Abstract
BACKGROUND
The Marin strain of Culex pipiens Say is a pyrethroid-resistant population that was collected in Marin County, California, in 2001 and subsequently maintained in the laboratory under regular permethrin exposure.
RESULTS
In this study, two genes, CpGSTd1 and CpGSTd2, encoding glutathione S-transferase (GST) were cloned from Cx. pipiens Marin. Phylogenetic analysis of the deduced amino acid sequences, CpGSTD1 and CpGSTD2, of these genes indicated that they belong to the Delta class of insect GSTs. The nucleotide and deduced amino acid sequences of CpGSTd1 and CpGSTd2 were 59% and 48% identical, respectively. CpGSTD1 and CpGSTD2 were expressed in Escherichia coli and purified by affinity chromatography. The recombinant GSTs exhibited unique selectivity towards the general GST substrates CDNB and DCNB, and also differed in their sensitivity to known inhibitors of GSTs. CpGSTD1 exhibited peroxidase activity with cumene hydroperoxide, while CpGSTD2 appeared to lack this activity. CpGSTD1 was able to metabolize DDT, while DDT metabolism by CpGSTD2 was not detectable. CpGSTD1 and CpGSTD2 showed no detectable metabolism of permethrin. Gene expression of CpGSTd1 and CpGSTd2 in Marin mosquitoes was elevated by about 2-fold in comparison to that found in a pyrethroid-sensitive mosquito strain.
CONCLUSION
Our results indicated that CpGSTD1 and CpGSTD2 have unique biochemical characteristics but they did not appear to play major roles in permethrin resistance in Marin mosquitoes.
Keywords: GST, glutathione S-transferase, Culex pipiens, mosquito, insecticide
1 INTRODUCTION
Members of the glutathione S-transferase (GST; EC 2.5.1.18) family of detoxification enzymes catalyze the conjugation of the tripeptide glutathione (GSH) to electrophilic centers of nonpolar compounds, making them more water soluble and thus easier to excrete from cells.1,2 GSTs are ubiquitous in both prokaryotes and eukaryotes, and play central roles in the detoxification of endogenous and xenobiotic compounds including drugs, herbicides, and insecticides. GSTs are also involved in the biosynthesis and intracellular transport of hormones, and protection against oxidative stress.2,3 In insects, the majority of GSTs are cytosolic dimeric enzymes, organized into six classes (Delta, Epsilon, Sigma, Theta, Omega, and Zeta) on the basis of their sequence similarity, chromosomal location and immunological properties.4 Generally, GSTs with greater than 40% sequence identity are grouped in the same class.
GSTs from the insect-specific Delta and Epsilon GST classes are associated with resistance to organophosphate insecticides through dearylation or dealkylation reactions,5,6 and organochlorine insecticides through dehydrochlorinase activity.7–9 Elevated GST activity and increased gst gene expression levels are reported in pyrethroid resistant insects including mosquitoes.10 Exposure to the pyrethroid deltamethrin can also increase or even decrease (depending on concentration) gst gene expression levels in locusts.11 GSTs, however, have not been shown to directly metabolize pyrethroid insecticides. In pyrethroid resistant brown planthoppers, Nilaparvata lugens, elevated GST activity protects against lipid peroxidation and other oxidative stresses that are induced by pyrethroid exposure.12 Insect GSTs may also function as binding proteins that sequester pyrethroids until they are metabolized by other detoxification enzymes such as carboxylesterases or P450 oxidases.13,14
Mosquitoes in the Culex pipiens complex are vectors of multiple human pathogens including West Nile virus,15 St. Louis encephalitis virus,16 Rift Valley fever virus,17 and Wuchereria bancrofti;18 as well as zoonotic pathogens including dog heartworm19 and avian malaria.20 Our ability to reduce the incidence of diseases vectored by Culex relies primarily on our ability to control them by habitat reduction and the use of biological and chemical insecticides.21 Monitoring and managing resistance to chemical insecticides is a critical component in our programs to maintain effective control of these disease vectors. Resistance to organochlorines,22 organophosphates,23,24 and more recently, pyrethroids is documented in Cx. pipiens sensu lato.25–27
Target site insensitivity and increased metabolic activity by detoxification enzymes (e.g., GSTs, carboxylesterases, and P450s) are key mechanisms of insecticide resistance.28,29 A clear understanding of the biochemical properties and expression profiles of detoxification enzymes such as GSTs is essential for our understanding of the roles that detoxification enzymes play in conferring resistance. In the case of GSTs, the presence of multiple cytosolic isozymes with similar physical and biochemical properties and overlapping substrate selectivity, makes it difficult to purify individual GSTs on the basis of biophysical techniques. A useful alternative is gene cloning, followed by recombinant protein expression, and subsequent biochemical characterization of the individual recombinant GSTs. In this study, we cloned two GST-encoding genes from pyrethroid resistant Cx. pipiens using degenerate PCR primers that targeted the conserved glutathione binding motif of GSTs. Two recombinant GSTs, CpGSTD1 and CpGSTD2, were expressed in Escherichia coli, purified, and characterized. In order to decipher their potential role(s) in insecticide resistance, the ability of CpGSTD1 and CpGSTD2 to metabolize permethrin and DDT was analyzed. The expression levels of CpGSTd1 and CpGSTd2 transcripts in permethrin resistant (i.e., Marin) and permethrin susceptible (i.e., CQ1) strains of Cx. pipiens were also compared. Our findings suggested that CpGSTd1 and CpGSTd2 encode two Delta class GSTs with unique biochemical characteristics. CpGSTD1 and CpGSTD2, however, appeared not to play major roles in permethrin resistance in Marin mosquitoes.
2 EXPERIMENTAL METHODS
2.1 Chemicals
1,1,1-Trichloro-2,2-bis(4-chlorophenyl)ethane (DDT), 1,1-dichloro-2,2-bis(4-chlorophenyl)ethane (DDE), 1-chloro-2,4-dinitrobenze (CDNB), 1,2-dichloro-4-nitrobenzene (DCNB), cumene hydroperoxide (CMHP), ethacrynic acid (EA) and S-hexylglutathione were purchased from Sigma-Aldrich Life Sciences (St. Louis, MO). Permethrin and lambda cyhalothrin were purchased from Chem Service (West Chester, PA). The reduced form of GSH was purchased from Fisher Scientific (Tustin, CA). Chalcone and 4-hydroxychalcone were purchased from Acros Organics (Geel, Belgium) and Fairfield Chemical (Blythewood, SC), respectively. Bivalent inhibitor30 was a generous gift from Dr. W. M. Atkins, Department of Medicinal Chemistry, University of Washington, Seattle, WA.
2.2 Mosquito colony and maintenance
A permethrin tolerant population of Cx. pipiens was originally collected in California (Marin County, San Rafael) in May 2001.26 This population was used to establish a permethrin resistant colony named Cx. pipiens var. molestus Marin by repeated exposure to permethrin (approximately an LC50 dose) as larvae at every five generations. In addition to permethrin resistance, the Marin colony exhibits resistance to DDT and lambda cyhalothrin.26 An insecticide susceptible population of Cx. quinquefasciatus named CQ1 was originally collected in Merced County, California, in the 1950s and subsequently maintained in the laboratory. Both Marin and CQ1 mosquitoes were reared under standard conditions with a 14:10 light:dark cycle and constant temperature of 27°C. The larvae were fed a diet of liver powder and ground rodent chow (LabDiet 5001, PMI Nutrition International, Brentwood, MO). Adults were provided constant access to a 10% (w:w) solution of sucrose.
