Abstract
Anopheline specimens were collected in 2011 by human landing catch, Shannon and CDC traps from the malaria endemic localities of Santa Rosa and San Pedro in Madre de Dios Department, Peru. Most specimens were either Anopheles (Nyssorhynchus) benarrochi B or An. (Nys.) rangeli, confirmed by polymerase chain reaction-restriction fragment length polymorphism-internal transcribed spacer 2 (PCR-RFLP-ITS2) and, for selected individuals, ITS2 sequences. A few specimens from Lupuna, Loreto Department, northern Amazonian Peru, were also identified as An. benarrochi B. A statistical parsimony network using ITS2 sequences confirmed that all Peruvian An. benarrochi B analyzed were identical to those in GenBank from Putumayo, southern Colombia. Sequences of the mtDNA COI BOLD region of specimens from all three Peruvian localities were connected using a statistical parsimony network, although there were multiple mutation steps between northern and southern Peruvian sequences. A Bayesian inference of concatenated Peruvian sequences of ITS2+COI detected a single clade with very high support for all An. benarrochi B except one individual from Lupuna that was excluded. No samples were positive for Plasmodium by CytB-PCR.
Introduction
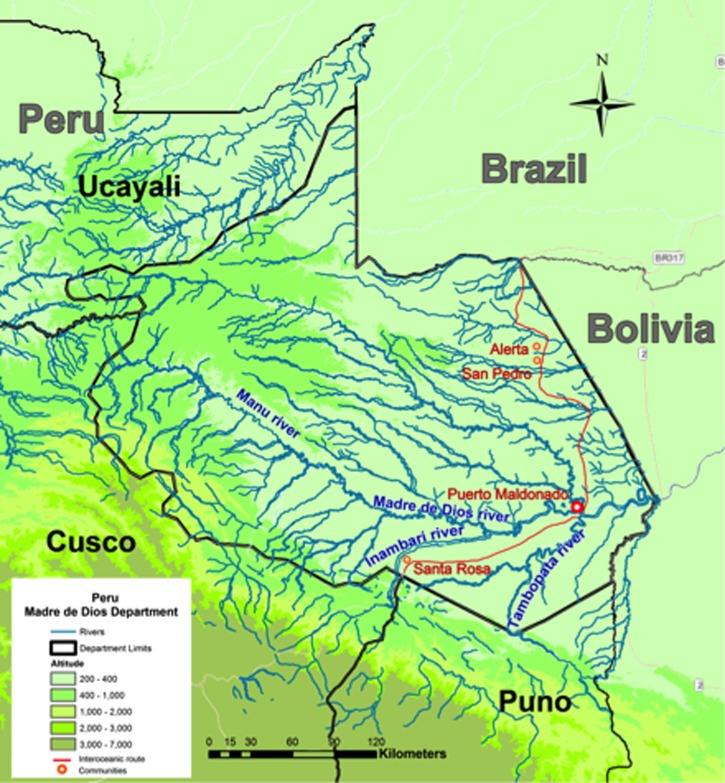
The majority of malaria cases in Peru are from the Amazonian departments of Loreto and Madre de Dios (MdD). MdD is located between 9°54′S, 72°29′W, and 13°22′S, 68°36′W and is part of the Southern Peruvian Amazon Basin (Figure 1). Of the 112,814 human inhabitants, 73% live in urban areas such as Puerto Maldonado (PM), the largest city1 (Figure 1). The main economic driver in MdD is gold mining, primarily in Bajo Pukiri (Delta-1) and Huepetuhe.2 According to the most recent census, these communities have 9,404 and 6,978 inhabitants, respectively, with 40% of the population involved in mining.1 Santa Rosa (SRA), along the Peru-Brazil Inter-Oceanic Highway 3 hours west of PM, is considered a new mining site (since 2009). The newly constructed highway, connecting Amazonian Brazil to coastal Peru, is also a source of significant migration and likely contributes to endemic malaria in MdD.2 SRA had 2,000 inhabitants in 2007 and at that time 55% of the population lived in rural areas. In general, mining areas lack electricity, potable water, and public sewage.1 Many of the workers who flock to this area are vulnerable migrants from other parts of Peru with no previous exposure to malaria. There are also small foci of Plasmodium vivax cases not far from the Bolivian border, in eastern MdD, near San Pedro (SPD), one of our research localities.
