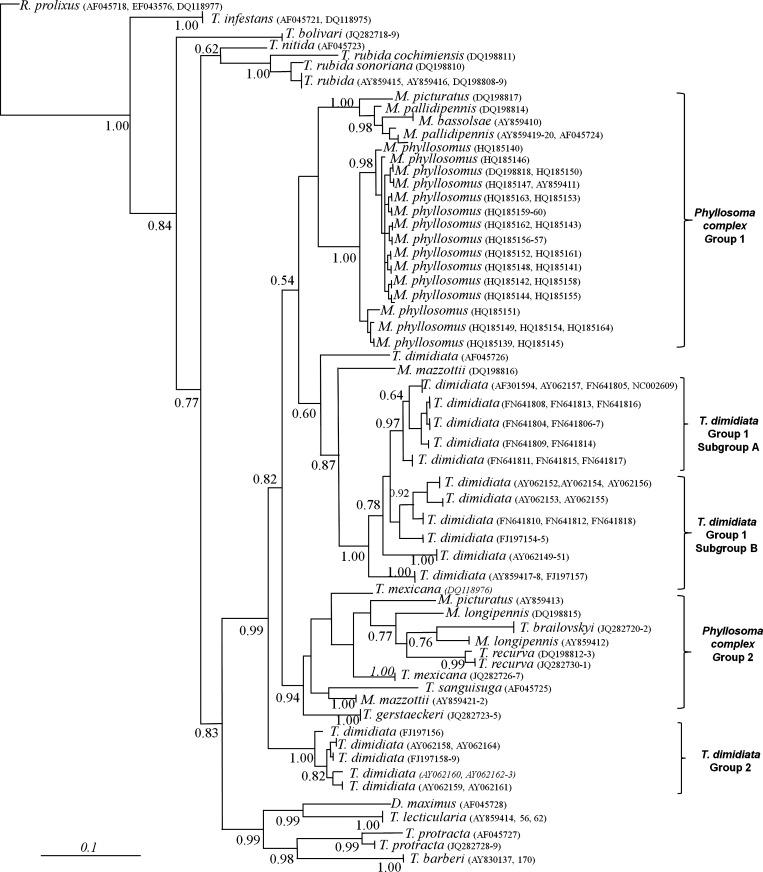
Figure 1.
Bayesian phylogenetic trees of triatomine insects with cytochrome B (cytB) sequences. Majority rule phylogram resulting from Bayesian analysis with the cytB data set under the Hasegawa, Kishino, and Yano + gamma distribution + proportion invariate model of evolution. Scale bar indicates nucleotide substitutions per site.
Using the ITS-2 marker, we found that T. bolivari, T. gerstaeckeri, M. pallidipennis, M. picturatus, M. mazzottii, T. recurva, and T. rubida populations showed high values of similarity (≥ 99%) and low diversity values (π ≤ 0.005, θ ≥ 0.7), in contrast with populations of T. protracta and T. mexicana, which showed high genetic differentiation (Table 1). As with cytB, T. dimidiata showed a similar grouping. The percentage of variable nucleotides between the subgroups (A and B) was 98.5% (7 of 459) and FSTA–B = 0.47. In contrast with the second group, which was integrated with 21 sequences, we observed 94% similarity (28 of 459) and FSTA–2 = 0.84 and 97.7% similarity (11 of 459) with FSTB–2 = 0.85 between subgroups A and B, respectively.
Microsatellite sequences.
Triatoma brailovskyi showed microsatellites identical to those of some species of the Phyllosoma complex ((AT)4TTT(AT)5). For T. bolivari, their microsatellites ((AT)4TTTAAA(AT)5) were more similar to those of T. rubida ((AT)4TTTATT(AT)5), a species that belongs to the Rubrofasciata complex. Triatoma mexicana had three microsatellites, two from the samples collected in Guanajuato ((AT)3TTTATA(AT)4 and (AT)5T(AT)6) and one from Hidalgo ((AT)5TTT(AT)6). Triatoma protracta had two microsatellites sequences, one from Sonora ((AT)3TTTTATAA(AT)4) and one from Chihuahua ((AT)3TTTTATAA(AT)4). T. recurva microsatellites sequences (AT)4TTTATT(AT)5 were more similar to T. bolivari microsatellites; only two mutations were observed.
Phylogenetic analysis.