2.3 Molecular cloning of CpGSTd1 and CpGSTd2
Total RNA was isolated from 100 4th instar Marin larvae at generation 78 following field collection (see26) using an RNeasy kit (Qiagen, Valencia, CA), according to manufacturer’s instructions. The permethrin resistance ratio of the Marin strain was not determined at generation 78. However, at generation 57 the Marin strain showed a resistance ratio of 126 when compared with the CQ1 strain (comparison of LC90 values), and we believe that a similar resistance ratio was maintained at generation 78. Messenger RNA (mRNA) was isolated from total RNAs using a Nucleotrap Midi kit (Clontech, Mountain View, CA) according to manufacturer’s instructions. First strand cDNAs were generated from 1 μg of mRNA using MMLV reverse transcriptase (Clontech). Two degenerate primers, GSTi and GSTv (Table 1), corresponding to the conserved glutathione binding domain were designed following ClustalW alignment31 of known GST sequences from Anopheles gambiae32 and Drosophila melanogaster.33 The GSTi and GSTv primers were used for 3′-RACE (Clontech) using KOD HotStart Polymerase (Promega, Madison, WI), which generated roughly 600 nts of sequence that corresponded to the 3′ ends and 3′ untranslated regions (UTRs) of two potential GSTs. These sequences were used to design two gene specific primers, GST3prime1 and GST3prime2 (Table 1), for 5′-RACE to generate sequences corresponding to the 5′ UTRs and 5′ ends of the two putative GSTs. The full-length coding sequences of the two putative GSTs were amplified using the primer pairs GST1F/GST1R and GST2F/GST2R (Table 1). These primer pairs incorporated HindIII restriction endonuclease sites at the ends of the putative GSTs for cloning into a bacterial expression vector as described below. The amplicons were inserted into the cloning vector pCR2.1-TOPO (Invitrogen, Carlsbad, CA) and the sequences of the inserts were confirmed in both directions. Analysis of the deduced amino acid sequences of the inserts by BLAST34 indicated that they were likely Delta class GSTs, thus the coding sequences were named CpGSTd1 and CpGSTd2.
Table 1.
Primers used for cloning and expression analysis of CpGSTd1 and CpGSTd2
| Primer name | Sequence | Targeta |
|---|---|---|
| GSTi | AAYCCNCARCAYNNNATHCCNAC | |
| GSTv | AAYCCNCARCAYNNNGTNCCNAC | |
| GST3prime1 | TCACTTTCCTCCCAGGAACTTGGC | |
| GST3prime2 | TTAGTGCTTAACATCAACAAAGTACG | |
| GST1F | CCCCAAGCTTCGATGGATTTCTACTACCTGCC | |
| GST1R | GGGGAAGCTTCACTTTCCTCCCAGGAAC | |
| GST2F | CCCCAAGCTTCGATGAATCTTTATCACATGGAAC | |
| GST2R | GGGGAAGCTTTAGTGCTTAACATCAACAAAG | |
| CpGSTd1F | CTGAACCTTAAGCTGACCAACCTG | CpGSTd1 |
| CpGSTd1R | GTTCTTCTTGCACCGCTCCAAC | |
| CpGSTd2F | GTGCCAGTCGGTGCGGCTTCT | CpGSTd2 |
| CpGSTd2R | CGTACTTGGCCAAATCAACTCCG | |
| RPS7F | CAGGCCATCCTGGAGCTGGAG | RPS7 |
| RPS7R | CGGGAACTCGAACGTGACGTC |
Target gene for expression analysis.
2.4 Phylogenetic analysis of CpGSTd1 and CpGSTd2
The deduced amino acid sequences of CpGSTd1 and CpGSTd2 were aligned with GST sequences of An. gambiae32 using the ClustalW program.31 The alignment was then used to construct a phylogenetic tree by the Neighbor-Joining algorithm provided in the MEGA4 software package.35
2.5 Bacterial expression and purification of recombinant CpGSTD1 and CpGSTD2
Functionally active insect GSTs have been commonly expressed using bacterial expression systems.14,36,37 In order to express recombinant proteins encoded by CpGSTd1 and CpGSTd2, these sequences were excised by HindIII digestion from the pCR2.1-TOPO vector containing these sequences (see above) and ligated into the HindIII site of the bacterial expression vector pLP-PROTet-6xHN (Clontech). These constructs placed the coding sequences under the tight regulation of the anhydrotetracycline inducible PL promoter and expressed a recombinant protein with an N-terminal histidine tag. Recombinant pLB-PROTet-6xHN vectors carrying either CpGSTd1 or CpGSTd2 were transformed into BL21 Pro cells (Clontech) for recombinant protein expression. Transformed BL21 Pro cells (250 culture volume) were grown in Luria Broth (LB) medium as recommended by the manufacturer (Clontech) until an OD600 of 0.6 was reached. At this point, anhydrotetracycline was added to the culture (final concentration of 100 ng ml−1) to induce expression of the recombinant GST and the culture was grown for another 4 hours.
BL21 Pro cells expressing CpGSTD1 or CpGSTD2 were precipitated by centrifugation for 20 min at 3,000g. Following centrifugation, the supernatant was discarded and the pellet was resuspended in 10 ml of BugBuster reagent (EMD Biosciences, San Diego, CA) and incubated for 10 min at room temperature. The mixture was then centrifuged for 20 min at 8,600g, and the supernatant was transferred to a new centrifuge tube. The supernatant was then diluted 1:1 with TALON equilibration and wash buffer (Clontech) containing 5 mM imidazole. This solution was loaded onto a column containing 1 ml of prewashed and equilibrated TALON Metal Affinity Resin (Clontech). Following protein loading, the column was washed first with 10 ml of TALON equilibration and wash buffer, containing 10 mM imidazole, then 10 ml of TALON equilibration and wash buffer containing 25 mM imidazole. Finally, the bound proteins were eluted with 10 ml of TALON equilibration and wash buffer containing 150 mM imidazole. The elutant was desalted and simultaneously concentrated into 0.1 M sodium phosphate buffer, pH 6.5, using a 10 kiloDalton cutoff Centricon filter (Millipore, Billerica, MA).
2.6 Biochemical characterization of CpGSTD1 and CpGSTD2
GST activity was measured as described previously38 using the general GST substrates CDNB or DCNB with reduced GSH serving as co-substrate in clear 96-well plates. Stock solutions of CDNB and DCNB were prepared in ethanol, while reduced GSH was dissolved in 0.1 M sodium phosphate buffer. Each 300 μl reaction contained 100 ng of CpGSTD1 or 140 ng of CpGSTD2, 1 mM CDNB or DCNB, 5 mM reduced GSH, and 1.67% (v:v) ethanol in 0.1 M sodium phosphate buffer. Protein concentrations were determined with Bradford’s reagent (Bio-Rad, Hercules, CA) using bovine serum albumin (BSA) as a standard. Following a 5 min preincubation, the enzyme reaction was initiated by the addition of CDNB or DCNB and reduced GSH to each well. The conjugation of GSH with CDNB or DCNB was measured at 15 sec intervals at a wavelength of 340 nm (A340) for 5 min using a Spectra Max M2 spectrophotometer (Molecular Devices, Sunnyvale, CA). Wells containing substrate (CDNB or DCNB and reduced GSH) in buffer but no enzyme served as reference blanks. The effect of pH on GST activity was determined at 30°C in 0.1 M sodium phosphate buffer, pH 5.5 to pH 8.0 at 0.5 pH unit intervals. The effect of temperature on GST activity was determined in 0.1 M sodium phosphate buffer, pH 6.5 for CpGSTD1 and pH 7.5 for CpGSTD2, and incubation temperatures from 24°C to 44°C at 2°C intervals. All of the assays using CDNB or DCNB as substrates were performed in quadruplicate and repeated three times.
