Abstract
The cat flea, Ctenocephalides felis, is an inefficient vector of the plague bacterium (Yersinia pestis) and is the predominant off-host flea species in human habitations in the West Nile region, an established plague focus in northwest Uganda. To determine if C. felis might serve as a Y. pestis bridging vector in the West Nile region, we collected on- and off-host fleas from human habitations and used a real-time polymerase chain reaction-based assay to estimate the proportion of off-host C. felis that had fed on humans and the proportion that had fed on potentially infectious rodents or shrews. Our findings indicate that cat fleas in human habitations in the West Nile region feed primarily on domesticated species. We conclude that C. felis is unlikely to serve as a Y. pestis bridging vector in this region.
Introduction
Plague is a rare but highly virulent zoonotic disease.1 The etiologic agent, Yersinia pestis, circulates primarily in enzootic cycles between rodents or shrews and their fleas.2 Humans are most susceptible to infection during epizootics; when a large number of infected rodents die, their infectious fleas must seek new hosts, and these fleas may act as “bridging vectors” to humans.2 Control methods aimed at reducing the incidence of human plague often target these vectors.3 To serve as a bridging vector, a flea species must be capable of transmitting Y. pestis, it must feed on potentially infected zoonotic hosts, and it must feed on humans.
The West Nile region is an established plague focus in northwest Uganda.4 More than 2,400 suspect human plague cases were reported in this region between 1999 and 2011.5 Although research has not determined exactly where humans are most likely to encounter infectious fleas in Uganda, studies in other regions suggest that human exposure occurs most often in the domestic or peridomestic environment.6,7 Researchers generally identify rat flea species, Xenopsylla brasiliensis, Xenopsylla cheopis, and Dinopsyllus lypusus, as the primary bridging vectors for Y. pestis in East Africa.8–10 Some studies, however, have implicated the human flea, Pulex irritans, as a potentially important Y. pestis vector in regions where it is the predominant host-seeking (off-host) flea species in human habitations, although it is unclear to what extent its postulated role in human plague outbreaks derives from an ability to spread the bacterium from one person to another versus an ability to serve as a bridging vector from rodent hosts to humans.10–14 Recent investigations have found that the cat flea, Ctenocephalides felis, comprises the majority of off-host fleas captured in huts in the West Nile region,15,16 however relatively little is known about its potential role as a Y. pestis vector in this plague focus.
Cat fleas in East Africa are most often identified as C. felis strongylus based on morphological characteristics15,17,18; recent molecular studies, however, have found no evidence for the existence of C. felis subspecies.19,20 Ctenocephalides felis strongylus may therefore share biological characteristics, including feeding behavior and transmission efficiency, with other C. felis subspecies, including Ctenocephalides felis felis. Worldwide, C. felis is associated with a wide variety of hosts including cats, dogs, livestock and wild mammals, including rodents.21–27 The cat flea also feeds readily on humans.26,28–30 In the West Nile region, this species occasionally infests rodents, including the black rat (Rattus rattus), which is abundant in the domestic environment and susceptible to Y. pestis infection.8,15,31 Further implicating the cat flea as a potential bridging vector, Eisen and others15 recently showed that C. felis is capable of early-phase transmission of Y. pestis.
Here, we sought to investigate the potential for C. felis to serve as a Y. pestis bridging vector in the West Nile region. Specifically, we aimed to identify vertebrate blood meals in off-host C. felis collected in huts in this region as a means to determine the proportion of blood meals taken from humans and the proportion taken from potential rodent or shrew reservoirs of Y. pestis. We also sought to determine what flea species infested small mammals captured in the same huts.
Materials and Methods
Study area.
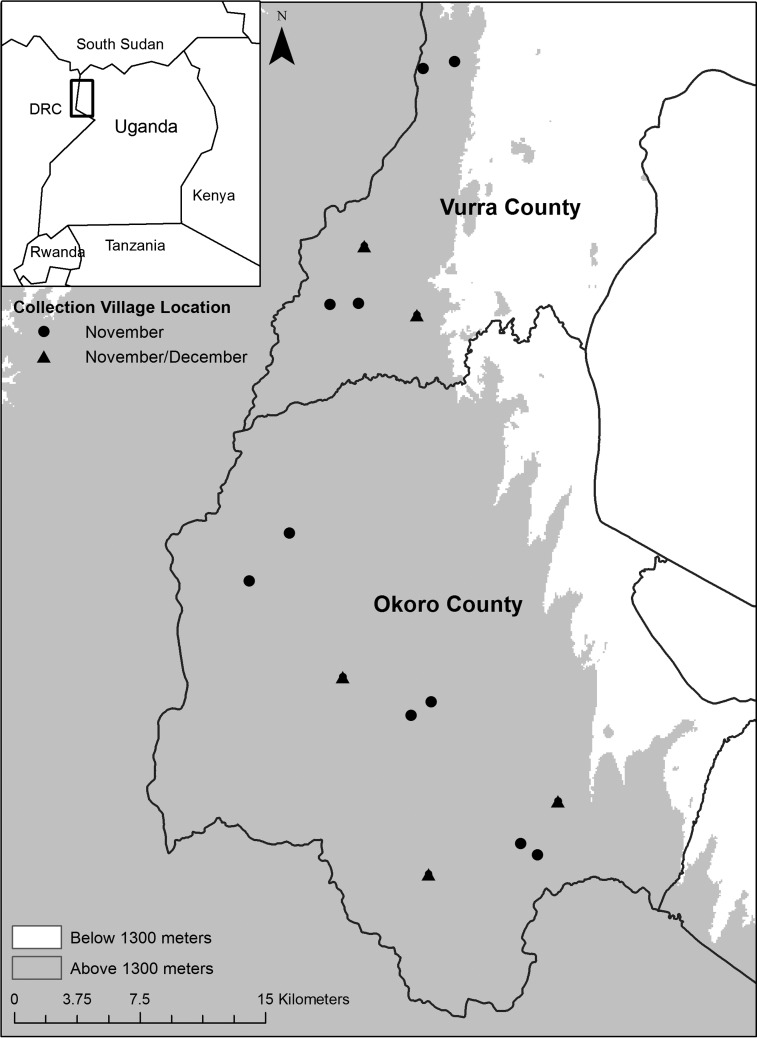
We collected fleas and small mammals in Vurra and Okoro, contiguous counties located along the Democratic Republic of Congo (DRC) border in the West Nile region of Uganda (Figure 1). The Rift Valley escarpment roughly bisects these counties, and most human plague cases are reported from villages located west of the escarpment at elevations above 1,300 m.32 The western highlands are characterized by lush vegetation, fertile soil, numerous water sources, and highly fragmented land use associated with subsistence farming.32,33 This sub-humid region experiences heavy, reliable rains from late August through November and a less reliable rainy season between March and June.32,33 We randomly selected 100 huts in each of 15 villages west of the escarpment for flea and small mammal collections in November 2009. We conducted a second collection 22 days later in five of the villages (Figure 1). Although most human plague cases are reported from this region between September and January,5,32 our study occurred during an inter-epizootic period; there were no confirmed human plague cases in Vurra or Okoro County between March 2009 and October 2011 (CDC, unpublished data). We collected all fleas and rodents inside homes typical of this area: square or round huts constructed of mud bricks and waddle with thatched grass roofs. Residents had often smeared a mixture of mud and bovine feces on their dirt floors and walls that had dried to create a hard surface.33
Figure 1.