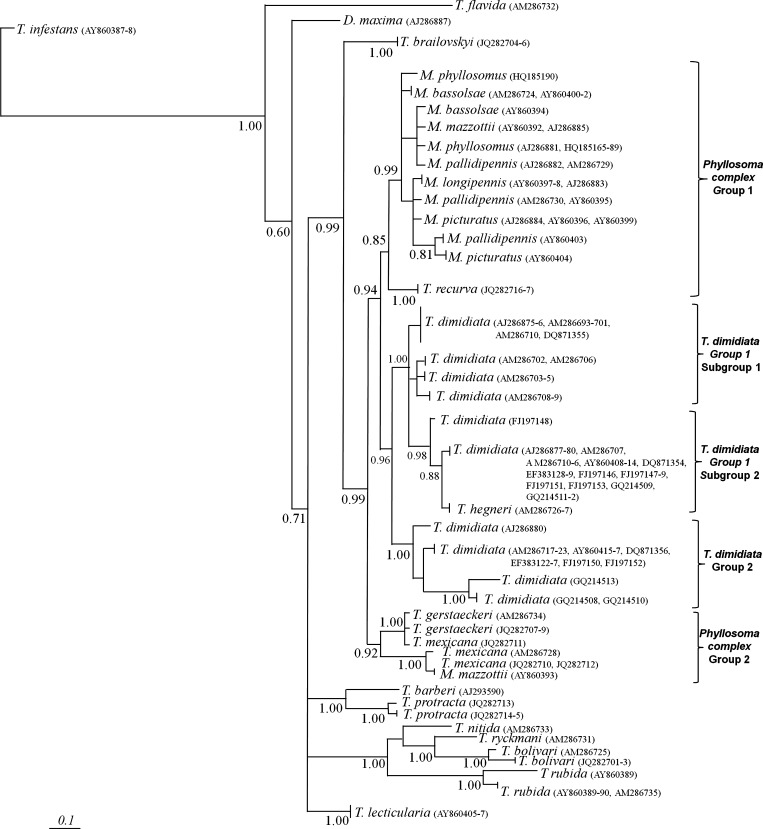
Similarity between phylogenetic reconstructions using Bayesian inference with the cytB (Figure 1) and ITS-2 sequences (Figure 2) was observed. Phylogenetic analyses of Phyllosoma and Dimidiata complex species and T. brailovskyi, T. gerstaeckeri, and T. recurva showed a strong clade with 0.99 of posterior probability. In this major clade, there are four subgroups: T. dimidiata group 1 (with two subgroups), T. dimidiata group 2, and Phyllosoma complex groups 1 and 2.
Figure 2.
Bayesian phylogenetic trees of triatomine insects with internal transcribed spacer (ITS-2). Majority rule phylogram resulting from Bayesian analysis with the ITS-2 data set under the general time reversible + gamma distribution model of evolution. Values at nodes indicate percentage of the posterior probability. Scale bar indicates nucleotide substitutions per site.
Triatoma bolivari showed a clear relationship with T. rubida, with posterior probability values of 77% for cytB and 100% for ITS-2. T. protracta and T. barberi showed high similarity with T. bolivari.
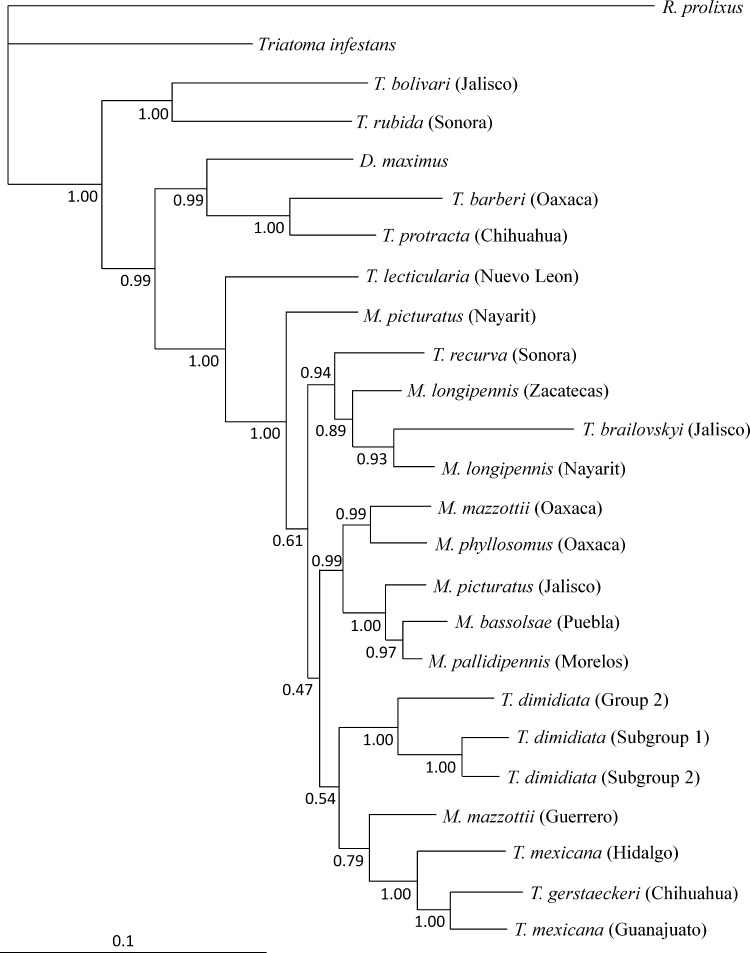
To increase support for clades of the separate trees and to obtain a better evolutionary correlation, a third phylogenetic tree was built to align cytB and ITS-2 for each species. Only sequences that had similar numbers of basepairs and geographic origins for both markers were used. A total of 1,053 characteristics were analyzed, with approximately 20% informative sites. Meccus longipennis, T. mexicana, and M. picturatus from different geographic origins were distributed into separate clades. Among the other species, the genetic relationships were similar to the topology for particulars trees (Figure 3).