Figure 1.
Map of southern Amazonian Peru. The main city is Puerto Maldonado. Research sites are San Pedro, north of Puerto Maldonado, and Santa Rosa to the southwest. Major river systems are also depicted. Many of the gold mining workers come from the nearby departments of Cusco and Puno.
MdD ranks fourth in number of malaria cases in Peru (25,677 between 2005 and 2010) and all the cases during this time were P. vivax.3,4 According to the Regional Health Directorate, Delta-1 accounts for ∼70% of all cases and the remaining ones are mostly from Huepetuhe.5 SRA reported 113 malaria cases in 2011, a 47% increase in a single year. In general in MdD, malaria cases increase during the rainy season (December to February) however transmission takes place yearlong. According to data from the Dirección Generalde Epidemiologia-Ministerio de Salud de Peru (DGE-MINSA), the number of P. vivax cases declined from 2008 (3,700) to 2011 (1,750). No Plasmodium falciparum cases have been registered in MdD since 2007 (Table 1). Some of this reduction may be attributed to the distribution of insecticide-treated bed nets through the Global Fund-supported PAMAFRO from 2006 to 2011. With the recent termination of the Global Fund, funding for malaria control measures is threatened, therefore monitoring high malaria risk regions of Peru remains important.
Table 1.
Confirmed and suspected (=total) malaria cases from Madre de Dios and Loreto Departments, Peru, 2008–2011*
| 2008 | 2009 | 2010 | 2011 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TOTAL | P. vivax | P. falciparum | TOTAL | P. vivax | P. falciparum | TOTAL | P. vivax | P. falciparum | TOTAL | P. vivax | P. falciparum | |
| Loreto | 25,163 | 20,565 | 4,598 | 26,006 | 22,031 | 3,975 | 11,504 | 9,208 | 2,296 | 11,663 | 9,198 | 2,465 |
| MdD | 4,489 | 4,489 | 0 | 2,151 | 2,151 | 0 | 3,041 | 3,041 | 0 | 1,750 | 1,750 | 0 |
Data from http://www.minsa.gob.pe/.
The port city of Iquitos in Loreto was the focus of a remarkable surge of malaria cases in the 1990s.6,7 This trend, blamed on ecological changes that facilitated the westward spread of the primary malaria vector, Anopheles (Nyssorhynchus) darlingi,8,9 has continued as frequent outbreaks, mostly in the surrounding rural villages. Here, most people who become ill with malaria are involved in agriculture, fishing, or timber extraction.2 Plasmodium vivax cases in Loreto have decreased overall during the most recent 4-year period, from 20,565 (2008) to 9,198 (2011). Similarly, P. falciparum cases have declined from 4,598 (2008) to 0 (2011) (Table 1). The Pan American Health Organization (PAHO) 2008 map of malaria transmission in endemic areas of South America (http://new.paho.org/hq/index.php?option-com.content&task-view&id-2459&Itemid-2049) shows that parts of MdD range from very high (where our research site, SRA, is located) to low (where SPD is located), whereas Loreto is an area of intermediate risk.
In MdD, near the Peru-Bolivia border, both An. darlingi and Anopheles benarrochi have been shown to be abundant,10 and An. benarrochi has been hypothesized to be an important regional vector.11 Despite being more frequently collected in western Loreto and Ucayali than An. darlingi (71% versus 24%), An. benarrochi was found to be infected with either P. falciparum or P. vivax at much lower rates.11 These data are consistent with a study in southern Colombia that found despite An. benarocchi B's anthropophily and high prevalence (66.1% of 2,445 anophelines tested), no individual specimen was positive for Plasmodium by ELISA.12,13 Researchers have investigated seasonal species diversity in a distinctive ecological setting near Iquitos, collecting mosquitoes using a human landing catch (HLC) method. Not surprisingly, An. darlingi was the most abundant species in all site types, however, interestingly, An. benarrochi was quite abundant in a rural area, but not in the forest, forest village, or periurban sites.14 It has also been reported that both An. benarrochi s.s. and An. benarrochi B occur in Peru, and that Anopheles rangeli, a species in the Oswaldoi Group of subgenus Nyssorhynchus, is a important local or regional vector in southern Colombia.13 The major objective of this study was to verify the molecular identity of selected species, and then focus on An. benarrochi samples collected in MdD, compare them with those from San José de Lupuna (LUP) in Loreto, Peru and from other South American localities (by GenBank), and test them for infection with Plasmodium using CytB-polymerase chain reaction (PCR).
