Summary
KsgA, a universally conserved small ribosomal subunit (SSU) rRNA methyltransferase, has recently been shown to facilitate a checkpoint within the ribosome maturation pathway. Under standard growth conditions removal of the KsgA checkpoint has a subtle impact on cell growth, yet upon overexpresssion of RbfA, a ribosome maturation factor, KsgA becomes essential. Our results demonstrate the requirement of KsgA, in the presence of excess RbfA, for both the incorporation of ribosomal protein S21 to the developing SSU, and for final maturation of SSU rRNA. Also, when SSU biogenesis is perturbed by an imbalance in KsgA and RbfA, a population of 70S-like particles accumulate that are compositionally, functionally and structurally distinct from mature 70S ribosomes. Thus, our work suggests that KsgA and RbfA function together and are required for SSU maturation, and that additional checkpoints likely act to modulate malfunctional 70S particle formation in vivo.
Keywords: KsgA, RbfA, rRNA processing, ribosome biogenesis, translation initiation
Introduction
Proper ribosome biogenesis is of fundamental importance to assure the production and integrity of translationally active ribosomes; yet studies of ribosome biogenesis have lagged considerably behind those of the mature, fully-formed ribosome. Characterization of the bacterial ribosomal subunit biogenesis cascade has been hindered by the presence of redundant and parallel pathways [see (Connolly & Culver, 2009, Shajani et al., 2011)]. Recent data indicates that SSU biogenesis factors may act as blocks to translation initiation (Xu et al., 2008, Connolly et al., 2008, Strunk et al., 2011). Specifically, KsgA, a bacterial SSU rRNA methyltransferase (Helser et al., 1972) and biogenesis factor (Connolly et al., 2008), has been shown to compete with Initiation Factor 3 (IF3) and the large ribosomal subunit (LSU) for SSU association; therefore, a model has been proposed whereby KsgA binding prevents the incorporation of immature SSUs in the translating population of ribosomes by blocking the association of IF3 (Xu et al., 2008, Connolly et al., 2008). IF3 is the first translation initiation factor to bind the SSU during formation of the pre-initiation complex (Petrelli et al., 2001). Additionally, IF3 plays a role in start codon selection making its appropriate binding critical for the fidelity of translation initiation (Hartz et al., 1989, Sussman et al., 1996, Seshadri et al., 2009). Therefore, KsgA binding and release acts as a checkpoint during SSU biogenesis and acts to separate assembly intermediates from functional SSUs. (Connolly et al., 2008, Campbell & Brown, 2008).
The cellular role of KsgA is of particular interest as it appears to be universally conserved in both form and function (Klootwijk et al., 1972, Noon et al., 1998, Steege et al., 1982, Van Buul et al., 1984, O'Farrell et al., 2006, O'Farrell et al., 2008). KsgA and homologs dimethylate two adjacent adenosines near the 3’ end of SSU rRNA (residues A1518 and A1519 in E. coli) (Helser et al., 1972). Previous studies demonstrated that overexpression of a catalytically inactive KsgA (KsgA E66A) inhibits most 70S ribosome formation and results in accumulation of a population of SSUs physically associated with KsgA E66A (Connolly et al., 2008). These data suggest that release of KsgA from the developing SSU is greatly stimulated by methylation and that release is required for association of IF3 and/or LSUs with SSUs (Xu et al., 2008, Boehringer et al., 2012). In addition, overexpression of KsgA E66A also results in a profound SSU biogenesis defect, suggesting that impairing the ability of KsgA to methylate and dissociate from the SSU prevents late biogenesis events, including final maturation of 16S rRNA (Connolly et al., 2008) (K.C. and G.M.C., in preparation). Recent structural studies show that Dim1 (the eukaryotic homolog of KsgA), along with other assembly factors, bind eukaryotic ribosomes in similar positions to translation initiation factors (Strunk et al., 2011). Thus a general role for several SSU biogenesis factors appears to be restricting interactions of immature SSUs with other cellular components important for translation.
While yeast Dim1p is essential for rRNA processing, and therefore growth (Lafontaine et al., 1994), KsgA, like many bacterial ribosome biogenesis factors, is not essential in E. coli under standard growth conditions (Connolly & Culver, 2009). Mild phenotypes are often observed when a single E. coli ribosome biogenesis factor is perturbed, likely due to the apparent redundancy of factors and functions in bacterial ribosome biogenesis [see (Connolly & Culver, 2009)]. One exception is when the SSU biogenesis protein Ribosome Binding Factor A, RbfA, is deleted; this strain exhibits a significant growth defect, due to impaired SSU biogenesis, even under standard growth conditions (Dammel & Noller, 1995, Xia et al., 2003). Despite this and an understanding that RbfA is a cold shock protein (Jones & Inouye, 1996), the role for RbfA in SSU biogenesis has remained enigmatic. It has been speculated that RbfA plays a role in structural transitions that facilitate SSU maturation (Datta et al., 2007); however, no enzymatic activity has been assigned to RbfA. A GTPase, RsgA/YjeQ has been implicated in removing RbfA from SSUs (Goto et al., 2010). Cryo-EM studies revealed that, when bound to mature SSUs, RbfA appears to displace helix 44 and 45 of 16S rRNA, elements associated with the decoding process (Datta et al., 2007). Moreover, helix 44 has been shown to be a primary binding site for KsgA (Xu et al., 2008) and the loop of helix 45 contains the two adenosines methylated by KsgA (Helser et al., 1972). Recent cryo-EM data for KsgA bound to SSUs also suggests that it perturbs the structure of helix 44 (Boehringer et al., 2012). Thus binding of RbfA or KsgA alter the structure of similar regions of the SSU. Taken together these data suggest a link between appropriate assembly of functional regions of SSUs and the functions of RbfA and KsgA.
In the present study the SSU biogenesis cascade is investigated using a combination of deletion of ksgA and overexpression of RbfA. Under standard growth conditions, deletion of ksgA or overexpression of RbfA is well tolerated (Connolly et al., 2008, Dammel & Noller, 1995), yet when RbfA is overexpressed in the ksgA deletion background (ΔksgA) the result is a dramatic loss of viability, even at standard growth temperatures. Further examination of this strain revealed a large population of particles sedimenting near 70S even when RbfA is expressed and growth is undetectable. These 70S-like particles were shown to be immature, in terms of rRNA processing and association of ribosomal protein S21, and to have different conformations from mature SSUs in complex with LSUs. S21 has been implicated in translation initiation by exposing the anti-Shine-Dalgarno (SD) region of 16S rRNA and thus functioning in mRNA recruitment and appropriate start site selection during translation initiation (van Duin et al., 1972, Backendorf et al., 1981). Consistent with these findings, decreased fidelity of translation initiation and overall translational capacity are observed in this strain. Our work suggests that in E. coli there are several critical SSU biogenesis checkpoints prior to formation of initiation competent SSUs; KsgA methylation and release, as well as S21 association and rRNA maturation are all linked and important events late in SSU maturation as well as in translation initiation. Thus events associated with KsgA and RbfA function appear to play critical roles segregating immature SSUs from mature, initiation-competent SSU particles.
