Abstract
Protein degradation is a fundamental cellular process, the genomic control of which is incompletely understood. The advent of transgene-coded reporter proteins has enabled the development of C. elegans into a model for studying this problem. The regulation of muscle protein degradation is surprisingly complex, integrating multiple signals from hypodermis, intestine, neurons and muscle itself. Within the muscle, degradation is executed by separately regulated autophagy-lysosomal, ubiquitin-proteasome and calpain-mediated systems. The signal-transduction mechanisms, in some instances, involve modules previously identified for their roles in developmental processes, repurposed in terminally differentiated muscle to regulate the activities of pre-formed proteins. Here we review the genes, and mechanisms, which appear to coordinately control protein degradation within C. elegans muscle. We also consider these mechanisms in the context of development, physiology, pathophysiology and disease models.
Introduction
Since the demonstration that it was possible to isolate and characterize mutant lines of C. elegans with abnormal movement,1 it has been shown that these abnormalities can arise both from defects in nerves and in muscle. Characterization of mutants with structural defects in muscle has revealed how the sarcomeres, which power muscle contraction, are established2,3 and these general principles of assembly and function appear to be conserved across several metazoan species.4 Recently it was shown that calpain mediated protein degradation is required for establishment and maintenance of C. elegans sarcomeres, giving rise to the suggestion that this proteolytic system normally functions to allow proper coordinate growth of muscle and hypodermis.5 This work came from a long-term program of study aimed at forming an integrated picture of the control of general cytosolic protein degradation in muscle, rather than from the more mainstream study of sarcomere assembly and maintenance. Here we review how the control of general cytosolic protein degradation has been studied, and the genes that appear to control this biochemical process that can be studied in C. elegans muscle. We conclude by considering the roles of these mechanisms in development, physiology, pathophysiology and some disease models.
Control of the Main Proteolytic Systems Present in Muscle
The control of muscle protein synthesis and degradation is an area of broad biomedical interest due to the public health costs associated with muscle pathologies, particularly loss of muscle mass.6 There is abundant evidence in mammals that catabolism of muscle protein is used to provide amino acids to other tissues in time of need, but there is currently no direct evidence that amino acids from muscle protein catabolism are efficiently reutilized by other tissues in C. elegans.
Four main proteolytic systems that exist in muscle (the lysosomes, proteasomes, calpains and caspases) are key to the control of human muscle size. Which proteolytic system is ″most important″ likely depends on what proteins are being considered and on what (patho)physiologic conditions are being considered. Both proteasomes and lysosomes are thought to be capable of the degradation of general proteins trafficked to the proteases. In the case of the proteasomes, most proteins are made available to the protease active sites on the proteasome interior via a combination of trafficking to the proteasome itself and unfolding of the protein and feeding of the peptide into the catalytic core of the proteasome.7 In the case of the lysosomes, proteins require encapsulation within a lipid vesicle (e.g., autophagosome or endosome), which is then trafficked to lysosomes, where the encapsulated protein can be unfolded and degraded.8 In contrast, the calpains9 and caspases10 are generally thought to be capable of general degradation of proteins in the immediate vicinity of the proteases. Furthermore, calpains are probably better able to attack proteins in supramolecular complexes than are proteasomes or the autophagy-lysosomal system. Unlike the proteasomes and lysosomes, which are constitutively active, calpains and caspases are believed to exist in a default inactive state with triggers such as elevated calcium and/or membrane disruption resulting in activation of the protease and degradation of proteins near to the catalytic site. Despite our knowledge of these proteolytic systems we know relatively little of how the myriad of extra-muscular (and intramuscular) signals that have been suggested to control protein degradation in man11,12 act to control bulk trafficking of proteins to the lysosomes or proteasomes, or activation of calpains or caspases to induce bulk degradation of proteins.
Transgenic Proteins Provide a Visible Phenotype for Studying Protein Degradation
Study of the control of protein degradation has historically lagged behind study of the control of protein synthesis. This lag is largely for technical reasons; for example, the difficulty of linking the sub-cellular process of degradation to more overt phenotypes and the difficulty in uncoupling protein synthesis from degradation. The development of transgenes for studying the control of muscle protein synthesis13 has also enabled the use of transgene-coded proteins as tools for studying the control of muscle protein degradation.14 The C. elegans transgene ccIs55 is a translational fusion of a 5′-terminal portion of unc-54 (myosin heavy chain B) fused to E. coli lacZ and was developed to understand the enhancer elements that control unc-54 expression.13 This transgene produces an enzymatically active β-galactosidase that also contains an N-terminal segment of UNC-54. The transgenic protein is expressed specifically in body-wall and vulval muscles and accumulates throughout development, as assayed by both activity (activity assays) and amount (Western blots).14 The myosin moiety is insufficient to support assembly into filaments, so the fusion protein is a soluble tetramer in muscle cytosol. The protein, surprisingly, remains stable (e.g., is neither synthesized nor degraded) in the cytosol for 72–96 h post-adulthood in well fed worms,14 showing that this foreign protein does not induce an unfolded protein response.15,16 The stability of this reporter protein suggested that one could use it in a fashion similar to the use of traditional tracer techniques, where C. elegans muscle is loaded with marked protein during development and the loss of marked protein indicates that protein degradation is occurring. Indeed, starvation, a condition well appreciated to trigger muscle protein degradation across species, results in ubiquitinylation and degradation of the reporter protein.14 Since the reporter is ubiquitinylated and degradation is blocked when worms are treated with a proteasome inhibitor,17 it appears that starvation triggers proteasome mediated degradation of muscle protein in C. elegans. This was confirmed by subsequent studies, which showed that the endogenous proteins arginine kinase and adenylate kinase and a transgenic GFP expressed in body wall muscle are all also degraded upon starvation,18 at about the same rate as the myosin-LacZ reporter protein. Thus, the loss of this transgenically expressed marker protein can be used as an indicator that degradation, not normally observed in well fed adult worms, has been initiated (NB this does not imply that the proteasomes or lysosomes are inactive in well fed adult worms; this implies that the transgenic protein is not trafficked to the proteasomes or lysosomes in well fed adult worms). The ability to visually observe loss of a normally stable protein in muscle provided a phenotype that has now been used to identify many genes that appear to control the initiation of degradation in C. elegans muscle.5,17,19-23
While this review focuses on genes that appear to control the initiation of degradation as assayed by aberrant degradation of normally stable transgenic proteins, several other transgenic proteins can also be used to visually observe other features of protein degradation. These include Ubiquitin GFP fusions which can inform on inhibition of the proteasome,24 a Ubiquitin Dendra2 fusion which can be used to monitor rates of proteasome mediated degradation,25 and LGG-126 or LGG-227 GFP fusions which can be used to monitor accumulation of autophagosomes.
