Abstract
We evaluated Trypanosoma cruzi infection in 397 wild Mepraia gajardoi specimens from five coastal localities in northern Chile by detection of minicircle DNA by polymerase chain reaction. The wild capture sites were classified into two ecotopes: a fully wild ecotope (ecotope 1) and a wild ecotope near human dwellings (ecotope 2). Infection rates varied between 11% and 27%. Minicircle hybridization assays showed that T. cruzi lineages Tc II and Tc VI were commonly detected in ecotope 1 and ecotope 2, respectively. These results are discussed in the context of insect proximity to human dwellings, the alimentary profile of Mepraia sp., T. cruzi lineages detected in the past in the same disease-endemic area circulating in humans, and Triatoma infestans.
Chagas disease is a human parasitic disease in the Western Hemisphere that is caused by Trypanosoma cruzi, and transmitted by blood-sucking insects of the subfamily Triatominae (Hemiptera: Reduviidae).1 The taxon T. cruzi is divided into six discrete typing units or lineages (Tc I–Tc VI).2 In T. cruzi, mitochondrial DNA or kinetoplast DNA is composed of maxicircles and minicircles. At least 12 maxicircles are present per cell, and they contain mitochondrial genes. The other components are minicircles (10,000–20,000 copies/cell). Trypanosoma cruzi minicircles are 1.4 kb in size and are composed of four conserved regions and four hypervariable intercalated regions.3 Clones of T. cruzi have specific minicircle classes, which define each lineage.4
In Chile, the vectors Triatoma infestans and Mepraia spinolai propagate T. cruzi in domestic/peridomestic and wild/peridomestic habitats, respectively.1,5 However, Chile has eliminated domestic transmission of T. infestans.6 Mepraia spinolai is frequently found in corrals of domestic animals, on stony hills, and in rock crevices of arid and semiarid zones of central Chile.7 Mepraia gajardoi is a genetically related triatomine described as a separate species from M. spinolai. It has similar feeding behavior and is distributed along the northern coast of Chile between 18°S and 21°S.8,9 Blood meal analyses from an insular population of M. spinolai from the southern Pacific Ocean coast of Chile (26°S) showed that coastal populations feed on seabirds, marine mammals, and reptiles.10
The purpose of this study was to determine T. cruzi genotypes in M. gajardoi from wild ecotopes of northern Chile. To meet this objective, we quantified T. cruzi infection on M. gajardoi from two wild ecotopes in northern Chile by detection of minicircle DNA by polymerase chain reaction (PCR)11 and genotyping by means of minicircle hybridization tests. The information generated by this study will be used when evaluating risk factors for human populations who are adjacent to sampling sites.
Mepraia gajardoi (stage III nymphs and adults) were collected during the austral spring-summer of 2009–2011 from coastal zones in northern Chile. Capture sites were subsequently classified into two ecotopes according to their distance from human dwellings Table 1: a fully wild ecotope (ecotope 1) that included Corazones (Arica City), Vitor, Camarones, and a wild ecotope near human dwellings (ecotope 2) that included Rio Seco and San Marcos. Study localities in northern Chile are shown in Figure 1. Study sites have a coastal desert climate with less than 2 mm of annual precipitation.12 These sites are extremely arid, have low plant cover, and include beaches with a mixture of rocks, pebbles, cobblestones, and sand. Lizards, sea birds, and wild rodents inhabit the collecting sites.13 Insects were collected from noon to 4:00 pm by two persons at each site. Captured bugs were transported to the laboratory and kept separately inside a climate chamber at 27°C with a relative humidity of 70% a 14:10 hour light:dark photoperiod. Triatomines were fed on Mus musculus to maintain the infective status of T. cruzi.14
Table 1.