Results
Growth defects are associated with overexpression of RbfA in ΔksgA
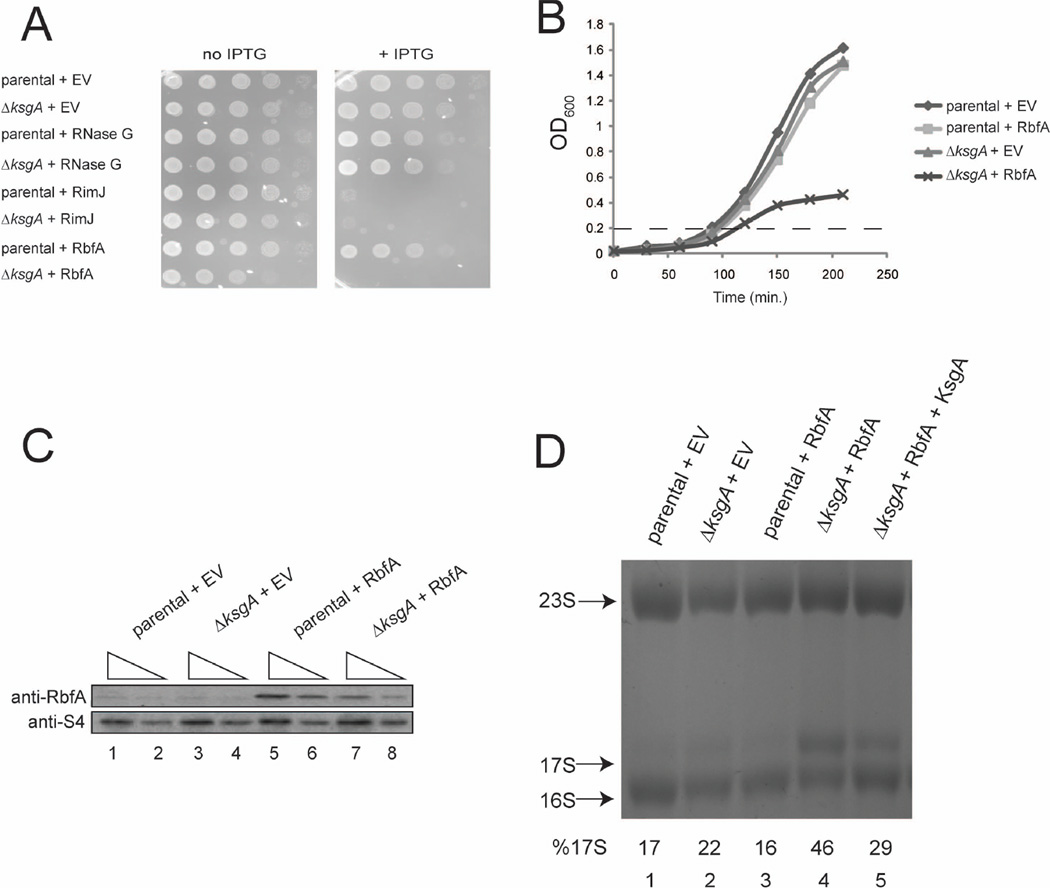
Although the function of many E. coli ribosome biogenesis factors has eluded understanding, previous studies identified a role for KsgA as a checkpoint within the bacterial SSU biogenesis cascade (Connolly et al., 2008). By extension, we sought to understand the integrated function of KsgA with other SSU biogenesis factors by identifying phenotypes associated with an imbalance in biogenesis factor concentration. To this end, a directed approach using overexpression of known biogenesis factors in the absence of the KsgA checkpoint (ΔksgA strain) was initiated. For some factors, such as RNase G, a SSU maturation endonuclease (Li et al., 1999), no growth phenotype is observed upon RNase G overexpression when compared to growth of strains (parental or ΔksgA) harboring empty vector (EV) (Figure 1A). Overexpression of other factors resulted in drastic growth defects, as was the case for RimJ, regardless of the presence or absence of KsgA (Figure 1A). Overexpression of RbfA, a SSU biogenesis factor of unknown function (Dammel & Noller, 1995, Bylund et al., 1998), was well tolerated in the parental strain (similarly to RNase G) while a severe growth defect in the ΔksgA strain was observed (Figure 1A). Overexpression of RbfA was also specifically toxic in kasugamycin resistant strains harboring a null mutation in ksgA (ksgA-ksgAR) and a mutation resulting in a catalytically inactive KsgA form (KsgA-G45S) that is otherwise expressed from the endogenous chromosomal locus (ksgA-ca) (Supplemental Figure 1A and 1B and data not shown). Thus, a severe growth phenotype is observed when RbfA is overexpressed in the absence of the KsgA protein, or its methylation activity.
Figure 1. Overexpression of RbfA is specifically toxic in the absence of KsgA.
A) Overnight E. coli cultures were plated in 10-fold serial dilutions on medium with or without inducer (IPTG). Parental and ΔksgA strains harbored empty vector or vectors for IPTG-inducible overexpression of RNaseG, RimJ or RbfA. B) Growth of the parental or ΔksgA strains containing either empty vector (EV) or overexpression vector for RbfA. The dashed line indicates the OD600 at which inducer was added. C) Western detection using an anti-RbfA (top panel) or an anti-r-protein S4 (bottom panel) antibodies of two serial dilutions (10 and 5 ugs total protein) of total protein from the same strains shown in B. D) Ethidium bromide stained agarose gel of total RNA from strains as in B, with the addition of a ΔksgA strain harboring overexpression vectors for both RbfA and KsgA.
Next, we were interested in understanding the underlying changes that are associated with this dramatic defect in growth. To this end, ΔksgA harboring a plasmid overexpressing RbfA (ΔksgA + RbfA) was grown in liquid culture. Following ~5 doublings (see Material and Methods) overexpression was initiated (Figure 1B, dotted line) and growth is arrested in ΔksgA + RbfA cells while parental cells harboring EV or an RbfA overexpression vector, and ΔksgA cells harboring EV, maintained normal growth (Figure 1B). Upon overexpression, enhanced levels of RbfA are observed in both the parental and ΔksgA strains, compared to endogenous levels (Figure 1C, compare lanes 1–4 with lanes 5–8). Moreover, the expression of RbfA is greater in the parental suggesting that the toxic phenotype cannot be simply due to total levels of RbfA. The observation that extra RbfA protein is specifically toxic when the KsgA checkpoint is absent, suggests that the KsgA methyl-mediated switch and RbfA functions are linked.
Given that both KsgA and RbfA are involved in ribosome biogenesis, we next examined if intermediates of the ribosome biogenesis pathway accumulate during overexpression of RbfA in ΔksgA. Following 1 hour of growth in the presence of inducer rRNA species were examined from whole cell lysate (Figure 1D). While mature 16S rRNA is the dominant SSU species in both the parental and ΔksgA strains harboring empty vector (EV) (Figure 1D, lanes 1 and 2), an immature 16S rRNA precursor, 17S rRNA (Supplemental Figure 1C), accumulates when RbfA is overexpressed in ΔksgA but not the parental strain (Figure 1D, lanes 3 and 4). Simultaneous overexpression of RbfA and KsgA reduces accumulation of 17S rRNA (Figure 1D, lane 5) indicating that RbfA toxicity is due to the lack of KsgA. Simultaneous overexpression of RbfA and KsgA does not greatly reduce cellular levels of overexpressed RbfA (Supplemental Figure 1D). These data indicate that additional copies of RbfA lead to changes in 16S rRNA maturation when KsgA is absent.
Changes in ribosomal populations are associated with simultaneous perturbation in KsgA and RbfA levels
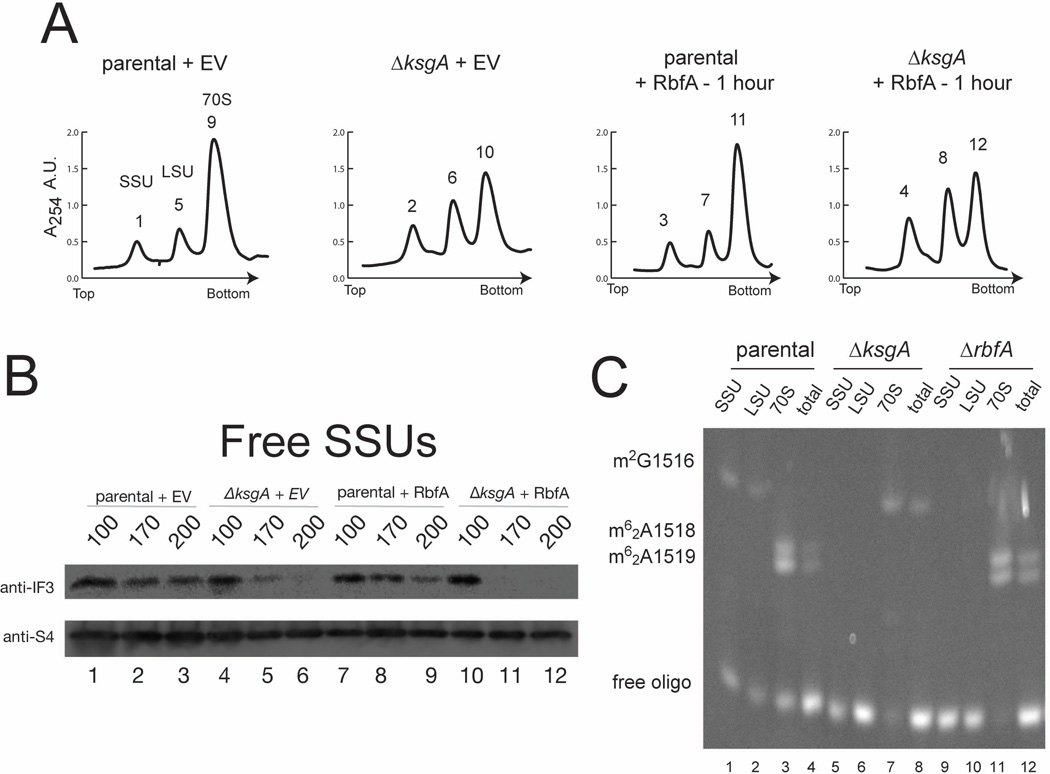
Analysis of cellular ribosomal particles allows the propensity of SSUs to bind LSUs and form ribosomes to be assessed. In both the parental strain harboring EV (strains harboring EV will be referred to as control) and overexpressing RbfA, the majority of SSUs are associated with LSUs as 70S ribosomes (Figure 2A). Overexpression of RbfA in the parental strain has little impact on A254 traces and only minimal changes in A254 traces are observed when RbfA is overexpressed in the ΔksgA strain (Figure 2A). This latter result is somewhat surprising given the extreme toxicity and impaired SSU rRNA maturation when RbfA is overexpressed in ΔksgA (Figure 1D). In contrast, when KsgA-E66A, a catalytically inactive, biogenesis defective form of KsgA is overexpressed, 70S ribosomes substantially decrease in the ΔksgA and parental strains (Connolly et al., 2008) (Supplemental Figure 2A). These results support the hypothesis that RbfA and KsgA have distinct yet coordinated roles in SSU biogenesis.
Figure 2. Ribosomes are altered in strains overexpressing RbfA but lacking KsgA.