Proteasome-Mediated Muscle Protein Degradation is Opposed by Acetylcholine from Neurons
The degradation of LacZ observed in response to starvation was noted to proceed in a temporal and cell specific fashion with degradation first observed in 63 posterior body-wall muscles then the 8 vulval muscles and finally the 32 anterior body-wall muscles at the head.17 The correlation of this pattern of degradation with sources of innervation suggested that neuronal input was controlling starvation induced degradation in body-wall muscle. Indeed, it was shown that temperature sensitive cha-1 mutants, which rapidly lose mobility at 25°C through failure to produce acetylcholine, began LacZ degradation when adults were acutely placed at non-permissive temperature. This degradation could be blocked by treatment with the acetylcholine (ACh) agonist Levamisole or with proteasome inhibitors.17 Furthermore, degradtaion occurred more rapidly in starved animals that released less ACh (unc-17 or unc-13) or had mutations in the ACh receptor (unc-29 or unc-38) and Levamisole failed to protect starved animals from muscle protein degradation when they contained a mutation in the ACh receptor (lev-1). Together, these results provided the first in vivo demonstration of a signaling system that regulated muscle protein degradation (Table 1A and Fig. 1) and cross tissue regulation of muscle protein degradation (Table 3).
Table 1. Genes identified as regulators of muscle protein degradation.
| (A) Genes contributing to acetylcholine control of proteasome mediated degradation17 | |
|---|---|
|
cha-1 |
Choline acetyltransferase |
|
unc-17 |
ACh vesicular transporter |
|
unc-13 |
Regulator of neurotransmitter release |
|
lev-1 |
AChR (non-α subunit) |
|
unc-29 |
AChR (non-α subunit) |
| unc-38 | AChR (α subunit) |
| (B) Genes contributing to Ras-MAPK control of autophagic degradation19 | |
|---|---|
|
let-60 |
Ras |
|
gap-1 |
Ras GTPase activating protein |
|
gap-2 |
Ras GTPase activating protein |
|
mpk-1 |
MAPK |
|
mek-2 |
MEK |
|
lin-45 |
Raf |
| gla-3 | TIS11 like zinc finger containing protein20 |
| (C) Genes contributing to FGF control of autophagic degradation upstream of Ras21 | |
|---|---|
|
clr-1 |
Tyrosine phosphatase |
|
egl-15 |
FGFR |
|
egl-17 |
FGF |
|
let-756 |
FGF |
|
sem-5 |
GRB2 |
| soc-2 | Leucine rich repeat protein |
| (D) Genes contributing to insulin-like control of autophagic degradation upstream of Raf22 | |
|---|---|
|
daf-2 |
IGFR |
|
age-1 |
PI3K |
|
pdk-1 |
3-phosphoinositide-dependent kinase 1 |
|
daf-18 |
PTEN |
| akt-1 | Akt |
| (E) Genes contributing to control of calpain-mediated degradation5 | |
|---|---|
|
pat-2 |
Integrin (α subunit) |
|
pat-3 |
Integrin (β subunit) |
|
pat-4 |
Integrin-linked kinase |
|
pat-6 |
α-parvin |
|
tln-1 |
Talin |
|
zyx-1 |
Zyxin |
|
atn-1 |
α-actinin |
|
cdc-42 |
Rho GTPase |
|
uig-1 |
Cdc-42 guanine nucleotide exchange factor |
|
deb-1 |
Vinculin |
|
unc-52 |
Perlecan |
|
unc-82 |
Kinase |
|
unc-97 |
PINCH-1 |
| unc-112 | Kindlin-2 |
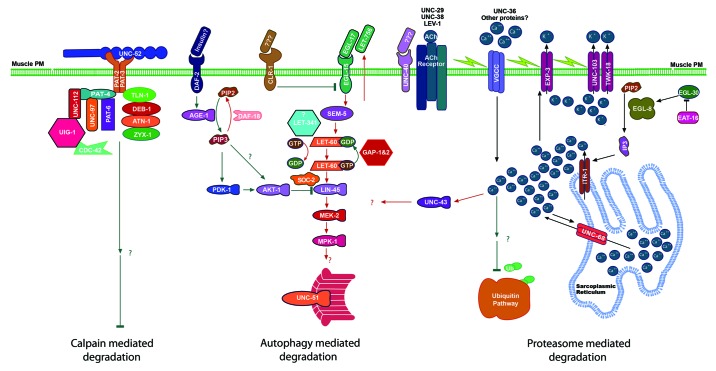
Figure 1. Putative model of the integrated control of cytosolic muscle protein degradation. Left: calpain mediated degradation is opposed by integrin attachment complex binding to perlecan in the basement membrane.5 Middle: insulin-like signaling opposes22 constitutive, autocrine28 FGF signaling19,21 to modulate autophagic degradation. Right: intra-cellular calcium levels23 as controlled by plasma membrane depolarization17,23 act to inhibit proteasome mediated degradation.
Table 3. Cross tissue regulation of muscle protein degradation.