Lineages of Trypanosoma cruzi in northern Chile*
| Location | Coordinates | Distance to human dwellings (km) | Tc I | Tc II | Tc V | Tc VI | ND | No. infected vectors | No. vectors analyzed |
|---|---|---|---|---|---|---|---|---|---|
| Corazones | 18°28′47″S, 70°19′27″W | 0.080–0.100 | 2 | 3 | 3 | 1 | 0 | 8 | 77 |
| Vitor | 18°45′45″S, 70°20′34″W | 45 | 2 | 19 | 2 | 0 | 0 | 19 | 151 |
| Camarones | 19°12′16″S, 70°16′08″W | 0.050–0.070 | 2 | 4 | 0 | 0 | 3 | 9 | 95 |
| Total for ecotope 1 | 6 | 26 | 5 | 1 | 3 | 36 | 323 | ||
| Rio Seco | 21°00′6″S, 70°9′52″W | 0.010–0.030 | 3 | 1 | 3 | 7 | 0 | 12 | 46 |
| San Marcos | 21°6′56″S, 70°7′30″W | 0.005–0.010 | 1 | 2 | 1 | 6 | 0 | 8 | 28 |
| Total for ecotope 2 | 4 | 3 | 4 | 13 | 0 | 20 | 74 |
ND = not determined.
Figure 1.
A, Map of northern Chile showing triatomine collection localities. B, Caleta Vitor (locality in ecotope 1). Note the presence of abundant stony areas and the absence of sand. C, San Marcos (locality in ecotope 2). Note the abundant stony area. Several fishermen's dwellings are seen.
For parasitologic analyses, we obtained fecal samples after feeding.15 Intestinal contents were mixed with 200 μL of phosphate-buffered saline, centrifuged at 10,000 × g, and frozen at −20°C until the PCR was conducted. Negative samples were evaluated for inhibitors. Each PCR included positive and negative controls. A 330-basepair product indicated a positive result.
For genotyping, DNA blot analyses were performed by using 10 μL of each PCR product. Four T. cruzi clones (sp 104 cl 1, CBB cl 3, NR cl 3, and V195 cl 1), corresponding to Tc I, Tc II, Tc V and Tc VI, respectively, were used to generate lineage-specific probes. Construction of minicircle probes and radiolabeling was performed as described.4 The PCR products were subjected to electrophoresis, transferred onto Hybond N+ nylon membranes (Amersham, Piscataway, NJ), and cross-linked by ultraviolet light for DNA fixation. After transferring PCR products, four membranes were pre-hybridized for at least 2 hours at 55°C. Each membrane was then hybridized with a lineage-specific probe labeled with 32P (1 × 106 cpm/membrane). After hybridization, membranes were washed under high stringency conditions and then exposed in the Molecular Imager FX (Bio-Rad Laboratories, Hercules, CA). Genotype distribution between ecotopes 1 and 2 was compared using by using chi-square and Fisher exact tests with Bonferroni adjustment when required.16
Overall, 397 M. gajardoi insects (mainly stage III and IV nymphs) were captured (ecotope 1: Corazones, n = 77; Camarones, n = 95; Vitor, n = 151 and ecotope 2: Rio Seco, n = 46; San Marcos, n = 28). Typical ecologic habitats of ecotope 1 and ecotope 2 are shown in Figure 1. The PCR analyses detected 56 insects positive for T. cruzi in the four localities, 36 from ecotope 1 (11.1%), and 20 from ecotope 2 (27.0%). Results from genotyping indicated that M. gajardoi is mainly infected with Tc II (n = 26), in ecotope 1, followed by Tc I (n = 6), Tc V (n = 5), Tc VI (n = 1), and unknown lineages (n = 3). In contrast, Tc VI (n = 13) was the most prevalent lineage in ecotope 2, followed by Tc I (n = 4), Tc V (n = 4), and Tc II (n = 3).