A) Sucrose sedimentation analysis of ribosomes (70S) and small and large ribosomal subunits (SSU and LSU, respectively) from parental and ΔksgA strains harboring empty vector (EV), and parental and ΔksgA strains both overexpressing RbfA for 1 hour. Peaks are labeled for later analysis (see Figure 5A and B). B) Western analysis for IF3 (anti-IF3) and S4 (anti-S4) association to SSUs that were purified though sucrose gradients containing 100, 170 or 200 mM NH4Cl prior to analysis. C) Primer extension analysis of SSU, LSU, 70S ribosome and total RNA from parental, ΔksgA, and ΔrbfA strains. 5’ Cy5 labeled primer initiates transcription at position C1521 (5’-TAAGGAGGTATCCAACCGCAG-3’) and stops indicate m2G1516, m62A1518 and m62A1519 modifications as labeled.
The large 70S population in the ΔksgA + RbfA strain was surprising given the significant impact on growth. Generally, subunit joining is governed by initiation complex formation (Kaczanowska & Ryden-Aulin, 2007). Initiation Factor 3 (IF3) is the first initiation factor to bind free SSUs (Milon et al., 2012), and to sequester this population from associating with LSUs prior to start site selection and initiator tRNA binding. One possible explanation for the presence of 70S particles (Figure 2A) that cannot support growth (see Figure 1 A and B) in the ΔksgA + RbfA strain could be impaired IF3 binding. Thus, association of IF3 to free SSUs in the parental and ΔksgA strains +/− RbfA overexpression was analyzed. While the proportion of immature and mature SSU rRNA is similar between control parental and ΔksgA strains (Figure 1D), the association of IF3 with free SSUs is substantially reduced in the ΔksgA control strain (Figure 2 compare lanes 1–3 with lanes 4–6); the association is further reduced when RbfA is overexpressed in ΔksgA strains (Figure 2B, compare lanes 4–6 with 10–12). These results suggest that KsgA and/or the methylations catalyzed by KsgA play an important role in transitioning SSUs to the translationally active form and that in the absence of KsgA excess RbfA further perturbs this process.
The requirement for KsgA when RbfA is overexpressed suggests that functions of RbfA and KsgA are linked. We next wanted to determine if RbfA was required for KsgA activity. Therefore rRNA was isolated from parental, ΔksgA, and ΔrbfA ribosomal particles and primer extension was performed to assess A1518 and A1519 methylation in SSU and 70S rRNA as well as total RNA. While rRNA isolated from free SSUs from each strain lack methylations at A1518 and A1519 (Figure 2C, lanes 1,5, and 9), these modifications are apparent on rRNA isolated from 70S ribosomes and total RNA from parental and ΔrbfA strains (Figure 2C, lanes 3 and 11, and 4 and 12, respectively). As expected the modifications are not present on rRNA isolated from ribosomes in ΔksgA. These data indicate that RbfA is not required for methylation of A1518 and A1519 by KsgA and may imply that RbfA acts downstream of KsgA methylation.
Ribosome subunit association is less stable when RbfA is overexpressed in ΔksgA
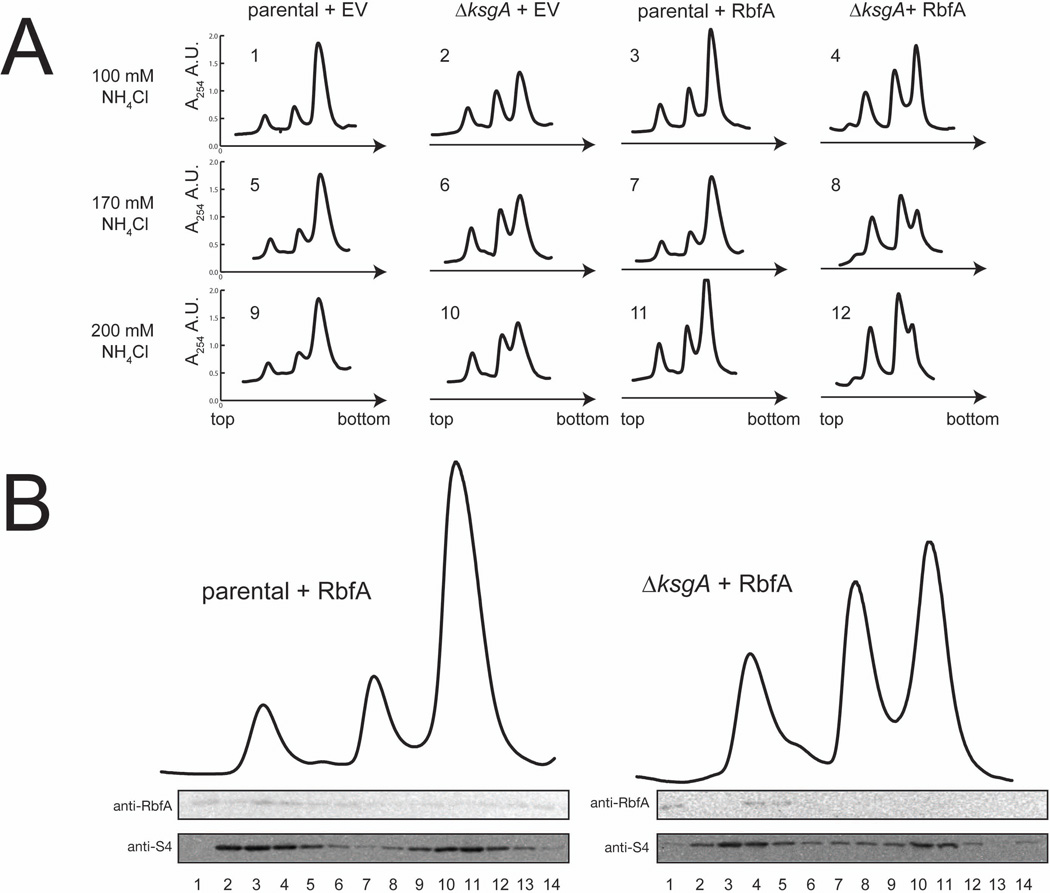
Given that the ribosomal particles formed when RbfA is overexpressed in the ΔksgA strain contain a significant level of 17S rRNA and that decreased affinity for IF3 is observed for the SSUs formed in this strain, the stability of the 70S particles was examined. The 70S ribosomes produced in parental and ΔksgA control strains and the RbfA overexpressing parental strain do not readily dissociate upon exposure to increasing salt concentrations (Figure 3A). However, 70S particles produced in the RbfA overexpressing ΔksgA strain dissociate as NH4Cl concentrations are increased from 100 mM to 200 mM (Figure 3A). As a result, there are a greater proportion of SSUs and LSUs in the free form at 200 mM NH4Cl in the biogenesis compromised strain compared to the controls (Figure 3A). These data suggest that when RbfA is overexpressed in the absence of the KsgA checkpoint, rRNA maturation and possibly other SSU biogenesis events are bypassed resulting in formation of less stable ribosome-like particles that are not competent to support growth.
Figure 3. Ribosomes produced when RbfA is overexpressed in ΔksgA exhibit changes in subunit dissociation but not in RbfA localization.
A) Ribosomes and ribosomal subunits from parental and ΔksgA strains harboring empty vector (parental + EV and ΔksgA + EV, respectively) and parental and ΔksgA strains overexpressing RbfA (parental + RbfA and ΔksgA + RbfA, respectively) were resolved by sucrose sedimentation in buffer containing 100, 140 or 200 mM NH4Cl. Ribosomes and ribosomal subunits used for later analysis were isolated from gradients containing 100 mM NH4Cl. B) Western analysis of total protein isolated from sucrose gradient fractions corresponding to ribosomal particles from parental and ΔksgA strains overexpressing RbfA using antibodies recognizing RbfA and S4 (as control).
RbfA dissociates from pre-30S particles prior to association with 50S subunits
RbfA binds at a juncture of the head and body on the interface side of the SSU, a position that would be occupied by the LSU in the 70S ribosome (Datta et al., 2007); therefore if RbfA were to remain bound to SSUs, it could result in less stable subunit association and thus explain the observed dissociation differences in the 70S particles formed when RbfA is overexpressed in the absence of KsgA compared to controls. We assessed the presence of RbfA in different ribosomal particle fractions from the parental and ΔksgA strains following RbfA overexpression (Figure 3B). In the parental strain RbfA comigrates with SSUs but does not appear to significantly associate with 70S ribosomes (Figure 3B, compare lanes 2–4 with 10–12), consistent with cryo-EM positioning of RbfA at the subunit interface (Datta et al., 2007). When fractions from sucrose sedimentation gradients were collected from the ΔksgA + RbfA strain, RbfA again comigrates with free SSUs but does not appear to be associated with 70S ribosomes (Figure 3B, compare lanes 2–5 with 10–12). These data suggest that RbfA does not remain bound to SSUs in 70S-like complexes and thus changes in dissociation properties must have another cause.