| Tissue | Signal | Effect |
|---|---|---|
| Intestine |
H+ |
Inhibits autophagic degradation? |
| Muscle |
LET-756 |
Promotes autophagic degradation |
| Muscle/Hypodermis |
UNC-52 |
Inhibits calpain-mediated degradation |
| Nerve |
ACh |
Inhibits proteasome-mediated degradation |
| Nerve |
Contact ? |
Inhibits autophagic degradation of GABAA |
| Nerve |
VPR-1, ligand for CLR-1? |
Promotes autophagic degradation? |
| Unknown, Nerve? | Ligand for DAF-2 | Inhibits autophagic degradation |
Autophagic Muscle Protein Degradation is Coordinately Controlled by Growth Factors
A temperature sensitive let-60 (Ras) gain-of-function mutation was also found to induce degradation in adults acutely placed at non-permissive temperature.19 In contrast to the degradation induced by starvation or in cha-1 mutants, degradation observed in temperature sensitive let-60 gain-of-function mutants was not affected by proteasome inhibitors. Additionally while mutations in the LET-60 Ras effectors LIN-45 (Raf), MEK-2 or MPK-1 suppressed the degradation in let-60 mutants they did not suppress the degradation induced by starvation or in cha-1 mutants. These results suggested that a second signaling system (Ras-Raf-MEK-MAPK) was regulating muscle protein degradation and doing so via a different proteolytic pathway, which later was identified as autophagy (Table 1B and Fig. 1). Additional experiments showed that mutational activation of the LET-60 effector LIN-45 was also sufficient to induce degradation, as was expression of an active form of MPK-1.19 This, coupled with the separate observation that the MPK-1 interacting protein GLA-3, which appears to be a negative regulator of MPK-1 signaling, also appears to regulate muscle protein degradation,20 demonstrated that Ras-MAPK was regulating muscle protein degradation. Surprisingly, gain of function mutations in the EGF receptor homolog LET-23, which signals to the Ras-Raf-MEK-MAPK module during vulval development29 did not provoke degradation.19 Rather, it was later shown that activation of the EGL-15 FGF receptor by loss of function mutation in clr-1, a negative regulator of EGL-15, caused degradation and that this degradation was dependent upon EGL-15 and its ligands EGL-17 and LET-756;21 the two ligands apparently act redundantly. Since reduction of function mutations in sem-5, soc-2, let-60, lin-45, mek-2 and mpk-1 all suppressed clr-1 induced degradation, and as reduction of function of clr-1 resulted in EGL-15 dependent phosphorylation of MPK-1, it was evident that EGL-15 was the upstream receptor controlling LET-60 regulation of muscle protein degradation (Table 1C and Fig. 1). The recent demonstration that VPR-1 appears to be secreted from neurons and to bind to and inhibit CLR-130 suggests that nerves may have multiple mechanisms by which they modulate protein degradation (Table 3), but this role of VPR-1 for control of protein degradation via CLR-1 remains to be demonstrated. Intriguingly, LET-756 is produced and secreted by body-wall muscle itself,28 suggesting that muscle may have a constitutive, autocrine loop by which FGFR-Ras-MAPK is poised to keep degradation on (Fig. 1 and Table 3). In addition to CLR-1 acting to prevent EGL-15 from causing constitutive degradation, signaling from DAF-2 (homolog of insulin/IGF-1 like receptor) via AGE-1, PDK-1, and AKT-1 acts, as it does in mammalian muscle,31 to oppose LIN-45 mediated protein degradation22 (Table 1D and Fig. 1). Specifically, temperature sensitive daf-2 or age-1 mutants degrade LacZ when shifted to non-permissive temperature as adults and this degradation is blocked in pdk-1 or akt-1 gain of function mutants as well as in soc-2, lin-45, mek-2 or mpk-1 loss of function mutants. Additionally, muscle specific expression of wild-type DAF-2 or AGE-1 is sufficient to block degradation in response to mutation of each respective gene, and MPK-1 is activated in response to AGE-1 inhibition with LY-294002 (as assayed by western blot using a pTpY-ERK antibody). Further experiments suggest that AKT-1 acts to phosphorylate and inhibit LIN-45 Raf such that LIN-45 serves to integrate signal from both EGL-15 and DAF-2.22 Consistent with this presumptive balancing of growth factor signals, increased signal downstream of DAF-2 (in daf-18 mutants) can prevent degradation when the FGF receptor is hyperactivated in clr-1 mutants. Finally, degradation in response to MPK-1 activation is blocked by mutation in the autophagy initiator UNC-51 and degradation in daf-2 mutants is blocked by N6,N6-dimethyladenosine treatment, suggesting that growth factor signals interact to control autophagic degradation in C. elegans muscle.
unc Mutants are an Enriched Gene Class for Potential Regulators of Muscle Protein Degradation
The discovery that genes that control muscle contraction, e.g., ACh signaling, also control protein degradation and the observation that long-term activation of autophagy via either increased EGL-15 or decreased DAF-2 signaling leads to a severe movement decline, raised the conjecture that other potential regulators of muscle protein degradation might be found among mutants with an uncoordinated (Unc) phenotype. This hypothesis was tested by treating fully developed adult animals with RNAi against 159 genes from the various classes of established muscle mutants.23 Much as drug treatments or acute shifts of mutants to non-permissive temperature can trigger protein degradation in adult worms, RNAi treatment of adults can also do so. Accordingly, 47 genes were identified as potential negative regulators of muscle protein degradation using this method and another three were identified by examination of dominant mutants.23 Using the results from past studies, RNAi knockdowns of these genes were further tested against mutants in the autophagy pathway (unc-51, mpk-1, daf-18) and in animals treated with proteasome inhibitor (this group also included the 3 dominant mutants). Fifteen genes appear to be potential new regulators of autophagy (Table 2A), ten appear to be potential new regulators of proteasome mediated degradation (Table 2B), and 25 RNAi treatments were not suppressed (Table 2C and unc-52, unc-82, unc-97 and unc-112 in Table 1E), suggesting regulators of other proteases, perhaps calpains and/or caspases. Note that in the clustering experiments restoration of muscle function was not assayed so that no conclusions about the causal role of protein degradation in the various muscle phenotypes can be made from these studies alone. Additionally, further studies are required to determine if the products of these genes are direct or indirect regulators of muscle protein degradation (for example it seems likely that the commonly used transformation marker rol-6 is an indirect regulator based upon its product being cuticular) as well as to place these genes into accurate signaling networks. However, several testable predictions can be made. For example, netrin receptors, which are known to be expressed in muscle to control synapse formation,32 and ion channels controlling calcium flux are good candidates to mediate ACh regulation of proteasomal degradation and UNC-43, CaMKII, could be controlling the effects of elevated intracellular calcium (Fig. 1). Importantly, the clustering methodology that combines RNAi knockdowns with mutational epistasis tests also allows for stratification of novel regulators of degradation into potentially known vs. unknown pathways, so that priority can be given either to uncovering novel players in established pathways or to constructing new pathways. Lastly, as the genes in Tables 1 and 2 all come from candidate gene screens it is quite likely that many additional mutations can impact muscle protein degradation (for example extrapolation of the screen of muscle mutants suggests at least 200 more await to be uncovered from among the genes for which RNAi produces a movement defect).
