Abstract
Spt10 is a putative acetyltransferase of Saccharomyces cerevisiae that directly activates the transcription of histone genes. Deletion of SPT10 causes a severe slow growth phenotype, showing that Spt10 is critical for normal cell division. To gain insight into the function of Spt10, we identified mutations that impair or improve the growth of spt10 null (spt10Δ) mutants. Mutations that cause lethality in combination with spt10Δ include particular components of the SAGA complex as well as asf1Δ and hir1Δ. Partial suppressors of the spt10Δ growth defect include mutations that perturb cell-cycle progression through the G1/S transition, S phase, and G2/M. Consistent with these results, slowing of cell-cycle progression by treatment with hydroxyurea or growth on medium containing glycerol as the carbon source also partially suppresses the spt10Δ slow-growth defect. In addition, mutations that impair the Lsm1-7−Pat1 complex, which regulates decapping of polyadenylated mRNAs, also partially suppress the spt10Δ growth defect. Interestingly, suppression of the spt10Δ growth defect is not accompanied by a restoration of normal histone mRNA levels. These findings suggest that Spt10 has multiple roles during cell division.
Keywords: Spt10, Spt21, histones, suppressors
The Saccharomyces cerevisiae Spt10 protein plays important roles in gene expression and growth. Mutations in the SPT10 gene have been identified in many different ways, including as suppressors of the transcriptional defects caused by Ty and Ty LTR insertion mutations (Fassler and Winston 1988; Natsoulis et al. 1991), suppressors of glucose repression of ADH2 (Denis and Malvar 1990), and suppressors of loss of an upstream activation sequence (Prelich and Winston 1993; Yamashita 1993). Several subsequent studies have demonstrated that Spt10 is a site-specific DNA binding protein that binds cooperatively at the regulatory regions of the four S. cerevisiae histone loci where it activates transcription (Dollard et al. 1994; Eriksson et al. 2005, 2011; Hess et al. 2004; Mendiratta et al. 2006, 2007; Xu et al. 2005). DNA binding is dependent upon both a zinc finger domain and an adjacent region required for cooperative binding (Mendiratta et al. 2006, 2007). Spt10 also plays a negative role in histone gene transcription, as it is required for repression of several histone loci outside of S phase (Sherwood and Osley 1991). An intriguing feature of the Spt10 amino acid sequence is a conserved acetyltransferase domain (Neuwald and Landsman 1997). Although this domain is required for Spt10 function (Hess et al. 2004), no acetyltransferase activity or acetyltransferase substrates have yet been identified for Spt10, despite efforts by several laboratories.
The SPT21 gene is functionally related to SPT10. Mutations in SPT21 were isolated in two of the same mutant selections as mutations in SPT10 (Natsoulis et al. 1991; Prelich and Winston 1993), including one large-scale selection that identified only these two genes (Natsoulis et al. 1991). In addition, mutations in SPT21 appear to cause the same pattern of histone locus transcription defects as do mutations in SPT10 (Dollard et al. 1994; Hess et al. 2004; Sherwood and Osley 1991). In vivo, Spt21 is also recruited to all four histone loci, and this recruitment is required for the recruitment of Spt10 during S-phase (Hess et al. 2004). Mutations in SPT10 and SPT21 share other phenotypes, including silencing defects (Chang and Winston 2011). Mutations have been identified in SPT10 that suppress the requirement for SPT21, suggesting that Spt21 is an accessory factor, required for optimal Spt10 function (Hess et al. 2004).
In addition to the close functional relationships between SPT10 and SPT21, obvious differences between them suggest that they do not always function together. There are three especially striking differences between the two. First, SPT10 is transcribed throughout the cell cycle, whereas SPT21 is transcribed only during S phase, at the same time as histone genes (Cho et al. 1998; Spellman et al. 1998). Second, a complete deletion of SPT10 (spt10Δ) causes a severe growth defect, whereas a complete deletion of SPT21 (spt21Δ) causes a only a mild growth defect (Natsoulis et al. 1994). Finally, mutations that suppress an spt21Δ mutation do not suppress spt10Δ and, in fact, sometimes cause lethality when combined with spt10Δ (Hess and Winston 2005). Taken together, the common and distinct phenotypes of spt10Δ and spt21Δ mutants suggest that Spt10 and Spt21 function together to regulate histone gene expression and that, in addition, Spt10 plays other roles that are critical for normal growth.