Figure 3.
Bayesian phylogenetic trees of triatomine insects using cytochrome B (cytB) and internal transcribed spacer 2(ITS-2) sequences. Majority rule phylogram resulting from Bayesian analysis with cytB and ITS-2 data set under the general time reversible + gamma distribution + proportion invariate model of evolution. Values at nodes indicate percentage of the posterior probability. Scale bar indicates nucleotide substitutions per site.
RNA secondary structure.
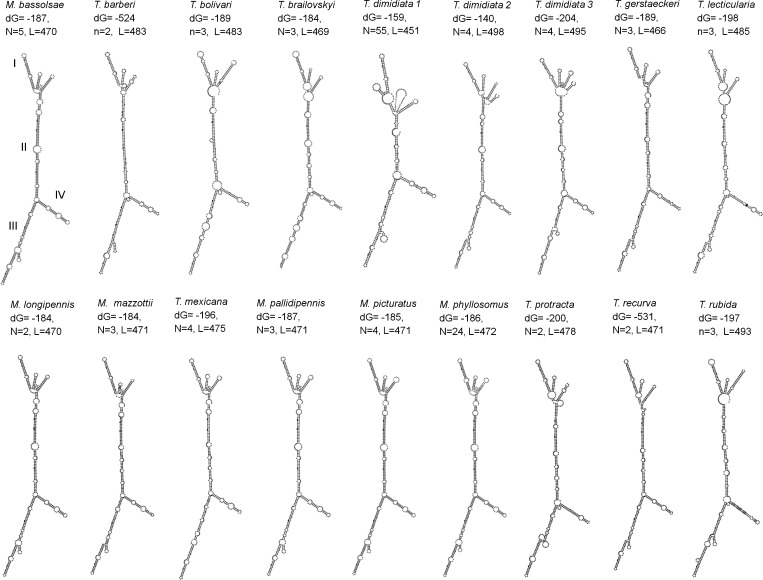
The ITS-2 RNA secondary structures obtained showed the lowest free energy structures, with values less than −184 kcal/mol denoting highly stable models. Four preserved domains were identified (I, II, III, and IV) by using consensus alignment and logo structure. All analyzed sequences had four domains and some modifications between hairpins and bulges in each domain. Meccus bassolsae, M. longipennis, M. mazzottii, M. pallidipennis, M. phyllosomus, M. picturatus, and T. gerstaeckeri had similar structures and few modifications in domains I and II. Triatoma mexicana showed high similarity between domains I, II and IV and modifications in domain III, and T. recurva showed similarity only in domain IV with the above mentioned species.
Triatoma dimidiata RNA secondary structures were constructed according to the sequences that the ITS-2 tree used for each group or subgroup: T. dimidiata subgroup A was constructed with 15 sequences, T. dimidiata subgroup B with 20 sequences, and T. dimidiata group 2 with 16 sequences. These groups showed three structures with modifications in domains I, II, and III and were similar in domain IV. Triatoma bolivari and T. rubida showed similarity within domains I and IV; T. barberi and T. protracta shared only domain I. In general, domains II and IV were the most conserved (Figure 4).
Figure 4.
RNA secondary structures of internal transcribed spacer 2 for species of triatomines. dG = entrophy energy; n = no. sequences used; L = no. basepairs.