Study area within a plague-endemic region of Uganda. Points indicate village locations where fleas and rodents were collected from huts in November 2009 (circles) or November and December 2009 (triangles) for this study.
Off-host flea collection.
We set one modified Kilonzo light trap34 to collect photosensitive fleas in each hut. The trap consisted of a flashlight suspended over a metal pan (25.4 cm diameter) containing 2% saline with a surfactant (1 drop/L Tween 80 detergent) to force trapped fleas to sink. We put fresh batteries in the flashlight for each night of collection. We applied petroleum jelly to the rim of the pan to prevent fleas from escaping. Residents turned the flashlights on at night and left them on overnight. The following day, we collected the fleas from each trap and stored them in 70% ethanol.
Small mammal and on-host flea collection.
Small mammals and their fleas were collected in the same huts on the same nights we collected off-host fleas as part of a previously described study.35 We placed two traps (48.3 × 17.1 × 17.1 cm, Tomahawk Trap Co., Tomahawk, WI) against the inside wall of each hut. The following day we sedated trapped animals by halothane inhalation and identified captures to species based on morphological measurements (e.g., length of body, tail, right hind foot, and ear)36; we combed each animal to recover on-host fleas and stored the fleas in 70% ethanol. Rodents were then released at the point of capture.
We identified all fleas to species using published keys,17,37,38 and we differentiated C. felis strongylus from the morphologically similar C. felis felis based on the relative lengths of the first two spines of the genal comb.26
Blood meal identification.
Real-time polymerase chain reaction (PCR) and sequencing.
We used a SYBR-Green I-based real-time PCR assay using a single primer set to amplify vertebrate DNA for identification by sequencing. The assay is described in detail elsewhere39; briefly, we used primers targeting regions of the 12S mitochondrial RNA gene that are conserved across vertebrate species but differ in C. felis. The primers flank a variable region of ∼100 nucleotides (nt), and each includes an M13 tag to facilitate sequencing.40 We ran all reactions in an Mx3005P thermal cycler (Stratagene, La Jolla, CA). A 5-minute initial denaturation was followed by 38 amplification cycles, and the final amplification cycle was immediately followed by a melting analysis cycle. We stored all PCR products at 4°C or −20°C until purification. Selected amplicons were purified using the QIAquick PCR Purification Kit (Qiagen, Valencia, CA) and sequenced immediately or stored at −20°C. We generated forward and reverse sequences as previously described,39 and we used DNASTAR Lasergene software (Madison, WI) to edit sequences and generate a single sequence for each sample. We manually removed the primer sequences from either end of the sequence for final analysis.
Analysis of DNA from known vertebrates.
To test the assay's ability to detect and differentiate potential vertebrate host species from the study area, we purchased citrated human (Homo sapiens), cat (Felis catus), dog (Canis lupus familiaris), goat (Capra hircus), and chicken (Gallus gallus) whole blood from a commercial vendor (Bioreclamation, Westbury, NY). These are all species previously observed in or within 10 m of huts in the West Nile region (CDC, unpublished data). We collected tail tissue or whole blood from the four small mammal species that predominate in domestic and peridomestic environments in our study area: black rat, Nile rat (Arvicanthis niloticus), multimammate mouse (Mastomys natalensis), and shrew (Crocidura species).15,31 Mammals were trapped in the West Nile region and identified to genus or species based on morphological measurements.36 Tail tissue was stored in 70% ethanol. Whole blood was absorbed on Nobuto strips (Advantec, Toyo Roshi Kaisha, Ltd., Tokyo, Japan) made of cellulose paper. The absorbed blood (∼100 μL per strip) was air dried and the strips were stored at ambient temperature. We extracted DNA from citrated whole blood and tail tissue using the DNeasy Blood and Tissue Kit and from dried blood samples using the QIAamp DNA Micro Kit (Qiagen) per the manufacturer's protocols. All DNA was stored at −80°C until PCR analysis. We subjected each sample to amplification and sequencing as described previously and aligned the nine sequences to verify that no two sequences were identical. We also used the basic local alignment search tool (BLAST) to identify similar mitochondrial sequences in the nucleotide database.
All procedures for animal handling were approved by the Animal Care and Use Committee at the Division of Vector-Borne Diseases, Centers for Disease Control and Prevention, Fort Collins, CO. Voucher specimens for small mammals were previously submitted to Makerere University (Kampala, Uganda) for verification of species identification.
Blood meal identification in off-host fleas.
We removed surface contaminants from each Kilonzo-trapped flea, homogenized it in calcium- and magnesium-free Dulbecco's phosphate buffered saline, and extracted DNA as previously described.39 Each set of extractions included a teneral (unfed), colony-reared C. felis (Heska Corporation, Loveland, CO) as a negative control. We used 10 μL eluted DNA per real-time PCR. Each real-time run included at least two no template control wells (water in place of template DNA). Rattus norvegicus DNA from a single stock stored −80°C in single-use aliquots served as a positive control and an inter-run calibrator as previously described.39 We ran all samples in duplicate.
To minimize the risk of cross-contamination, we conducted DNA isolation, PCR setup, and amplicon processing in three separate rooms. To limit human DNA contamination, we used certified DNA-free pipet tips and tubes for DNA isolation and PCR set-up. All surfaces and equipment were treated with 0.62–0.99% sodium hypochlorite or DNA Away (Molecular BioProducts, San Diego, CA) and rinsed with 70% reagent alcohol (Ricca Chemical Company, Arlington, TX) before and after use.
We considered a flea sample positive for vertebrate DNA if both replicates crossed the threshold within 38 cycles and had similar dissociation curves and a collective threshold cycle (Ct) value less than or equal to our previously established Ct cutoff of 36.39 We classified a sample as having no detectable vertebrate DNA if both replicates had Ct values greater than 36 or if the sample generated similar replicates with a collective Ct value greater than 36. We repeated any sample with dissimilar replicates. If a sample repeatedly yielded dissimilar replicates, we concluded that the assay could not reliably detect a blood meal in that sample. We chose a single replicate from each positive sample for sequencing and used BLAST to identify the blood meal source.
Estimating the true number of human blood-fed fleas and statistical analyses.
Given that our assay previously detected human DNA in 3 of 50 unfed fleas,39 we assumed that the total number of fleas with detectable human DNA in our study, h, included fleas that had actually consumed human blood, x, and samples that contained no detectable blood meal but had been contaminated with human DNA, c:
We estimated that ∼6% of fleas with no detectable blood meal, n, tested positive for contaminating human DNA:
The total number of fleas tested, t, included those with no detectable blood meal, those with a detectable human blood meal, and those with a detectable blood meal from a non-human vertebrate species, b:
Combining Equations 1–3 and solving for x:
We solved Equation 4 to estimate the number of field-collected fleas that had taken a human blood meal.
We used JMP software (SAS Institute Inc., Cary, NC) to calculate a Wilson score confidence interval41 for the percentage of C. felis with detectable vertebrate blood meals that had fed on wild rodents or shrews.