Table 2. Genes putatively identified as regulators of muscle protein degradation.
| (A) Genes that appear to control autophagic degradation23 | |
|---|---|
|
unc-32 |
Vacuolar proton pump |
|
aex-5 |
Kex2/subtilisin-like proprotein convertase |
|
egl-46 |
Insulinoma associated protein 2 |
|
sqt-1 |
Cuticle collagen |
|
unc-103 |
ERG-like K+ channel |
|
unc-116 |
Kinesin-1 heavy chain |
|
aex-3 |
RAB-3 guanine nucleotide exchange factor |
|
eat-16 |
Regulator of G protein signaling |
|
eat-20 |
Fibrillin |
|
egl-47 |
G protein coupled receptor |
|
exp-3 |
Calcium activated K+ channel |
|
sdc-1 |
Zinc finger protein |
|
aex-2 |
G protein coupled receptor |
|
pbo-4 |
Voltage gated Na+/H+ exchanger |
| unc-36 | Ca2+ channel (α2δ subunit) |
| (B) Genes that appear to control proteasome mediated degradation23 | |
|---|---|
|
unc-40 |
Netrin receptor |
|
unc-98 |
Zinc finger protein |
|
egl-6 |
FMRFamide G protein coupled receptor |
|
egl-8 |
PLC |
|
itr-1 |
IP3 receptor |
|
unc-5 |
Netrin receptor |
|
unc-42 |
Homeodomain transcription factor |
|
unc-61 |
Septin |
|
unc-85 |
Asf1 |
| twk-18 | TWK K+ channel |
| (C) Genes that appear to control protein degradation via unidentified proteases23 | |
|---|---|
|
ina-1 |
Integrin (α subunit) |
|
mup-2 |
Troponin T |
|
mup-4 |
Fibrillin |
|
unc-78 |
Actin-interacting protein |
|
vab-10 |
Spectraplakin |
|
egl-27 |
Atrophin 1 |
|
eat-6 |
Na+/K+ ATPase |
|
rol-6 |
Cuticle collagen |
|
unc-77 |
Voltage insensitive cation leak channel (α1 subunit) |
|
unc-120 |
Serum response factor |
|
unc-124 |
Innexin |
|
eat-3 |
Mitochondrial dynamin |
|
egl-3 |
Subtilisin-like proprotein convertase |
|
egl-13 |
Sox domain transcription factor |
|
rol-3 |
Kinase |
|
unc-24 |
Stomatin |
|
unc-73 |
Guanine nucleotide exchange factor |
|
unc-96 |
LIM domain protein |
|
unc-119 |
Synaptic vesicle protein |
|
unc-43 |
CaM kinase |
| unc-105 | Degenerin channel |
Calpain Mediated Degradation is Controlled By Integrin Attachment Complexes
Among the 25 genes identified as regulating an unknown protease were an integrin receptor, an integrin receptor ligand and several intracellular binding partners of integrin receptors. The prediction that integrin attachment complexes control muscle protein degradation was further tested, and it was determined that protein degradation was induced by RNAi knockdown of any one of 14 out of 15 members of the integrin-based muscle attachment complex.5 Further experiments showed that degradation also occurred in adults carrying temperature sensitive mutations in unc-52 or unc-112, and that this degradation was inhibited by treatment with calpain inhibitors or RNAi against any one of several calpain encoding genes. It was also shown that RNAi against these integrin attachment complex proteins results in physical disruption of the attachments and that when physical disruption is attenuated in dim-1 mutants, degradation does not occur (dim-133 encodes an immunoglobulin-like repeat protein that localizes around dense bodies). Together, these results suggest that physical disruption of integrin complexes, perhaps due to mechanical use via contraction/locomotion, results in activation of calpains and consequent protein degradation. Surprisingly, RNAi knockdown of various calpain encoding genes in wild-type adults results in muscle defects, and calpain activation results in degradation of at least one component of the attachment complex.5 It therefore appears that calpain activation upon disruption of integrin complexes is largely for repair and/or reassembly of the integrin complexes themselves, rather than for control of degradation of bulk muscle protein (which is observed under conditions of pathological non-attachment).
Activation of Pre-Existing Proteolytic Systems
The picture that emerges is that different proteolytic systems in muscle are responsible for degrading proteins in various subcellular ″compartments″ (cytosol, myofibrils, attachment complexes) under various conditions. The proteasomal and lysosomal proteases are constitutively active, but the access of substrate proteins is regulated. By contrast, the calpains appear to have a role in degrading proteins in the contractile structures under both normal and pathological conditions. The data in C. elegans muscle on all three systems show that protein degradation can be triggered in the absence of de novo protein synthesis, that is, by activation of pre-existing proteolytic or substrate-delivery systems. There may also be increased synthesis of the components of these catabolic systems (mpk-1 appears to control this in other cell types24), but if so, this is not absolutely required to initiate or maintain degradation.
Roles of These Systems in Development and Physiology
C. elegans is a multi-cellular animal with multiple tissues. Thus, muscle growth and/or shrinkage needs to be coordinated with the growth of other tissues to maintain functionally significant contacts with, for example, motor neurons and the hypodermis. Similarly, the energy and material needs for muscle growth need to be balanced against the other functions of muscle (e.g., contraction) and requirements of other tissues. It is likely that signals that regulate global protein synthesis and degradation (for example synthetic capacity or rate of degradation) do so as part of an integrated network of signals that promote growth and/or maintenance of organism-wide homeostasis.
Proteasome-mediated degradation of bulk cytosolic protein occurs when diminished cholinergic signal from motor neurons leads to decreased intramuscular calcium. Thus, it would appear that active signal from nerves to muscle promotes not only contraction, but also maintenance of appropriate levels of protein synthesis and degradation in muscle. The integrated control of contraction and degradation suggests that a primitive ″use it or lose it″ mechanism underlies this degradation. Additionally, hydrogen ions can signal from the gut to muscle to induce contraction during defecation34 and PBO-4, which initiates this proton-induced contraction, appears to regulate muscle protein degradation as well (Table 2A). This may suggest that ″use it or lose it″ mechanisms are not limited to nerve-muscle interactions, a conjecture supported by the role of integrin-based attachment complexes in hypodermis-muscle signaling. Together, these results suggest that depolarization of the muscle plasma membrane, which in turn affects calcium flux through voltage-gated channels, may be an ″emergent property″ of muscle that controls muscle protein degradation. This depolarization is apparently not simply responsive to a single signal from a single tissue (e.g., ACh from nerve) but rather may reflect the signal from multiple tissues. If this speculation is correct, the proteasome may be the key proteolytic system that responds to ion flux and it may be that proteins like calreticulin35 bridge the gap between calcium flux and proteasome activation.