Discussion
This study analyzed five species of Triatoma from Mexico and the United States for which little genetic and epidemiologic data are available: T. brailovskyi, T. bolivari, T. gerstaeckeri, T. recurva, and T. protracta. Triatoma gerstaeckeri and T. recurva have been associated with human dwellings and high rates of T. cruzi infections (90%). In contrast, T. bolivari, T. brailovskyi, and T. protracta are considered to be sylvatic species.5–7,10,23,34
The aim of this study was to determine genetic variation in populations and perform taxonomic analysis by using ITS-2 and cytB sequences of some triatomine species from North America, particularly in Phyllosoma, Dimidiata, and Rubrofasciata complexes. Although several studies have also focused on these aspects, we expanded our analysis to include important phenotypic characteristics such RNA secondary structures, which commonly determine similarity and evolutionary relationships between species. The secondary structures of ITS-1 and ITS-2 regions of RNA reportedly are involved in RNA ribosome maturation processing. Therefore, structures that retain a highly conserved configuration may be useful tools for analyzing this cryptic species.35
Using cytB sequences, we detected low values of intraspecific similarity (86–91%) and high values of nucleotide diversity (0.09–0.14) between the Phyllosoma complex species M. longipennis, M. mazzottii, T. mexicana, and M. picturatus from different geographic areas. For example, divergence levels for several T. infestans populations in South America did not exceed 2%36; Within T. rubida populations from Mexico and the United States, the percentage similarity was 87%, and within the T. recurva population, the percentage similarity was 92%.24 In contrast, T. brasiliensis populations showed high divergence.37
A species complex was proposed for T. rubida, T. recurva, and T. brasiliensis. Analysis of our data from the Phyllosoma complex suggests two hypotheses. The first hypothesis is that geographic fragmentation of these populations generated genetic variants that could indicate either a subspecies or a speciation process. The second hypothesis is that these differences are caused by classification problems related to morphologic plasticity or hybridization of different triatomines species, resulting in genetic introgression. Both hypotheses are feasible. For example, inconsistency in morphologic and genetic characteristics between sympatric species as a result of natural hybridization has been reported, and re-validation of the subspecies in this complex has been proposed.38–41 In areas with sympatric speciation, classification problems could be common, and occasionally morphologic variants are not genetically differentiated.
Unlike previous analyzes of T. dimidiata that focused on genetic grouping and correlation with geographic origin or subspecies of T. dimidiata,23,25 our intraspecific variation analysis formed two groups based on habits: the first group corresponds to insects with domestic and peridomestic habits and the second group corresponds to sylvatic insects. Data for both genetic markers analyzed were similar, and we observed the greatest variation between these two groups. According to the fixation index, the observed null interaction between the two groups and both markers (FST ≥ 0.81) suggests limited migrations. Similar results were reported by Tamay-Segovia and others, who analyzed two groups in a tropical area (sylvatic) and a mainly coastal distribution (peridomestic and domestic).26
Triatoma protracta also showed high variation between the two populations. In this regard, Ryckman proposed five subspecies,20 and Lent and Wygodzinsky proposed synonymy.9 No previous genetic studies have analyzed these populations. However, these types of studies are important because T. protracta populations have high variation, and genetic analyses can verify the taxonomic status of these populations.
We conducted few ITS-2 sequence-structure analyses because no comparative geographic data are available for some species. Triatoma bolivari, T. gerstaeckeri, and M. pallidipennis showed low intraspecific similitude between populations (99.3–99.4%), indicating homogeny that is independent of their geographic distributions. In this instance, data suggested that natural selection pressures are stable and do not favor the new variant generation. However, we suggest that these data should be confirmed by extensive population genetic studies using several markers and other genes.
The cytB sequences, RNA secondary structures of ITS-2 (with similarity between domains II, III and domain IV) and microsatellites sequences ((AT)4TTT(AT)7) grouped insects in T. dimidiata group 2 with species of the Phyllosoma complex. Triatoma dimidiata group 2 from sylvatic areas could be the origin of the entire Phyllosoma complex species and T. dimidiata group 1. The domestication process could be an important source of genetic variation between groups. Insects in the Phyllosoma complex from sylvatic areas should be analyzed to determinate this relationship. An additional important consideration is the taxonomic controversy, particularly in the inclusion or separation of T. dimidiata from the Phyllosoma complex, which could be complicated by genetic similarity between these groups (T. dimidiata group 2 and Phyllosoma complex group 2).
Previous taxonomic classification of T. brailovskyi morphologically related this species to the Dimidiata complex (T. dimidiata, T. hegneri, and T. gomeznunezi).22 We observed that T. brailovskyi was related to the Phyllosoma complex on the basis of analogous microsatellites ((AT)4TTT(AT)5) and a strong phylogenetic relationship with M. longipennis and T. mexicana as demonstrated by cytB and ITS-2 trees. However, T. brailovskyi had a different ITS-2 secondary structure and greater similarities with T. lecticularia (Lecticularia complex) in domain I, T. protracta (Protracta complex) in domain 2, and the Phyllosoma complex in domains III and IV. The phylogenetic tree consensus (cytB and ITS-2) and RNA structure suggest that T. brailovskyi could be related to T. recurva and M. longipennis from Zacatecas.