Ribosomal-like particles formed when RbfA is overexpressed in ΔksgA exhibit impaired translation
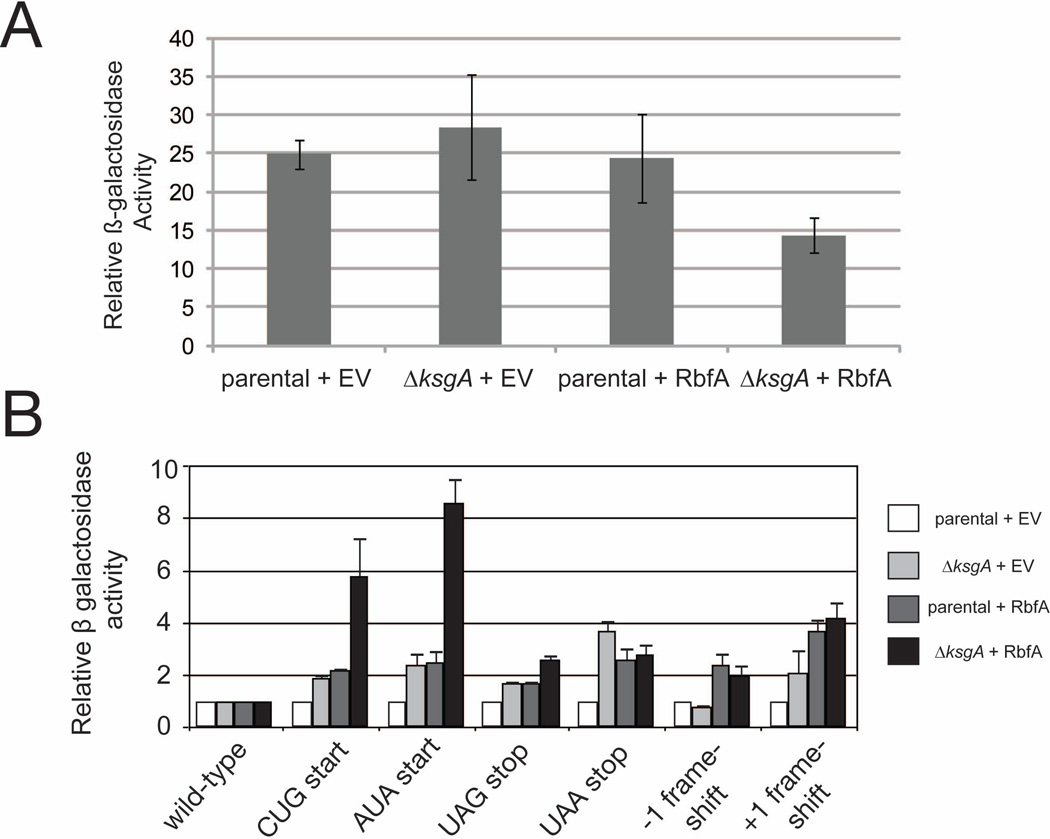
Given that the 70S particles formed in ΔksgA when RbfA is overexpressed have different dissociation properties from control ribosomes, we next sought to determine the translational capacity of these strains, shortly after induction of RbfA overexpression, using a β-galactosidase activity assay (O'Connor et al., 1997) (Figure 4A). While the parental and ΔksgA control strains and the parental strain overexpressing RbfA exhibit similar levels of β-galactosidase production, the ΔksgA strain overexpressing RbfA shows reduced levels of β-galactosidase production (Figure 4A). Therefore, while there is a large population of 70S-like particles observed in sucrose sedimentation experiments (Figure 2A), these particles exhibit reduced stability of association (Figure 3A) and translational capacity (Figure 4A). Moreover, these changes can be observed shortly after RbfA expression is induced.
Figure 4. Ribosomes produced when RbfA is overexpressed in the ΔksgA strain have altered translational properties.
A) Normalized β-galactosidase activity (in Miller units) was measured using a plasmid-bourne reporter construct (O'Connor et al., 1997) in parental and ΔksgA strains harboring empty vector or overexpression vector for RbfA and are expressed. B) Fidelity of translation at the initiation (abherent CUG and AUA start codons), termination (premature UAG and UAA stop codon) and elongation (−1 frameshift and +1 frameshift) stages was assayed in parental and ΔksgA strains harboring empty vector (parental + EV and ΔksgA + EV, respectively) and parental and ΔksgA strains overexpressing RbfA (parental + RbfA and ΔksgA + RbfA, respectively). Production of plasmid-encoded β galactosidase, assayed by calorimetric assay, requires read-through of the indicated aberrant codon (O'Connor et al., 1997).
To understand the change in translational output and the subsequent death, fidelity of translation initiation, elongation and termination were examined using a series of plasmids with aberrant start, stop or frameshift codons incorporated early in the coding sequence for a β-galactosidase reporter gene (O'Connor et al., 1997) (Figure 4B). While slight changes in read-through and frameshifting are observed in the ΔksgA control strains and the parental and ΔksgA strains overexpressing RbfA compared to the parental control strain, translation initiation is profoundly compromised in the ΔksgA strain one hour after RbfA induction (Figure 4B). It is likely that these defects arise as immature SSUs accumulate and non-functional 70S particles begin to form. These data further support that changes in translation initiation, subunit association, and translational capacity can occur rapidly when SSU biogenesis is compromised by this imbalance in factors.
Overexpression of RbfA in the absence of KsgA results in incorporation of immature SSUs in 70S-like complexes
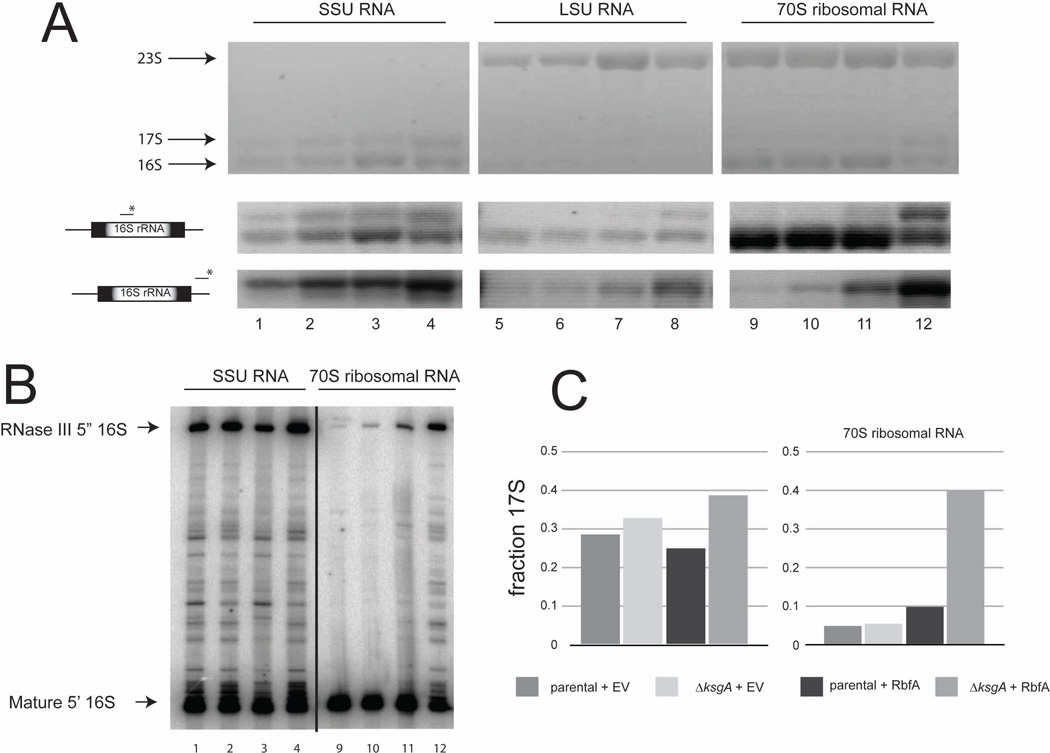
To further dissect the differences in the ΔksgA + RbfA strain compared to other strains the extent of rRNA maturation in specific ribosomal particles was examined (Figure 5A; Note: lane numbers in Figures 5A and 5B correspond to labeled peaks in Figure 2A). SSU rRNA is processed sequentially from a primary rRNA transcript that contains the mature rRNA sequences as well as transcribed spacer sequences (Srivastava & Schlessinger, 1990) (Supplemental Figure 1C) and the major SSU rRNA processing species and LSU rRNAs can be resolved by gel electrophoresis. Northern analysis with probes to mature 16S rRNA and the 3’ trailer confirm the identity of specific rRNA species (Figure 5A). The length of the 5’ leader was determined using primer extension analysis (Figure 5B). SSU rRNA extracted from control parental or ΔksgA 70S ribosomal particles were found to contain mostly mature 16S rRNA (Figure 5A, lanes 9 and 10). Surprisingly, while A254 traces from the ΔksgA strain overexpressing RbfA are similar to the ΔksgA control strain, these 70S-like particles are composed of a significant portion of 17S rRNA (Figure 5A, lane 12 and Figure 5C). In contrast, when KsgA-E66A, a catalytically inactive form of KsgA, which has also been shown to impair SSU biogenesis, is overexpressed, the population of 70S ribosomes substantially decreases in the ΔksgA strain and are more modestly reduced in the parental strain (Connolly et al., 2008) (Supplemental Figure 2A). Although there is only a small population of 70S ribosomes when KsgA-E66A is overexpressed in the absence of endogenous KsgA, these 70S particles contain immature SSU rRNA (Supplemental Figure 2B). We have previously observed a modest accumulation of 17S rRNA in 70S ribosomes produced in a strain with a mutation in r-protein S5 (Roy-Chaudhuri et al., 2010); this S5 mutant strain also exhibited decreased viability and translation fidelity (Roy-Chaudhuri et al., 2010). Thus in several strains carrying mutations that alter SSU biogenesis, immature SSU rRNA accumulates in 70S particles and this accumulation correlates with changes in function. These findings support the hypothesis that SSU rRNA maturation is linked to appropriate biogenesis and formation of functional ribosomes and that KsgA and RbfA are both involved in these processes in vivo.