The EGL-15 FGFR signal that promotes autophagic degradation is triggered, in part, by autocrine signaling through LET-756, suggesting that muscle is constitutively programmed to degrade soluble protein unless other, growth-permissive signals provide countervailing instructions. DAF-2 (insulin/IGF-1 like receptor), which restrains the autophagy promoted by EGL-15, receives signal from an unidentified ligand emanating from an as yet unidentified tissue. It is not yet clear what physiological information is conveyed by this signal, but the role of insulin/IGF in regulating cell growth in other organisms suggests that it may have a role in coordinating muscle growth with growth of other tissues. If this idea is correct, then it may be the case that the Ras-Raf-MEK-MAPK module serves in muscle as a central integrator of signals from other tissues, much as the polarization of the plasma membrane appears to do for proteasome based degradation. This idea further implies that autophagy may be the key proteolytic system that responds to growth factors; indeed, mutants deficient in autophagy are generally abnormal in growth regulation.36
The PAT-2/PAT-3 attachment complex, in addition to maintaining contacts between adjacent muscle cells, permits coordinated structures to be established in the body wall muscle and in hypodermis.2,37 Both tissues continue to grow, by hypertrophy, after the worm reaches adulthood, allowing coordinated longitudinal growth of the worm. Since the worm maintains an approximately constant axial ratio during post-adult growth, it would not be surprising if there were a feedback mechanism between this longitudinal growth and radial growth, which may be controlled in part by DAF-2. Thus, unlike proteasomal and autophagic degradation, calpain mediated degradation would appear to allow for fine-tuning of signals that regulate these other proteolytic systems.
The number of genes identified as having roles in regulating muscle proteolysis is vastly greater for C. elegans than for other metazoans, including man. These discoveries not only reflect the formidable experimental advantages available when studying this worm but also demonstrate, once again, how this worm can be a very good model for understanding the regulation of a complex phenotype. It may even be possible to map and quantitatively model genomic control of muscle protein degradation in C. elegans.
Relation to Pathophysiology and Disease Models
Elucidation of signaling systems that appear to regulate various proteolytic systems in muscle not only lends itself to the question of what role these systems play in normal development and physiology but also to the relationship to pathologies and disease states.
The initial experiments to validate the transgene-encoded LacZ and GFP as reporters of intramuscular proteolysis suggest that the proteasome system (Table 1A and Fig. 1) is important in regulating protein degradation in response to starvation and denervation. Recently it was shown that selenium exposure results in neurotoxicity with post-synaptic muscles receiving less ACh and displaying proteasome mediated degradation.38 This may represent a mechanism underlying functional decline in muscle that receives less innervation (for example aging human muscle) or in post-synaptic cells receiving less ACh signal from degenerating pre-synaptic cells (for example neurodegenerative diseases). However, denervated muscle has also been shown to induce accumulation of GABAA but not ACh receptors in autophagosomes.39 Thus it may be that both proteasome and autophagic degradation are induced by denervation with multiple inputs from nerves having been removed (Table 3). This highlights the difficulty of both assuming and demonstrating that a single pathway or system is modulated in pathology or underlies a disease. This problem is compounded by the fact that it may often be the amount of perturbation of a particular signaling system that is important.22,40 For example, if ACh signaling to muscle can control intracellular calcium levels and the amount, timing or spatial distribution regulates both proteasome and autophagic degradation (Fig. 1) as well as calpain mediated muscle degeneration41 then demonstration of the importance of a single system in modulating a disease process can prove quite difficult. Similarly, as signals may balance each other it is possible that gain/loss of signal through one pathway can compensate for loss/gain of signal in another counterpoised signal to prevent pathological degradation.22 Lastly, both the loss5 and gain of function of calpains41 in muscle results in pathologies. This highlights the difficult path to considering treatment options which may need to be individually tailored to restore or remove a specific amount of activity and which also may explain why depolarization of the plasma membrane is both ″good″ because it opposes aberrant proteasome mediated degradation17,23 and ″bad″ because it contributes to muscle cell degeneration41,42 and protein aggregation.43
In developing transgene-encoded LacZ and GFP as reporters of intramuscular proteolysis, care has been taken to show that these proteins are reasonably representative of ″normal″ muscle proteins,18 and neither evoke an unfolded protein response15,16 nor produce other cellular pathologies. However, the same protein products, when expressed at much higher levels than those used in the studies described above (as the result of higher transgene copy-number) often accumulate in aggregates or granules. This produces various levels of cytotoxicity and is of no interest in the context of normal cellular physiology, but represents a general caveat of using transgenic markers in C. elegans.
Others have chosen to express proteins associated with human pathological processes involving protein folding or aggregation, in order to develop C. elegans as a model for studying the pathophysiology, biochemistry and genetics of such diseases. These pathologies in humans usually manifest primarily in neurons (e.g., Alzheimer disease, Huntington disease, Parkinson disease).40 Nevertheless, the C. elegans models have often used expression in body-wall muscles, for reasons of experimental convenience:44 Muscle-specific promoters are strong and well-characterized, muscle cells are large to permit easier examination of transgene expression and product localization, and muscle cells are fully susceptible to RNA interference (RNAi), in contrast to neurons. Although many of these model proteins have also been expressed in neurons, we limit the discussion below to phenomena observed in muscle.