Our study is the first to demonstrate that Phyllosoma complex species have genetic differences between T. mexicana, M. longipennis, and M. picturatus; these species have at least two geographically differentiated variants. This finding is important because genetic diversity or phylogenetic relationships with other species or genera could also be different. Therefore, we propose that the Phyllosoma complex is divided into two paraphyletic genetic groups: group 1 is composed of M. bassolsae, M. longipennis, M. mazzottii, M. pallidipennis, M. phyllosomus, and M. picturatus, and group 2 is composed of T. brailovskyi, T. mexicana, T. gerstaeckeri, T. recurva, and other variants of M. mazzottii and M. picturatus. Phylogenetic, taxonomic, and paraphyletic characteristics of this complex need to carefully considered because of large genetic variations in some species such as M. mazzottii, M. longipennis, and M. picturatus.
RNA secondary structure analyzed in species of the Phyllosoma complex (T. mexicana, M. bassolsae, T. gerstaeckeri, M. longipennis, M. pallidipennis, M. picturatus, M. mazzottii, M. phyllosomus, and T. recurva) generally showed four domains with similar morphology and minimal variation in domains I, II, and IV. Domain IV seems to be characteristic of M. bassolsae, M. longipennis, M. mazzottii, M. pallidipennis, M. phyllosomus, and M. picturatus. The ITS-2 secondary structure correlation with minimal changes observed and results of phylogenetic analysis suggested close evolutionary relationships in the Phyllosoma complex.
Pfeiler and others used cytB and the cytochrome oxidase I gene to include T. recurva in the Phyllosoma complex.24 Our data for cytB and ITS-2 corroborates this inclusion. Moreover, T. recurva showed greater similarity to M. longipennis when cytB sequences were compared and with M. mazzottii when ITS-2 secondary structure was analyzed.
Triatoma species have been related morphologically to the Rubrofasciata complex (T. rubida and T. rubrofasciata) because of similar topology for ITS-2 and cytB and similarities in secondary structure of domains I and IV of RNA in T. bolivari and T. rubida.22,23 We have confirmed this relationship. Additional morphologic and genetic studies are needed to determine potential relationships between others species.
Finally, there is little data for triatomine groupings. Our analysis reflects the difficulty in separating these insects because of their high genetic variability. The continuous domestication process and fragmentation of habitats could be an important variation source. Therefore, grouping proposals demand careful consideration. These considerations are useful for establishing adequate vector control and for increasing biologic, taxonomic, and phylogenetic information for this these important species.
Footnotes
Financial support: This study was supported by the Consejo Nacional de Ciencia y Tecnología (grant no. 113985, Dirección General de Asuntos del Personal Académico, and Universidad Nacional Autónoma de México grant no. 229209 to Bertha Espinoza.
Authors' addresses: Bertha Espinoza, Patricia De La Torre, and Juan Perdo Laclette, Departamento de Inmunología, Instituto de Investigaciones Biomédicas, Universidad Nacional Autónoma de México, Circuito Escolar, Ciudad Universitaria, PC 04510, Distrito Federal, México, E-mails: besgu@biomedicas.unam.mx, dltorre@biomedicas.unam.mx, and laclette@servidor.unam.mx. Jose Alejandro Martínez-Ibarra, Área de Entomología Médica, Centro Universitario del Sur, Universidad de Guadalajara, Ciudad Guzmán, Jalisco, PC 49000, México, E-mail: aibarra@cusur.udg.mx. Guiehdani Villalobos, Departamento de Parasitología, Escuela de Ciencias Biológicas, Instituto Politécnico Nacional, Prolongación de Carpio y Plan de Ayala S/N, Col. Casco de Sto. Tomas, PC 11340, DF, México, E-mail: guiehda@yahoo.com.mx. Fernando Martínez-Hernández, Departamento de Ecología de Agentes Patógenos, Hospital General Doctor Manuel Gea González, Tlalpan 4800, Col. Sección XVI, PC 14080, Distrito Federal, México, E-mail: fherxyz@yahoo.com.