The kinetic constants of CpGSTd1 and CpGSTd2 were determined using varying concentrations (7.8 to 750 μM) of CDNB or DCNB, and a fixed concentration (5 mM) of reduced GSH at 30°C in 0.1 M sodium phosphate buffer (pH 6.5 for CpGSTD1 and pH 7.5 for CpGSTD2). In addition, varying concentrations of reduced GSH (39 μM to 5 mM) were used while keeping a constant concentration (750 μM) of CDNB. The assays were performed with 100 ng of CpGSTD1 or 140 ng of CpGSTD2. The kinetic constants were determined by linear regression using SigmaPlot software (Systat Software Inc., San Jose, CA).
The ability of chalcone, 4-hydroxychalcone, EA, bivalent inhibitor, and S-hexylglutathione to inhibit CpGSTD1 and CpGSTD2 was determined using CDNB (125 μM) and reduced GSH (5 mM) as substrates at 30°C in 0.1 M sodium phosphate buffer (pH 6.5 for CpGSTD1 and pH 7.5 for CpGSTD2). The inhibitors were preincubated with CpGSTD1 (100 ng) or CpGSTD2 (140 ng) for 5 min at 30°C prior to the addition of CDNB. The conjugation of GSH with CDNB was measured at 15 sec intervals at A340 for 5 min as described above. All of the inhibitors except S-hexylglutathione were dissolved in dimethyl sulfoxide (DMSO) to give a final concentration of 1% (v:v) DMSO. S-Hexylglutathione was dissolved in 0.1 M sodium phosphate buffer, pH 9.0. The half maximal inhibitory concentration (IC50) was determined using a range of concentrations (100 nM to 100 μM for chalcone and 4-hydroxychalcone, 3 nM to 30 μM for EA, 1 nM to 16 μM for bivalent inhibitor, and 16 μM to 4 mM for S-hexylglutathione) that included at least four concentrations above and four concentrations below the apparent IC50.
2.7 Peroxidase activity assay
Peroxidase activity of CpGSTD1 and CpGSTD2 was measured generally as described previously 12 in 96-well plates in a 200 μl reaction containing CpGSTD1 (1.4 μg) or CpGSTD2 (2.0 μg), varying concentrations (0.125 to 1.0 mM) of CMHP, 0.5 units of glutathione reductase (GSR, Sigma-Aldrich), 7.5 mM reduced GSH, 0.5 mM NADPH (Sigma-Aldrich), and 1% (v:v) ethanol in 0.1 M sodium phosphate buffer (pH 6.5 for CpGSTD1 and pH 7.5 for CpGSTD2). Stock solutions of CMHP were prepared in ethanol. For each assay, CpGSTD1 or CpGSTD2, GSR, reduced GSH, and NADPH were preincubated for 5 min at 25°C prior to the addition of CMHP. The oxidation of NADPH (resulting in a decrease in absorbance) was measured at 15 sec intervals at A340 for 5 min as described above. Wells lacking enzyme (CpGSTD1 or CpGSTD2) but containing all of the substrates served as blanks. The assays were performed in quadruplicate and repeated three times.
2.8 DDT dehydrochlorination assay
Dehydrochlorinase activity of CpGSTD1 and CpGSTD2 was determined generally as described previously7 in a 500 μl reaction containing 1 μM DDT, CpGSTD1 (10, 20 or 40 μg) or CpGSTD2 (10, 20 or 40 μg), 15 mM reduced GSH, and 1% (v:v) ethanol in 0.1 M sodium phosphate buffer (pH 6.5 for CpGSTD1 and pH 7.5 for CpGSTD2). Reactions containing heat inactivated (30 min at 70°C) CpGSTD1 or CpGSTD2 or BSA served as negative controls. Each reaction was run in triplicate in individual glass tubes (10×75 mm) and allowed to proceed for 2 h at 30°C with shaking. At the end of the 2 h incubation, the reaction mixture was extracted twice with 500 μl of methyl-tertiary-butyl ether (MTBE). The extracts were combined, dried under a gentle stream of nitrogen, resuspended in 50 μl of ethanol, and analyzed by HPLC using a reverse phase C18 column (Phenomenex, Torrance, CA). A gradient of acetonitrile (5 to 95%) and water containing 0.1% (v:v) TFA served as the mobile phase with a flow rate of 0.3 ml per min for 45 min. The retention times of DDT (18 ng in 10 μl) and DDE (16 ng in 10 μl) that were separated under identical conditions were used as standards to identify the substrates and potential products.
2.9 Permethrin metabolism assay
The ability of CpGSTD1 and CpGSTD2 to metabolize permethrin was determined as generally described by39,40 in a 500 μl reaction containing 1 μM permethrin, CpGSTD1 (40 or 60 μg) or CpGSTD2 (40 or 60 μg), 15 mM reduced GSH, and 1% (v:v) ethanol in 0.1 M sodium phosphate buffer (pH 6.5 for CpGSTD1 and pH 7.5 for CpGSTD2). Reactions containing heat inactivated (30 min at 70°C) CpGSTD1 or CpGSTD2 or BSA served as negative controls. Each reaction was run in triplicate in individual glass tubes (10×75 mm) and allowed to proceed for 2 h at 30°C with shaking. After the 2 h incubation, lambda cyhalothrin was added to the reaction mixture to a final concentration of 1 μM to serve as an internal standard. The mixture was then extracted twice with 500 μl of MTBE as described above. Following HPLC separation as described above, the area under the curve of the cis-permethrin, trans-permethrin, and lambda cyhalothrin peaks were determined. The decrease in area of cis- and trans-permethrin peaks relative to lambda cyhalothrin peak was taken to indicate the metabolism or sequestration of permethrin by the GSTs.
2.10 Semi-quantitative PCR
Semi-quantitative PCR was used to determine the expression levels of CpGSTd1 and CpGSTd2 in 4th instar larvae and 1 day old adults of Marin and CQ1 mosquitoes. Total RNA was isolated from a pool of 50 larvae or 50 adults using Trizol reagent (Invitrogen), according to the manufacturer’s instructions. Next mRNAs were isolated from total RNAs with oligo-dT cellulose using a Fast Track 2.0 kit (Invitrogen) according to the manufacturer’s protocol. The mRNAs (1.0 μg) were used as templates for the generation of first strand cDNAs as described above. Primers pairs (Table 1) were designed to amplify 500 bp-long regions of CpGSTd1, CpGSTd2 or the ribosomal protein S7 (RPS7) gene. The expression level of RPS7 from Cx. quinquefasciatus (GenBank #AF272670.1) was used as internal control for normalization. The PCRs were repeated three times in a 100 μl reaction containing 5 units of GoTaq polymerase (Promega), 0.8 μg of first strand cDNA template, and 3 μM of each primer for 17 to 33 cycles of: 95°C, 30 sec; 65°C, 30 sec; and 72°C, 30 sec. The PCR products were separated by 0.7% agarose gel electrophoresis and visualized by staining with ethidium bromide. The intensities of the DNA bands were quantified using ImageJ software (http://rsb.info.nih.gov/ij).