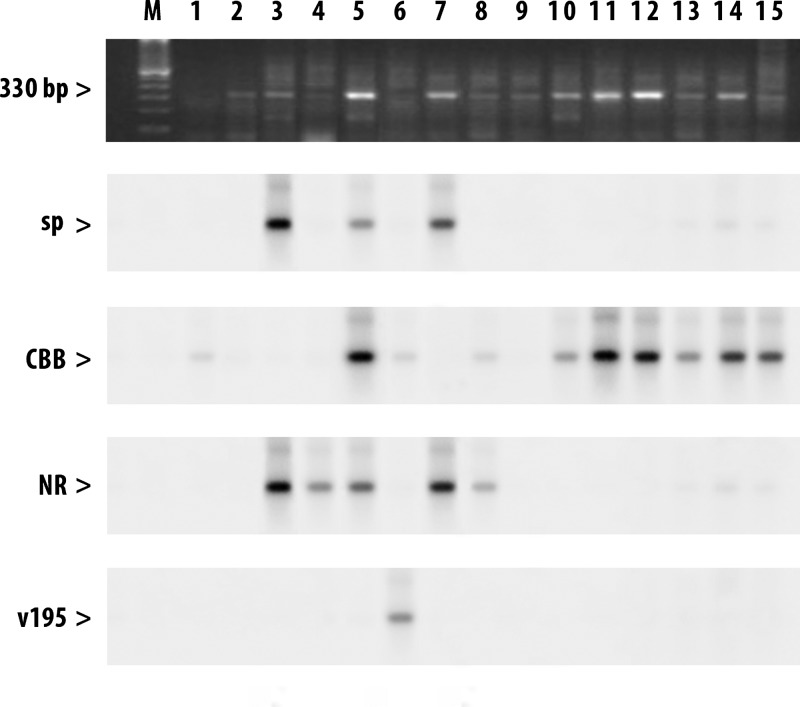
Statistical analyses indicated that Tc II is significantly more frequent in ecotope 1 than in ecotope 2 (P < 0.001), and Tc VI is more frequent in ecotope 2 than ecotope 1 (P < 0.001). The geo-reference of each triatomine locality and detailed genotyping results is shown in Table 1. Specifically, in locality-based analyses, results indicated that TcII is more common in Vitor than in Rio Seco (P < 0.001) and San Marcos (P < 0.01). The TcVI lineage is less common in Vitor than in Rio Seco (P < 0.001) and San Marcos (P < 0.001). Representative results of T. cruzi lineages of triatomine samples from ecotope 1 are shown in Figure 2. Some triatomine samples (3, 5, and 7) corresponded to mixed infections with two and up to three lineages. Other triatomine samples (2 and 9) are infected with unknown T. cruzi lineages.
Figure 2.
Trypanosoma cruzi amplicons from ecotope 1 stained with ethidium bromide. Hybridization profiles obtained with genotype-specific probes corresponding to TcI (sp104 cl1), TcII (CBB cl3), TcV (NR c13), and TcVI (v195 cl1). Lane M = molecular mass marker. A 330-basepair (bp) product indicates a positive result.
We showed that M. gajardoi in Chile is naturally infected with T. cruzi. Overall, the percentage of M. gajardoi nymphs infected was low (14%). There were few cases of mixed infections compared with those in M. spinolai, in which T. cruzi infection can reach up to 46.2% in some areas of central Chile, with up to half of the insects containing mixed infections.17 Our results indicate that infection would not be widely spread in the ecotopes studied. One explanation could be that blood donor vertebrates inhabiting coastal areas may be refractory or dead-end hosts for maintaining and propagating T. cruzi. Birds have an immune system with a potent complement system and are naturally resistant to T. cruzi infection.18,19 In all study localities, reptiles and marine birds are among the most frequent vertebrates found.