References
- 1.Zárate LG, Zárate RJ. A checklist of the Triatominae (Hemiptera: Reduviidae) of México. Int J Entomol. 1985;27:102–127. [Google Scholar]
- 2.Galvão C, Carcavallo R, Da Silva Rocha D, Jurberg J. A checklist of the current valid species of the subfamily Triatominae Jeannel, 1919 (Hemiptera: Reduviidae) and their geographical distribution with nomenclatural and taxonomic note. Zootaxa. 2003;202:1–36. [Google Scholar]
- 3.World Health Organization . Control of Chagas Disease Second Report of the WHO Expert Committee. Geneva: World Health Organization; 2002. [Google Scholar]
- 4.Clayton J. Chagas disease: pushing through the pipeline. Nature. 2010;465:S12–S15. doi: 10.1038/nature09224. [DOI] [PubMed] [Google Scholar]
- 5.Martínez A, Carcavallo RU, Pelaez D. Triatoma brailovskyi, nueva especie de triatominae de México. Chagas. 1984;1:72–75. [Google Scholar]
- 6.Carcavallo RU, Martínez A, Peláez D. Una nueva especie de Triatoma Laporte en México. Chagas. 1987;4:477–477. [Google Scholar]
- 7.Ramsey JM, Ordoñez R, Cruz-Celis A, Alvear A, Chávez V, López R, Pintor JR, Gama F, Carrillo S. Distribution of domestic triatominae and stratification of Chagas disease transmition in Oaxaca, Mexico. Med Vet Entomol. 2000;14:19–30. doi: 10.1046/j.1365-2915.2000.00214.x. [DOI] [PubMed] [Google Scholar]
- 8.Martínez-Ibarra JA, Martínez-Hernández F, Villalobos G, Vences-Blanco MO, Salazar-Schettino PM. Update on the distribution of Triatoma bolivari and Triatoma brailovskyi (Hemiptera: Reduviidae: Triatominae) in western Mexico. J Vector Ecol. 2010;35:432–434. doi: 10.1111/j.1948-7134.2010.00103.x. [DOI] [PubMed] [Google Scholar]
- 9.Lent H, Wygodzinsky P. Revision of de Triatominae (Hemiptera: Ruduviidae) and their significance as vector of Chagas disease. Bull Am Mus Nat Hist. 1979;163:125–520. [Google Scholar]
- 10.Vidal-Acosta V, Ibáñez S, Bernal R, Martínez-Campos C. Infección natural de chinches Triatominae con Trypanosoma cruzi asociada a la vivienda humana en México. Salud Publica Mex. 2000;42:496–503. [PubMed] [Google Scholar]
- 11.Martínez-Ibarra JA, Galaviz-Silva L, Lara-Campos C, Trujillo-García JC. Distribución de los triatominos asociados al domicilio humano en el municipio General de Terán, Nuevo León, México. Southwest Entomologist. 1992;17:261–265. [Google Scholar]
- 12.Licon-Trillo A. Infección de Triatoma recurva por Trypanosoma cruzi en um campamento minero de Urique, Chihuaha, México. Rev Salud Publica Med Nut. 2006;65:1–8. [Google Scholar]
- 13.Molina-Garza ZJ, Rosales-Encina JL, Galaviz-Silva L, Molina-Garza D. Prevalencia de Trypanosoma cruzi en triatominos silvestres de Nuevo León, México. Salud Publica Mex. 2007;49:37–44. doi: 10.1590/s0036-36342007000100006. [DOI] [PubMed] [Google Scholar]
- 14.Beard C, Pye G, Steurer FJ, Rodriguez R, Campman R, Townsend P, Ramsey J, Wirtz RA, Robinson LE. Chagas disease in a domestic transmission cycle, southern Texas. Emerg Infect Dis. 2003;9:103–105. doi: 10.3201/eid0901.020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood SF. Notes on the distribution and habits of Reduviid vectors of Chagas disease in the Southwestern United States. Pan-Pac Entomol. 1941;27:85–94. [Google Scholar]
- 16.Sullivan TD, McGregor T, Eads RB, Davis DJ. Incidence of Trypanosoma cruzi, Chagas, in Triatoma (Hemiptera, Reduviidae) in Texas. Am J Trop Med. 1949;29:453–458. doi: 10.4269/ajtmh.1949.s1-29.453. [DOI] [PubMed] [Google Scholar]
- 17.Eads RB, Trevino HA, Campos EG. Triatoma (Hemiptera: Reduviidae) infected with Trypanosoma cruzi in south Texas wood rat dens. Southwest Nat. 1963;8:38–42. [Google Scholar]
- 18.Burkholder JE, Allison TC, Kelly VP. Trypanosoma cruzi (Chagas) (Protozoa: Kinetoplastida) in invertebrate, reservoir, and human hosts of the Lower Rio Grande Valley of Texas. J Parasitol. 1980;66:305–311. [PubMed] [Google Scholar]
- 19.Paredes GEA, Valdez-Miranda J, Nogueda-Torres B, Alejandre-Aguilar R, Canett-Romero R. Vectorial importance of Triatominae bugs (Hemiptera: Reduviidae) in Guaymas, México. Rev Latinoam Microbiol. 2001;43:119–122. [PubMed] [Google Scholar]
- 20.Ryckman RE. Biosystematics and hosts of the Triatoma protracta complex in North America (Hemiptera: Reduviidae) (Rodentia: Cricetidae) University of California Publications in Entomology. 1962;27:93–240. [Google Scholar]
