Abstract
mRNA localization is a crucial mechanism for post-transcriptional control of gene expression used in numerous cellular contexts to generate asymmetric enrichment of an encoded protein. This process has emerged as a fundamental regulatory mechanism that operates in a wide range of organisms to control an array of cellular processes. Recently, significant advancements have been made in our understanding of the mechanisms that regulate several steps in the mRNA localization pathway. Here we discuss progress made in understanding localization element recognition, paying particular attention to the role of RNA structure. We also consider the function of mRNP granules in mRNA transport, as well as new results pointing to roles for the endocytic pathway in mRNA localization.
Introduction
Over 25 years ago, a new cellular regulatory mechanism was revealed with the observation that β-actin mRNA is asymmetrically distributed within the cytoplasm of Ascidian eggs [1]. As later demonstrated, the function of mRNA localization is the generation of polarized gene expression through local protein synthesis [2]. Since this discovery, and the realization of the importance of mRNA localization in many aspects of multi-cellular life, including embryonic development [3], cell motility [4], and synaptic plasticity [5], inroads have been made into understanding the mechanisms controlling this process. A combination of powerful genetic and biochemical approaches has led to the discovery of trans-acting protein factors that function in mRNA localization [6]. In addition, new cell biological techniques for imaging RNA movement in live cells have enabled unprecedented views of this process in a number of systems [7]. Moreover, a global analysis underscored the importance of mRNA localization during Drosophila development, with over 70% of the ~3,000 transcripts analyzed displaying distinct localization patterns [8]. We now know that mRNA localization is a mechanism used to spatially regulate gene expression that is conserved not only throughout the Eukarya, but also in the Bacteria, emphasizing its importance in all domains of life [9].
The ultimate goal of mRNA localization is the subsequent spatial restriction of protein expression (Box 1). Mechanisms to accomplish this include degradation of protein products outside of the specified region, entrapment of a freely diffusing mRNA at the site of local protein synthesis, or active transport of mRNAs to specific cellular destinations (with the two latter mechanisms requiring a static anchor for the mRNA [6]). Importantly, to faithfully restrict protein accumulation to one part of a polarized cell, an mRNA must remain translationally silent during active transport or diffusion; this topic has been reviewed recently [10] and will not be covered here. Instead, our focus will be on the cellular mechanisms used to identify, package, and anchor mRNAs that are actively transported to specific domains within the cell cytoplasm.
Box 1. Functions of mRNA localization in eukaryotes.
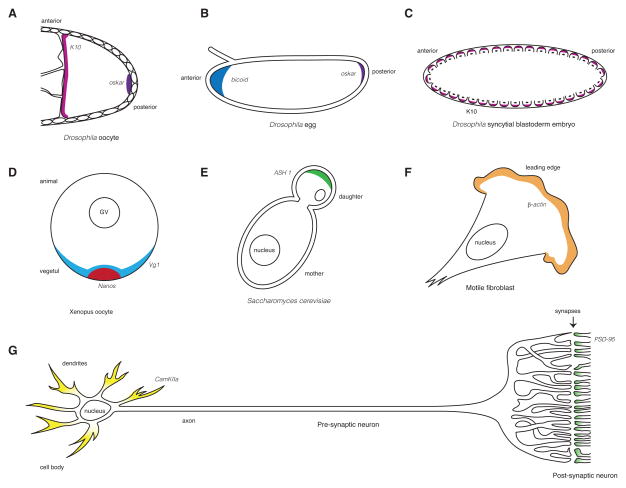
Cytoplasmic localization of mRNAs ultimately results in asymmetric distribution of protein expression, which can generate both cellular and developmental polarity. Some of the earliest examples of mRNA localization were identified in developmental systems. Early Drosophila development relies on maternally localized mRNAs, with bicoid mRNA at the anterior [55] and oskar mRNA at the posterior [56] providing spatial determinants that define the anterior-posterior axis (Fig. 1B). In Xenopus oocytes, selected transcripts are localized along the animal-vegetal axis of the oocyte during early oogenesis (Fig. 1D). Some of these mRNAs go on to specify the germ line [57], while others are involved in defining the primary germ layers, and thus are critical for patterning of the early embryo [58,59]. mRNA localization is also critical for generating cellular polarity. For example, in the yeast Saccharomyces cerevisiae ASH1 mRNA is localized to the tip of a budding daughter cell during mitosis (Fig. 1E; [12,60]). Local translation of Ash1p prevents mating-type switching, thus specifying daughter cell fate [12]. β-actin mRNA is localized to the leading edge of migrating fibroblasts (Fig. 1F) where local translation results in cytoskeletal remodeling necessary for cell movement [4]. mRNA localization is also critical for neuronal function [61]. Studies in Xenopus [62], Drosophila [63], and mammalian [64] neurons have shown that local protein synthesis is involved in neuronal growth during development, and in faithful synaptic function in the adult (Fig. 1G; [65]). These selected examples of mRNA localization represent some of the most widely studied model systems, with each offering unique advantages and distinct yet overlapping insights.