To determine if the distribution of fleas per hut and fleas per rat was consistent with a Poisson distribution, we compared actual and expected values for the number of rats and the number of huts infested with 0, 1, 2, 3, and 4 or more fleas and conducted a χ2 goodness-of-fit test.
Results
Small mammal and flea collections.
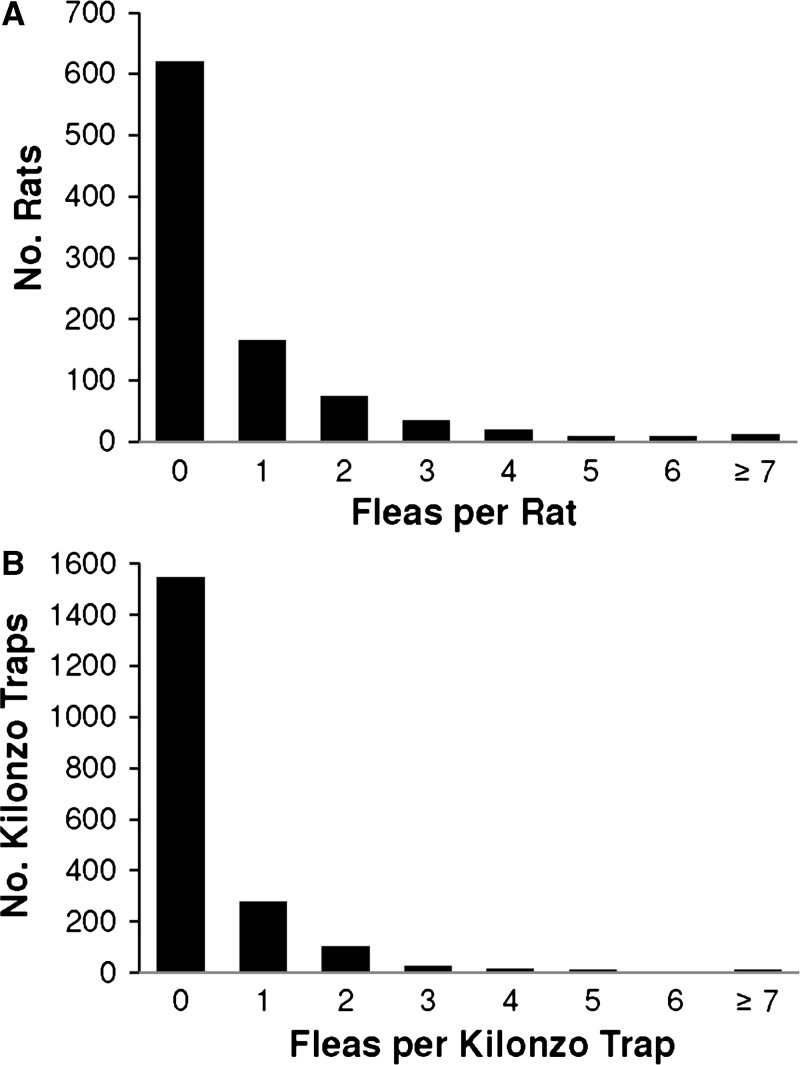
Rattus rattus (N = 944) was the only small mammal species collected in the huts during the 4,000 trap nights included in this study. We trapped at least one R. rattus in 577 of the 1,500 huts sampled in November and in 188 of the 500 huts sampled in December for an overall capture rate of 38.3% per hut per sampling occasion. We collected a total of 727 fleas from R. rattus, of which the majority (91.1%) were X. cheopis or X. brasiliensis (Table 1). By contrast, the majority (86.9%) of the 839 off-host fleas collected from modified Kilonzo light traps were C. felis (Table 1). All off-host C. felis were identified to subspecies as C. felis strongylus. The number of fleas per R. rattus ranged from 0 to 27, but more than 90% of the captures were infested with two or fewer fleas (Figure 2A). Among sampled huts that yielded off-host fleas (N = 455 of 2000), the number ranged from 1 to 23 fleas per hut per sampling occasion, although most Kilonzo traps yielded no more than 1 flea (Figure 2B). The χ2 goodness-of-fit analysis revealed that neither the distribution of fleas per rat (N = 944, λ = 0.77) nor the distribution of fleas per hut (N = 2000, λ = 0.42) was consistent with a Poisson distribution (P < 0.001). The number of rats and huts with zero fleas and the number of rats and huts with more than four fleas were higher than the expected Poisson values. The number of rats and huts with one flea were lower than the expected Poisson values.
Table 1.
Summary of on- and off-host flea captures, by species, in human habitations in Vurra and Okoro counties*
| Number of fleas (% of all fleas collected from the same source) | ||||||
|---|---|---|---|---|---|---|
| Flea source | Cf | Xb/Xc | Eg | Dl | Other† | Unknown‡ |
| Kilonzo traps (off-host) | 729 (86.9%) | 32 (3.8%) | 68 (8.1%) | 0 (0.0%) | 10 (1.2%) | 0 (0.0%) |
| Rattus rattus§ | 2 (0.3%) | 662 (91.1%) | 0 (0.0%) | 17 (2.3%) | 23 (3.2%) | 23 (3.2%) |
Fleas were collected using modified Kilonzo light traps (off-host) or by combing Rattus rattus trapped in huts.
Other off-host flea species: Tunga penetrans, Ctenocephalides spp. too damaged to identify to species. Other flea species collected from R. rattus: Xenopsylla spp. too damaged to identify to species, Ctenophthalmus calceatus cabirus, Afristivalius torvus Rothschild (syn. Stivalius torvus8).
“Unknown” includes fleas that were lost or too damaged to identify to genus.
Flea species collected from R. rattus were previously reported as part of a larger data set.35
Cf = Ctenocephalides felis; Xb/Xc = Xenopsylla brasiliensis and Xenopsylla cheopis; Eg = Echidnophaga gallinacea; Dl = Dinopsyllus lypusus.
Figure 2.
Distribution of fleas collected inside huts in Vurra and Okoro counties per (A) Rattus rattus and (B) modified Kilonzo light trap. The χ2 goodness-of-fit analysis revealed that neither the distribution of fleas per rat (N = 944, λ = 0.77) nor the distribution of fleas per hut (N = 2000, λ = 0.42) was consistent with a Poisson distribution (P < 0.001).
DNA amplification and sequencing from known vertebrate species.