In our study, we found higher infection rates in insects from ecotope 2, which is a wild area but near human dwellings compared with the ecotope 1, which is a fully wild area. This observation could be explained by the different feeding source of insects in ecotope 2, which probably includes more mammals (humans, domestic animals, and/or synanthropic rodents) than in ecotope 1. The potential importance of M. gajardoi and M. spinolai in transmitting T. cruzi as secondary vector cannot be overlooked because the domestic and wild transmission cycles may overlap, as occurs in other disease-endemic areas.20 Mepraia sp. has a patchy distribution,21 a home range of 47 meters2, and a maximum mobility distance of 12 meters.7 This information is relevant to ascertain overlapping between insect focus points and human dwellings.
Although the southern Pacific Ocean coast of Chile has been considered an area without active transmission of Chagas disease,10 our results indicate that some coastal populations of M. gajardoi are infected with T. cruzi and represent a threat for humans, such as fisherman and algae collectors, living in those areas. Lineages of T. cruzi found in M. gajardoi in Rio Seco and San Marcos differ substantially from those found in Corazones, Camarones, and Vitor. A high prevalence of Tc VI is found in ecotope 2, which is an identical lineage to the T. cruzi clone V195 isolated in this area and used as probe,22 and other T. cruzi stocks circulating in the highlands of San Pedro de Atacama at 23°S as described.23,24 Previous studies of T. cruzi lineages in northern Chile showed that Tc V was found in humans, whereas Tc I and Tc VI were found in T. infestans.25 The finding of Tc VI in ecotope 2 is similar to findings in northern Argentina, where this lineage is frequently found in humans, T. infestans, and dogs.20,26 In contrast, Tc I is the most prevalent lineage found in M. spinolai at 31°S.17 A high prevalence of Tc II was found in ecotope 1, which is the same lineage detected in Mepraia sp., in a coastal locality at 24°S.27
Different vertebrate hosts maintaining vector populations could explain the dissimilar geographic distribution of T. cruzi lineages in the two study ecotopes. These different host species in two types of ecotopes studied could transmit different T. cruzi lineages. Nevertheless, geographic distances between our sampling sites (ecotopes 1 and 2) may also explain the observed dissimilarity.
ACKNOWLEDGMENTS
We thank Marco Arenas and Iván Cordova for helping during fieldwork, and Alonso Parra of the Ministry of Health for assistance.
Footnotes
Financial support: This study was supported by grants 1085154 and 1120122 from Fondo Nacional de Desarrollo Cientifíco Tecnológico to A. Solari and grant 11090086 from Fondo Nacional de Desarrollo Cientifíco Tecnológico to C. Botto-Mahan.
Disclosure: None of the authors has any conflicts of interest.
Authors' addresses: Andrea Toledo, Fernanda Vergara, Ricardo Campos, Sylvia Ortiz, and Ximena Coronado, Programa Biología Celular y Molecular, Instituto de Ciencias Biomédicas, Facultad de Medicina, Universidad de Chile, Santiago, Chile, E-mails: sonu444@hotmail.com, efe.vale@hotmail.com, hemanxcore@gmail.com, sortiz@med.uchile.cl, and ecoronado@med.uchile.cl. Carezza Botto-Mahan, Departamento de Ciencias Ecológicas, Facultad de Ciencias, Universidad de Chile, Santiago, Chile, E-mail: cbotto@uchile.cl. Aldo Solari, Programa Biología Celular y Molecular, Instituto de Ciencias Biomédicas, Facultad de Medicina, e Instituto de Ciencias Biomédicas, Programa Biología Celular y Molecular, Universidad de Chile, Independencia 1027, Santiago, Chile, E-mail: asolari@med.uchile.cl.