Localization Elements: Structure, not sequence, gives a message direction
The importance of cis-acting signals within localizing transcripts has been widely documented in a number of systems [11–13]. These localization elements (LEs) are generally found within the 3′-untranslated region (UTR) of localized mRNAs, and function predominantly to engage trans-acting protein factors [14]. Recruited proteins, including molecular motors [15–17] and translational repressors [18,19], thus function to direct the correct spatial and temporal distribution of the mRNA and its encoded protein product. Indeed, numerous proteins and protein families have been shown to be involved in mRNA localization. Perhaps the most widely characterized of these are a family of RNA binding proteins that includes ZBP-1, which was first described as a critical regulator of β-actin mRNA localization in chick embryo fibroblasts [20]. Related proteins have been shown to be required for mRNA localization in Xenopus oocytes (Vg1RBP/Vera; [21,22]), local translation in Drosophila oocytes and synapses (IMP; [23]), and translational regulation in mammalian fibroblasts (IMP-1,2,3; [24]). However, the molecular mechanisms governing how these proteins act in mRNA localization remain unclear. Given the conservation of mechanism and involved protein factors [25], it is surprising that a concordant conservation of LE sequences has not been observed [26]. Instead, it has been proposed that LEs fold into conserved secondary and tertiary structures that are recognized by RNA-binding proteins [26–28].
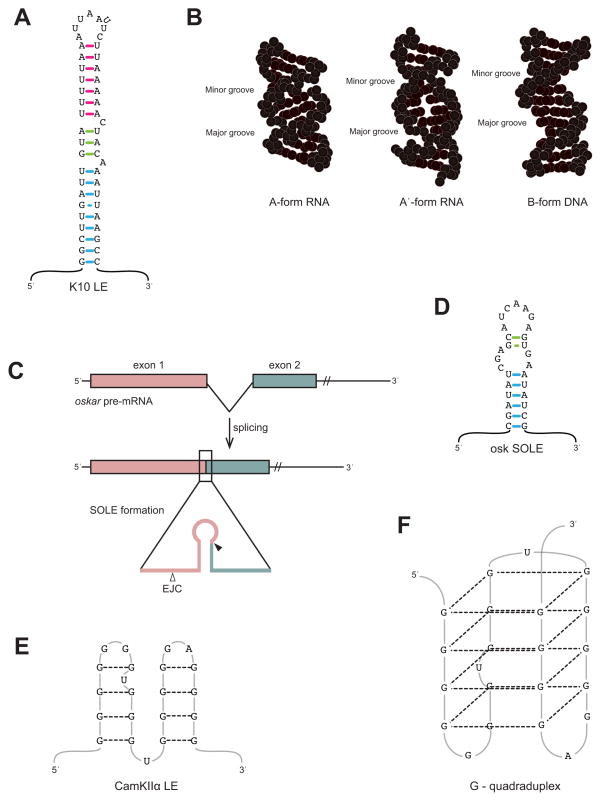
Until recently, this hypothesis was based on secondary-structure predictions, with stem-loop conformations as the likely candidate for defining an active LE [29]. However, in 2010 Bullock and colleagues succeeded in solving an NMR structure for the Drosophila K10 mRNA LE [30••], which localizes to the anterior of the oocyte and to the apical surface of the syncitial blastoderm in embryos (Fig. 1A, C; [31]). The 44-nucleotide K10 LE adopts a hairpin structure (Fig. 2A), but the helices do not conform to the expected A-form RNA. Instead, they form an A′-form helix, which, similar to B-form DNA, has a much wider major groove than A-form helices (Fig. 2B). In functional assays the widened major groove, the result of purine-purine base-stacking interactions, proved to be the key feature of the K10 LE [30••].
Figure 1. mRNA localization in Drosophila oocytes and embryos, and animal neurons.