Using our real-time PCR-based assay, we successfully amplified and sequenced the 12S rDNA molecular marker from nine vertebrate species found within the domestic or peridomestic environment in our study area. With the primer sequences trimmed from either end of the amplicon, the resulting sequence was 99–102 nt long. Using BLAST, we confirmed that seven of the test sequences were identical to mitochondrial sequences in GenBank from the expected species (A. niloticus, AF141259.2:3–101; C. hircus, HM623880.1:25–124; C. lupus familiaris, AB499817.1:513–611; F. catus, D28892.1:3–103; G. gallus, GU261719.1:1760–1861; H. sapiens, HQ700378.2:1097–1196; R. rattus, EU273707.1:514–613). GenBank did not contain the target sequence from M. natalensis, which was identified in our study using morphological characters found in taxonomic keys for this region of Africa.36 Our M. natalensis sequence (99 nt) shared 99% identity with corresponding sequences from two species in the same monophyletic group: Mastomys huberti (AF141282.2:3–101; 26C>T) and Mastomys erythroleucus (X85952.1:443–541; 465G>A).42 We isolated Crocidura sp. DNA from a shrew that had not been identified to species. The target sequence (99 nt) was not identical to any sequence in the GenBank database; it differed by one nucleotide from the corresponding sequence for Crocidura gueldenstaedtii (AF434825.1:447–545; 493A>T). Aligning the sequences from the nine species we tested revealed that each pair differed by at least seven nucleotides. None of the BLAST results from these nine species indicated that a different species from our study area had an identical target sequence.
Blood meal identification in off-host fleas.
Of the 729 C. felis collected in modified Kilonzo light traps, 694 undamaged fleas were tested. We detected vertebrate DNA in 151 (21.8%). Three samples contained vertebrate DNA from multiple sources as indicated by multiple melting peaks and/or sequence traces with overlapping fluorograms. In one case, repeated attempts to obtain reverse sequence for a mixed DNA sample failed, so we generated and analyzed a 75-nt consensus sequence from two forward sequence reactions. In the other two cases we were able to assemble 100-nt sequences from overlapping forward and reverse sequences. We aligned the mixed-sample sequences, including ambiguous base calls where overlapping fluorograms resulted in double peaks, with the target sequences from the Crocidura sp. trapped in our study area and from the three predominant domestic and peridomestic rodent species in our study area: black rat, Nile rat and multimammate mouse.15,31 The mixed-sample sequences differed from the rodent sequences by ≥ 5 nt and from the Crocidura sp. sequence by ≥ 3 nt. We then used the PHASE algorithm in DnaSP43 to infer haplotypes for each of the three mixed samples from the sequence data associated with all 151 samples containing detectable vertebrate DNA. The BLAST analysis of the inferred haplotypes suggested that two of the samples contained a combination of goat and human DNA, and the third sample contained DNA from a combination of human and cow (Bos sp.). Because these data indicated that these fleas had not fed on a rodent or shrew host, we did not pursue positive identification of each vertebrate DNA source. We did not include these three samples in our statistical analyses.
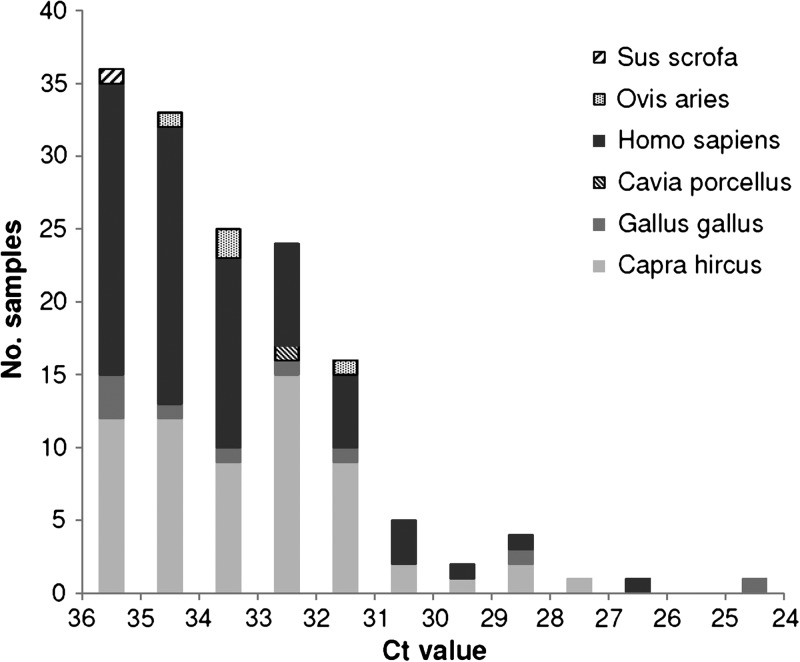
We identified a single vertebrate DNA source in 148 of the off-host C. felis samples. The Ct values associated with these samples ranged from 24.81 to 36.00, but samples tended to yield Ct values closer to rather than farther from the limit of detection (Figure 3). Seventy samples contained human DNA. The remaining samples contained DNA from various domesticated animals (Table 2). Using Equation 4 and the values listed in Table 3, we estimated that ∼35 of the 70 field-collected C. felis samples in which our assay detected human DNA had actually taken a blood meal from a human host.
Figure 3.
Distribution of threshold cycle threshold (Ct) values associated with field-collected Ctenocephalides felis in which our real-time PCR-based assay detected and identified vertebrate DNA (N = 148).
Table 2.
Vertebrate DNA identified in off-host Ctenocephalides felis collected inside huts in Vurra and Okoro counties
| Vertebrate (species) | No. detected (% of total with detectable vertebrate DNA) |
|---|---|
| Human (Homo sapiens) | 70* (47%) |
| Goat (Capra hircus) | 63 (43%) |
| Chicken (Gallus gallus) | 9 (6%) |
| Sheep (Ovis aries) | 4 (3%) |
| Guinea Pig (Cavia porcellus) | 1 (< 1%) |
| Pig (Sus scrofa) | 1 (< 1%) |
Our assay has a 6% false positive rate for human DNA.39 Using Equation 4 and the values listed in Table 3, we estimated that ∼35 of the samples with detectable human DNA represent human blood-fed fleas.
Table 3.
Terms used in Equation 4 to estimate the number of fleas testing positive for human DNA that had actually consumed human blood
| Variable | Definition | Value* |
|---|---|---|
| h | No. fleas testing positive for human DNA | 70 |
| b | No. fleas containing detectable DNA from non-human vertebrates | 78 |
| t | Total number of fleas tested | 691 |
Values do not include three samples containing mixed DNA.
We detected non-human vertebrate DNA in 78 samples. None of them contained DNA from a rat or shrew species. Thus, even if all of the samples testing positive for human DNA contained contaminating DNA, the proportion of detectable C. felis blood meals taken from rat species was 0/78. From this we estimate that no more than 4.7% (upper 95% confidence limit) of off-host C. felis infesting huts in Vurra and Okoro counties during the collection period had fed on rats or other wild rodent or shrew species.
Discussion
Consistent with previous studies,15,16 we found that C. felis is the predominant off-host flea species in human habitations in the West Nile region of Uganda. Laboratory studies have shown that this species is capable of transmitting Y. pestis, but estimated transmission efficiency is very low (0.57%).15 To determine if C. felis might serve as a bridging vector in this region, we investigated C. felis infestation of small mammals trapped in human habitations in Vurra and Okoro counties, and we estimated the proportion of off-host cat fleas with a detectable blood meal that had fed on potentially infectious wild hosts. Consistent with previous studies,15,31 we observed very low C. felis infestation rates on rats trapped in huts (< 0.5%). Results from our blood meal analysis indicate that the off-host cat fleas had not fed on potentially infected wild hosts; they had fed primarily on non-rodent domesticated species.