Mosquitoes were collected in SRA, using HLC, Shannon traps, and Centers for Disease Control and Prevention (CDC) light traps, alternating 12 h (18 h–6 h) and 4 h (18 h–22 h) nights. In SPD, collections were done using only Shannon traps for logistical reasons. In Loreto, mosquitoes were collected from LUP on the Nanay River with HLC and Shannon traps, using the same collection schedule (Table 1). In this report, we present only the samples of An. benarrochi that were collected in LUP February 24, April 4, and April 6, 2011 for comparative purposes. Identifications and biological details of other anopheline species from northern Amazonian Peru will be reported elsewhere. Mosquitoes were identified in the field,15–17 and then stored in individual tubes with silica gel. The DNA extraction was performed at the UCSD-PRISMA Laboratory, Iquitos, from the head+thorax portion of each specimen following the manufacturer's instructions (Qiagen DNA blood tissue extraction kit, Valencia, CA). Aliquots of the head+thorax DNA were used for the Plasmodium Cyt-B-PCR protocol18 for detection of parasites in Iquitos, and for the molecular identification of the anopheline species at the Wadsworth Center in Albany, New York. The latter procedure was undertaken with the PCR-fragment length polymorphism-internal transcribed spacer 2 (RFLP-ITS2) using the restriction endonucleases Alu I and BsrB I as previously described.19 Amplification of the ITS2 followed the published protocol.20 After standard PCR reactions, both strands were sequenced at the Applied Genomic Technologies Core (Wadsworth Center) on an ABI PRISM 3700 automated DNA sequencer (Life Technologies, Applied Biosystems, Carlsbad, CA). The BOLD (Barcode of Life Database) region of the COI gene was amplified using the universal primers.21 Sequencher 4.1 (Gene Codes Corps., Ann Arbor, MI) was used for automatic sequence alignment into contigs and proofreading sequences files; subsequently, all sequences were aligned with Clustal W and then compiled in MEGA5.0522 Unique sequences were submitted to GenBank using Sequin v12.30 (http://www.ncbi.nlm.nih.gov/Sequin/).
To infer the haplotype relationships within the ITS2 and BOLD data sets, we performed the median-joining network algorithm,23 available in NETWORK v 4.5.1.0 (www.fluxus-engineering.com), which combines the topology of a minimum spanning tree with a parsimonious search for the missing haplotypes. Because this algorithm was designed for non-recombining molecules (i.e., mitochondrial DNA [mtDNA]), we analyzed ITS2 using a statistical parsimony network24 at the 90% confidence level with TCS v 1.21 software.25 We also conducted a Bayesian inference of the concatenated (ITS2+COI) sequences using MrBayes 3.0.426 with An. rangeli as the outgroup.27 The settings were two simultaneous runs of the Markov Chain Monte Carlo for 20 million generations, sampling every 2,000 and discarding the first 25% as burn-in.
Although there was only a single Shannon trap collection (two nights) in SPD, An. benarrochi B was relatively abundant (11 of 13 specimens). Two individuals of An. rangeli were also confirmed molecularly (Table 2). In SRA, despite low densities, the most commonly collected species was An. rangeli (13 specimens confirmed by PCR-RFLP-ITS2 of 15 tested). There were two confirmed An. benarrochi B specimens collected in January 2011. Generally, for An. triannulatus, morphological identification is not problematic, therefore we did not molecularly confirm the two specimens collected in September (Table 2). Four specimens of An. rangeli, collected in January, were initially identified as Anopheles oswaldoi. Misidentification based on adult female morphology is common among An. nuneztovari s.l., An. rangeli, An. oswaldoi s.l., and An. benarrochi s.l. and other closely related species and can compromise vector control strategies12,13,19,28,29; in contrast, ITS2 is generally reliable.27,30,31 All specimens collected were tested for the presence of Plasmodium using CytB-PCR but none were positive. However, the sample sizes were small, and as the overall average prevalence of Plasmodium in neotropical vectors is around 1%, this was not an unexpected result. Overall, because of anthropophily and relative abundance, it appears that An. benarrochi B and (or) An. rangeli have the potential to be involved in malaria transmission in our study sites in MdD, but additional collections are needed to investigate this further.