A. K10 mRNA (magenta) localizes to the anterior and oskar mRNA to the posterior (purple) of a developing Drosophila oocyte. The oocyte is depicted on the right, surrounded by somatic follicle cells. B. bicoid mRNA (blue) localizes to the anterior and oskar (purple) mRNA to the posterior of the Drosophila egg. C. K10 mRNA (magenta) is found at the apical side of the blastoderm nuclei (black), which are partially separated from one another by invaginated cell membranes. D. Nanos (red) and Vg1 (pale blue) localize to the vegtal pole of the Xenopus oocyte utilizing two alternative localization pathways referred to as the early (active in stages I and II of oogenesis) and late (active in stages II–IV) pathways, respectively. The oocyte depicted is in stage IV of oogenesis. E. ASH1 mRNA (green) localizes to the distal tip of a budding daughter cell during mitosis in the yeast Saccharomyces cerevisiae. F. β-actin mRNA (orange) localizes to the leading edge of motile fibroblast. G. CamKIIα mRNA (yellow) localizes to the dendrites of a neuron, whereas PSD-95 mRNA (pale green) is found at the post-synaptic side of a synapse.
Figure 2. Structural motifs involved in mRNA localization.
A. Secondary structure of the K-10 LE. The three helical sections of the hairpin are shown with colored base-pairs (proximal blue, medial green, and distal pink). A flipped out U is present in the loop. This structural representation is based upon Figure 1 of [30••]. B. Depiction of A-form RNA, A′-form RNA, and B-form DNA helices. The K-10 LE stem folds into A′-form helices, in which the size of the major groove is similar to that of B-form DNA. This is in contrast to the small major groove found in the more common A-form RNA helix. This image is again based upon Figure 1 of [30••]. C. The SOLE is formed after splicing of the first intron in osk pre-mRNA [32••]. Exon 1 is shown as pink and exon 2 is green in both the pre-mRNA (top) and the spliced product (below). The inset shows the exon 1 (pink) and exon 2 (green) sequences within the SOLE and the position of the EJC (open arrowhead), 20–24nts upstream of the splice junction (filled arrowhead). D. The oskar SOLE is predicted to form a hairpin, with the proximal and distal stems shown with blue and green basepairs, respectively. E. Secondary structure of the CamKIIα LE in mammals, as depicted in [37•]. F. CamKIIα LE folds into a G-quadruplex through hydrogen-bond interactions indicated by dashed lines.
For the Drosophila oskar transcript, which is localized to the posterior pole of the developing oocyte and egg (Fig. 1A, B), a cis-LE has, until very recently, evaded identification [32••]. While the osk 3′-UTR was known to play a role [33], splicing of the first intron in the osk pre-mRNA and deposition of the exon-junction complex (EJC) are also required for osk localization [34]. Ghosh et al. [32••] have now reconciled these data into a model whereby splicing results in the formation of a spliced oskar localization element (SOLE); the two juxtaposed exons form a hairpin structure that directs osk mRNA localization (Fig. 2C, D). Interplay between the SOLE and the EJC also appears to be important, but the underlying mechanism is not yet clear. These findings underscore the importance of structural elements in mRNA localization, and also emphasize the complexity of identifying them through comparative computational analyses.
While short LEs can fold into discrete stem-loop structures, other much longer LEs have the potential to form complex structural conformations. With the discovery [35] that Fragile X Mental Retardation Protein (FMRP) is required for dendritic localization (Fig. 1G) of a number of mRNAs, came a renewed interest in the RNA-recognition potential of this protein. FMRP shows a propensity for binding intramolecular G-quadruplex structures [36], but a biological role for this interaction had yet to be shown. Subramanian et al. tested this interaction by examining the putative G-quadruplexes in the 3′-UTRs of two post-synaptically-localizing mRNAs, PSD-95 and CamKIIa (Figs. 1G, 2E-F; [37•]). They found that the structural conformation of the G-quadruplex is essential for mRNA localization, and that this activity was independent of sequence information. In addition, the G-quadruplex motif was found in almost a third of all neurite-targeted mRNAs, suggesting its function as a general LE.
Although the importance of structural motifs in LE function is gaining significant support, it remains a challenge to predict novel mRNA secondary and tertiary structures, especially in the context of longer localization signals (such as the 340-nt LE in Xenopus Vg1 mRNA [13]). However, this year saw the publication of the first real test of RNA tertiary structure prediction, RNA-Puzzles [38•]. Similar to the contribution of the CASP competition to in silico protein structure prediction [39], the goal of RNA-Puzzles is to improve RNA tertiary structure prediction through applying various computer programs to RNAs of known structure. The success of this project in coming years will be of great interest to the mRNA localization field, and has the potential to improve our ability to identify structural LEs.