We verified that our SYBR Green real-time PCR-based assay allows detection and differentiation of vertebrate DNA from at least nine different species found in or near huts in the West Nile region. This includes black rats, Nile grass rats, multimammate mice, and shrews, the predominant domestic and peridomestic small mammal species in our study area.15,31 Using this assay, we detected vertebrate DNA in approximately one-fifth of off-host C. felis. We may have observed this low proportion because many off-host fleas had either not taken a blood meal or had not fed recently and had only thoroughly-digested remnants of blood meals in their guts. Using microscopy, we sought to determine if field-collected fleas had fed. However, the flea samples were stored in ethanol and had developed cloudy, gray midguts, which made it impossible to reliably distinguish blood-fed from unfed specimens. In laboratory studies, our assay detected human, rat, and goat DNA in artificially fed C. felis held alive up to 72 hours post feeding, but detection decreased as the time between blood feeding and collection increased.39 A previous study found that the amount of blood consumed was the principle limiting factor for amplification of triatomine blood meal DNA by PCR,44 therefore it is also possible that we failed to detect vertebrate DNA in smaller fleas that had taken smaller blood meals. A study in Anopheles gambiae found, however, that there was no significant difference in blood meal detection in mosquitoes held for 0–32 hours post feeding that had consumed blood meals categorized as either small or large based on gravimetric analysis.45
Because we were unable to distinguish blood-fed from unfed specimens by microscopy, we tested every off-host flea for vertebrate DNA using our real-time PCR assay. Given that our assay may fail to detect vertebrate DNA in some blood-fed fleas, however, we cannot conclude that fleas with no detectable vertebrate DNA were unfed. For future studies, it might be beneficial to adapt a catalytic assay like the phenolphthalein (Kastle-Meyer) test to detect heme in homogenized samples.46 Only samples that tested positive for heme, indicating that they had consumed a blood meal, would be subjected to real-time PCR analysis. Not only would this reduce the number of real-time PCR reactions required, it would reduce the number of false positives associated with unfed fleas contaminated with human DNA. This would, in turn, increase the probability that fleas testing positive for human DNA had actually consumed human blood.
Although we cannot make any conclusions about the proportion of field-collected fleas that had actually taken a blood meal, our assay was designed to minimize bias toward detection of any one species. Others have reported that amplicon size can affect the sensitivity of PCR-based blood meal assays.47,48 By using a single primer set that targets highly conserved regions of the 12S mitochondrial gene and thus generates similar amplicons from all vertebrate species, we hoped to achieve similar sensitivity to blood from different hosts. There is evidence that fleas with catholic feeding habits take similar sized blood meals from different vertebrate species, even if they prefer one species over another49; therefore, differences in blood meal size should not bias our assay toward detection of any particular vertebrate, although host species may affect the rate of blood digestion.50 Laboratory studies found no significant difference in our assay's sensitivity to human, rat and goat blood at any of seven time points post feed.39 Therefore, we used data from those samples with detectable vertebrate DNA to estimate the proportion of all off-host C. felis blood meals from each host.
A significant proportion of the off-host C. felis had fed on domesticated animals, particularly goats and, to a lesser extent, chickens. This is consistent with a 2006 observational survey in the West Nile region, which found that these were the two domesticated species most likely to be present inside or within 10 m of human habitations (CDC, unpublished data). Previous studies in other parts of East Africa have found domesticated species including goats, pigs, sheep, cats, dogs, and cows infested with C. felis.17,21 None of the non-rodent domesticated species we detected is likely to serve as a source of Y. pestis infection. To infect a feeding flea, a host must generally develop an overwhelming bacteremia (> 106 cfu/mL), which is often fatal.51,52 Chickens are resistant to Y. pestis infection.53 Pigs are susceptible to infection but do not show any obvious signs of disease54; they are highly unlikely to develop the high bacteremia required to infect feeding fleas. The German Plague Commission found evidence of active infection in experimentally infected sheep and goats, but the animals all recovered.55 Others have found that sheep succumb to Y. pestis infection when inoculated intravenously.56 There is limited evidence, however, of naturally acquired infection or plague-associated deaths in domesticated sheep or goats. Based on serological observations and flea infestation data from domesticated animals in Tanzanian villages where plague was active, Kilonzo25 concluded that goats and sheep were not involved in plague epidemiology in that region. One report from Libya suggested that a small number of human plague cases may have been associated with killing or skinning sick goats and sheep, but there was no indication of transmission from these species by fleas. Even in this case the authors “Regard the goat as a sentinel animal . . . not necessarily the immediate cause of cases in humans.”57
Guinea pigs are highly susceptible to Y. pestis infection; they have been used in laboratory studies of plague since the early 20th century.58 Contact with infected guinea pigs or their fleas has been cited as a potential source of human Y. pestis infection at a zoo in India59 and in Ecuador.60,61 Guinea pigs were observed in < 2% of huts included in a 2006 survey of the West Nile region (CDC, unpublished data). We did not collect data on the percentage of huts included in our study that were raising guinea pigs, however, so we cannot determine the extent to which guinea pig abundance contributed to the low proportion of guinea pig blood meals detected in off-host C. felis. Given the cat flea's low transmission efficiency, transmission by this species from an infected guinea pig to a human is likely only if C. felis feed frequently on guinea pigs in human habitations where this domesticated species is abundant. In addition, guinea pigs are unlikely to become a source of infection unless they are also infested with flea species that feed regularly on rats and could thus transmit Y. pestis from a wild host. Kilonzo21 observed that domesticated guinea pigs were poor hosts of fleas in the Lushoto district of Tanzania and unlikely to play a large role in plague epidemiology there. Liston59 noted, however, that although X. cheopis rarely feeds on guinea pigs, some guinea pigs kept at the zoological gardens of Bombay were heavily infested with X. cheopis a few days after dead rats were found near their cage. The infested guinea pigs died of plague. Further study is needed to determine what role guinea pigs might play in plague epidemiology in the West Nile region.
Our results indicate that C. felis in human habitations in the West Nile region feed very infrequently on potentially infectious wild hosts. Though we trapped at least one R. rattus in 38.3% of huts, we did not detect R. rattus DNA in any of the off-host C. felis collected concurrently. Only 2 of the 727 fleas collected from the rats were C. felis. Other studies have indicated that other small mammals including Nile rats, multimammate mice, and shrews inhabit the domestic and peridomestic environments in this region,15,31 but we did not detect DNA from any of these species in off-host C. felis either. We estimate that not more than 4.7% of off-host C. felis infesting huts during the collection period had fed on R. rattus or other wild hosts that could serve as a source of Y. pestis.
Using a modification of a plague model based on Macdonald's equation,51,62 Eisen and others15 estimated that at least 20 cat fleas per host would be required for a focal Y. pestis infection to give rise to a secondary infection. This estimate assumes that all cat fleas are feeding on a single host. To serve as a bridging vector, a flea must first feed on a potentially infectious zoonotic host and then on a human host. If not more than 4.7% of off-host C. felis infesting huts feed on a potentially infectious wild host, the estimated number of cat fleas required for a focal infection (in a rodent or shrew) to give rise to a secondary infection increases more than 20-fold. Given that our assay detects contaminating human DNA in ∼6% of samples tested, we estimate that ∼35 of the off-host C. felis in which we detected human DNA had fed on a human. Notably, this is less than the number of goat blood meals we detected. Our data thus suggest that although cat fleas may be feeding on humans in this region, they take the majority of their blood meals from domesticated animals that are unlikely to play a significant role in perpetuating transmission of Y. pestis. This finding further increases the estimated number of cat fleas required for a focal infection in a zoonotic host to give rise to a secondary infection in a human. We conclude that C. felis is highly unlikely to serve as a bridging vector for Y. pestis in the West Nile region.