Table 2.
Identities of anopheline specimens from San Pedro and Santa Rosa, Madre de Dios, 2011*
| Site | Date | No. | Trap | Morph. Id. | PCR-RFLP Id. | ITS2 | COI |
|---|---|---|---|---|---|---|---|
| SPD | 2/1 | 5 | SHA | nuneztovari | benarrochi B | benarrochi B | benarrochi B |
| SPD | 2/1 | 4 | SHA | benarrochi | benarrochi B | benarrochi B | benarrochi B |
| SPD | 2/2 | 2 | SHA | benarrochi | benarrochi B | benarrochi B | benarrochi B |
| SPD | 2/2 | 2 | SHA | rangeli | rangeli | ||
| SRA | 1/24 | 1 | CDC | oswaldoi | rangeli | ||
| SRA | 1/24 | 3 | SHA | oswaldoi | rangeli | ||
| SRA | 1/27 | 1 | SHA | rangeli | benarrochi B | benarrochi B | benarrochi B |
| SRA | 1/27 | 1 | HLC | rangeli | benarrochi B | benarrochi B | benarrochi B |
| SRA | 3/22 | 1 | SHA | Ano. | NT | ||
| SRA | 5/27 | 1 | SHA | benarrochi | NT | ||
| SRA | 9/27 | 5 | SHA | rangeli | rangeli | ||
| SRA | 9/27 | 1 | SHA | trinkae | rangeli | ||
| SRA | 9/27 | 1 | SHA | benarrochi | NT | ||
| SRA | 9/27 | 2 | SHA | triannulatus | NT | ||
| SRA | 9/27 | 1 | SHA | Nys. | NT | ||
| SRA | 9/28 | 1 | SHA | trinkae | rangeli | ||
| SRA | 9/28 | 1 | SHA | rangeli | rangeli | ||
| SRA | 11/15 | 1 | SHA | Nys. | rangeli |
SPD = San Pedro; SRA = Santa Rosa; No. = number of specimens; SHA = Shannon trap; CDC = CDC light trap; HLC = human landing catch, Morph. Id. = morphological identification; PCR-RFLP Id. = PCR-RFLP-ITS2, see text; ITS2 = rDNA internal transcribed spacer 2 sequence; COI = mtDNA COI sequence; Ano. = Anopheles; Nys. = Nyssorhynchus; NT= not tested.
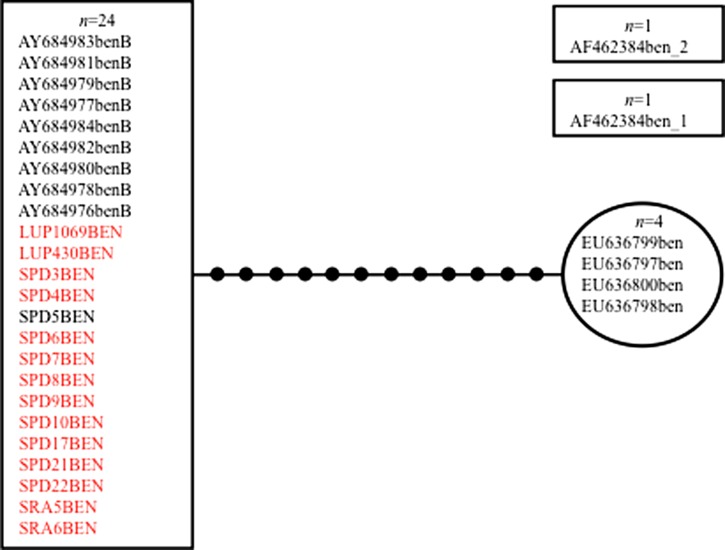
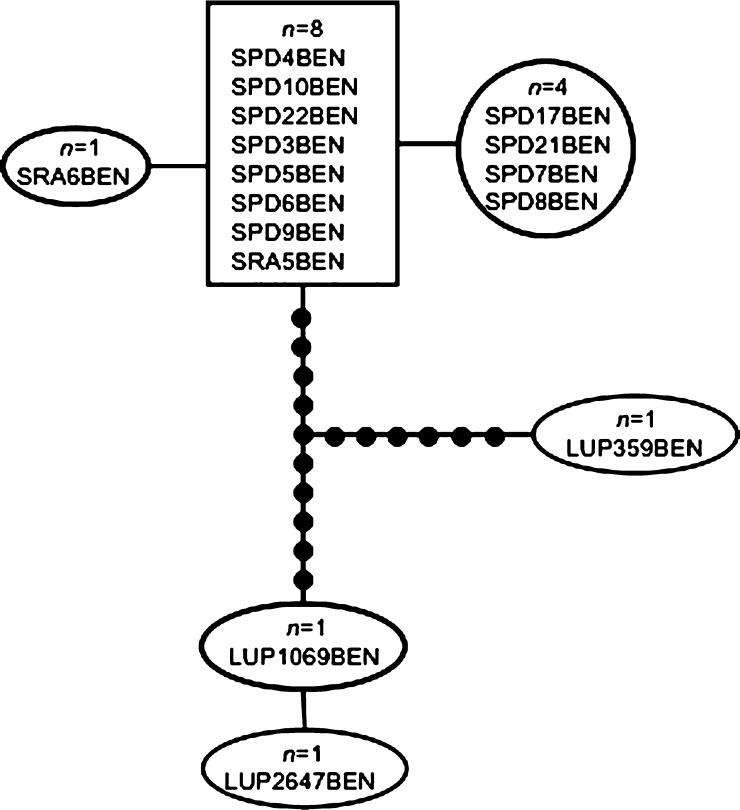
The ITS2 sequences of An. benarrochi from LUP, SPD, and SRA, Peru (GenBank accession no. JX173490) were all 448 bp in length, and the statistical parsimony network (SPN) at 90%, grouped them as a single haplotype (Figure 2), together with nine An. benarrochi B sequences from southern Colombia (GenBank; Table 3). They were 12 mutation steps away from An. benarrochi from Acre state, Brazil,31 and could not be connected with the GenBank samples from Rondonia, Brazil,27 suggesting