Localizing mRNAs can go it alone
An increasingly accepted model to explain motor-dependent RNA transport proposes that, after LE recognition, multiple localizing mRNA species associate to form a localization-competent mRNP. The resulting large RNP granule (Box 2), containing multiple mRNAs, could then recruit molecular motors, thus transporting many mRNAs in a single trip [40]. Potentially, different mRNA species with similar LEs could be loaded into the same mRNP and co-transported to the same cellular location; an idea that was validated in yeast using fluorescent protein-tethering to track movement of ASH1 and IST2 mRNAs in vivo [41•]. While co-transport is also evident in other systems, recent studies using quantitative high-resolution microscopy have challenged the view that mRNP granule formation is obligatory for transport.
Box 2. Cytoplasmic mRNP granules.
mRNA, from transcription to degradation, is associated with a vast array of RNA-binding proteins in ribonucleoprotein particles or mRNPs. These associations are far from random, with specific mRNP configurations observable in different cellular circumstances. Several such mRNPs—termed granules due to their large size—have been intensely studied, including processing-bodies (P-bodies [66]), stress granules [67], and GW bodies [68]. These cytoplasmic granules are composed of overlapping sets of proteins and mRNAs, with some apparently common functions. For example, in both stress granules and P-bodies mRNAs can be stored in a translationally inactive state to be subsequently returned to translation [69], or degraded [66]. RNA-protein interactions and the formation of mRNP granules are crucial events in the life of an mRNA, and localizing mRNAs are no exception. Several localized mRNAs have been shown through both biochemical and cell biological techniques to form granule-like structures, including Drosophila oskar [46•] and Xenopus Vg1 [70]. How granule formation may aid in localization is unclear, although translational repression, protection of the mRNA from the degradation machinery, and transport of multiple mRNA species in a single localizing particle, are all attractive, and not mutually exclusive, hypotheses.
In neurons, synaptic plasticity and neuronal growth are dependent on local synthesis of proteins from a variety of transported mRNAs [26]. That these mRNAs would localize in distinct granules containing only one type of mRNA makes sense, as the spatio-temporal distribution throughout the neuron is different for each protein. This idea was tested by Kiebler and co-workers, who showed that MAP2 and CamKIIα mRNAs were transported in distinct particles [42•], and that these particles contained only one or a few mRNA molecules [43•]. Similar results were reported by Batish et al., who used high-resolution in situ hybridization to assess neuronal transport particles [44•]. Interestingly, this study also showed that mRNAs containing the same recognition motifs within their LEs were no more likely than those with apparently dissimilar LEs to co-localize. In the Drosophila blastoderm embryo as well, formation of large granules containing multiple RNAs does not appear to be required for localization. Amrute-Nayak and Bullock [45•] differentially labeled K10 LE RNA (Fig. 1C) to determine whether more than one mRNA assembled into a single localizing particle. No co-localization of red- and green-labeled RNAs was observed in embryos, indicating that the injected RNAs do not co-assemble into the same transport particle [45•].
Although it is apparent that some mRNAs localize singly, others are transported in large granules containing several transcripts; a prominent example of this is seen in osk mRNA transport in the Drosophila oocyte (Fig. 1A). In this instance, formation of granules of 50–80S in size is proposed to function in translational repression of osk during transport to the posterior pole of the oocyte [46•]. Similarly, in the Xenopus oocyte Vg1 mRNA has been observed to localize to the vegetal pole in granules, and recent data suggest that VegT is co-transported in these particles (T. Wood, personal communication). These examples, together with the data for ASH1 and IST2 RNAs in yeast [41•], indicate that co-transport of localized mRNAs in RNP granules has potential biological importance, thus raising the question: What controls whether an mRNA is transported in a large granule or alone? Cellular context likely plays an important role, where RNA-binding proteins could alternatively promote or prevent the formation of higher order structures. Indeed, in neurons, the RNA-binding protein Staufen appears to control the number of mRNAs in a localizing mRNP [43•] suggesting that the composition of granules, whether small or large, is tightly regulated. However, information within the localizing mRNA itself must be considered. For example, mRNAs that are localized in a group might contain multiple copies of a motif that would allow for mRNP formation by multi-domain RNA-binding proteins, protein-protein, or even inter-molecular RNA-RNA interactions (as has been suggested for osk [47]). Further complicating matters, these mechanisms need not function in a mutually exclusive manner, with a network of cis- and trans-factors potentially working together.