Figure 5. SSU rRNA maturation defects are associated with RbfA overexpression in ΔksgA.
A) RNA isolated from sucrose sedimentation peaks shown in Figure 2 was analyzed by ethidium bromide staining (top panel) and northern analysis using an internal 16S rRNA (middle panel) and 3’ trailer probe (bottom panel). Lane number corresponds to peak labeling in Figure 2. B) Primer extension analysis of the 5’ end of SSU rRNA from RNA as in (A). Primer that initiates extension at position 50 of 16S rRNA was used. C) Quantitation of 17S rRNA fraction of SSU rRNA in (A).
Ribosome-like particles produced when RbfA is overexpressed in the absence of KsgA have reduced levels of ribosomal protein S21
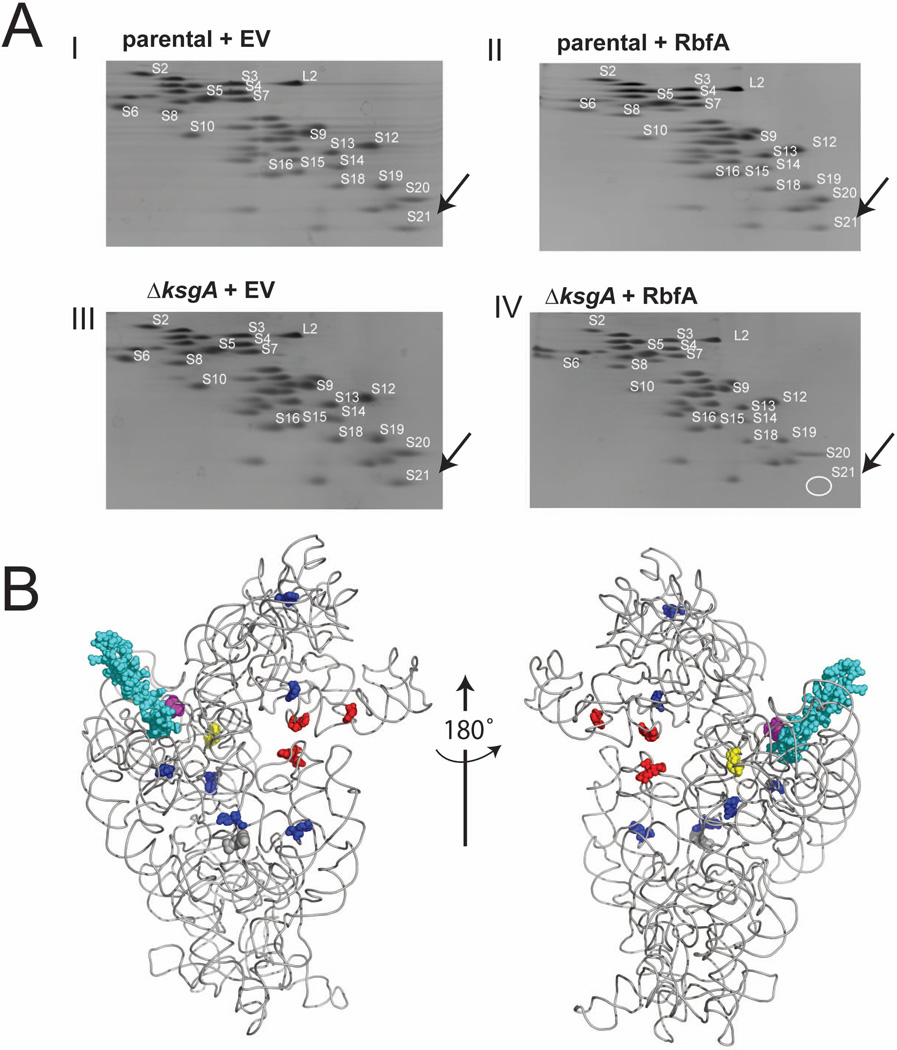
To further understand the changes in the 70S-like particles the ribosomal protein (r-protein) content of 70S or 70S-like particles was examined using 2D gel electorphoresis (Figure 6A I-IV). While the r-protein content is globally similar for each strain (Figure 6A, I-IV), r-protein S21 is underrepresented in 70S-like particles isolated from the ΔksgA + RbfA strain (Figure 6A, IV). The mature 3’ end of 16S rRNA is adjacent to S21 (see Figure 6B) and given this proximity to the 3’ trailer sequence it seems possible that S21 does not stably associate prior to 3’ end maturation. The reduction in S21 was particularly interesting given that S21 plays a role in SSU and LSU joining and anti-Shine-Dalgarno availability during translation initiation (van Duin et al., 1972). Our findings suggest that structural requirements for appropriate translation initiation are not met during SSU biogenesis when RbfA is overexpressed in the absence of KsgA.
Figure 6. 70S-like particles exhibit reduced r-protein S21 and altered structure.
A) Ribosomal proteins from sucrose gradient purified 70S ribosomes were extracted from I) parental + EV, II) parental + RbfA, III) ΔksgA + EV and IV) ΔksgA + RbfA (see Figure 2A-D for representative profiles) and resolved on 2-D urea-acid gels (Geyl et al., 1981). SSU r-proteins are labeled in white according to previous studies (Geyl et al., 1981) with LSU r-protein L2 (1 ug added post-purification to all samples as a loading control) labeled to show relative abundance of LSU and SSU r-protein. This circle indicates the area where S21 should run in this system. B) Solvent exposed (left) and interface (right) sides of the SSU X-ray crystal structure (Schuwirth et al., 2005) (pdb: 2AVY) are pictured with r-protein S21 shown in cyan. The 3’ end of 16S rRNA is shown in purple and methylations catalyzed by KsgA are shown in yellow. These figures were produced using Pymol (www.pymol.org). DMS probing (Supplemental Figure 3) reveals altered reactivity of residues both proximal (residues C518, C519 A1055 and A1046; highlighted in red) and distal (residues highlighted in blue) to the translational active site.
Next, we assessed structural differences between 70S(-like) particles using chemical probing of functional (ΔksgA + empty vector: control) and mal-functional (ΔksgA + RbfA) ribosomes (Figure 6B and Supplemental Figure 3). We identified a number of residues with altered reactivity between the two strains near functional regions of the SSU. In particular, residues C518, C519, A1055 and A1046 (Figure 6B, red, Supplemental Figure 3) reside in a region that is directly involved in translation. Residues A7, A546, A872, A889, C1059 and C1149 are peripheral to the active site of the ribosome but may indicate an altered conformation that results in less stable ribosomes (Figure 6B). Interestingly these residues appear to be distally located to S21 (Figure 6B), which is highly underrepresented in 70S-like particles from the ΔksgA + RbfA strain. These data suggest there are significant differences between 70S and 70S-like ribosomal particles. However, the overall similarity, in terms of structure and composition of these particles, indicates that proper biogenesis has occurred to a given point in the cascade. Taken together, the altered SSU conformation of 70S-like particles and impaired fidelity of translation initiation suggest that structural requirements are not achieved when there is an imbalance of KsgA and RbfA.
Discussion
The balance of ribosome biogenesis factors KsgA and RbfA appears to be required for appropriate ribosome maturation and function and ultimately for cell growth. Traditionally bacterial ribosome biogenesis has been difficult to study due to redundancy in this cascade, thus we still have an incomplete view of the steps required for the formation of the bacterial ribosome [see (Connolly & Culver, 2009)]. Given the intricate dynamics of ribosome biogenesis factor association, previous studies have taken advantage of the deleterious effects of overexpression of ribosome biogenesis factors to parse out assembly events (Connolly et al., 2008, Campbell & Brown, 2008). This current study used overexpression of RbfA in the absence of the KsgA checkpoint to examine properties of ribosome biogenesis that are perturbed when the KsgA checkpoint is removed and the ratio of SSUs to biogenesis factors is disrupted. The KsgA checkpoint is important for preventing premature incorporation of SSU particles in the translating population (Connolly et al., 2008, Xu et al., 2008); RbfA is a SSU biogenesis factor of unknown function, but whose deletion leads to a significant growth defect (Dammel & Noller, 1993, Dammel & Noller, 1995). Previously, it was demonstrated that even minor incorporation of immature SSU rRNA into 70S populations compromises ribosome function (Roy-Chaudhuri et al., 2010). These ribosomes with mutations in r-protein S5 have altered structures near the 5’ end of 16S rRNA, which is the region of 16S rRNA that RbfA genetic interacts with (Dammel & Noller, 1995). These findings suggest that when additional pressure is applied to the SSU biogenesis cascade (as in the case of RbfA overexpression), the importance of the KsgA checkpoint is amplified. Other studies are also consistent with our observations of a link between RbfA and KsgA function in vivo; work investigating genetic interactions of ribosome biogenesis factors revealed that deletion of both rsgA and ksgA resulted in an exacerbated growth defect when compared to strains where either gene was singly deleted (Campbell & Brown, 2008). RsgA has been shown to be important for the removal of RbfA from the developing SSU (Goto et al., 2010). We have observed that overexpression of RsgA in the ksgA deletion strain yields a similar growth phenotype as observed with RbfA overexpression (Supplemental Figure 4). Thus the toxicity of excess production of SSU biogenesis factors in ΔksgA extends beyond RbfA to other related factors (Supplemental Figure 4). These studies also provide further support that the roles of KsgA and RbfA are linked in vivo.