that these Rondonia sequences may represent a distinct species. However, the TCS program would not run at 95% for the ITS2 sequences because the 24 sequences from Colombia and Peru were all a single haplotype. The SPN and the median-joining network of the mtDNA COI BOLD region of An. benarrochi B (632 bp) were identical; only the SPN is shown (Figure 3) (GenBank accession nos. JX170904–909). In this case, the three southern Peruvian haplotypes are single mutation steps apart, and form a compact group, whereas the three northern Peruvian haplotypes are separated from each other by 13–14 steps, and differ from the southern haplotypes by 11–12 steps. Of note is that for both network analyses of COI sequences at 95% the three LUP samples were not connected to those from MdD. We could not compare the Peruvian samples with any from GenBank because none are publically available. We were able to sequence only one specimen of An. benarrochi for both ITS2 and the BOLD region from LUP, sample 1069 (Figures 2 and 3). Because of the relatively large number of mutation steps separating this specimen from the southern An. benarrochi for the BOLD region (Figure 3), and because the three LUP sequences did not connect at 95%, we conducted a Bayesian inference using the concatenated data set (ITS2+COI) to test the hypothesis that the northern and southern Peruvian An. benarrochi are in the same clade. These results (Figure 4) show that the single LUP (northern) specimen does not group with the southern ones. Clearly, this warrants additional investigation. It is possible that the northern Peruvian An. benarrochi represent a distinctive lineage but this requires additional collections to increase the sample sizes, and further analysis.
Figure 2.
TCS 90% Statistical Parsimony Network: Anopheles benarrochi B ITS 2 (448 bp). All samples from SRA, SPD, and LUP, Peru group as one haplotype with An. benarrochi B samples from Colombia. Samples from Acre state, Brazil grouped 12 mutation steps away as a single haplotype. Samples from Rondonia, Brazil are not connected to the network. Details in Table 3.
Table 3.