The functional consequences of mRNA localization in granules are also important. First, the transport of multiple mRNAs in a single particle results in a greater net polarization of the transcript using fewer molecular motors. Secondly, granule formation has been linked to translational control [46•], thus coordinately controlling protein expression of multiple mRNAs. The converse situation could also be beneficial, however, since individual transport allows for transcript-specific regulation. These alternatives are perhaps best exemplified by comparing neurons, where synaptic translation of single mRNAs is tightly controlled, with oocytes, where large numbers of mRNAs are localized and translated later at specific developmental time points. Further analysis in such different cell types may shed some light on the mechanisms that orchestrate assembly of transport mRNPs, be they large or small.
mRNA anchoring and the endocytic pathway
Regardless of how an actively transported mRNA reaches its cellular destination, mechanisms must be in place to anchor the mRNA in order to prevent it from diffusing back into the bulk cytoplasm. This critical feature of the mRNA localization pathway remains one of its least understood parts. However, a recent report has implicated the endocytic pathway in anchoring of the Drosophila pole plasm through an interaction with posteriorly-localized osk mRNA [48•]. Oskar protein had previously been shown to stimulate endocytosis upon localization, an activity that when disrupted resulted in diffusion of pole plasm [49]. The molecular culprit behind this activity, it appears, is Mon2, a conserved protein that is associated with the Golgi apparatus and the endosomal compartment. While Mon2 is dispensable for osk-induced endosomal cycling and maintenance of the polarized microtubule cytoskeleton, it is critical for the formation of the long actin projections that are necessary for anchoring pole plasm components [48•]. This induction of actin remodeling requires the activity of three proteins previously shown to regulate actin dynamics, Cappuccino, Spire, and the small GTPase Rho [50], as well as additional as yet uncharacterized regulators [48•]. Thus, Mon2 links posterior osk mRNA localization and anchoring of pole plasm with the endocytic pathway, the activity of which is in itself posteriorly polarized [48•].
The involvement of the endocytic pathway in the anchoring of a localized RNA is perhaps not unexpected. ESCRT-II (endosomal sorting complex required for transport-II) was previously shown to play a role in bicoid mRNA localization at the anterior pole of the Drosophila embryo [51]. However it is not yet clear how ESCRT-II promotes bicoid accumulation at the anterior of the embryo, although it has been shown to be distinct from its role in endosomal cycling [51]. In the pathogenic yeast Ustiglio maydis, various mRNAs are localized in a microtubule-dependent manner as the cell creates invasive filaments [52]. This process was recently linked to the endocytic pathway, with mRNPs observed to “hitch-hike” onto endosomes. Both constituents are then transported through an association with molecular motors towards the tip of the growing filament [53•]. These data hint at the idea that mRNA localization pathways can intersect with other cellular processes, such as endosomal sorting, and may utilize shared machinery (albeit in distinct ways). Indeed, a diverse array of functions, including cytokinesis, cell migration, and the generation of polarity, rely on the endocytic pathway, leading to the emerging hypothesis that endosomal sorting acts as a multi-purpose platform for a variety of cellular activities [54].
Conclusions and Future Directions
From recognition to anchoring the life of a localizing mRNA is a complex and still rather mysterious one. The recent elucidation of bone fide structural elements in the 3′-UTR of several mRNAs has opened a new chapter in the delineation of LEs, one that the field has long been working towards. It will be exciting to see whether advances in computational approaches will contribute to the discovery of additional structured LEs. It will also be important to tease apart the significance of RNP granule formation in mRNA localization. With the recent data in some systems highlighting the fact that mRNAs can travel alone, comparison of mRNP assembly in multiple cell types will be critical. And, as has been the case for some time, understanding the mRNA anchoring process remains a particular challenge. Recent studies aimed at understanding the involvement of endocytic pathway components offer a tantalizing glimpse into the overlap of cellular trafficking mechanisms. Understanding precisely how these pathways interact represents a particularly exciting area of future investigation in the field of mRNA localization.