Although it is unclear how overexpression of RbfA in the absence of KsgA prevents final SSU rRNA maturation and association of S21. The observation that S21 was vastly underrepresented in the 70S peak when RbfA is overexpressed in the ΔksgA strain also lends insight into the functional deficit of the 70S-like particles. S21 has been implicated as a critical component of initiation competent SSUs (Held et al., 1974, van Duin et al., 1972, Van Duin & Wijnands, 1981). Interestingly, S21 is required for in vitro translation of MS2 mRNA but not for translation of poly-U RNA transcripts (Van Duin & Wijnands, 1981). The changes in translation initiation specificity that we observe in the ΔksgA stain overexpressing RbfA (Figure 4B) are reminiscent of these reported differences. Thus our findings are consistent with suggestions that S21 is important for canonical translation initiation, which requires interaction between the Shine-Dalgarno (SD) of mRNA and anti-SD at the 3‘ end of 16S rRNA (Van Duin & Wijnands, 1981). In fact SSUs lacking S21, but otherwise mature, are unable to bind a SD mimic (Backendorf et al., 1981) (KC and GMC, unpublished results). Futhermore, S21 inhibits methylation by KsgA suggesting that methylation precedes the association of S21 (Thammana & Held, 1974). In these 70S-like particles, approximately one-half of the SSU rRNA is mature but S21 is substantially reduced, suggesting that the absence of S21 is the critical element for appropriate translation initiation. It is still unclear if the structural differences we observe (see Figure 6B and Supplemental Figure 3) are due to lack of methylation, impaired SSU rRNA maturation and/or the association of S21. Also, it is difficult to determine if the changes in initiation and overall translational capacity (see Figure 4) is sufficient to cause the observed toxicity or if this is an initial response that is followed by more catastrophic changes. However, the aggregate effect observed in the ΔksgA strain overexpressing RbfA is the formation of a large population of 70S-like ribosomes that are not competent to support growth. These findings suggest that perturbation of specific combinations of E. coli SSU biogenesis factors can be lethal [also see (Campbell & Brown, 2008)] and thus supports the hypotheses that this process could be used for antimicrobial development.
Do these 70S-like particles represent a normal stage in SSU biogenesis? The accumulation of a “70S-like” population of ribosomes in cells where the normal ribosome biogenesis cascade is perturbed raises a number of hypotheses regarding their functional significance. These particles could represent an accumulation of nonfucntional (as a result of improper biogenesis) ribosomes, aspecific state of sequestration of immature subunits, or on path intermediates of the ribosome biogenesis pathway.
One hypothesis is that these particles are aggregates that form when SSU biogenesis is stalled. This sequestration of particles in non-functional complexes could be a means of halting assembly and thereby translation until conditions improve. This would be analogous to conversation of ribosomes into non-functional dimers in stationary phase (Wada et al., 1990). Similar to our findings, structural studies suggest that the anti-SD/SD helix cannot form in these dimers and that translation initiation is thus impaired (Polikanov et al., 2012). These particles are therefore not actively recycled but are “stored” until translational requirements increase thus requiring an increased population of mature subunits. This would allow a rapid increase in functional ribosomes in response to changing conditions.
Another hypothesis would suggest that the association of SSU and LSUs occurs during the ribosome biogenesis cascade and there are data from the literature which support the existence of a 70S-like particle that is formed during SSU biogenesis (see Figure 7). First, it has been suggested that final maturation of 16S rRNA occurs after subunit association; pulse-chase experiments indicated that 17S rRNA was incorporated into 70S(-like) particles in wild-type strains (Mangiarotti et al., 1974). No mechanism for resolving such particles has been proposed but our data suggest that IF3 could fulfill such a role. Generally, IF3 keeps free SSUs and LSUs, which have a high affinity for one another, separated until the appropriate initiation complex has formed on the SSU (Dallas & Noller, 2001, Petrelli et al., 2001). The decreased binding of IF3 to pre-SSUs observed in this work could account for changes in association of pre-SSUs with LSUs when biogenesis is perturbed in the ΔksgA strain overexpressing RbfA. A mutant infC (the gene coding for IF3) allele, infC135, is especially toxic when combined with deletion of ksgA, further supporting an in vivo link between ribosome biogenesis and the translation initiation system (Seshadri et al., 2009). Some of the regions of the 70S-like particles from ΔksgA + RbfA that are structurally altered (see Figure 6B and Supplemental Figure 3) are sites targeted by IF3 in directed probing experiments (Dallas & Noller, 2001); thus changes in IF3 affinity could be linked to late biogenesis events and to the mechanism of resolution of the 70S-like particles. Understanding the timing of SSU rRNA maturation and S21 association also could be critical. The lack of S21 binding until after final processing and release would insure that these precursor molecules do not bind mRNA by preventing exposure of the anti-SD sequence (Figure 7C).
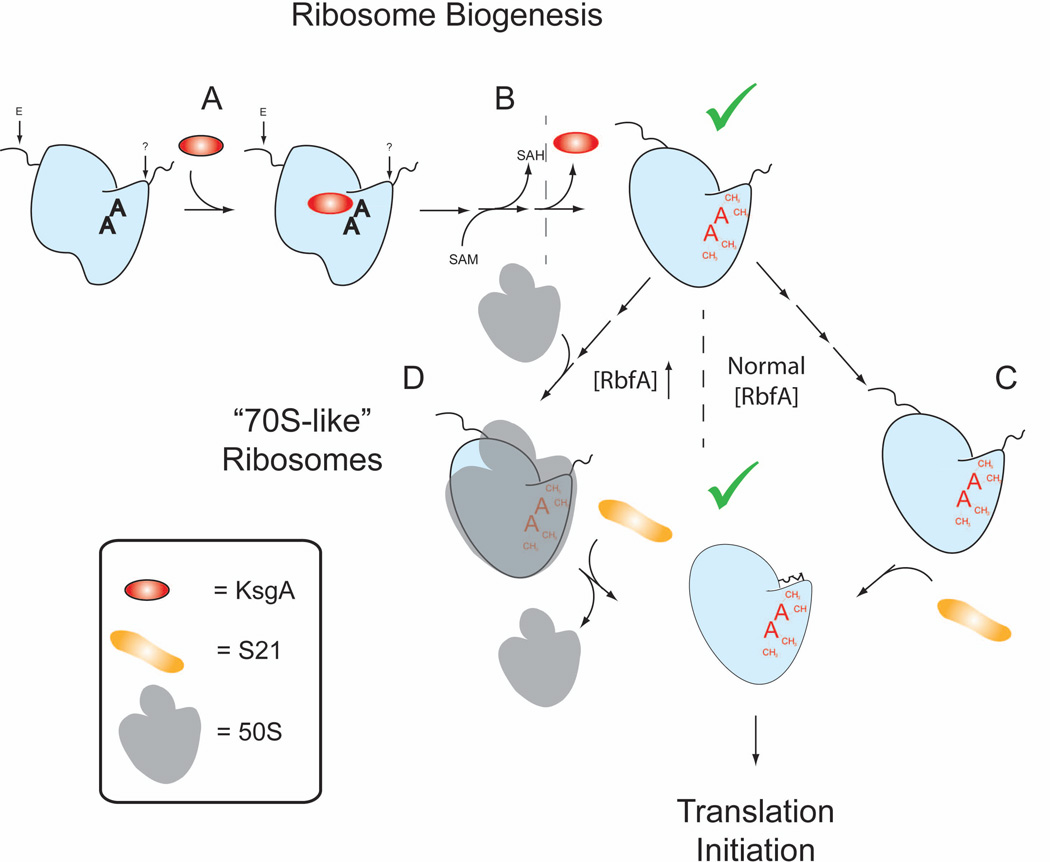
Figure 7. A multi-stage checkpoint system at work during SSU maturation and incorporation into functional ribosomes.
A) KsgA associates with a SSU biogenesis intermediate and once a certain level of maturation has occurred and B) KsgA methylates A1518 and A1519 residues of 16S rRNA thus prompting its release and serving as a checkpoint (green checkmark) within the SSU maturation cascade (Connolly et al., 2008). Following methylation by KsgA two competing pathways facilitate an additional checkpoint in SSU maturation prior to incorporation into the translation cycle. C) In one pathway association of S21 exposes the anti Shine-Dalgarno region of 16S rRNA thus making SSUs competent for translation initiation. D) In another pathway, which is induced by excess RbfA, 50S subunits associate with immature SSUs to produce “70S-like” particles, which are then either act as a repository for nearly mature SSUs and would under appropriate conditions be further processed into mature particles.
One final hypothesis predicts that association with LSUs is a structural checkpoint for SSU biogenesis and that only if the requirements of this checkpoint could be met would final processing/maturation occur. In eukaryotic systems, ‘80S-like’ particles composed of LSUs and immature SSUs have been observed (Lebaron et al., 2012, Strunk et al., 2012). Moreover, links between translation initiation factors and these 80S-like particles have been revealed (Strunk et al., 2012). It appears that the observations we have made in the ΔksgA + RbfA strain are consistent with a link between translation initiation and the final stages of SSU maturation. Taken together, our data reveal that KsgA methylation, SSU rRNA maturation, S21 association, RbfA function and LSU joining likely represent a detailed system that prevents incorporation of immature SSUs or malformed SSUs from entering in the translation cycle.