Link45 first expressed a 42-residue β-amyloid peptide (Aβ), derived from human amyloid precursor protein, in worm muscle under the direction of the unc-54 (myosin heavy chain B) promoter. Because these transgenic worm lines used either extrachromosomal arrays or integrants derived from such arrays, it seems likely that the transgene was present at high copy number. The overexpressed Aβ peptide produced visible aggregates in the muscle cells, promoted accumulation of autophagic vesicles, and caused progressive paralysis.46 The Aβ peptide level was reduced, and vesicular accumulation and paralysis were relieved, by a reduction of function mutation in the DAF-2 insulin/IGF receptor.46 These ameliorating effects were blocked by RNAi knockdown of components (BEC-1, ATG-7) of the autophagic pathway. Conversely, Aβ toxicity was enhanced by RNAi knockdown of lysosomal aspartyl proteases (especially ASP-6) or of a putative lysosomal proton pump VHA-15. These observations implied a model in which autophagy, negatively regulated by signaling from the insulin/IGF pathway, can serve as a major mechanism for preventing accumulation of toxic protein aggregates in muscle. It is not resolved whether the role of autophagy is to clear large aggregates once they are formed, or to dispose of smaller aggregates before they can become large and toxic.40 These scenarios cannot be easily distinguished because the effects of unconditional mutations or RNAi cannot be applied acutely after aggregates have accumulated. It is noteworthy that the pro-degradation effect of tuning down IGFR signaling is similar to that observed22 using soluble reporters in the muscle cytosol (Table 1D and Fig. 1).
Visible aggregates were also produced upon expression of a GFP fusion47 or a YFP fusion44 of α-synuclein (α-syn) in muscle from the unc-54 promoter. α-syn is an aggregation-prone protein that is probably in equilibrium between folded and unfolded form,48-50 which may enable it to have some chaperone-like properties.51 Some α-syn mutations enhance the accumulation of aggregates, which are associated with the Lewy bodies of autosomal-dominant familial forms of Parkinson disease. In an effort to identify genes that might be involved in controlling α-syn aggregation, two reverse-genetic screens were performed using RNAi:
van Ham et al.44 used a genome-wide screen to find genes whose normal function opposed the early formation of α-syn::YFP fusion protein inclusions. The screen identified 80 genes whose knockdown enhanced aggregate accumulation, among which there was a high representation of genes involved in membrane or vesicle trafficking, but a notable absence of genes involved in either the ubiquitin-proteasome or the autophagic pathways.
Hamamichi et al.47 screened 868 candidate genes, chosen for possible relationship to known Parkinson pathways, for RNAi enhancement of early α-syn::GFP aggregation in a genetic background sensitized to reduce aggregation. This screen turned up genes involved in vesicle trafficking, as well as a few that potentially function (but perhaps do not function exclusively) in autophagy or in the ubiquitin-proteasome pathway.
Although these two screens were in many respects superficially similar, they identified nearly non-overlapping sets of genes.52
A conceptually similar RNAi screen was performed to identify genes whose knockdown enhanced aggregation of a Q35::YFP construct in muscle.53 This screen identified a number of proteasome components and chaperones, as well as a significant number of genes involved in mRNA processing and protein biosynthesis. However, there was again relatively little overlap with the gene set identified in the α-syn studies.44,47
The study of Prahlad and Morimoto54 showed that accumulation of aggregation-prone proteins (Q44::GFP and a mutant paramyosin) in muscle could be regulated cell-nonautonomously, through action of the neurosecretory thermosensory neurons. These same thermosensory neurons regulate the heat-shock response.55 Mutations (gcy-8 or ttx-3) that affect these neurons suppressed both the accumulation of aggregates and motility defects. It was conjectured54 that the thermosensory neurons might be involved, directly or indirectly, in the secretion of insulin-like peptides. If this is correct, then reduced levels of insulin-like peptides might lead to reduced stimulation of muscle DAF-2 insulin/IGF receptors and consequently increased intramuscular autophagy.22,46 It is important to note that the RNAi screens targeted at muscle-expressed proteins would not have uncovered the role of neurons in controlling intramuscular proteostasis, since C. elegans neurons are generally insensitive to RNAi.56,57
An alternative to insulin mediated control of autophagy is offered by the study of Garcia et al.43 They showed that accumulation of aggregation-prone Q35::GFP was exacerbated when pre-synaptic signaling was disrupted such that muscle was ‘overstimulated’. Specifically, loss of GABA signal or gain of ACh signal resulted in increased numbers of aggregates. It remains to be determined if the presumptive calcium modulation of proteasome and/or autophagic degradation (Table 2A and B and Fig. 1) underlies the increased aggregation.
In addition to these disease-specific models, aggregation can be studied utilizing the attachment of a 16-residue “degron” (degradation-promoting peptide) to the C-terminus of an otherwise normal GFP.58 This degron targets the attached protein to the proteasome in mammalian cells,59 but also produces toxic perinuclear aggregates when expressed in either mammalian cells or in C. elegans muscles.58 Overexpression of AIP-1, a positive regulator of the proteasome, reduces accumulation of both GFP::degron and Aβ peptide, and protects against Aβ toxicity.60 Lastly, it is possible to take a combinatorial approach by combining different models to attempt to look for commonalities.61
Studies of toxic proteins provide intriguing hints that various proteolytic processes may help alleviate toxic effects in muscle, but no consistent picture of which route to proteolysis is the most significant. This may reflect the biochemical reality that different proteins in different states or stages of aggregation are susceptible to somewhat different combinations of proteolytic processes. Furthermore, the balance between these proteolytic processes might well be modulated by factors (including the toxic aggregates) that alter the expression level or activities of the proteolytic pathway components. We should probably avoid the temptation to make sweeping generalizations and content ourselves for now with the power of C. elegans to point the way to specific intervention points for each kind of abnormal protein associated with a human pathology.
Acknowledgments
Thanks to Chris Link for discussions around the various protein aggregation models. The bulk of the work presented in Tables 1 and 2 and in Figure 1 was funded by grants to L.A.J. from the NSF (MCB-0542355 and MCB-0918031) and to N.J.S. from the NIH (AR-054342) and the MRC (G0801271). Figure 1 was generated using Biocarta’s open source pathway master template.