Acknowledgments
We thank the members of the Mowry Lab for helpful discussion throughout the writing of this manuscript and Andrew Long for assistance with figures. We would also like to apologize to our colleagues in the field of mRNA localization whose work we were unable to reference due to space limitations. Our work on this topic is supported by Public Health Service grant GM071049 from the National Institute of General Medicine to K.L.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jeffery WR, Tomlinson CR, Brodeur RD. Localization of actin messenger RNA during early ascidian development. Dev Biol. 1983;99:408–417. doi: 10.1016/0012-1606(83)90290-7. [DOI] [PubMed] [Google Scholar]
- 2.Tomlinson CR, Bates WR, Jeffery WR. Development of a muscle actin specified by maternal and zygotic mRNA in ascidian embryos. Dev Biol. 1987;123:470–482. doi: 10.1016/0012-1606(87)90404-0. [DOI] [PubMed] [Google Scholar]
- 3.Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nusslein-Volhard C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawrence JB, Singer RH. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 1986;45:407–415. doi: 10.1016/0092-8674(86)90326-0. [DOI] [PubMed] [Google Scholar]
- 5.Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- 6.Shahbabian K, Chartrand P. Control of cytoplasmic mRNA localization. Cell Mol Life Sci. 2012;69:535–552. doi: 10.1007/s00018-011-0814-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu B, Piatkevich KD, Lionnet T, Singer RH, Verkhusha VV. Modern fluorescent proteins and imaging technologies to study gene expression, nuclear localization, and dynamics. Curr Opin Cell Biol. 2011;23:310–317. doi: 10.1016/j.ceb.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. Translation-independent localization of mRNA in E. coli. Science. 2011;331:1081–1084. doi: 10.1126/science.1195691. [DOI] [PubMed] [Google Scholar]
- 10.Lasko P. Translational control during early development. Prog Mol Biol Transl Sci. 2009;90:211–254. doi: 10.1016/S1877-1173(09)90006-0. [DOI] [PubMed] [Google Scholar]
- 11.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 13.Mowry KL, Melton DA. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science. 1992;255:991–994. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- 14.Macdonald PM. mRNA localization: assembly of transport complexes and their incorporation into particles. Curr Opin Genet Dev. 2011;21:407–413. doi: 10.1016/j.gde.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Betley JN, Heinrich B, Vernos I, Sardet C, Prodon F, Deshler JO. Kinesin II mediates Vg1 mRNA transport in Xenopus oocytes. Curr Biol. 2004;14 :219–224. doi: 10.1016/j.cub.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 16.Bobola N, Jansen RP, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- 17.Schnorrer F, Bohmann K, Nusslein-Volhard C. The molecular motor dynein is involved in targeting swallow and bicoid RNA to the anterior pole of Drosophila oocytes. Nat Cell Biol. 2000;2:185–190. doi: 10.1038/35008601. [DOI] [PubMed] [Google Scholar]
- 18.Gu W, Deng Y, Zenklusen D, Singer RH. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 2004;18:1452–1465. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilhelm JE, Hilton M, Amos Q, Henzel WJ. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J Cell Biol. 2003;163:1197–1204. doi: 10.1083/jcb.200309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deshler JO, Highett MI, Schnapp BJ. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276:1128–1131. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- 22.Havin L, Git A, Elisha Z, Oberman F, Yaniv K, Schwartz SP, Standart N, Yisraeli JK. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev. 1998;12:1593–1598. doi: 10.1101/gad.12.11.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boylan KL, Mische S, Li M, Marques G, Morin X, Chia W, Hays TS. Motility screen identifies Drosophila IGF-II mRNA-binding protein--zipcode-binding protein acting in oogenesis and synaptogenesis. PLoS Genet. 2008;4:e36. doi: 10.1371/journal.pgen.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262–1270. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macdonald PM. mRNA localization: assembly of transport complexes and their incorporation into particles. Curr Opin Genet Dev. 21:407–413. doi: 10.1016/j.gde.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.dos Santos G, Simmonds AJ, Krause HM. A stem-loop structure in the wingless transcript defines a consensus motif for apical RNA transport. Development. 2008;135:133–143. doi: 10.1242/dev.014068. [DOI] [PubMed] [Google Scholar]