Experimental Procedures
Growth Experiments
BW2113 (parental) and JW0050 (∆ksgA) strains (Baba et al., 2006) harboring either empty pQE-80 vector (Qiagen) or pQE-80 vector with cloned rbfA (pQE-RbfA), rimJ or rng according to manufacturer’s protocol were grown overnight in the presence of 100 µg/mL ampicillin in LB media until saturated in culture. For dilution plating experiments overnight culture was diluted in 10-fold increments and plated on LB plates containing 100 µg/mL ampicillin or 100 µg/mL ampicillin and 1mM IPTG. Plates were incubated at 37 °C for ~7–8 hours. For growth in liquid media saturated culture was diluted to an OD600 of 0.02 and grown with shaking (200 r.p.m.). Time points were taken during growth and growth curves were generated by measuring optical density at 600 nm (OD600) in a standard spectrophotometer. Overexpression of both KsgA and RbfA simultaneously was done by transforming ASKA ksgA plasmid (Kitagawa et al., 2005) in ∆ksgA harboring pQE-RbfA. This growth was performed as described above with the exception that 34 µg/mL chloramphenicol was added to the media.
Cells grown for sucrose sedimentation profiles were grown in this same manner with the exception that they were harvested one hour following the addition of inducer by chilling cells on ice, pelleting cells, and resuspending cells in 1/100 volume polysome lysis buffer [10mM Tris-HCl (pH 7.8) and 15 mM MgCl2] containing 1 mg/ml lysozyme (Sigma) and protease inhibitor (Roche). The cells were then ‘quick froze’ in liquid nitrogen and stored at −80 °C.
RNA Extraction and Analysis
RNA was extracted from cell lysate or sucrose gradient fractions as described previously (Connolly et al., 2008). 10 µg total RNA (Figure 1D), 1 µg SSU rRNA, 2 µgs LSU rRNA and 3 µgs 70S rRNA (Figure 2 G) were loaded on a 2% agarose gel containing formaldehyde (Sambrook J., 1989). RNA was transferred to Genescreen® membrane according to manufacturer’s protocol. Membrane was then probed with oligos [mature 16S - (5’-CGCATTTCACCGCTACA-3’) and 3’ trailer - (5’-GCACTGCAAAGTACGCTTC)], labeled with T4 Polynucleotide Kinase (New England Biolabs) according to manufacture’s protocol, using UltraHyb-Oligo (Ambion®) Buffer System as described by manufacturer. Quantification of bands was performed using Quantity One software (BioRad).
Sucrose Sedimentation Profiles
Sucrose sedimentation profiles were performed essentially as described (Connolly et al., 2008) except centrifuge time was reduced from 17 hours to 15 hours. In addition, cells were treated with Complete Protease Inhibitor Cocktail Tablets (Roche) according to manufacture’s instructions during harvest and with 10 U DNase I (Fermentas) prior to clearing of the lysate. For ribosome stability experiments (Figure 3A), sucrose sedimentation profiles were performed exactly as described above with the exception of the NH4Cl was altered as described in the figure.
β-galactosidase Activity Assay
β-galactosidase activity assays were performed as described (O'Connor et al., 1997). βgalactosidase values are given in Miller units. Miller units are defined as follows: 1 Miller unit = 1000 * [OD420 - (1.75 - OD550)]/(t * v * OD600) where OD420 = the absorbance of ο-nitrophenol, (1.75 - OD550) = scatter from cell debris at OD420, t = reaction time (mins), v = volume of culture assayed (mLs) and OD600 = cell density. The OD600 refers to the number of cells in the culture as it correlates with the colony forming units. This experiment therefore calculates the translational output (βgal expression) / # of cells. β-galactosidase activity was measured in strains harboring mutations in the AUG start codon (CUG start and AUA start), an upstream stop codon (UAG stop and UAA stop) and −1 and +1 frameshifts and normalized to expression of WT β-galactosidase activity in each strain (Figure 3B). Normalized β-galactosidase activity values are expressed in Miller units and plotted relative to the parental control strain.
β-galactosidase activity assays were also analyzed using the wild-type control and normalizing according to previously described methods (O'Connor et al., 1997) to show total translational capacity (Figure 4A).
Protein extraction and analysis
Peaks corresponding to 70S ribosome particles were collected and concentrated using 3 kD cut-off Centricons (Pall®) according to manufacturers instructions. Following concentration, 1 initial volume sucrose gradient buffer was added and concentration was repeated 4 times to allow complete exchange of buffer. 1/10 volume 1M MgCl2 and 2 volumes glacial acetic acid was added and incubated with shaking at 4 °C for 45 min. Sample was then centrifuged for 30 min at 13 K at 4 °C and supernatant was dialyzed against 1 L 2% glacial acetic acid for 18 hours with 3 buffer exchanges before being dried under vacuum. Dried samples were resuspended in Geyl loading buffer (Geyl et al., 1981) prior to further analysis.
Protein gels
1-D Tris-Tricine gels were prepared and ran as described (Schagger, 2006). Gels were transferred to PVDF membrane (Bio-Rad) according to manufacture’s protocol. Blot was probed using antibodies recognizing RbfA (Figure 1D) or IF3 (Figure 2I). An antibody recognizing r-protein S4 was used at a loading control in western experiments. Incubation of membrane with each antibody was followed by detection with a goat anti-rabbit secondary antibody as described previously for S3 antibody (Connolly et al., 2008).
A previously reported 2-D gel system (Geyl et al., 1981) was modified as described below. First dimension 4% gels were cast using glass capillary tubes and set up according manufacture’s protocol using the Mini-PROTEAN® 2-D Electrophoresis Cell and 70 µgs extracted protein loaded on the top of each gel. One-half volume of Geyl loading dye with 1–2 drops 0.5mg/ml basic fucshin per 10 mLs was added and the first dimension was ran with reported buffer system (Geyl et al., 1981) at 200V until the dye just exited the tube. The gel was extruded from the capillary tube and placed on 18% 1 mm slab gels (Bio-Rad, mini-gel casting system). A small layer of Geyl loading dye with basic fucshin was loaded on top and the gels were again ran according as previously reported (Geyl et al., 1981) until the dye just exited the gel. Gels were stained with Coomassie R-250 in 15% formaldehyde.
Primer Extension Experiments
Primer Extension experiments were performed as described (Stern et al., 1988) using primer 5’-TCGCCGCTACTGGGGAA-3’ (Figure 5B). Flourecent primer extensions were performed as above with the exception that 110 µM set dNTPs were used for the extension reaction and extension products were resolved using a 20% denaturing gel. Visualization of bands was performed using the BioRad Versa Doc system according to manufacturer’s protocol.
Isolation of ksgA mutant alleles
Wild-type BW2113 (parental) strain was grown in LB media and plated on increasing concentrations of kasugamycin (20–2,000 ug/mL) with growth in non selective liquid media between each plating. The ksgA allele was PCR amplified from kasugamycin resistant clones (up to 2,000 ug/mL) and sequenced. The ksgA-ksgR strain had a frameshift mutation near the start of the ksgA gene as follows: 5’ -nucleotide postion 60 from start - TCTCAACGATCAG - 3’ to 5’ -nt 60 from start - TCTCACGATCAG - 3’. The ksgA-ca allele had the following single nucleotide substitution: 5’ -nucleotide postion 126 from start - GTCGAAATCGGCCCCGGTCTG - 3’ to 5’ -nt 126 from start - GTCGAAATCAGCCCCGGTCTG - 3’. This mutation results in a serine substitution at the conserved G45 position, a residue that directly contacts the SAM cofactor (O'Farrell et al., 2004) likely resulting in a fully expressed (Supplemental Figure 1B) but catalytically inactive KsgA form.
Supplementary Material
Acknowledgments
The authors would like to thank Drs. Eric Phizicky, Scott Butler, and Yitao Yu for helpful discussions during manuscript preparation, as well as, Drs. Zhili Xu and Biswajoy Roy-Chaudhuri for help with technical aspects of the experiments presented in this work. The authors would also like to thank Deepika Calidas and Hiram Lyon for critical reading of this manuscript. This work was funded by NIH Grants GM062432 (to G.M.C.) as well as the T32 G068411 NIH Training Grant in Cellular, Biochemical and Molecular Sciences.
Footnotes
Author contributions
KMC performed experiments. KMC and GMC designed experiments and analyzed results and wrote the paper.