References
- 1.Lent H, Wygodzinsky P. Revision of the triatominae (Hemiptera: Reduviidae) and their significance as vectors of Chagas disease. Bull Am Mus Nat Hist. 1979;163:130–138. [Google Scholar]
- 2.Zingales B, Andrade SG, Briones MR, Campbell DA, Chiari E, Fernandes O, Guhl F, Lage Silva E, Macedo AM, Machado CR, Miles MA, Romanha AJ, Sturm N, Tibayrenc M, Schijman AG. A new consensus for Trypanosome cruzi intraspecific nomenclature second revision meeting recommend TcI to TcVI. Mem Inst Oswaldo Cruz. 2009;104:1051–1054. doi: 10.1590/s0074-02762009000700021. [DOI] [PubMed] [Google Scholar]
- 3.Avila HA, Simpson L. Organization and complexity of minicircle-encoded guide RNAs in Trypanosoma cruzi. RNA. 1995;1:939–947. [PMC free article] [PubMed] [Google Scholar]
- 4.Veas F, Breniere SF, Cuny G, Brengues C, Solari A, Tibayrenc M. General procedure to construct highly specific kDNA probes for clones of Trypanosoma cruzi for sensitive detection by polymerase chain reaction. Cell Mol Biol. 1991;37:73–84. [PubMed] [Google Scholar]
- 5.Lent H, Jurberg J, Galvão C. Revalidação do gênero Mepraia, Mazza, Gajardo & Jorg, 1940 (Hemiptera, Reduviidae, Triatominae) Mem Inst Oswaldo Cruz. 1994;89:347–352. [Google Scholar]
- 6.WHO Chagas disease: interruption of transmission in Chile. Wkly Epidemiol Rec. 2000;75:10–12. [Google Scholar]
- 7.Botto-Mahan C, Cattan PE, Canals M, Acuña M. Seasonal variation in the home range and host availability of the blood-sucking insect Mepraia spinolai in wild environment. Acta Trop. 2005;95:160–163. doi: 10.1016/j.actatropica.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Frías DA, Henry AA, González CR. Mepraia gajardoi: a new species of Triatominae (Hemiptera: Reduviidae) from Chile and its comparison with Mepraia spinolai. Rev Chil Hist Nat. 1998;71:177–188. [Google Scholar]
- 9.Campos R, Botto-Mahan C, Coronado X, Catala S, Solari A. Phylogenetic relationships of the Spinolai complex and other Triatomini based on mitochondrial DNA sequences (Hemiptera: Reduvidae) Vector Borne Zoonotic Dis. 2013;13:73–76. doi: 10.1089/vbz.2011.0954. [DOI] [PubMed] [Google Scholar]
- 10.Sagua H, Araya J, González J, Neira I. Mepraia spinolai in the southeastern Pacific Ocean coast (Chile): first insular record and feeding pattern on the Pan de Azúcar Island. Mem Inst Oswaldo Cruz. 2000;95:167–170. doi: 10.1590/s0074-02762000000200006. [DOI] [PubMed] [Google Scholar]
- 11.Wincker P, Britto C, Pereira JB, Cardoso MA, Oeleman O, Morel CM. Use of a simplified polymerase chain reaction procedure to detect Trypanosoma cruzi in blood samples from chronic chagasic patients in a rural endemic area. Am J Trop Med Hyg. 1994;51:771–777. doi: 10.4269/ajtmh.1994.51.771. [DOI] [PubMed] [Google Scholar]
- 12.di Castri F, Hajek ER. Bioclimatología de Chile. Santiago, Chile: Ediciones de la Universidad Católica de Chile; 1976. [Google Scholar]
- 13.Ortiz JC. Estudios comparativos de algunas poblaciones de Tropidurus de la costa Chilena. Ann Mus His Nat. 1980;13:267–280. [Google Scholar]
- 14.Kollien A, Schaub G. Development of Trypanosoma cruzi after starvation and feeding of the vector: a review. Tokai J Exp Clin Med. 1999;23:335–340. [PubMed] [Google Scholar]