To gain insight into other possible roles for Spt10, we have screened for both enhancers and suppressors of the spt10Δ growth defect. The identification of mutations that cause lethality when combined with spt10Δ suggests that Spt10 has overlapping roles with the SAGA coactivator complex. In addition, Spt10 appears to be functionally related to Asf1, the Hir complex, and the Caf-1 complex, whose functions are connected in histone gene regulation, transcriptional silencing, and chromatin assembly (Amin et al. 2012; Eriksson et al. 2012; Kaufman et al. 1998; Sutton et al. 2001). The identification of partial suppressors of the spt10Δ growth defect suggests that Spt10 plays important roles throughout the cell cycle. In support of the idea that these functions are independent of the role of Spt10 as an activator of histone gene transcription, suppressors of the spt10Δ growth defect do not reverse the defects in histone gene transcription.
Materials and Methods
Yeast strains, media, and crosses
All S. cerevisiae strains (Table 1) are GAL2+ derivatives of the S288C background (Winston et al. 1995). Capital letters denote wild-type genes, lowercase letters denote mutant alleles, and Δ indicates a complete open reading frame deletion. To construct spt10Δ haploids, the open reading frame of SPT10 was first replaced with the LEU2 gene or a kanamycin resistance marker in a diploid strain. Then, plasmid pFW217 (SPT10-URA3-CEN) was used to transform the diploid to Ura+, followed by sporulation of the diploid to obtain haploids with the spt10Δ mutation and pFW217. Whenever possible, spt10Δ strains were grown in the presence of pFW217 to minimize selection for spontaneous growth suppressors. Then, the spt10Δ phenotypes were tested after growth on medium with 5-fluoroorotic acid (5-FOA) to select for cells that had lost pFW217. For the nap1Δ::kanMX, hsl1Δ::kanMX, mih1Δ::kanMX, swe1Δ::kanMX, and pat1Δ::kanMX alleles, a 2.4-kb cassette was amplified by polymerase chain reaction (PCR) from genomic DNA isolated from the corresponding deletion set strain (Giaever et al. 2002), then used to transform a wild-type strain. The cassette contains a replacement of the entire open reading frame with a kanamycin resistance marker. The cln3Δ::HIS3, lsm1Δ::natMX, and bck2Δ::hphMX alleles were generated by PCR-mediated disruption of the entire open reading frame (Goldstein and McCusker 1999). All deletions were confirmed by PCR. The cdc28-T18A Y19F allele was generated by digesting p433 (a generous gift from A. Amon) with EcoRI and using the fragment containing the cdc28-T18A Y19F allele and the URA3 marker to transform a wild-type strain. The URA3 gene was then replaced with the KanMX drug resistance cassette of pRS400. Media, basic yeast techniques, mating, sporulation, and tetrad dissection were as previously described (Rose et al. 1990). Crosses to test double mutant lethality generally contained one parent with an spt10Δ mutation and also carrying plasmid pFW217 (SPT10-URA3-CEN). Double-mutant lethality was assayed by replica plating the spore colonies to 5-FOA plates to determine whether strains that had lost pFW217 were viable.
Table 1. S. cerevisiae strains used in this study.