Footnotes
Previously published online: www.landesbioscience.com/journals/worm/article/20465
References
- 1.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moerman DG, Williams BD. Sarcomere assembly in C. elegans muscle (January 16, 2006), WormBook, ed. The C. elegansResearch Community, WormBook, /10.1895/wormbook.1.81.1,www.wormbook.org [DOI] [PMC free article] [PubMed]
- 3.Qadota H, Benian GM. Molecular structure of sarcomere-to-membrane attachment at M-Lines in C. elegans muscle. J Biomed Biotechnol. 2010;2010:864749. doi: 10.1155/2010/864749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sparrow J, Hughes SM, Segalat L. Other model organisms for sarcomeric muscle diseases. Adv Exp Med Biol. 2008;642:192–206. doi: 10.1007/978-0-387-84847-1_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Etheridge T, Oczypok EA, Lehmann S, Fields BD, Shephard F, Jacobson LA, et al. Calpains mediate integrin attachment complex maintenance of adult muscle in Caenorhabditis elegans. PLoS Genet. 2012;8:e1002471. doi: 10.1371/journal.pgen.1002471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.American Academy of Orthopaedic Surgeons, ed. The Burden of Musculoskeletal Diseases in the United States. Rosemont, IL: American Academy of Orthopaedic Surgeons, 2008. [Google Scholar]
- 7.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807–19. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 8.Dice JF. Lysosomal Degradation of Proteins. eLS: John Wiley & Sons, Ltd., 2007. http://www.els.net[10.1002/9780470015902.a0000646.pub2]
- 9.Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 10.Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem. 2011;80:1055–87. doi: 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- 11.Szewczyk NJ, Jacobson LA. Signal-transduction networks and the regulation of muscle protein degradation. Int J Biochem Cell Biol. 2005;37:1997–2011. doi: 10.1016/j.biocel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 12.Atherton PJ, Szewczyk NJ. Regulation of muscle proteostasis via extramuscular signals. In: Adams JD, Parker KK, eds. Extracellular and Intracellular Signaling: Royal Society of Chemistry, 2011. [Google Scholar]
- 13.Fire A, Waterston RH. Proper expression of myosin genes in transgenic nematodes. EMBO J. 1989;8:3419–28. doi: 10.1002/j.1460-2075.1989.tb08506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zdinak LA, Greenberg IB, Szewczyk NJ, Barmada SJ, Cardamone-Rayner M, Hartman JJ, et al. Transgene-coded chimeric proteins as reporters of intracellular proteolysis: starvation-induced catabolism of a lacZ fusion protein in muscle cells of Caenorhabditis elegans. J Cell Biochem. 1997;67:143–53. [PubMed] [Google Scholar]
- 15.McCracken AA, Brodsky JL. A molecular portrait of the response to unfolded proteins. Genome Biol. 2000;1:REVIEWS1013. doi: 10.1186/gb-2000-1-2-reviews1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–89. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 17.Szewczyk NJ, Hartman JJ, Barmada SJ, Jacobson LA. Genetic defects in acetylcholine signalling promote protein degradation in muscle cells of Caenorhabditis elegans. J Cell Sci. 2000;113:2003–10. doi: 10.1242/jcs.113.11.2003. [DOI] [PubMed] [Google Scholar]
- 18.Fostel JL, Benner Coste L, Jacobson LA. Degradation of transgene-coded and endogenous proteins in the muscles of Caenorhabditis elegans. Biochem Biophys Res Commun. 2003;312:173–7. doi: 10.1016/j.bbrc.2003.09.248. [DOI] [PubMed] [Google Scholar]
- 19.Szewczyk NJ, Peterson BK, Jacobson LA. Activation of Ras and the mitogen-activated protein kinase pathway promotes protein degradation in muscle cells of Caenorhabditis elegans. Mol Cell Biol. 2002;22:4181–8. doi: 10.1128/MCB.22.12.4181-4188.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kritikou EA, Milstein S, Vidalain PO, Lettre G, Bogan E, Doukoumetzidis K, et al. C. elegans GLA-3 is a novel component of the MAP kinase MPK-1 signaling pathway required for germ cell survival. Genes Dev. 2006;20:2279–92. doi: 10.1101/gad.384506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szewczyk NJ, Jacobson LA. Activated EGL-15 FGF receptor promotes protein degradation in muscles of Caenorhabditis elegans. EMBO J. 2003;22:5058–67. doi: 10.1093/emboj/cdg472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szewczyk NJ, Peterson BK, Barmada SJ, Parkinson LP, Jacobson LA. Opposed growth factor signals control protein degradation in muscles of Caenorhabditis elegans. EMBO J. 2007;26:935–43. doi: 10.1038/sj.emboj.7601540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shephard F, Adenle AA, Jacobson LA, Szewczyk NJ. Identification and functional clustering of genes regulating muscle protein degradation from amongst the known C. elegans muscle mutants. PLoS One. 2011;6:e24686. doi: 10.1371/journal.pone.0024686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu G, Rogers J, Murphy CT, Rongo C. EGF signalling activates the ubiquitin proteasome system to modulate C. elegans lifespan. EMBO J. 2011;30:2990–3003. doi: 10.1038/emboj.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamer G, Matilainen O, Holmberg CI. A photoconvertible reporter of the ubiquitin-proteasome system in vivo. Nat Methods. 2010;7:473–8. doi: 10.1038/nmeth.1460. [DOI] [PubMed] [Google Scholar]
- 26.Meléndez A, Tallóczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–91. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 27.Alberti A, Michelet X, Djeddi A, Legouis R. The autophagosomal protein LGG-2 acts synergistically with LGG-1 in dauer formation and longevity in C. elegans. Autophagy. 2010;6:622–33. doi: 10.4161/auto.6.5.12252. [DOI] [PubMed] [Google Scholar]
- 28.Bülow HE, Boulin T, Hobert O. Differential functions of the C. elegans FGF receptor in axon outgrowth and maintenance of axon position. Neuron. 2004;42:367–74. doi: 10.1016/S0896-6273(04)00246-6. [DOI] [PubMed] [Google Scholar]
- 29.Aroian RV, Sternberg PW. Multiple functions of let-23, a Caenorhabditis elegans receptor tyrosine kinase gene required for vulval induction. Genetics. 1991;128:251–67. doi: 10.1093/genetics/128.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han SM, Tsuda H, Yang Y, Vibbert J, Cottee P, Lee SJ, et al. Secreted VAPB/ALS8 major sperm protein domains modulate mitochondrial localization and morphology via growth cone guidance receptors. Dev Cell. 2012;22:348–62. doi: 10.1016/j.devcel.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rommel C, Clarke BA, Zimmermann S, Nuñez L, Rossman R, Reid K, et al. Differentiation stage-specific inhibition of the Raf-MEK-ERK pathway by Akt. Science. 1999;286:1738–41. doi: 10.1126/science.286.5445.1738. [DOI] [PubMed] [Google Scholar]