Source of 2011 Peruvian Anopheles benarrochi field specimens and GenBank ITS2 and COI sequences analyzed for molecular confirmation of An. benarrochi
| Locality | Department/state | Country | N | Coordinates | COI accession no. | Source | |
|---|---|---|---|---|---|---|---|
| ITS2* | COI | ||||||
| SPD | Madre de Dios | Peru | 11 | 11 | 11°43.220′S, 69°10.588′W | JX170907-08 | This study |
| SRA | Madre de Dios | Peru | 2 | 2 | 12°55.457′S, 70°18.201′W | JX170909 | This study |
| LUP | Loreto | Peru | 2 | 3 | 03°44.591′S, 73°19.615′W | JX170904-06 | This study |
| Granada† | Acre | Brazil | 4 | – | 09°41′03.5″S, 67°08′05.3″W | EU636797-EU636800 | 31 |
| Puerto Asís | Putumayo | Colombia | 9 | – | 00°31′N, 66°31′W | AY684976-AY684983 | 12 |
| São Miguel | Rondônia | Brazil | 2 | – | 08°49′S, 63°54′W | AF462383, AF462384 | 27 |
All ITS2 sequences were identical (a single haplotype), GenBank accession no. JX173490.
Acrelandia.
ITS2 = internal transcribed spacer 2; COI = mtDNA COI sequence; SPD = San Pedro; SRA = Santa Rosa; LUP = Lupuna.
Figure 3.
90% SPN: An. benarrochi Folmer COI (632 bp).
Figure 4.
Bayesian inference of concatenated ITS2+COI Peruvian An. benarrochi B sequences. An. rangeli is the outgroup. 1 = Bayesian posterior probability for the clade including all An. benarrochi B sequences except LUP1069.
ACKNOWLEDGMENTS
For help with field work in Lupuna, Loreto Department, we thank Eliseo Ramirez, Jose Manuel Reyna, Victor Pacaya, David Arimuya, and Hercules Maytahuari. We appreciate the support of the communities of Santa Rosa and San Pedro (Madre de Dios Department) and Lupuna (Loreto Department). For collaboration and facilitating logistics in Madre de Dios and Loreto we thank the Direccion Regional de Salud (DIRESA, Puerto Maldonado, Madre de Dios) and the Direccion Regional de Salud (DIRESA, Iquitos, Loreto). We are particularly grateful to Carlos Manrique (DIRESA, Madre de Dios) for orienting us at the beginning of this project and suggesting San Pedro and Santa Rosa as potential localities for our study.
Disclaimer: The views expressed in this article are those of the authors only and do not necessarily reflect the official policy or position of the Navy, Department of Defense, or the U.S. Government. Several authors of this manuscript are active duty military service members or employees of the U.S. Government. This work was prepared as part of their duties. Title 17 U.S.C. § 105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.’ Title 17 U.S.C. § 101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person's official duties.
Footnotes
Financial support: Financial support was provided by U.S. National Institutes of Health (NIH) grant U19 AI089681 to JMV and NIH grant R01AI54139 to JEC. The participation of AGL in this project is sponsored by the training grant NIH/FIC 2D43 TW007393 awarded to NAMRU-6 by the Fogarty International Center of the U.S. National Institutes of Health.
Authors' addresses: Jan E. Conn and Sara A. Bickersmith, Griffin Laboratory, The Wadsworth Center, NYSDOH, Slingerlands, NY, E-mails: jconn@wadsworth.org and sab19@notes.health.state.ny.us. Marta Moreno and Joseph M. Vinetz, Division of Infectious Diseases, Department of Medicine, University of California San Diego, La Jolla CA, E-mails: mmorenoleirana@ucsd.edu and jvinetz@ad.ucsd.edu. Marlon Saavedra, Associacion Benefica PRISMA, Iquitos, Peru, E-mail: msaavedraromero75@gmail.com. Elisabeth Knoll, Department of Biomedical Sciences, School of Public Health, State University of New York-Albany, Albany, NY, E-mail: eknoll@albany.edu. Roberto Fernandez and Roxanne G. Burrus, Department of Entomology, U.S. Naval Medical Research Unit No.6 (NAMRU-6), Avenida Venezuela Cuadra 36 s/n, Callao 2, Peru, E-mails: Roberto.Fernandez@med.navy.mil and Roxanne.Burrus@med.navy.mil. Hubert Vera and Esteban Rivera, DIRESA, Avenida Ernesto Rivero, 475, Puerto Maldonado, Madre de Dios, Peru, E-mails: hubvera@hotmail.com and estebanriveracarrera@gmail.com. Andres G. Lescano and Juan Francisco Sanchez, Department of Parasitology, U.S. Naval Medical Research Unit No.6 (NAMRU-6), Avenida Venezuela Cuadra 36 s/n, Callao 2, Peru, E-mails: willy.lescano@med.navy.mil and Juan.F.Sanchez@med.navy.mil.