- 15.Schenome H. Xenodiagnosis. Mem Inst Oswaldo Cruz. 1999;94:289–294. doi: 10.1590/s0074-02761999000700052. [DOI] [PubMed] [Google Scholar]
- 16.Sokal R, Rohlf FJ. Biometry: The Principles and Practice of Statistics in Biological Research. New York: WH Freeman and Company; 1995. [Google Scholar]
- 17.Coronado X, Rozas M, Botto-Mahan C, Ortiz S, Apt W, Cattan P, Solari A. Molecular epidemiology of Chagas disease in the wild transmission cycle: the evolution in the sylvatic vector Mepraia spinolai from an endemic area of Chile. Am J Trop Med Hyg. 2009;81:656–659. doi: 10.4269/ajtmh.2009.09-0053. [DOI] [PubMed] [Google Scholar]
- 18.Kierszenbaum F, Ivanyi J, Budzko DB. Mechanisms of natural resistance to trypanosomal infection. Role of complement in avian resistance to Trypanosoma cruzi infection. Immunol. 1976;30:1–6. [PMC free article] [PubMed] [Google Scholar]
- 19.Teixeira ARL, Nascimento RJ, Sturm NR. Evolution and pathology in Chagas disease: a review. Mem Inst Oswaldo Cruz. 2006;101:463–491. doi: 10.1590/s0074-02762006000500001. [DOI] [PubMed] [Google Scholar]
- 20.Cardinal MV, Lauricella MA, Ceballos LA, Lanati L, Marcet PL, Levin MJ, Kitron U, Gürtler RE, Schijman AG. Molecular epidemiology of domestic and sylvatic Trypanosoma cruzi infection in rural northwestern Argentina. Int J Parasitol. 2008;38:1533–1543. doi: 10.1016/j.ijpara.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ordenes H, Ehrenfeld M, Cattan PE, Canals M. Infección tripano-triatomina de Triatoma spinolai en una zona de riesgo epidemiológico. Rev Med Chil. 1996;124:1053–1057. [PubMed] [Google Scholar]
- 22.Solari A, Wallace A, Ortiz S, Venegas J, Sanchez G. Biological characterization of Trypanosoma cruzi stocks from Chilean insect vectors. Exp Parasitol. 1998;89:312–322. doi: 10.1006/expr.1998.4289. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez J, Muñoz S, Ortiz S, Anacona D, Salgado S, Galleguillos M, Neira I, Sagua H, Solari A. Biochemical, immunological, and biological characterization of Trypanosoma cruzi populations of the Andean north of Chile. Exp Parasitol. 1995;81:125–135. doi: 10.1006/expr.1995.1100. [DOI] [PubMed] [Google Scholar]
- 24.Torres JP, Ortiz S, Muñoz S, Solari A. Trypanosoma cruzi isolates from Chile are heterogeneous and composed of mixed populations when characterized by schizodeme and southern analyses. Parasitol. 2004;128:161–168. doi: 10.1017/s0031182003004475. [DOI] [PubMed] [Google Scholar]
- 25.Arribada AC, Apt W, Aguilera X, Solari A, Arribada AM, Sandoval J. Cardiopatía chagásica en la primera región de Chile. Estudio clínico, epidemiológico y parasicológico. Rev Med Chil. 1990;118:846–854. [PubMed] [Google Scholar]
- 26.Diez C, Lorenz V, Ortiz S, Gonzalez V, Racca A, Bontempi I, Manattini S, Solari A, Marcipar I. Genotyping of Trypanosoma cruzi sublineages in human samples from a northeast Argentina area by hybridization with DNA probes and specific polymerase chain reaction (PCR) Am J Trop Med Hyg. 2010;82:67–73. doi: 10.4269/ajtmh.2010.09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Botto-Mahan C, Sepulveda M, Vidal M, Acuña M, Ortiz S, Solari A. Trypanosoma cruzi infection in the sylvatic kissing bug Mepraia gajardoi from the Chilean southern Pacific Ocean coast. Acta Trop. 2008;105:166–169. doi: 10.1016/j.actatropica.2007.11.003. [DOI] [PubMed] [Google Scholar]