3 RESULTS AND DISCUSSION
3.1 Molecular cloning and phylogenetic analysis of CpGSTd1 and CpGSTd2
A major aim of this study was to determine whether permethrin resistance in Cx. pipiens could be attributed to qualitative or quantitative changes in one or more GST isozymes. At the time we undertook this study, the genome sequence of Cx. quinquefasciatus was unavailable and GST-encoding gene sequences from Culex were still uncharacterized. Thus, we took a shotgun approach based on 3′- and 5′-RACE to identify gst genes using two degenerate primers, GSTi and GSTv, that were designed to bind the GSH binding motif of GSTs.41 Unfortunately, this strategy only generated two GST-encoding sequences, CpGSTd1 (GenBank #JN251103) and CpGSTd2 (GenBank #JH251104), from Marin mosquitoes. CpGSTd1 and CpGSTd2 encoded proteins of 212 and 215 amino acid residues that were predicted to belong to the Delta GST class. The 5′- and 3′-UTRs of CpGSTd1 were 96 and 118 nts long, respectively. A putative polyadenylation sequence (AATAAA) was found 66 nts downstream of the stop codon TGA. Similarly, the 5′- and 3′-UTRs of CpGSTd2 were 77 and 57 nts long, respectively; and a putative polyadenylation sequence was found 12 nts downstream of the stop codon TAA.
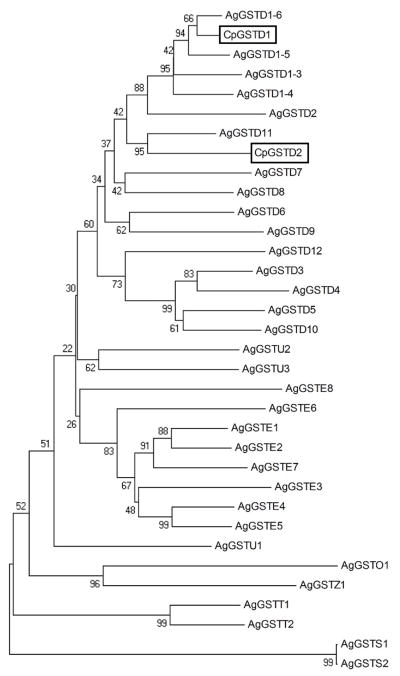
The nucleotide and deduced amino acid sequences of CpGSTd1 and CpGSTd2 showed 59% and 48% identity, respectively. Phylogenetic analyses clustered CpGSTD1 and CpGSTD2 in the same clade within Delta class GSTs from An. gambiae (Fig. 1). CpGSTD1 showed 34% (with AgGSTD10) to 84% (with AgGSTD1-6) identity with Delta class GSTs from An. gambiae. CpGSTD2 also showed the lowest (34%) identity with AgGSTD10 and 57% identity with AgGSTD11. Homologs of AgGSTD1-636 have been identified in Anopheles dirus14 and Cx. quinquefasciatus.42 Genome analyses of An. gambiae32 and Cx. quinquefasciatus42 indicate that there are four possible splice variants of GSTd1. CpGSTd1 and GSTd1-1 of Cx. quinquefasciatus show 97% and 98% identity at the nucleotide and deduced amino acid sequences levels. This exceptionally high identity is consistent with an earlier observation based on morphology and enzyme electrophoretic profiles that concluded that Cx. pipiens and Cx. quinquefasciatus interbreed and act as single species in California.43
Figure 1.
Phylogenetic relationship of CpGSTD1 and CpGSTD2 with GSTs from Anopheles gambiae.32 The deduced amino acid sequences of CpGSTD1 and CpGSTD2 were aligned with amino acid sequences of GSTs from An. gambiae using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2). The alignment was then used to construct a phylogenetic tree by the Neighbor-Joining method using the MEGA4 software package.35 Bootstrap values from a set of 500 replicates are listed at the nodes.
3.2 Enzyme kinetic characterization of CpGSTD1 and CpGSTD2
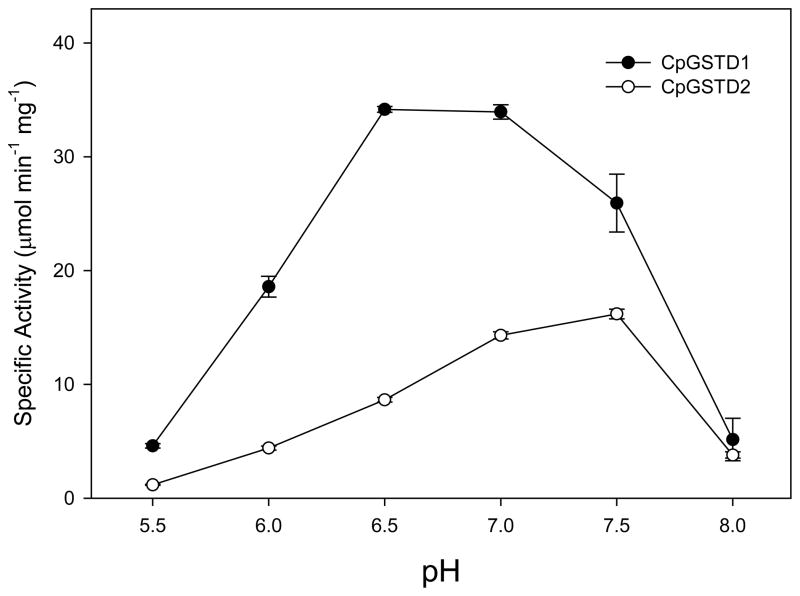
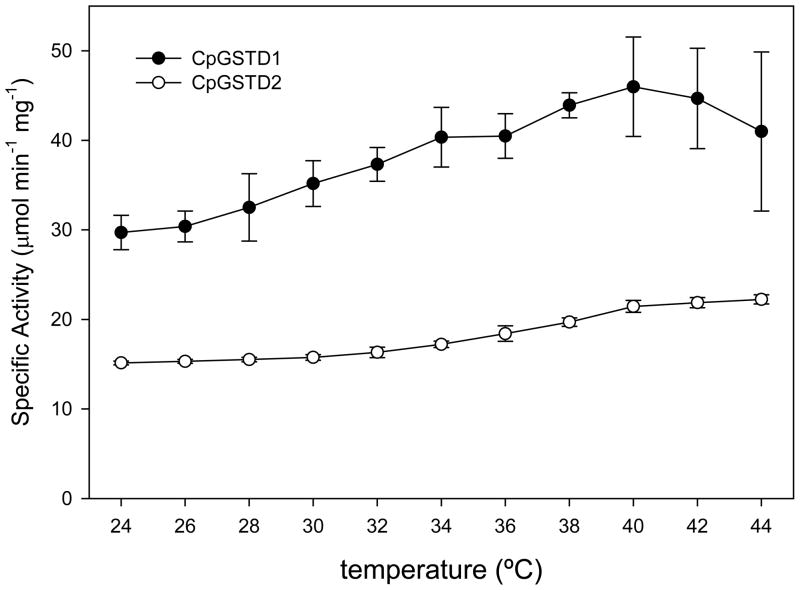
CpGSTD1 and CpGSTD2 were affinity purified using a histidine tag binding column and initially characterized with the colorimetric substrates CDNB and DCNB. CpGSTD1 turned over CDNB about 230-fold faster than DCNB, whereas CpGSTD2 turned over CDNB about 18-fold faster than DCNB (Table 2). CpGSTD1 turned over CDNB about 3-fold faster than CpGSTD2. The catalytic efficiency or specificity (kcat/KM) of both CpGSTD1 and CpGSTD2 was at least 100-fold higher with CDNB than with DCNB. CpGSTD1 showed the highest activity for CDNB at pH 6.5 and pH 7.0 with significant reductions in activity at pH 5.5 and pH 8.0 (Fig. 2). CpGSTD2 showed the highest activity for DCNB at pH 7.5 (Fig. 2). Both CpGSTD1 and CpGSTD2 showed a wide range of thermal stability (Fig. 3). CpGSTD1 showed approximately 30% higher activity at 40°C than at 24°C. The kinetic parameters of CpGSTD1 and CpGSTD2 were similar to Delta class GSTs from An. dirus,14 An. gambiae,36 and D. melanogaster.37 For example, the catalytic efficiency ratios (i.e., kcat/KM) of CpGSTD1, AgGSTD1-636 and AdGSTD1-114 for CDNB are 1.7 × 105, 7.9 × 105, and 4.8 × 104 M−1 s−1, respectively.