References
- 1.Instituto Nacional de Estadística e Informática (INEI) XI Censo de Poblacion y VI de Vivienda. 2007. http://www.inei.gob.pe/ Available at. Accessed June 6, 2012.
- 2.da Silva-Nunes M, Moreno M, Conn JE, Gamboa D, Abeles S, Vinetz JM, Ferreira MU. Amazonian malaria: asymptomatic human reservoirs, diagnostic challenges, environmentally driven changes in mosquito vector populations, and the mandate for sustainable control strategies. Acta Trop. 2012;121:281–291. doi: 10.1016/j.actatropica.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.December 24, 2011 epidemiological surveillance report in Peru Bol Epidemiol. 2011;20:1080–1091. [Google Scholar]
- 4.Pan American Health Organization . Report on the Situation of Malaria in the Americas, 2008. Washington, DC: Pan American Health Organization; 2010. [Google Scholar]
- 5.Regional Health Office of Madre de Dios Results of malaria control and interventions in migrant populations 2011 [Google Scholar]
- 6.Aramburu Guarda J, Asayag CR, Witzig R. Malaria reemergence in the Peruvian Amazon region. Emerg Infect Dis. 1999;5:209–215. doi: 10.3201/eid0502.990204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roper MH, Torres RS, Goicochea CG, Andersen EM, Guarda JS, Calampa C, Hightower AW, Magill AJ. The epidemiology of malaria in an epidemic area of the Peruvian Amazon. Am J Trop Med Hyg. 2000;262:247–256. doi: 10.4269/ajtmh.2000.62.247. [DOI] [PubMed] [Google Scholar]
- 8.Fernández R, Carbajal F, Quintana J, Chauca H, Watts DM. Presence of A. (N.) darlingi (Diptera: Culicidae), in the region surrounding Iquitos, Loreto-Peru. Bol Soc Peruana Enferm Inf Trop. 1996;5:10–20. [Google Scholar]
- 9.Vittor AY, Gilman RH, Tielsch J, Glass G, Shields T, Lozano WS, Pinedo-Cancino V, Patz JA. The effect of deforestation on the human-biting rate of Anopheles darlingi, the primary vector of Falciparum malaria in the Peruvian Amazon. Am J Trop Med Hyg. 2006;74:3–11. [PubMed] [Google Scholar]
- 10.Tineo ET, Medina CA, Fallaque SC, Chavez CL, Quispe FS, Mercado AM, Zevallos GJ, Leon CW, Palomino SM. Geographical distribution and seasonal biting behavior of Anopheles (Nyssorhynchus) darlingi Root 1926 in Peru-Bolivia frontier localities, Madre de Dios, Peru. Rev Peru Med Exp Salud Publica. 2003;20:78–83. [Google Scholar]
- 11.Flores-Mendoza C, Fernándezc R, Escobedo-Vargasc KS, Vela-Perezac Q, Schoelerbc GB. Natural Plasmodium infections in Anopheles darlingi and Anopheles benarrochi (Diptera: Culicidae) from eastern Peru. J Med Entomol. 2004;41:489–494. doi: 10.1603/0022-2585-41.3.489. [DOI] [PubMed] [Google Scholar]
- 12.Ruiz F, Quinones ML, Erazo HF, Calle DA, Alzate JF, Linton YM. Molecular differentiation of Anopheles (Nyssorhynchus) benarrochi and An. (N.) oswaldoi from southern Colombia. Mem Inst Oswaldo Cruz. 2005;100:155–160. doi: 10.1590/s0074-02762005000200008. [DOI] [PubMed] [Google Scholar]
- 13.Quiñones ML, Ruiz F, Calle DA, Harbach RE, Erazo HF, Linton YM. Incrimination of Anopheles (Nyssorhynchus) rangeli and An. (Nys.) oswaldoi as natural vectors of Plasmodium vivax in southern Colombia. Mem Inst Oswaldo Cruz. 2006;101:617–623. doi: 10.1590/s0074-02762006000600007. [DOI] [PubMed] [Google Scholar]