It should be noted that our study was conducted within a single 2-month period, and flea prevalence may vary between seasons. Amatre and others31 reported significant differences between flea loads on some wild rodent species, including R. rattus, trapped in the West Nile region during different 2-month collection periods between January and August of 2006. Regardless of season, however, a tiny proportion of on-host fleas were C. felis.31 Although the number of C. felis infesting huts might vary, we would not expect to find that a significantly higher proportion of off-host C. felis had fed on potentially infectious wild hosts during different seasons.
Our findings suggest that efforts to control C. felis are unlikely to impact Y. pestis transmission to humans in Vurra and Okoro counties. Efforts to prevent human plague cases in this plague-endemic region should remain focused on controlling the fleas that feed on potential zoonotic hosts, particularly X. cheopis and X. brasiliensis. During inter-epizootic periods, such as the period during which we conducted this study, these fleas are most likely to be found on rodent hosts, so flea-control measures that target on-rodent fleas are more likely to limit human exposure to Y. pestis than measures targeting only off-host fleas.
Our study does suggest, however, that the off-host C. felis found in huts in the West Nile region are biting humans. This is consistent with previous reports that cat fleas readily feed on people.26,28–30 This flea species may transmit pathogens other than Y. pestis, including Rickettsia typhi, Rickettsia felis, and Bartonella species.63–66 Further study is required to determine whether cat fleas are likely to transmit these pathogens to humans in Uganda. In addition, insofar as C. felis is a nuisance biter in this region, vector control efforts, including those that target plague, may be more appealing to residents of the West Nile region if they eliminate C. felis in addition to those species most likely to transmit pathogens.
ACKNOWLEDGMENTS
We thank J. Stovall, K. Boroughs, and H. Brown for technical assistance, and K. MacMillan for help with flea identification and figures.
Footnotes
Authors' addresses: Christine B. Graham, Jeff N. Borchert, Karen A. Boegler, Sean M. Moore, Kenneth L. Gage, and Rebecca J. Eisen, Division of Vector-Borne Diseases, Centers for Disease Control and Prevention, Fort Collins, CO, E-mails: cgraham@cdc.gov, gqx1@ug.cdc.gov, kje5@cdc.gov, kfy0@cdc.gov, klg0@cdc.gov, and dyn2@cdc.gov. William C. Black IV, Microbiology, Immunology, and Pathology Department, Colorado State University, Fort Collins, CO, E-mail: william.black@colostate.edu. Linda A. Atiku and Joseph T. Mpanga, Uganda Virus Research Institute, Entebbe, Uganda, E-mails: l_atikupraise@yahoo.com and joe1ug@msn.com.
References
- 1.Butler T. Plague into the 21st Century. Clin Infect Dis. 2009;49:736–742. doi: 10.1086/604718. [DOI] [PubMed] [Google Scholar]
- 2.Gage KL, Kosoy MY. Natural history of plague: perspectives from more than a century of research. Annu Rev Entomol. 2005;50:505–528. doi: 10.1146/annurev.ento.50.071803.130337. [DOI] [PubMed] [Google Scholar]
- 3.Gratz NG. Control of plague transmission. In: Dennis DT, Gage KL, Gratz N, Poland JD, Tikhomirov E, editors. Plague Manual: Epidemiology, Distribution, Surveillance and Control. Geneva: World Health Organization; 1999. pp. 97–134. [Google Scholar]
- 4.Borchert JN, Mach JJ, Linder TJ, Ogen-Odoi A, Angualia S. Invasive rats and bubonic plague in northwest Uganda. In: Witmer GW, Pitt WC, Fagerstone A, editors. Managing Vertebrate Invasive Species: Proceedings of an International Symposium. Fort Collins, CO: USDA/APHIS Wildlife Services, National Wildlife Research Center; 2007. pp. 283–293. [Google Scholar]
- 5.Moore SM, Monaghan A, Griffith KS, Apangu T, Mead PS, Eisen RJ. Improvement of disease prediction and modeling through the use of meteorological ensembles: human plague in Uganda. PLoS ONE. 2012;7:e44431. doi: 10.1371/journal.pone.0044431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tikhomirov E. Epidemiology and distribution of plague. In: Dennis DT, Gage KL, Gratz N, Poland JD, Tikhomirov E, editors. Plague Manual: Epidemiology, Distribution, Surveillance and Control. Geneva: World Health Organization; 1999. pp. 11–42. [Google Scholar]
- 7.Craven RB, Maupin GO, Beard ML, Quan TJ, Barnes AM. Reported cases of human plague infections in the United States, 1970–1991. J Med Entomol. 1993;30:758–761. doi: 10.1093/jmedent/30.4.758. [DOI] [PubMed] [Google Scholar]
- 8.Hopkins GHE. Report on Rats, Fleas and Plague in Uganda. Uganda: Government Printer of Uganda; 1949. [Google Scholar]
- 9.Kilonzo BS. Observations on the epidemiology of plague in Tanzania during the period 1974–1988. East Afr Med J. 1992;69:494–499. [PubMed] [Google Scholar]
- 10.Gratz NG. Rodent reservoirs and flea vectors of natural foci of plague. In: Dennis DT, Gage KL, Gratz N, Poland JD, Tikhomirov E, editors. Plague Manual: Epidemiology, Distribution, Surveillance and Control. Geneva: World Health Organization; 1999. pp. 63–96. [Google Scholar]
- 11.Laudisoit A, Leirs H, Makundi RH, Van Dongen S, Davis S, Neerinckx S, Deckers J, Libois R. Plague and the human flea, Tanzania. Emerg Infect Dis. 2007;13:687–693. doi: 10.3201/eid1305.061084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kilonzo BS, Makundi RH, Mbise TJ. A decade of plague epidemiology and control in the western Usambara Mountains, north-east Tanzania. Acta Trop. 1992;50:323–329. doi: 10.1016/0001-706x(92)90067-8. [DOI] [PubMed] [Google Scholar]
- 13.Guiguen C, Beaucournu JC. Presence of Pulex irritans L. (Siphonaptera) in Burundi, plague risk area. Bull Soc Pathol Exot. 1979;72:481–486. [PubMed] [Google Scholar]
- 14.Karimi Y, Farhang-Azad A. Pulex irritans, a human flea in the plague infection focus at General Mobutu Lake (formerly Lake Albert): epidemiologic significance. Bull World Health Organ. 1974;50:564–565. [PMC free article] [PubMed] [Google Scholar]