Table 2.
Enzyme kinetic properties of CpGSTD1 and CpGSTD2
| CpGSTD1 | CpGSTD2 | |||
|---|---|---|---|---|
|
| ||||
| CDNB | DCNB | CDNB | DCNB | |
| Vmax (μmol min−1 mg−1) | 57 ± 1 | 0.19 ± 0.01 | 18 ± 0.2 | 0.96 ± 0.02 |
| KM (μM) | 140 ± 10 | 170 ± 10 | 30 ± 2 | 220 ± 10 |
| KM of GSH (μM) | 820 ± 60 | n.d.a | 270 ± 20 | n.d. |
| kcat (s−1) | 23.3 | 0.1 | 7.0 | 0.4 |
| kcat/KM (M−1 s−1) | 1.7 × 105 | 4.7 × 102 | 2.3 × 105 | 1.7 × 103 |
n.d.: not determined.
Figure 2.
The effect of pH on the activity of CpGSTD1 and CpGSTD2. The reaction was performed in 100 mM sodium phosphate buffer (pH 5.5 to pH 8.0 at 0.5 pH unit intervals), containing 100 ng of CpGSTD1 (●) or 140 ng of CpGSTD2 (○), 5 mM GSH, and 1 mM CDNB at 30°C. The error bars represent the standard deviation of the mean of three separate experiments.
Figure 3.
The effect of temperature on the activity of CpGSTD1 and CpGSTD2. The reaction was performed in 100 mM sodium phosphate buffer, pH 6.5 for CpGSTD1 or pH 7.5 for CpGSTD2, containing 100 ng of CpGSTD1 (●) or 140 ng of CpGSTD2 (○), 5 mM GSH, and 5 mM CDNB at varying temperatures (24°C to 44°C at 2°C intervals). The error bars represent the standard deviation of the mean of three separate experiments.
In some insects, GST isozymes that function as Se-independent peroxidases are believed to aid in cellular anti-oxidant defense by reducing organic hydroperoxides within membranes and lipoproteins.44 Similarly, in pyrethroid resistant brown planthoppers elevated GST levels reduce mortality by limiting pyrethroid induced lipid peroxidation.12 The role of GSTs as antioxidants that aid in insecticide resistance, however, has not been established in mosquitoes. CpGSTD1 and CpGSTD2 showed dramatic differences in their ability to metabolize CMHP. CpGSTD1 reduced CMHP at a rate of 1.6 μmol min−1 mg−1, whereas, CpGSTD2 showed no detectable activity towards CMHP under conditions where a rate of at least 0.1 μmol min−1 mg−1 could be detected with CpGSTD1. Recombinant Delta class GSTs from An. gambiae (e.g., AgGSTD1-5 and AgGSTD1-6)36 and D. melanogaster (e.g., DmGSTD1 and DmGSTD21)45 also show peroxidase activity (0.13 to 0.98 μmol min−1 mg−1 range) against CMHP. These findings suggest that some Delta class GSTs may play a role in cellular anti-oxidative defense against oxidative damage induced by DDT or pyrethroids.
Homologs of CpGSTD1 that are found in An. gambiae36 and An. dirus14 are able to dehydrochlorinate DDT to DDE at a rate of 4 to 8 nmol per mg protein over an incubation period of 2 h. Similarly, HPLC analysis showed that CpGSTD1 metabolized DDT to DDE at a rate of 6.4 ± 0.6 nmol per mg protein over a period of 2 h. In contrast to these Delta class GSTs, recombinant Epsilon class GSTs from An. gambiae46 and Ae. aegypti8 dehydrochlorinate DDT at roughly 400- to 600-fold greater rates suggesting that in mosquitoes Delta class GSTs do not play a significant role in the dehydrochlorination of DDT to DDE. Delta class GSTs, however, may play roles in binding to DDT prior to metabolism by Epsilon class GSTs or other detoxification enzymes.13,14 In contrast to CpGSTD1, CpGSTD2 showed no metabolism of DDT under conditions where a rate of at least 0.1 to 1 nmol of DDE formed per mg of CpGSTD1 over a period of 2 h could be detected.
Although elevated GST activity and gene expression levels are found in pyrethroid resistant insects, there is no direct evidence to show that GSTs directly metabolize pyrethroids. Our HPLC analyses indicated that neither CpGSTD1 nor CpGSTD2 are able to metabolize permethrin. Specifically, we found no difference in the area under the curve of peaks corresponding to cis- and trans-permethrin (relative to the lambda cyhalothrin internal standard) following a 2 h-long incubation with CpGSTD1 or CpGSTD2 in comparison to incubation under identical conditions with heat inactivated CpGSTD1 or CpGSTD2, respectively, or BSA (Table 3). Interestingly, however, in a related study by our laboratory (Huang et al., manuscript in preparation) we found that CpGSTD1 is able to metabolize fluorescent permethrin-like substrates (both cis- and trans-isomers) at rates up to 150 nmol min−1 mg−1. A chemical mechanism involving a GST-catalyzed thiolysis reaction is proposed in this reaction.
Table 3.
Potential metabolism of cis- and trans-permethrin by CpGSTD1 or CpGSTD2a
| Protein | Ratio of the area under the curve relative to lambda cyhalothrin
|
|
|---|---|---|
| cis-permethrin | trans-permethrin | |
| CpGSTD1 | 0.47 ± 0.02 | 0.24 ± 0.01 |
| CpGSTD1, heat inactivated | 0.49 ± 0.03 | 0.27 ± 0.02 |
| CpGSTD2 | 0.51 ± 0.03 | 0.29 ± 0.02 |
| CpGSTD2, heat inactivated | 0.53 ± 0.04 | 0.26 ± 0.02 |
| BSA | 0.50 ± 0.01 | 0.25 ± 0.01 |
HPLC analysis was used to quantify cis- and trans-permethrin following a 2 h incubation at 30°C. These data indicated that there was no metabolism of cis- or trans-permethrin by CpGSTD1 or CpGSTD2 under conditions where a rate of 7 pmol min−1 mg−1 could have been reliably detected.