- 28.Mickleburgh I, Chabanon H, Nury D, Fan K, Burtle B, Chrzanowska-Lightowlers Z, Hesketh J. Elongation factor 1alpha binds to the region of the metallothionein-1 mRNA implicated in perinuclear localization--importance of an internal stem-loop. RNA. 2006;12:1397–1407. doi: 10.1261/rna.2730106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chartrand P, Meng XH, Singer RH, Long RM. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol. 1999;9:333–336. doi: 10.1016/s0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- 30••.Bullock SL, Ringel I, Ish-Horowicz D, Lukavsky PJ. A′-form RNA helices are required for cytoplasmic mRNA transport in Drosophila. Nat Struct Mol Biol. 2010;17:703–709. doi: 10.1038/nsmb.1813. This study details the first NMR structure of a localization element. The 44nt K10 LE was shown to fold into an A′-form helix by virtue of purine base-stacking interactions, and this structure is required for localization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serano TL, Cohen RS. A small predicted stem-loop structure mediates oocyte localization of Drosophila K10 mRNA. Development. 1995;121:3809–3818. doi: 10.1242/dev.121.11.3809. [DOI] [PubMed] [Google Scholar]
- 32••.Ghosh S, Marchand V, Gaspar I, Ephrussi A. Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat Struct Mol Biol. 2012;19:441–449. doi: 10.1038/nsmb.2257. The oskar LE is formed after the removal of a 3′-UTR intron, thus forming the spliced oskar localization element or SOLE. These data reconcile much of the previous work in the field, showing a requirement for the spliceosomal machinery in the formation of the hairpin SOLE. [DOI] [PubMed] [Google Scholar]
- 33.Kim-Ha J, Webster PJ, Smith JL, Macdonald PM. Multiple RNA regulatory elements mediate distinct steps in localization of oskar mRNA. Development. 1993;119:169–178. doi: 10.1242/dev.119.1.169. [DOI] [PubMed] [Google Scholar]
- 34.Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- 35.Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 37•.Subramanian M, Rage F, Tabet R, Flatter E, Mandel JL, Moine H. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011;12:697–704. doi: 10.1038/embor.2011.76. The authors provide biological data implicating the formation of G-quadruplex structures in neurite-localized mRNAs. The role of FMRP in this process is also addressed, through its direct binding to the G-quadruplex structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Cruz JA, Blanchet MF, Boniecki M, Bujnicki JM, Chen SJ, Cao S, Das R, Ding F, Dokholyan NV, Flores SC, et al. RNA-Puzzles: a CASP-like evaluation of RNA three-dimensional structure prediction. RNA. 2012;18:610–625. doi: 10.1261/rna.031054.111. This publication documents the first test of computational approaches to RNA tertiary structure prediction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moult J, Pedersen JT, Judson R, Fidelis K. A large-scale experiment to assess protein structure prediction methods. Proteins. 1995;23:ii–v. doi: 10.1002/prot.340230303. [DOI] [PubMed] [Google Scholar]
- 40.Kato Y, Nakamura A. Roles of cytoplasmic RNP granules in intracellular RNA localization and translational control in the Drosophila oocyte. Dev Growth Differ. 2011 doi: 10.1111/j.1440-169X.2011.01314.x. [DOI] [PubMed] [Google Scholar]
- 41•.Lange S, Katayama Y, Schmid M, Burkacky O, Brauchle C, Lamb DC, Jansen RP. Simultaneous transport of different localized mRNA species revealed by live-cell imaging. Traffic. 2008;9:1256–1267. doi: 10.1111/j.1600-0854.2008.00763.x. These authors used live cell imaging techniques and RNA-tethering of fluorescent proteins to observed mRNA localization in yeast. Their data suggest that different mRNA species can localize in the same mRNP. [DOI] [PubMed] [Google Scholar]
- 42.Tubing F, Vendra G, Mikl M, Macchi P, Thomas S, Kiebler MA. Dendritically localized transcripts are sorted into distinct ribonucleoprotein particles that display fast directional motility along dendrites of hippocampal neurons. J Neurosci. 2010;30:4160–4170. doi: 10.1523/JNEUROSCI.3537-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikl M, Vendra G, Kiebler MA. Independent localization of MAP2, CaMKIIalpha and beta-actin RNAs in low copy numbers. EMBO Rep. 2011;12 :1077–1084. doi: 10.1038/embor.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44•.Batish M, van den Bogaard P, Kramer FR, Tyagi S. Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci U S A. 2012;109:4645–4650. doi: 10.1073/pnas.1111226109. These three references report on the phenomenon of mRNAs localizing in individual particles, not in large mRNPs containing multiple mRNAs. In all instances high-resolution imaging is used to track particular mRNAs in neurons, and point to the importance of specific post-transcriptional regulation of these mRNAs, especially at synapses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Amrute-Nayak M, Bullock SL. Single-molecule assays reveal that RNA localization signals regulate dynein-dynactin copy number on individual transcript cargoes. Nat Cell Biol. 2012;14:416–423. doi: 10.1038/ncb2446. This study uses sophisticated in vitro imaging techniques to analyze motor-dependent movement of mRNAs along microtubules. In vivo experiments also showed that K10 mRNA is preferentially transported alone, not in large mRNP granules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46•.Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. Biochemical analyses of osk RNPs revealed that granule formation is required for translational repression of the mRNA. This paper therefore suggests that Bruno is capable of orchestrating multiple mechanisms of translational control through recruitment of Cup and granule formation. [DOI] [PubMed] [Google Scholar]