- 15.Eisen RJ, Borchert JN, Holmes JL, Amatre G, Van Wyk K, Enscore RE, Babi N, Atiku LA, Wilder AP, Vetter SM, Bearden SW, Montenieri JA, Gage KL. Early-phase transmission of Yersinia pestis by cat fleas (Ctenocephalides felis) and their potential role as vectors in a plague-endemic region of Uganda. Am J Trop Med Hyg. 2008;78:949–956. [PubMed] [Google Scholar]
- 16.Borchert JN, Eisen RJ, Holmes JL, Atiku LA, Mpanga JT, Brown HE, Graham CB, Babi N, Montenieri JA, Enscore RE, Gage KL. Evaluation and modification of off-host flea collection techniques used in northwest Uganda: laboratory and field studies. J Med Entomol. 2012;49:210–214. doi: 10.1603/me11045. [DOI] [PubMed] [Google Scholar]
- 17.Hopkins GHE. Annotated and illustrated keys to the known fleas of East Africa. Uganda J. 1947;11:133–190. [Google Scholar]
- 18.Lewis RE. Notes on the geographical distribution and host preferences in the order Siphonaptera. 1. Pulicidae. J Med Entomol. 1972;9:511–520. doi: 10.1093/jmedent/9.6.511. [DOI] [PubMed] [Google Scholar]
- 19.Vobis M, D'Haese J, Mehlhorn H, Mencke N, Blagburn B, Bond R, Denholm I, Dryden MW, Payne P, Rust MK, Schroeder I, Vaughn MB, Bledsoe D. Molecular phylogeny of isolates of Ctenocephalides felis and related species based on analysis of ITS1, ITS2 and mitochondrial 16S rDNA sequences and random binding primers. Parasitol Res. 2004;94:219–226. doi: 10.1007/s00436-004-1201-x. [DOI] [PubMed] [Google Scholar]
- 20.Mehlhorn H, D'Haese J, Vobis M, Mencke N. No molecular indications for the occurrence of subspecies in the cat flea Ctenocephalides felis (Siphonaptera: Ctenocephalidae) Entomol Gen. 2005;27:295–301. [Google Scholar]
- 21.Kilonzo BS. Studies on determining the involvement of domestic animals in plague epidemiology in Tanzania. Tanzanian Veterinary Bulletin. 1980;2:37–44. [Google Scholar]
- 22.Dipeolu OO, Ayoade GO. Various hosts of Ctenocephalides felis strongylus. Vet Q. 1982;4:191–192. doi: 10.1080/01652176.1982.9693863. [DOI] [PubMed] [Google Scholar]
- 23.Fagbemi BO. Effect of Ctenocephalides felis strongylus infestation on the performance of West African dwarf sheep and goats. Vet Q. 1982;4:92–95. doi: 10.1080/01652176.1982.9693846. [DOI] [PubMed] [Google Scholar]
- 24.Rust MK, Dryden MW. The biology, ecology, and management of the cat flea. Annu Rev Entomol. 1997;42:451–473. doi: 10.1146/annurev.ento.42.1.451. [DOI] [PubMed] [Google Scholar]
- 25.Kilonzo BS, Gisakanyi ND, Sabuni CA. Involvement of dogs in plague epidemiology in Tanzania. Serological observations in domestic animals in Lushoto District. Scand J Infect Dis. 1993;25:503–506. doi: 10.3109/00365549309008533. [DOI] [PubMed] [Google Scholar]
- 26.Kaal JF, Baker K, Torgerson PR. Epidemiology of flea infestation of ruminants in Libya. Vet Parasitol. 2006;141:313–318. doi: 10.1016/j.vetpar.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 27.Psaroulaki A, Antoniou M, Papaeustathiou A, Toumazos P, Loukaides F, Tselentis Y. First detection of Rickettsia felis in Ctenocephalides felis fleas parasitizing rats in Cyprus. Am J Trop Med Hyg. 2006;74:120–122. [PubMed] [Google Scholar]
- 28.Pet'ko B. Domestic cats as a source of human flea infestations in towns. Cesk Epidemiol Mikrobiol Imunol. 1993;42:190–191. [PubMed] [Google Scholar]
- 29.Woods ME, Montenieri JA, Eisen RJ, Zeidner NS, Borchert JN, Laudisoit A, Babi N, Atiku LA, Enscore RE, Gage KL. Identification of flea blood meals using multiplexed real-time polymerase chain reaction targeting mitochondrial gene fragments. Am J Trop Med Hyg. 2009;80:998–1003. [PubMed] [Google Scholar]
- 30.Hunter KW, Jr, Campbell AR, Sayles PC. Human infestation by cat fleas, Ctenocephalides felis (Siphonaptera: Pulicidae), from suburban raccoons. J Med Entomol. 1979;16:547. doi: 10.1093/jmedent/16.6.547. [DOI] [PubMed] [Google Scholar]
- 31.Amatre G, Babi N, Enscore RE, Ogen-Odoi A, Atiku LA, Akol A, Gage KL, Eisen RJ. Flea diversity and infestation prevalence on rodents in a plague-endemic region of Uganda. Am J Trop Med Hyg. 2009;81:718–724. doi: 10.4269/ajtmh.2009.09-0104. [DOI] [PubMed] [Google Scholar]
- 32.Winters AM, Staples JE, Ogen-Odoi A, Mead PS, Griffith K, Owor N, Babi N, Enscore RE, Eisen L, Gage KL, Eisen RJ. Spatial risk models for human plague in the West Nile region of Uganda. Am J Trop Med Hyg. 2009;80:1014–1022. [PubMed] [Google Scholar]
- 33.Borchert JN, Enscore RE, Eisen RJ, Atiku LA, Owor N, Acayo S, Babi N, Montenieri JA, Gage KL. Evaluation of rodent bait containing imidacloprid for the control of fleas on commensal rodents in a plague-endemic region of northwest Uganda. J Med Entomol. 2010;47:842–850. doi: 10.1603/me09221. [DOI] [PubMed] [Google Scholar]
- 34.Kilonzo BS. Geneva: World Health Organization; 1977. A simple light trap for field collection of adult fleas: studies on its efficiency and suitability in north-east Tanzania. WHO/VBC/77.673, 11. [Google Scholar]
- 35.Borchert JN, Eisen RJ, Atiku LA, Delorey MJ, Mpanga JT, Babi N, Enscore RE, Gage K. Efficacy of indoor residual spraying using lambda-cyhalothrin for controlling non-target vector fleas (Siphonaptera) on commensal rats in a plague endemic region of northwest Uganda. J Med Entomol. 2012;49:1027–1034. doi: 10.1603/me11230. [DOI] [PubMed] [Google Scholar]
- 36.Delany MJ. The Rodents of Uganda. Kettering Northamptonshire: The George Press; 1975. [Google Scholar]
- 37.Haselbarth E. Siphonaptera. In: Zumpt F, editor. The Arthropod Parasites of Vertebrates in Africa South of the Sahara (Ethiopia region) Johannesburg: South African Institute of Medical Research; 1966. pp. 117–212. [Google Scholar]
- 38.Smit FGAM. Siphonaptera (fleas) In: Smith KGV, editor. Insects and Other Arthropods of Medical Importance. London: British Museum of Natural History; 1973. pp. 325–371. [Google Scholar]