3.3 Inhibition of CpGSTD1 and CpGSTD2
The ability of known GST inhibitors and novel compounds to inhibit CpGSTD1 and CpGSTD2 was analyzed using CDNB and reduced GSH as substrates. Chalcone and 4-hydroxychalcone showed IC50 values in the low μM range against both CpGSTD1 and CpGSTD2 (Table 4). Similarly, chalcone shows relatively poor inhibition of GST-2 and GST-1b of Aedes aegypti (IC50 of 165 μM and 166 μM, respectively).38 In contrast to chalcone and 4-hydroxychalcone, EA and its derivative bivalent inhibitor showed more than 100-fold greater potency against CpGSTD1 (IC50 of 110 nM and 20 nM, respectively, Table 4). On the other hand, CpGSTD2 was relatively poorly inhibited by EA and bivalent inhibitor (IC50 of 2.5 μM and 2.4 μM, respectively, Table 4). Chalcone, EA, and their derivatives have a ketone moiety that forms a conjugate with GSH through a GST catalyzed Michael addition reaction.47 This reaction is thermodynamically more favorable in comparison to the conjugation of CDNB to GSH via an addition-substitution reaction.47 Thus, chalcone, EA, and their derivatives can function to deplete GSH. In addition, Miyamoto et al.48,49 found that some chalcone derivatives not only deplete GSH, but that the resulting conjugate can also inhibit mammalian GSTs by mechanisms that involve substrate competition and competitive inhibition. S-Hexylglutathione is another class of GST inhibitor that occupies the G-site of GSTs thus preventing the binding of GSH.50 S-Hexylglutathione was a good inhibitor of CpGSTD2 (IC50 of 240 nM) but a dramatically less potent inhibitor of CpGSTD1 (Table 4). Although the amino acid sequence comparisons indicated that both CpGSTD1 and CpGSTD2 are Delta class GSTs, these GSTs showed unique inhibition profiles suggesting that their substrate preferences may also be unique.
Table 4.
Inhibition (IC50) of CpGSTD1 and CpGSTD2a
| Compound | Structure | IC50 (μM)b
|
|
|---|---|---|---|
| CpGSTD1 | CpGSTD2 | ||
| chalcone |
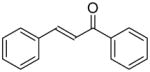
|
12.1 ± 0.4 | 4 ± 0.2 |
| 4-hydroxy chalcone |
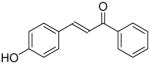
|
16 ± 1 | 5 ± 0.2 |
| ethacrynic acid (EA) |
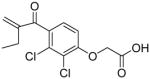
|
0.11 ± 0.01 | 2.5 ± 0.1 |
| bivalent inhibitor |
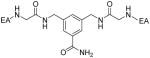
|
0.02 ± 0.001 | 2.4 ± 0.1 |
| S-hexyl glutathione |
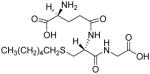
|
68 ± 3 | 0.24 ± 0.01 |
GST activity was determined using CDNB and reduced GSH as described in the Materials and Methods.
The values reported are the mean ± S.D. of three experiments.
3.4 Gene expression of CpGSTd1 and CpGSTd2 in Marin and CQ1 mosquitoes
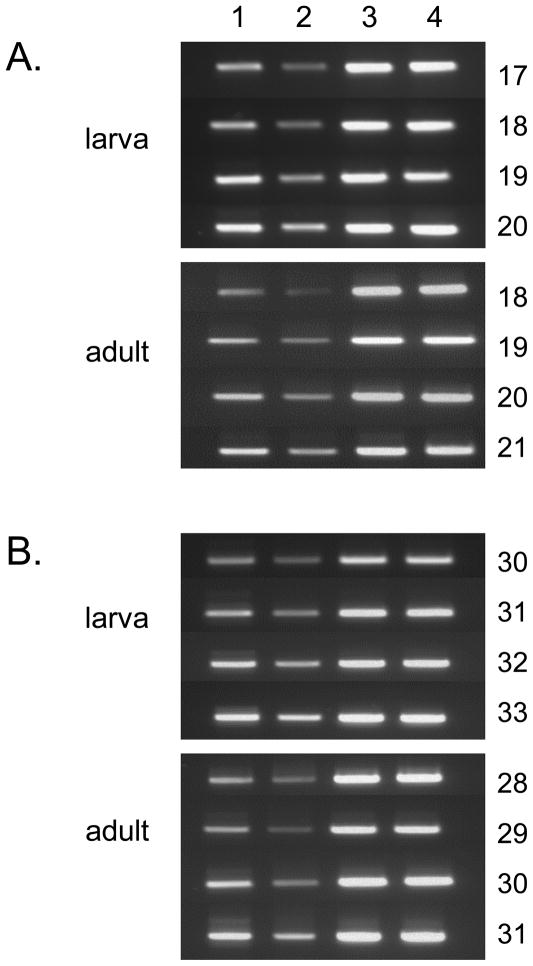
Previous gene expression analyses by quantitative PCR and microarray techniques have shown elevated gst gene expression levels in DDT and permethrin resistant An. gambiae10,32 and Ae. aegypti.8 We were therefore interested in determining the relative expression levels of CpGSTd1 and CpGSTd2 in insecticide resistant Marin and insecticide susceptible CQ1 mosquitoes. Semi-quantitative analysis from three separate experiments indicated that the expression of CpGSTd1 was 2.1 ± 0.2 fold higher in both larvae and adults of Marin mosquitoes in comparison to that in CQ1 mosquitoes, whereas the expression of CpGSTd2 was 1.8 ± 0.1 and 2.5 ± 0.4 fold higher in larvae and adults, respectively, of Marin mosquitoes (Fig. 4). Similarly, increased expression rates (3.88- and 2.36-fold higher) of an Epsilon class GST (AgGSTe2) are found in An. gambiae that are permethrin and DDT resistant, respectively.10 In contrast to these findings, microarray analysis showed no difference in GSTd1 expression between DDT resistant JPal-per and DDT susceptible Ogasawara strains of Cx. quinquefasciatus.42 In addition, the expression of CpGSTd1 in both larvae and adults of Marin mosquitoes appeared to be significantly higher than that of CpGSTd2 as suggested by the lower number of PCR cycles (roughly 20 cycles v. 30 cycles) that were required to clearly amplify the CpGSTd1 target.
Figure 4.
Semiquantitative PCR analysis of the expression of CpGSTd1 (A) and CpGSTd2 (B) in larvae and adults of the Marin (pyrethroid resistant) and CQ1 (pyrethroid susceptible) mosquitoes. The template for the semiquantitative PCR was first strand cDNA (0.8 μg) that was generated from total RNA isolated from 4th instar larvae or adults of Marin (columns 1 and 3) or CQ1 (columns 2 and 4) mosquitoes. The semiquantitative PCR was performed using primers that amplified 500 bp-long regions of CpGSTd1 (columns 1 and 2 in A), CpGSTd2 (columns 1 and 2 in B), and the internal standard RPS7 (columns 3 and 4 in both A and B). The number of PCR cycles that were used for amplification is shown to the right of each panel. The PCR reaction was repeated three times for each primer pair; the figure shows an example of typical results.