- 21.Reisenman CE, Lawrence G, Guerenstein PG, Gregory T, Dotson E, Hildebran JG. Infection of kissing bugs with Trypanosoma cruzi, Tucson Arizona, USA. Emerg Infect Dis. 2010;16:400–405. doi: 10.3201/eid1603.090648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carcavallo R, Jurberg J, Lent H, Noireau F, Galvão C. Phylogeny of the Triatominae (Hemiptera: Reduviidae). Proposals for taxonomic arrangements. Entomol Vector. 2000;7:1–101. [Google Scholar]
- 23.Bargues MD, Klisiowicz DR, Gonzalez-Candelas F, Ramsey JM, Monroy C, Ponce C, Salazar-Schettino PM, Panzera F, Abad-Franch F, Sousa OE, Schofield CJ, Dujardin JP, Guhl F, Mas-Coma S. Phylogeography and genetic variation of Triatoma dimidiata, the main Chagas disease vector in Central America, and its position within the genus Triatoma. Plos Negl Trop Dis. 2008;2:1–19. doi: 10.1371/journal.pntd.0000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeiler E, Bitler BG, Ramsey JM, Palacios-Cardiel C, Markow TA. Genetic variation population structure and phylogenetic relationships of Triatoma rubida and Triatoma recurva (Hemiptera: Reduviidae: Triatominae) from the Sonoran Desert, insect vectors of the Chagas disease parasite Trypanosoma cruzi. Mol Phylogenet Evol. 2006;41:435–446. doi: 10.1016/j.ympev.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Dorn PL, Calderon C, Melgar S, Moguel B, Solorzano E, Dumonteil E, Rodas A, de la Rua N, Garnica R, Monroy C. Two distinct Triatoma dimidiata (Latreille, 1811) Taxa are found in sympatry in Guatemala and Mexico. Plos Negl Trop Dis. 2009;3:e393. doi: 10.1371/journal.pntd.0000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamay-Segovia P, Alejandre-Aguilar R, Martínez F, Zavala-Días de la Serna F, De la Torre P, Laclette JP, Blum-Domínguez C, Espinoza B. Two Triatoma dimidiata clades (Chagas disease vector) associated with different habitats in southern Mexico and Central America. Am J Trop Med Hyg. 2008;78:472–478. [PubMed] [Google Scholar]
- 27.Dumonteil E, Tripet F, Ramirez-Sierra MJ, Payet V, Lanzaro G, Menu F. Assessment of Triatoma dimidiata dispersal in the Yucatan Peninsula of Mexico by morphometry and microsatellite markers. Am J Trop Med Hyg. 2007;76:930–937. [PubMed] [Google Scholar]
- 28.Herrera-Aguilar M, Be-Barragan LA, Ramirez-Sierra MJ, Tripet F, Dorn P, Dumonteil E. Identification of a large hybrid zone between sympatric sibling species of Triatoma dimidiata in the Yucatan peninsula, Mexico, and its epidemiological importance. Infect Genet Evol. 2009;9:1345–1351. doi: 10.1016/j.meegid.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Martínez F, Villalobos G, Cevallos AM, de la Torre P, Laclette JP, Alejandre-Aguilar R, Espinoza B. Taxonomic study of the Phyllosoma complex and other triatomine (Insecta: Hemiptera: Reduviidae) species of epidemiological importance in the transmission of Chagas disease: using ITS-2 and mtcyt B sequences. Mol Phylogenet Evol. 2006;41:279–287. doi: 10.1016/j.ympev.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 30.Marcilla A, Bargues MD, Ramsey JM, Magallon-Gastelum E, Salazar-Schettino PM, Abad-Franch F, Dujardin JP, Schofield CJ, Mas-Coma S. The ITS-2 of the nuclear rDNA as a molecular marker for papulations, species, and phylogenetic relationships in triatominae (Hemiptera: Reduviidae), vector of Chagas disease. Mol Phylogenet Evol. 2001;18:136–142. doi: 10.1006/mpev.2000.0864. [DOI] [PubMed] [Google Scholar]
- 31.Lyman DF, Monteiro FA, Escalante AA, Cordon-Rosales C, Wesson DM, Dujardin JP, Beard CB. Mitochondrial DNA sequence variation among triatomine vectors of Chagas disease. Am J Trop Med Hyg. 1999;60:377–386. doi: 10.4269/ajtmh.1999.60.377. [DOI] [PubMed] [Google Scholar]
- 32.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorodkin J, Heyer LJ, Brunak S, Stormo GD. Displaying the information contents of structural RNA alignments: the structure logos. Comput Appl Biosci. 1997;13:583–586. doi: 10.1093/bioinformatics/13.6.583. [DOI] [PubMed] [Google Scholar]