- 14.Reinbold-Wasson DD, Sardelis MR, Jones JW, Watts DM, Fernandez R, Carbajal F, Pecor JE, Calampa C, Klein TA, Turrel MJ. Determinants of Anopheles seasonal distribution patterns across a forest to periurban gradient near Iquitos, Peru. Am J Trop Med Hyg. 2012;86:459–463. doi: 10.4269/ajtmh.2012.11-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forattini OP. Entomologica Medica. Volume 1. Sao Paulo, Fac. Hig. Saude; Sao Paulo, Brazil: 1962. [Google Scholar]
- 16.Faran ME, Linthicum KJ. A handbook of the Amazonian species of Anopheles (Nyssorhynchus) (Diptera: Culicidae) Mosq Syst. 1981;13:1–81. [Google Scholar]
- 17.Consoli RA, Lourenço-de-Oliveira RL. Major mosquito vectors in Brazil. Fundação Oswaldo Cruz; Editora Fiocruz: 1994. [Google Scholar]
- 18.Hasan AU, Suguri S, Sattabongkot J, Fujimoto C, Amakawa M, Harada M, Ohmae M. Implementation of a novel PCR based method for detecting malaria parasites from naturally infected mosquitoes in Papua New Guinea. Malar J. 2009;8:182. doi: 10.1186/1475-2875-8-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matson R, Tong Rios C, Banda Chavez C, Gilman RH, Florin D, Lopez Sifuentes V, Cardenas Greffa R, Peñataro Yori P, Fernandez R, Velasquez Portocarrero D, Vinetz JM, Kosek M. Improved molecular technique for the differentiation of neotropical anopheline species. Am J Trop Med Hyg. 2008;78:492–498. [PMC free article] [PubMed] [Google Scholar]
- 20.Li C, Wilkerson RC. Intragenomic rDNA ITS2 variation in the neotropical Anopheles (Nyssorhynchus) albitarsis complex (Diptera: Culicidae) J Hered. 2007;98:51–59. doi: 10.1093/jhered/esl037. [DOI] [PubMed] [Google Scholar]
- 21.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial Cytochrome c Oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 22.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 24.Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 26.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 27.Marrelli MT, Floeter-Winter F, Malafronte RS, Tadei WP, Lourenço-de-Oliveira R, Flores-Mendoza C, Marinotti O. Amazonian malaria vector anopheline relationships interpreted from ITS2 rDNA sequences. Med Vet Entomol. 2005;19:208–218. doi: 10.1111/j.0269-283X.2005.00558.x. [DOI] [PubMed] [Google Scholar]
- 28.Cienfuegos AV, Gómez GF, Córdoba LA, Luckhart S, Conn JE, Correa MM. Design and evaluation of methods based on PCR-RFLP of ITS2 to identify mosquitoes of Anopheles species (Diptera: Culicidae) from the Pacific coast of Colombia. Rev Biomed. 2008;19:35–44. [Google Scholar]
- 29.Cienfuegos AV, Rosero DA, Naranjo N, Luckhart S, Conn JE, Correa MM. Evaluation of a PCR-RFLP-ITS2 assay for discrimination of Anopheles species in northern and western Colombia. Acta Trop. 2011;118:128–135. doi: 10.1016/j.actatropica.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marrelli MT, Sallum MAM, Marinotti O. The second internal transcribed spacer of nuclear ribosomal DNA as a tool for Latin American taxonomy: a critical review. Mem Inst Oswaldo Cruz. 2006;101:817–832. doi: 10.1590/s0074-02762006000800002. [DOI] [PubMed] [Google Scholar]
- 31.Sallum MAM, Marrelli MT, Nagaki SS, Laporta GZ, Dos Santos CLS. Insight into Anopheles (Nyssorhychus) (Diptera: Culicidae) species from Brazil. J Med Entomol. 2008;45:970–981. doi: 10.1603/0022-2585(2008)45[970:iiandc]2.0.co;2. [DOI] [PubMed] [Google Scholar]