4. CONCLUSIONS
A major goal of this study was to determine if overexpressed GSTs from insecticide resistant Cx. pipiens play qualitative or quantitative roles in pyrethroid resistance. To achieve this goal, we cloned and expressed CpGSTD1 and CpGSTD2 from pyrethroid resistant Marin larvae. CpGSTD1 and CpGSTD2 appeared to belong to the Delta GST class on the basis of their deduced amino acid sequences. The kinetic properties of CpGSTD1 with CDNB and DCNB were similar to other known Delta class mosquito GSTs. CpGSTD1 was able to dehydrochlorinate DDT and metabolize CMHP at rates that are similar to those of other Delta class GSTs from mosquitoes and fruit fly. CpGSTD1 and CpGSTD2 showed unique inhibition profiles. HPLC analysis indicated that CpGSTD1 and CpGSTD2 were unable to metabolize or sequester permethrin. Taken together, these findings suggested that Delta class GSTs do not play a major direct role in permethrin resistance in Culex.
Acknowledgments
We thank Julie Christianson and Rory McAbee for assistance with rearing of the mosquitoes. We thank W. M. Atkins for providing bivalent inhibitor. This work was partially funded by grants from the National Institute of Allergy and Infectious Diseases (#R01 AI068855) to A. J. Cornel and National Institute of Environmental Health Sciences (#R01 ES002710) to B. D. Hammock. B. D. Hammock is a senior fellow of the American Asthma Society.
References
- 1.Salinas AE, Wong MG. Glutathione S-transferases - a review. Curr Med Chem. 1999;6:279–309. [PubMed] [Google Scholar]
- 2.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Ann Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 3.Cnubben NHP, Rietjens IMCM, Wortelboer H, van Zanden J, van Bladeren PJ. The interplay of glutathione-related processes in antioxidant defense. Environ Toxicol Pharmacol. 2001;10:141–152. doi: 10.1016/s1382-6689(01)00077-1. [DOI] [PubMed] [Google Scholar]
- 4.Ranson H, Hemingway J. Mosquito glutathione transferase. In: Sies H, Packer L, editors. Glutathione transferases and gamma-glutamyl transpeptidases. Academic Press; New York: 2005. pp. 226–239. [Google Scholar]
- 5.Chiang FM, Sun CN. Glutathione transferase isozymes of diamondback moth larvae and their role in the degradation of some organophosphorus insecticides. Pestic Biochem Physiol. 1993;45:7–14. [Google Scholar]
- 6.Enayati AA, Ranson H, Hemingway J. Insect glutathione transferases and insecticide resistance. Insect Mol Biol. 2005;14:3–8. doi: 10.1111/j.1365-2583.2004.00529.x. [DOI] [PubMed] [Google Scholar]
- 7.Prapanthadara LA, Hemingway J, Ketterman AJ. Partial purification and characterization of glutathione S-transferases involved in DDT resistance from the mosquito Anopheles gambiae. Pestic Biochem Physiol. 1993;47:119–133. [Google Scholar]
- 8.Lumjuan N, McCarroll L, Prapanthadara LA, Hemingway J, Ranson H. Elevated activity of an Epsilon class glutathione transferase confers DDT resistance in the dengue vector, Aedes aegypti. Insect Biochem Mol Biol. 2005;35:861–871. doi: 10.1016/j.ibmb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Lumjuan N, Rajatileka S, Changsom D, Wicheer J, Leelapat P, Prapanthadara L, Somboon P, Lycett G, Ranson H. The role of the Aedes aegypti Epsilon glutathione transferases in conferring resistance to DDT and pyrethroid insecticides. Insect Biochem Mol Biol. 2011;41:203–209. doi: 10.1016/j.ibmb.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 10.David JP, Strode C, Vontas J, Nikou D, Vaughan A, Pignatelli PM, Louis C, Hemingway J, Ranson H. The Anopheles gambiae detoxification chip: a highly specific microarray to study metabolic-based insecticide resistance in malaria vectors. PNAS. 2005;102:4080–4084. doi: 10.1073/pnas.0409348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin G, Jia M, Liu T, Xuan T, Zhu KY, Guo Y, Ma E, Zhang J. Identification and characterisation of ten glutathione S-transferase genes from oriental migratory locust, Locusta migratoria manilensis (Meyen) Pest Manag Sci. 2011;67:697–704. doi: 10.1002/ps.2110. [DOI] [PubMed] [Google Scholar]
- 12.Vontas JG, Small GJ, Hemingway J. Glutathione S-transferases as antioxidant defence agents confer pyrethroid resistance in Nilaparvata lugens. Biochem J. 2001;357:65–72. doi: 10.1042/0264-6021:3570065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kostaropoulos I, Papadopoulos AI, Metaxakis A, Boukouvala E, Papadopoulou-Mourkidou E. Glutathione S-transferase in the defence against pyrethroids in insects. Insect Biochem Mol Biol. 2001;31:313–319. doi: 10.1016/s0965-1748(00)00123-5. [DOI] [PubMed] [Google Scholar]
- 14.Jirajaroenrat K, Pongjaroenkit S, Krittanai C, Prapanthadara LA, Ketterman AJ. Heterologous expression and characterization of alternatively spliced glutathione S-transferases from a single Anopheles gene. Insect Biochem Mol Biol. 2001;31:867–875. doi: 10.1016/s0965-1748(01)00032-7. [DOI] [PubMed] [Google Scholar]
- 15.Sardelis MR, Turell MJ, Dohm DJ, O’Guinn ML. Vector competence of selected North American Culex and Coquillettidia mosquitoes for West Nile virus. Emerg Infect Dis. 2001;7:1018–1022. doi: 10.3201/eid0706.010617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai TF, Smith GC, Happ CM, Kirk LJ, Jakob WL, Bolin RA, Francy DB, Lampert KJ. Surveillance of St. Louis encephalitis virus vectors in Grand Junction, Colorado, in 1987. J Am Mosq Control Assoc. 1989;5:161–165. [PubMed] [Google Scholar]
- 17.Meegan JM. The Rift Valley fever epizootic in Egypt 1977–1978. 1. Description of the epizootic and virological studies. Trans Roy Soc Trop Med Hyg. 1979;73:618–623. doi: 10.1016/0035-9203(79)90004-x. [DOI] [PubMed] [Google Scholar]
- 18.Farid HA, Hammad RE, Hassan MM, Morsy ZS, Kamal IH, Weil GJ, Ramzy RMR. Detection of Wuchereria bancrofti in mosquitoes by the polymerase chain reaction: a potentially useful tool for large-scale control programmes. Trans Roy Soc Trop Med Hyg. 2001;95:29–32. doi: 10.1016/s0035-9203(01)90322-0. [DOI] [PubMed] [Google Scholar]
- 19.Lai CH, Tung KC, Ooi HK, Wang JS. Competence of Aedes albopictus and Culex quinquefasciatus as a vector of Dirofilaria immitis after blood meal with different microfilarial density. Vet Parasitol. 2000;90:231–237. doi: 10.1016/s0304-4017(00)00242-9. [DOI] [PubMed] [Google Scholar]
- 20.Atkinson CT, Dusek RJ, Woods KL, Iko WM. Pathogenicity of avian malaria in experimentally-infected Hawaii Amakihi. J Wildlife Dis. 2000;36:197–204. doi: 10.7589/0090-3558-36.2.197. [DOI] [PubMed] [Google Scholar]