- 32.Alexander M, Chan KK, Byrne AB, Selman G, Lee T, Ono J, et al. An UNC-40 pathway directs postsynaptic membrane extension in Caenorhabditis elegans. Development. 2009;136:911–22. doi: 10.1242/dev.030759. [DOI] [PubMed] [Google Scholar]
- 33.Rogalski TM, Gilbert MM, Devenport D, Norman KR, Moerman DG. DIM-1, a novel immunoglobulin superfamily protein in Caenorhabditis elegans, is necessary for maintaining bodywall muscle integrity. Genetics. 2003;163:905–15. doi: 10.1093/genetics/163.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beg AA, Ernstrom GG, Nix P, Davis MW, Jorgensen EM. Protons act as a transmitter for muscle contraction in C. elegans. Cell. 2008;132:149–60. doi: 10.1016/j.cell.2007.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee D, Singaravelu G, Park BJ, Ahnn J. Differential requirement of unfolded protein response pathway for calreticulin expression in Caenorhabditis elegans. J Mol Biol. 2007;372:331–40. doi: 10.1016/j.jmb.2007.06.071. [DOI] [PubMed] [Google Scholar]
- 36.Aladzsity I, Tóth ML, Sigmond T, Szabó E, Bicsák B, Barna J, et al. Autophagy genes unc-51 and bec-1 are required for normal cell size in Caenorhabditis elegans. Genetics. 2007;177:655–60. doi: 10.1534/genetics.107.075762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Gally C, Labouesse M. Tissue morphogenesis: how multiple cells cooperate to generate a tissue. Curr Opin Cell Biol. 2010;22:575–82. doi: 10.1016/j.ceb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 38.Estevez AO, Mueller CL, Morgan KL, Szewczyk NJ, Teece L, Miranda-Vizuete A, et al. Selenium induces cholinergic motor neuron degeneration in Caenorhabditis elegans. Neurotoxicology. 2012 doi: 10.1016/j.neuro.2012.04.019. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rowland AM, Richmond JE, Olsen JG, Hall DH, Bamber BA. Presynaptic terminals independently regulate synaptic clustering and autophagy of GABAA receptors in Caenorhabditis elegans. J Neurosci. 2006;26:1711–20. doi: 10.1523/JNEUROSCI.2279-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen E, Dillin A. The insulin paradox: aging, proteotoxicity and neurodegeneration. Nat Rev Neurosci. 2008;9:759–67. doi: 10.1038/nrn2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joyce PI, Satija R, Chen M, Kuwabara PE. The Atypical Calpains: Evolutionary Analyses and Roles in Caenorhabditis elegans Cellular Degeneration. PLoS Genet. 2012;8:e1002602. doi: 10.1371/journal.pgen.1002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mariol MC, Ségalat L. Muscular degeneration in the absence of dystrophin is a calcium-dependent process. Curr Biol. 2001;11:1691–4. doi: 10.1016/S0960-9822(01)00528-0. [DOI] [PubMed] [Google Scholar]
- 43.Garcia SM, Casanueva MO, Silva MC, Amaral MD, Morimoto RI. Neuronal signaling modulates protein homeostasis in Caenorhabditis elegans post-synaptic muscle cells. Genes Dev. 2007;21:3006–16. doi: 10.1101/gad.1575307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Ham TJ, Thijssen KL, Breitling R, Hofstra RM, Plasterk RH, Nollen EA. C. elegans model identifies genetic modifiers of alpha-synuclein inclusion formation during aging. PLoS Genet. 2008;4:e1000027. doi: 10.1371/journal.pgen.1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Link CD. Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1995;92:9368–72. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Florez-McClure ML, Hohsfield LA, Fonte G, Bealor MT, Link CD. Decreased insulin-receptor signaling promotes the autophagic degradation of beta-amyloid peptide in C. elegans. Autophagy. 2007;3:569–80. doi: 10.4161/auto.4776. [DOI] [PubMed] [Google Scholar]
- 47.Hamamichi S, Rivas RN, Knight AL, Cao S, Caldwell KA, Caldwell GA. Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson’s disease model. Proc Natl Acad Sci U S A. 2008;105:728–33. doi: 10.1073/pnas.0711018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartels T, Choi JG, Selkoe DJ. α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature. 2011;477:107–10. doi: 10.1038/nature10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eliezer D, Kutluay E, Bussell R, Jr., Browne G. Conformational properties of alpha-synuclein in its free and lipid-associated states. J Mol Biol. 2001;307:1061–73. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- 50.Wang W, Perovic I, Chittuluru J, Kaganovich A, Nguyen LT, Liao J, et al. A soluble α-synuclein construct forms a dynamic tetramer. Proc Natl Acad Sci U S A. 2011;108:17797–802. doi: 10.1073/pnas.1113260108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim TD, Paik SR, Yang CH, Kim J. Structural changes in alpha-synuclein affect its chaperone-like activity in vitro. Protein Sci. 2000;9:2489–96. doi: 10.1110/ps.9.12.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teschendorf D, Link CD. What have worm models told us about the mechanisms of neuronal dysfunction in human neurodegenerative diseases? Mol Neurodegener. 2009;4:38–51. doi: 10.1186/1750-1326-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nollen EA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, et al. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci U S A. 2004;101:6403–8. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prahlad V, Morimoto RI. Neuronal circuitry regulates the response of Caenorhabditis elegans to misfolded proteins. Proc Natl Acad Sci U S A. 2011;108:14204–9. doi: 10.1073/pnas.1106557108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prahlad V, Cornelius T, Morimoto RI. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–4. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–7. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 57.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–12. doi: 10.1016/S0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 58.Link CD, Fonte V, Hiester B, Yerg J, Ferguson J, Csontos S, et al. Conversion of green fluorescent protein into a toxic, aggregation-prone protein by C-terminal addition of a short peptide. J Biol Chem. 2006;281:1808–16. doi: 10.1074/jbc.M505581200. [DOI] [PubMed] [Google Scholar]
- 59.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–5. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 60.Hassan WM, Merin DA, Fonte V, Link CD. AIP-1 ameliorates beta-amyloid peptide toxicity in a Caenorhabditis elegans Alzheimer’s disease model. Hum Mol Genet. 2009;18:2739–47. doi: 10.1093/hmg/ddp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jensen LT, Møller TH, Larsen SA, Jakobsen H, Olsen A. A new role for laminins as modulators of protein toxicity in Caenorhabditis elegans. Aging Cell. 2012;11:82–92. doi: 10.1111/j.1474-9726.2011.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]