- 39.Graham CB, Black WC, IV, Boegler KA, Montenieri JA, Holmes JL, Gage KL, Eisen RJ. Combining real-time polymerase chain reaction using SYBR Green I detection and sequencing to identify vertebrate bloodmeals in fleas. J Med Entomol. 2012;49:1442–1452. doi: 10.1603/me12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN. Universal primer cocktails for fish DNA barcoding. Mol Ecol Notes. 2007;7:544–548. [Google Scholar]
- 41.Agresti A, Coull BA. Approximate is better than “exact” for interval estimation of binomial proportions. Am Stat. 1998;52:119–126. [Google Scholar]
- 42.Wilson DE, Reeder DM. Mammal Species of the World: A Taxonomic and Geographic Reference. Baltimore, MD: The Johns Hopkins University Press; 2005. [Google Scholar]
- 43.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 44.Mota J, Chacon JC, Gutiterrez-Cabrera AE, Sanchez-Cordero V, Wirtz RA, Ordoez R, Panzera F, Ramsey JM. Identification of blood meal source and infection with Trypanosoma cruzi of Chagas disease vectors using a multiplex cytochrome b polymerase chain reaction assay. Vector Borne Zoonotic Dis. 2007;7:617–627. doi: 10.1089/vbz.2007.0106. [DOI] [PubMed] [Google Scholar]
- 45.Mukabana WR, Takken W, Seda P, Killeen GF, Hawley WA, Knols BGJ. Extent of digestion affects the success of amplifying human DNA from blood meals of Anopheles gambiae (Diptera: Culicidae) Bull Entomol Res. 2002;92:233–239. doi: 10.1079/BER2002164. [DOI] [PubMed] [Google Scholar]
- 46.Gurtler RE, Oneto ML, Cecere MC, Castanera MB, Canale DM. A simple method to identify triatomine (Hemiptera: Reduviidae) feces in sensing devices used in vector surveillance programs. J Med Entomol. 2001;38:147–152. doi: 10.1603/0022-2585-38.2.147. [DOI] [PubMed] [Google Scholar]
- 47.Kirstein F, Gray JS. A molecular marker for the identification of the zoonotic reservoirs of Lyme borreliosis by analysis of the blood meal in its European vector Ixodes ricinus. Appl Environ Microbiol. 1996;62:4060–4065. doi: 10.1128/aem.62.11.4060-4065.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kent RJ, Norris DE. Identification of mammalian blood meals in mosquitoes by a multiplexed polymerase chain reaction targeting cytochrome b. Am J Trop Med Hyg. 2005;73:336–342. [PMC free article] [PubMed] [Google Scholar]
- 49.Krasnov BR, Korine C, Burdelova NV, Khokhlova IS, Pinshow B. Between-host phylogenetic distance and feeding efficiency in hematophagous ectoparasites: rodent fleas and a bat host. Parasitol Res. 2007;101:365–371. doi: 10.1007/s00436-007-0480-4. [DOI] [PubMed] [Google Scholar]
- 50.Krasnov BR, Sarfati M, Arakelyan MS, Khokhlova IS, Burdelova NV, Degen AA. Host specificity and foraging efficiency in blood-sucking parasite: feeding patterns of the flea Parapulex chephrenis on two species of desert rodents. Parasitol Res. 2003;90:393–399. doi: 10.1007/s00436-003-0873-y. [DOI] [PubMed] [Google Scholar]
- 51.Lorange EA, Race BL, Sebbane F, Hinnebusch BJ. Poor vector competence of fleas and the evolution of hypervirulence in Yersinia pestis. J Infect Dis. 2005;191:1907–1912. doi: 10.1086/429931. [DOI] [PubMed] [Google Scholar]
- 52.Eskey CR, Haas VH. Plague in the Western Part of the United States. Washington, DC: U.S. Government Printing Office; 1940. Public Health Bulletin 254. [Google Scholar]
- 53.Meyer KF. Immunity in plague; a critical consideration of some recent studies. J Immunol. 1950;64:139–163. [PubMed] [Google Scholar]
- 54.Marshall JD, Jr, Harrison DN, Murr JA, Cavanaugh DC. The role of domestic animals in the epidemiology of plague. 3. Experimental infection of swine. J Infect Dis. 1972;125:556–559. doi: 10.1093/infdis/125.5.556. [DOI] [PubMed] [Google Scholar]
- 55.Bannerman WB, Kapadia RJ. XXVII. Report on experiments undertaken to discover whether the common domestic animals of India are affected by plague. J Hyg (Lond) 1908;8:209–220. doi: 10.1017/s0022172400003272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Lien-teh. Plague: A Manual for Medical and Public Health Workers. Shanghai, China: The Mercury Press; 1936. [Google Scholar]
- 57.Christie AB, Chen TH, Elberg SS. Plague in camels and goat: their role in human epidemics. J Infect Dis. 1980;141:724–726. doi: 10.1093/infdis/141.6.724. [DOI] [PubMed] [Google Scholar]
- 58.Pollitzer R. Plague. World Health Organization Monograph Series. Geneva: World Health Organization; 1954. [Google Scholar]
- 59.Liston WG. Plague, rats and fleas. J Bombay Nat Hist Soc. 1905;16:253–274. [Google Scholar]
- 60.Gabastou JM, Proano J, Vimos A, Jaramillo G, Hayes E, Gage K, Chu M, Guarner J, Zaki S, Bowers J, Guillemard C, Tamayo H, Ruiz A. An outbreak of plague including cases with probable pneumonic infection, Ecuador, 1998. Trans R Soc Trop Med Hyg. 2000;94:387–391. doi: 10.1016/s0035-9203(00)90114-7. [DOI] [PubMed] [Google Scholar]
- 61.Macchiavello A. Epidemiology of plague in Ecuador. Am J Public Health Nations Health. 1943;33:807–811. doi: 10.2105/ajph.33.7.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eisen RJ, Bearden SW, Wilder AP, Montenieri JA, Antolin MF, Gage KL. Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc Natl Acad Sci USA. 2006;103:15380–15385. doi: 10.1073/pnas.0606831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elston DM, Do H. What's eating you? Cat flea (Ctenocephalides felis), Part 1: clinical features and role as a disease vector. Cutis. 2010;85:231–236. [PubMed] [Google Scholar]
- 64.Azad AF, Radulovic S, Higgins JA, Noden BH, Troyer JM. Flea-borne rickettsioses: ecologic considerations. Emerg Infect Dis. 1997;3:319–327. doi: 10.3201/eid0303.970308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schriefer ME, Sacci JB, Taylor JP, Higgins JA, Azad AF. Murine typhus: updated roles of multiple urban components and a 2nd typhus like Rickettsia. J Med Entomol. 1994;31:681–685. doi: 10.1093/jmedent/31.5.681. [DOI] [PubMed] [Google Scholar]
- 66.Sackal C, Laudisoit A, Kosoy M, Massung R, Eremeeva ME, Karpathy SE, Van Wyk K, Gabitzsch E, Zeidner NS. Bartonella spp. and Rickettsia felis in fleas, Democratic Republic of Congo. Emerg Infect Dis. 2008;14:1972–1974. doi: 10.3201/eid1412.080610. [DOI] [PMC free article] [PubMed] [Google Scholar]