- 47.Jambor H, Brunel C, Ephrussi A. Dimerization of oskar 3′ UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. RNA. 2011;17 :2049–2057. doi: 10.1261/rna.2686411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka T, Kato Y, Matsuda K, Hanyu-Nakamura K, Nakamura A. Drosophila Mon2 couples Oskar-induced endocytosis with actin remodeling for cortical anchorage of the germ plasm. Development. 2011;138:2523–2532. doi: 10.1242/dev.062208. [DOI] [PubMed] [Google Scholar]
- 49•.Tanaka T, Nakamura A. The endocytic pathway acts downstream of Oskar in Drosophila germ plasm assembly. Development. 2008;135:1107–1117. doi: 10.1242/dev.017293. This is the most recent example of endocytic components playing important roles in the anchoring of localized mRNAs at their destination. [DOI] [PubMed] [Google Scholar]
- 50.Qualmann B, Kessels MM. New players in actin polymerization--WH2-domain-containing actin nucleators. Trends Cell Biol. 2009;19:276–285. doi: 10.1016/j.tcb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 51.Irion U, St Johnston D. bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature. 2007;445:554–558. doi: 10.1038/nature05503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Becht P, Konig J, Feldbrugge M. The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J Cell Sci. 2006;119:4964–4973. doi: 10.1242/jcs.03287. [DOI] [PubMed] [Google Scholar]
- 53•.Baumann S, Pohlmann T, Jungbluth M, Brachmann A, Feldbrugge M. Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J Cell Sci. 2012 doi: 10.1242/jcs.101212. In this paper the role of the endocytic pathway is examined in active mRNA transport, with localized mRNAs recognizing endosomes and trafficking with them along microtubules rather than being recognized as discrete cargo themselves. [DOI] [PubMed] [Google Scholar]
- 54.Gould GW, Lippincott-Schwartz J. New roles for endosomes: from vesicular carriers to multi-purpose platforms. Nat Rev Mol Cell Biol. 2009;10:287–292. doi: 10.1038/nrm2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Driever W, Nusslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988;54:83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- 56.Kim-Ha J, Smith JL, Macdonald PM. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell. 1991;66:23–35. doi: 10.1016/0092-8674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- 57.Heasman J, Quarmby J, Wylie CC. The mitochondrial cloud of Xenopus oocytes: the source of germinal granule material. Dev Biol. 1984;105:458–469. doi: 10.1016/0012-1606(84)90303-8. [DOI] [PubMed] [Google Scholar]
- 58.Melton DA. Translocation of a localized maternal mRNA to the vegetal pole of Xenopus oocytes. Nature. 1987;328:80–82. doi: 10.1038/328080a0. [DOI] [PubMed] [Google Scholar]
- 59.Zhang J, Houston DW, King ML, Payne C, Wylie C, Heasman J. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell. 1998;94:515–524. doi: 10.1016/s0092-8674(00)81592-5. [DOI] [PubMed] [Google Scholar]
- 60.Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- 61.Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 13:308–324. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32 :1013–1026. doi: 10.1016/s0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- 63.Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 64.Davis L, Banker GA, Steward O. Selective dendritic transport of RNA in hippocampal neurons in culture. Nature. 1987;330:477–479. doi: 10.1038/330477a0. [DOI] [PubMed] [Google Scholar]
- 65.Martin KC, Barad M, Kandel ER. Local protein synthesis and its role in synapse-specific plasticity. Curr Opin Neurobiol. 2000;10:587–592. doi: 10.1016/s0959-4388(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 66.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nover L, Scharf KD, Neumann D. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol Cell Biol. 1983;3:1648–1655. doi: 10.1128/mcb.3.9.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol. 2006;71 :513–521. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- 70.Lewis RA, Mowry KL. Ribonucleoprotein remodeling during RNA localization. Differentiation. 2007;75:507–518. doi: 10.1111/j.1432-0436.2007.00159.x. [DOI] [PubMed] [Google Scholar]