- 21.Curtis CF. Pesticides and their application for the control of vectors and pests of public health importance. World Health Organization; 2006. Report No. WHO/CDS/NTD/WHOPES/GCDPP/2006.1. [Google Scholar]
- 22.Amin AM, Hemingway J. Preliminary investigation of the mechanisms of DDT and pyrethroid resistance in Culex quinquefasciatus Say (Diptera, Culicidae) from Saudi Arabia. Bull Entomol Res. 1989;79:361–366. [Google Scholar]
- 23.Curtis CF, Pasteur N. Organophosphate resistance in vector populations of the complex of Culex pipiens L (Diptera, Culicidae) Bull Entomol Res. 1981;71:153–161. [Google Scholar]
- 24.Wirth MC, Marquine M, Georghiou GP, Pasteur N. Esterase A2 and esterase B2 in Culex quinquefasciatus (Diptera, Culicidae) - role in organophosphate resistance and linkage. J Med Entomol. 1990;27:202–206. doi: 10.1093/jmedent/27.2.202. [DOI] [PubMed] [Google Scholar]
- 25.Hemingway J, Ranson H. Insecticide resistance in insect vectors of human disease. Annu Rev Entomol. 2000;45:371–391. doi: 10.1146/annurev.ento.45.1.371. [DOI] [PubMed] [Google Scholar]
- 26.McAbee RD, Kang KD, Stanich MA, Christiansen JA, Wheelock CE, Inman AD, Hammock BD, Cornel AJ. Pyrethroid tolerance in Culex pipiens pipiens var molestus from Marin County, California. Pest Manag Sci. 2004;60:359–368. doi: 10.1002/ps.799. [DOI] [PubMed] [Google Scholar]
- 27.Liu HQ, Cupp EW, Micher KM, Guo AG, Liu NN. Insecticide resistance and cross-resistance in Alabama and Florida strains of Culex quinquefaciatus. J Med Entomol. 2004;41:408–413. doi: 10.1603/0022-2585-41.3.408. [DOI] [PubMed] [Google Scholar]
- 28.Hemingway J, Hawkes NJ, McCarroll L, Ranson H. The molecular basis of insecticide resistance in mosquitoes. Insect Biochem Mol Biol. 2004;34:653–665. doi: 10.1016/j.ibmb.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 29.Liu N, Xu Q, Zhu F, Zhang L. Pyrethroid resistance in mosquitoes. Insect Sci. 2006;13:159–166. [Google Scholar]
- 30.Maeda DY, Mahajan SS, Atkins WM, Zebala JA. Bivalent inhibitors of glutathione S-transferase: the effect of spacer length on isozyme selectivity. Bioorg Med Chem Lett. 2006;16:3780–3783. doi: 10.1016/j.bmcl.2006.04.041. [DOI] [PubMed] [Google Scholar]
- 31.Thompson JD, Higgins DG, Gibson TJ. Clustal-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding YC, Ortelli F, Rossiter LC, Hemingway J, Ranson H. The Anopheles gambiae glutathione transferase supergene family: annotation, phylogeny and expression profiles. BMC Genomics. 2003;4:35. doi: 10.1186/1471-2164-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tu CPD, Akgul B. Drosophila glutathione S-transferases. In: Sies H, Packer L, editors. Glutathione transferases and gamma-glutamyl transpeptidases. Academic Press; New York: 2005. pp. 204–226. [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 36.Ranson H, Prapanthadara LA, Hemingway J. Cloning and characterization of two glutathione S-transferases from a DDT-resistant strain of Anopheles gambiae. Biochem J. 1997;324:97–102. doi: 10.1042/bj3240097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawicki R, Singh SP, Mondal AK, Benes H, Zimniak P. Cloning, expression and biochemical characterization of one Epsilon class (GST-3) and ten Delta class (GST-1) glutathione S-transferases from Drosophila melanogaster, and identification of additional nine members of the Epsilon class. Biochem J. 2003;370:661–669. doi: 10.1042/BJ20021287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grant DF, Dietze EC, Hammock BD. Glutathione S-transferase isozymes in Aedes aegypti - purification, characterization, and isozyme-specific regulation. Insect Biochem. 1991;21:421–433. [Google Scholar]
- 39.Abernathy CO, Casida JE. Pyrethroid insecticides: esterase cleavage in relation to selective toxicity. Science. 1973;179:1235–1236. doi: 10.1126/science.179.4079.1235. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura Y, Sugihara K, Sone T, Isobe M, Ohta S, Kitamura S. The in vitro metabolism of a pyrethroid insecticide, permethrin, and its hydrolysis products in rats. Toxicol. 2007;235:176–184. doi: 10.1016/j.tox.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 41.Chen LQ, Hall PR, Zhou XYE, Ranson H, Hemingway J, Meehan EJ. Structure of an insect Delta class glutathione S-transferase from a DDT-resistant strain of the malaria vector Anopheles gambiae. Acta Crystallogr, Sect D: Biol Crystallogr. 2003;59:2211–2217. doi: 10.1107/s0907444903018493. [DOI] [PubMed] [Google Scholar]
- 42.Kasai S, Komagata O, Okamura Y, Tomita T. Alternative splicing and developmental regulation of glutathione transferases in Culex quinquefasciatus Say. Pestic Biochem Physiol. 2009;94:21–29. [Google Scholar]
- 43.Cornel AJ, McAbee RD, Rasgon J, Stanich MA, Scott TW, Coetzee M. Differences in extent of genetic introgression between sympatric Culex pipiens and Culex quinquefasciatus (Diptera: Culicidae) in California and South Africa. J Med Entomol. 2003;40:36–51. doi: 10.1603/0022-2585-40.1.36. [DOI] [PubMed] [Google Scholar]
- 44.Parkes TL, Hilliker AJ, Phillips JP. Genetic and biochemical analysis of glutathione S-transferase in the oxygen defense system of Drosophila melanogaster. Genome. 1993;36:1007–1014. doi: 10.1139/g93-134. [DOI] [PubMed] [Google Scholar]
- 45.Tang AH, Tu CPD. Biochemical characterization of Drosophila glutathione S-transferases D1 and D21. J Biol Chem. 1994;269:27876–27884. [PubMed] [Google Scholar]
- 46.Ortelli F, Rossiter LC, Vontas J, Ranson H, Hemingway J. Heterologous expression of four glutathione transferase genes genetically linked to a major insecticide-resistance locus from the malaria vector Anopheles gambiae. Biochem J. 2003;373:957–963. doi: 10.1042/BJ20030169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Awasthi S, Srivastava SK, Ahmad F, Ahmad H, Ansari GAS. Interactions of glutathione S-transferase pi with ethacrynic acid and its glutathione conjugate. Biochim Biophys Acta. 1993;1164:173–178. doi: 10.1016/0167-4838(93)90245-m. [DOI] [PubMed] [Google Scholar]
- 48.Miyamoto T, Silva M, Hammock BD. Inhibition of epoxide hydrolases and glutathione S-transferases by 2-substituted, 3-substituted, and 4-substituted derivatives of 4′-phenylchalcone and its oxide. Arch Biochem Biophys. 1987;254:203–213. doi: 10.1016/0003-9861(87)90096-8. [DOI] [PubMed] [Google Scholar]
- 49.Miyamoto T, Yamamoto I. Glutathione conjugates as the activated form of chalcones for glutathione S-transferase inhibition. J Pestic Sci. 1994;19:53–58. [Google Scholar]
- 50.Lyon RP, Hill JJ, Atkins WM. Novel class of bivalent glutathione S-transferase inhibitors. Biochemistry. 2003;42:10418–10428. doi: 10.1021/bi0346188. [DOI] [PubMed] [Google Scholar]