- 34.Magallón-Gastélum E, Magdaleno-Peñaloza NC, Katthain-Duchateau G, Trujillo-Contreras F, Lozano-Kasten FJ, Hernández-Gutiérrez R. Distribución de los vectores de la enfermedad de Chagas (Hemiptera: Reduviidae: Triatominae), en el estado de Jalisco, México. Rev Biomed. 1998;9:151–157. [Google Scholar]
- 35.Beiggi S, Piercery-Normore MD. Evolutions of ITS ribosomal RNA secondary structures in fungal and alga symbionts of selected of Cladinia sect Cladonia (Cladiniaceae, Ascomycotina) J Mol Evol. 2007;64:528–542. doi: 10.1007/s00239-006-0115-x. [DOI] [PubMed] [Google Scholar]
- 36.Monteiro FA, Perez R, Panzera F, Dujardin JP, Galvao C, Rocha D, Noireau F, Schofield C, Beard CB. Mitochondrial DNA variation of Triatoma infestans populations and its implication on the specific status of T. melanosoma. Mem Inst Oswaldo Cruz. 1999;94:229–238. doi: 10.1590/s0074-02761999000700037. [DOI] [PubMed] [Google Scholar]
- 37.Monteiro FA, Donnelly M, Beard C, Costa J. Nested clade and phylogeographic analyses of the Chagas disease vector Triatoma brasiliensis in northeast Brazil. Mol Phylogenet Evol. 2004;32:46–56. doi: 10.1016/j.ympev.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 38.Martínez-Ibarra JA, Morales-Corona Z, Moreno-Ruiz MG. Híbridos naturales fértiles entre especies del complejo Meccus phyllosomus (Heteroptera: Reduviidae) in Jalisco México. Entomol Mex. 2005;4:313–319. [Google Scholar]
- 39.Martínez-Ibarra JA, Grant-Guillén Y, Morales-Corona ZY, Haro-Rodríguez S, Ventura-Rodríguez LV, Nogueda-Torres B, Bustos-Saldaña R. Importance of species of triatominae (Heteroptera: Reduviidae) in risk of transmission of Trypanosoma cruzi in western Mexico. J Med Entomol. 2008;45:476–482. doi: 10.1603/0022-2585(2008)45[476:iosoth]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 40.Brenière SF, Bosseno MF, Magallón-Gastelúm E, Castillo-Ruvalcaba EG, Soto-Gutiérrez M, Montaño-Luna EC, Tejeda-Basulto J, Mathieu-Daudè F, Walter A, Lozano-Kasten F. Peridomestic colonization of Triatoma longipennis (Hemiptera, Reduviidae) and Triatoma barberi (Hemiptera, Reduviidae) in a rural community with active transmission of Trypanosoma cruzi in Jalisco State, Mexico. Acta Trop. 2007;101:249–257. doi: 10.1016/j.actatropica.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 41.Martínez-Hernández F, Martínez-Ibarra JA, Catalá S, Villalobos G, de la Torre P, Laclette JP, Alejandre-Aguilar R, Espinoza B. Natural crossbreeding between sympatric species of the Phyllosoma complex (Insecta: Hemiptera: Reduviidae) indicate the existence of only one species with morphologic and genetic variations. Am J Trop Med Hyg. 2010;82:74–82. doi: 10.4269/ajtmh.2010.09-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blandon-Naranjo M, Zuriaga MA, Azofeifa G, Zeledon R, Bargues MD. Molecular evidence of intraspecific variability in different habitat-related populations of Triatoma dimidiata (Hemiptera: Reduviidae) from Costa Rica. Parasitol Res. 2010;106:895–905. doi: 10.1007/s00436-010-1762-9. [DOI] [PubMed] [Google Scholar]
- 43.Dorn PL, Monroy C, Curtis A. Triatoma dimidiata (Latreille, 1811): a review of its diversity across its geographic range and the relationship among populations. Infect Genet Evol. 2007;7:343–352. doi: 10.1016/j.meegid.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Dotson EM, Beard CB. Sequence and organization of the mitochondrial genome of the Chagas disease vector, Triatoma dimidiata. Insect Mol Biol. 2001;10:205–215. doi: 10.1046/j.1365-2583.2001.00258.x. [DOI] [PubMed] [Google Scholar]
- 45.Marcilla A, Bargues MD, Ramsey JM, Magallon-Gastelum E, Salazar-Schettino PM, Abad-Franch F, Dujardin JP, Schofield CJ, Mas-Coma S. The ITS-2 of the nuclear rDNA as a molecular marker for populations, species, and phylogenetic relationships in Triatominae (Hemiptera: Reduviidae), vector of Chagas Disease. Mol Phylogenet Evol. 2001;18:136–142. doi: 10.1006/mpev.2000.0864. [DOI] [PubMed] [Google Scholar]
- 46.Villalobos G, Martínez-Hernández F, de la Torre P, Laclette JP, Espinoza B. Entomological indices, feeding sources, and molecular identification of Triatoma phyllosoma (Hemiptera: Reduviidae) one of the main vectors of Chagas disease in the Istmo de Tehuantepec, Oaxaca, Mexico. Am J Trop Med Hyg. 2011;85:490–497. doi: 10.4269/ajtmh.2011.10-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]