Bibliography
- 1.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006–2008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backendorf C, Ravensbergen CJ, Van der Plas J, van Boom JH, Veeneman G, Van Duin J. Basepairing potential of the 3' terminus of 16S RNA: dependence on the functional state of the 30S subunit and the presence of protein S21. Nucleic Acids Res. 1981;9:1425–1444. doi: 10.1093/nar/9.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehringer D, O'Farrell HC, Rife JP, Ban N. Structural insights into methyltransferase KsgA function in 30S ribosomal subunit biogenesis. J Biol Chem. 2012;287:10453–10459. doi: 10.1074/jbc.M111.318121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bylund GO, Wipemo LC, Lundberg LA, Wikstrom PM. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli . J Bacteriol. 1998;180:73–82. doi: 10.1128/jb.180.1.73-82.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell TL, Brown ED. Genetic interaction screens with ordered overexpression and deletion clone sets implicate the Escherichia coli GTPase YjeQ in late ribosome biogenesis. J Bacteriol. 2008;190:2537–2545. doi: 10.1128/JB.01744-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connolly K, Culver G. Deconstructing ribosome construction. Trends Biochem Sci. 2009;34:256–263. doi: 10.1016/j.tibs.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connolly K, Rife JP, Culver G. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol Microbiol. 2008;70:1062–1075. doi: 10.1111/j.1365-2958.2008.06485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell. 2001;8:855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 9.Dammel CS, Noller HF. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 1993;7:660–670. doi: 10.1101/gad.7.4.660. [DOI] [PubMed] [Google Scholar]
- 10.Dammel CS, Noller HF. Suppression of a cold-sensitive mutation in 16S rRNA by overexpression of a novel ribosome-binding factor, RbfA. Genes Dev. 1995;9:626–637. doi: 10.1101/gad.9.5.626. [DOI] [PubMed] [Google Scholar]
- 11.Datta PP, Wilson DN, Kawazoe M, Swami NK, Kaminishi T, Sharma MR, Booth TM, Takemoto C, Fucini P, Yokoyama S, Agrawal RK. Structural aspects of RbfA action during small ribosomal subunit assembly. Mol Cell. 2007;28:434–445. doi: 10.1016/j.molcel.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geyl D, Bock A, Isono K. An improved method for two-dimensional gel-electrophoresis: analysis of mutationally altered ribosomal proteins of Escherichia coli . Mol Gen Genet. 1981;181:309–312. doi: 10.1007/BF00425603. [DOI] [PubMed] [Google Scholar]
- 13.Goto S, Kato S, Kimura T, Muto A, Himeno H. RsgA releases RbfA from 30S ribosome during a late stage of ribosome biosynthesis. EMBO J. 2010 doi: 10.1038/emboj.2010.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartz D, McPheeters DS, Gold L. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev. 1989;3:1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- 15.Held WA, Nomura M, Hershey JW. Ribosomal protein S21 is required for full activity in the initiation of protein synthesis. Mol Gen Genet. 1974;128:11–22. doi: 10.1007/BF00267291. [DOI] [PubMed] [Google Scholar]
- 16.Helser TL, Davies JE, Dahlberg JE. Mechanism of kasugamycin resistance in Escherichia coli . Nat New Biol. 1972;235:6–9. doi: 10.1038/newbio235006a0. [DOI] [PubMed] [Google Scholar]
- 17.Jones PG, Inouye M. RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol Microbiol. 1996;21:1207–1218. doi: 10.1111/j.1365-2958.1996.tb02582.x. [DOI] [PubMed] [Google Scholar]
- 18.Kaczanowska M, Ryden-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 20.Klootwijk J, van den Bos RC, Planta RJ. Secondary methylation of yeast ribosomal RNA. FEBS Lett. 1972;27:102–106. doi: 10.1016/0014-5793(72)80419-8. [DOI] [PubMed] [Google Scholar]
- 21.Lafontaine D, Delcour J, Glasser AL, Desgres J, Vandenhaute J. The DIM1 gene responsible for the conserved m6(2)Am6(2)A dimethylation in the 3'-terminal loop of 18 S rRNA is essential in yeast. J Mol Biol. 1994;241:492–497. doi: 10.1006/jmbi.1994.1525. [DOI] [PubMed] [Google Scholar]
- 22.Lebaron S, Schneider C, van Nues RW, Swiatkowska A, Walsh D, Bottcher B, Granneman S, Watkins NJ, Tollervey D. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat Struct Mol Biol. 2012;19:744–753. doi: 10.1038/nsmb.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Pandit S, Deutscher MP. RNase G (CafA protein) and RNase E are both required for the 5' maturation of 16S ribosomal RNA. EMBO J. 1999;18:2878–2885. doi: 10.1093/emboj/18.10.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangiarotti G, Turco E, Ponzetto A, Altruda F. Precursor 16S RNA in active 30S ribosomes. Nature. 1974;247:147–148. doi: 10.1038/247147a0. [DOI] [PubMed] [Google Scholar]
- 25.Milon P, Maracci C, Filonava L, Gualerzi CO, Rodnina MV. Real-time assembly landscape of bacterial 30S translation initiation complex. Nat Struct Mol Biol. 2012;19:609–615. doi: 10.1038/nsmb.2285. [DOI] [PubMed] [Google Scholar]
- 26.Noon KR, Bruenger E, McCloskey JA. Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus. J Bacteriol. 1998;180:2883–2888. doi: 10.1128/jb.180.11.2883-2888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Connor M, Thomas CL, Zimmermann RA, Dahlberg AE. Decoding fidelity at the ribosomal A and P sites: influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res. 1997;25:1185–1193. doi: 10.1093/nar/25.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Farrell HC, Pulicherla N, Desai PM, Rife JP. Recognition of a complex substrate by the KsgA/Dim1 family of enzymes has been conserved throughout evolution. RNA. 2006;12:725–733. doi: 10.1261/rna.2310406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Farrell HC, Scarsdale JN, Rife JP. Crystal structure of KsgA, a universally conserved rRNA adenine dimethyltransferase in Escherichia coli . J Mol Biol. 2004;339:337–353. doi: 10.1016/j.jmb.2004.02.068. [DOI] [PubMed] [Google Scholar]
- 30.O'Farrell HC, Xu Z, Culver GM, Rife JP. Sequence and structural evolution of the KsgA/Dim1 methyltransferase family. BMC Res Notes. 2008;1:108. doi: 10.1186/1756-0500-1-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrelli D, LaTeana A, Garofalo C, Spurio R, Pon CL, Gualerzi CO. Translation initiation factor IF3: two domains, five functions, one mechanism? EMBO J. 2001;20:4560–4569. doi: 10.1093/emboj/20.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polikanov YS, Blaha GM, Steitz TA. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012;336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy-Chaudhuri B, Kirthi N, Culver GM. Appropriate maturation and folding of 16S rRNA during 30S subunit biogenesis are critical for translational fidelity. Proc Natl Acad Sci U S A. 2010;107:4567–4572. doi: 10.1073/pnas.0912305107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, F FE, Maniatis T. Molecular Cloning: A Laboratory Manual. second edition. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Schagger H. Tricine-SDS-PAGE. Nat Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 36.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 37.Seshadri A, Dubey B, Weber MH, Varshney U. Impact of rRNA methylations on ribosome recycling and fidelity of initiation in Escherichia coli. Mol Microbiol. 2009;72:795–808. doi: 10.1111/j.1365-2958.2009.06685.x. [DOI] [PubMed] [Google Scholar]
- 38.Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu Rev Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 39.Srivastava AK, Schlessinger D. Mechanism and regulation of bacterial ribosomal RNA processing. Annu Rev Microbiol. 1990;44:105–129. doi: 10.1146/annurev.mi.44.100190.000541. [DOI] [PubMed] [Google Scholar]
- 40.Steege DA, Graves MC, Spremulli LL. Euglena gracilis chloroplast small subunit rRNA. Sequence and base pairing potential of the 3' terminus, cleavage by colicin E3. J Biol Chem. 1982;257:10430–10439. [PubMed] [Google Scholar]
- 41.Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 42.Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL, 3rd, Karbstein K, Skiniotis G. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strunk BS, Novak MN, Young CL, Karbstein K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150:111–121. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sussman JK, Simons EL, Simons RW. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol Microbiol. 1996;21:347–360. doi: 10.1046/j.1365-2958.1996.6371354.x. [DOI] [PubMed] [Google Scholar]
- 45.Thammana P, Held WA. Methylation of 16S RNA during ribosome assembly in vitro. Nature. 1974;251:682–686. doi: 10.1038/251682a0. [DOI] [PubMed] [Google Scholar]
- 46.Van Buul CP, Hamersma M, Visser W, Van Knippenberg PH. Partial methylation of two adjacent adenosines in ribosomes from Euglena gracilis chloroplasts suggests evolutionary loss of an intermediate stage in the methyl-transfer reaction. Nucleic Acids Res. 1984;12:9205–9208. doi: 10.1093/nar/12.23.9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Duin J, van Knippenberg PH, Dieben M. Functional heterogeneity of the 30S ribosomal subunit of Escherichia coli. II. Effect of S21 on initiation. Mol Gen Genet. 1972;116:181–191. doi: 10.1007/BF00582227. [DOI] [PubMed] [Google Scholar]
- 48.Van Duin J, Wijnands R. The function of ribosomal protein S21 in protein synthesis. Eur J Biochem. 1981;118:615–619. doi: 10.1111/j.1432-1033.1981.tb05563.x. [DOI] [PubMed] [Google Scholar]
- 49.Wada A, Yamazaki Y, Fujita N, Ishihama A. Structure and probable genetic location of a"ribosome modulation factor" associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc Natl Acad Sci U S A. 1990;87:2657–2661. doi: 10.1073/pnas.87.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia B, Ke H, Shinde U, Inouye M. The role of RbfA in 16S rRNA processing and cell growth at low temperature in Escherichia coli. J Mol Biol. 2003;332:575–584. doi: 10.1016/s0022-2836(03)00953-7. [DOI] [PubMed] [Google Scholar]
- 51.Xu Z, O'Farrell HC, Rife JP, Culver GM. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat Struct Mol Biol. 2008;15:534–536. doi: 10.1038/nsmb.1408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.