| Name | Genotype |
|---|---|
| FY2191 | MATa spt10Δ201::HIS3 lys2-128δ ura3-52 his3Δ200 leu2Δ1 + pFW217 (SPT10-URA3-CEN) |
| FY2915 | MATa hsl7-gs65f::Tn3-LEU2 spt10Δ201::HIS3 lys2-128δ ura3-52 his3Δ200 leu2Δ1 |
| FY2916 | MATa hsl7-gs63f::Tn3-LEU2 spt10Δ201::HIS3 lys2-128δ ura3-52 his3Δ200 leu2Δ1 |
| FY2917 | MATa lsm1-68f::Tn3-LEU2 spt10Δ201::HIS3 lys2-128δ ura3-52 his3Δ200 leu2Δ1 |
| FY2918 | MATa asf1-69c::Tn3-LEU2 spt10Δ201::HIS3 lys2-128δ ura3-52 his3Δ200 leu2Δ1 |
| FY2919 | MATa asf1-57b::Tn3-LEU2 spt10Δ201::HIS3 lys2-128δ ura3-52 his3Δ200 leu2Δ1 |
| FY2920 | MATa ydr333c-710a::Tn3-LEU2 spt10Δ201::HIS3 lys2-128δ ura3-52 his3Δ200 leu2Δ1 |
| FY2921 | MATa dbf2-719a::Tn3-LEU2 spt10Δ201::HIS3 lys2-128δ ura3-52 his3Δ200 leu2Δ1 |
| FY2922 | MATa lea1-719d::Tn3-LEU2 spt10Δ201::HIS3 lys2-128δ ura3-52 his3Δ200 leu2Δ1 |
| FY2923 | MATα spt10Δ::LEU2 can1Δ::STE2pr-HIS3 lys2-128d ura3Δ0 his3Δ1 or Δ200 leu2Δ0 lyp1Δ or LYP1 + pFW217 (SPT10-URA3-CEN) |
| FY2200 | MATa lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2924 | MATa spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2925 | MATa spt8-302::LEU2 spt10Δ::kanMX lys2-128δ or LYS2-173R2 ura3-52 leu2Δ1 trp1Δ63 + pFW217 (SPT10-URA3-CEN) |
| FY2926 | MATa spt20Δ200::ARG4 spt10Δ::LEU2 lys2-128δ or LYS2-173R2 ura3Δ0 or -52 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2927 | MATα gcn5Δ::HIS3 spt10Δ::LEU2 ura3Δ0 or ura3-52 his3Δ200 leu2Δ0 or leu2Δ1 his3Δ200 + pFW217 (SPT10-URA3-CEN) |
| FY2928 | MATa ubp8Δ::kanMX4 spt10Δ::LEU2 lys2-128δ or LYS2-173R2 ura3Δ0 or -52 his3Δ200 leu2Δ0 or leu2Δ1 arg4-12 + pFW217 (SPT10-URA3-CEN) |
| FY2482 | MATα spt21Δ::kanMX lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2929 | MATa (hta2-htb2)Δ::URA3 hhf2Δ::LEU2 ura3-52 his3Δ200 leu2Δ1 |
| FY2930 | MATa hsl7Δ::HIS3 spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2931 | MATa nap1Δ::kanMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2932 | MATa bck2Δ::hphMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2933 | MATa lsm1Δ::natMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2934 | MATa hsl7Δ::HIS3 ura3Δ0 his3Δ200 leu2Δ0 |
| FY2935 | MATa nap1Δ::kanMX lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2936 | MATa bck2Δ::hphMX lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2937 | MATa lsm1Δ::natMX lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2938 | MATα spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2939 | MATa hsl7Δ::HIS3 nap1Δ::kanMX spt10Δ::LEU2 ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2940 | MATa hsl7Δ::HIS3 bck2Δ::hphMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2941 | MATa hsl7Δ::HIS3 lsm1Δ::natMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2942 | MATa nap1Δ::kanMX bck2Δ::hphMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2943 | MATa nap1Δ::kanMX lsm1Δ::natMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2944 | MATa bck2Δ::hphMX lsm1Δ::natMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2945 | MATa hsl7Δ::HIS3 nap1Δ::kanMX bck2Δ::hphMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2946 | MATa hsl7Δ::HIS3 nap1Δ::kanMX lsm1Δ::natMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2947 | MATa hsl7Δ::HIS3 bck2Δ::hphMX lsm1Δ::natMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2948 | MATa nap1Δ::kanMX bck2Δ::hphMX lsm1Δ::natMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2949 | MATa hsl7Δ::HIS3 nap1Δ::kanMX bck2Δ::hphMX lsm1Δ::natMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY1856 | MATα lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2950 | MATα hsl7Δ::HIS3 spt10Δ::LEU2 ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2951 | MATa hsl1Δ::kanMX4 spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2952 | MATa mih1Δ::kanMX4 spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2953 | MATa swe1Δ::kanMX4 spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2954 | MATa hsl7Δ::HIS3 swe1Δ::kanMX4 spt10Δ::LEU2 ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2955 | MATa hsl1Δ::kanMX4 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2956 | MATa mih1Δ::kanMX4 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2957 | MATa swe1Δ::kanMX4 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2958 | MATa cdc28-T18A Y19F:kanMX lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2959 | MATa cdc28-T18A Y19F:kanMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2960 | MATa hsl7Δ::HIS3 cdc28-T18A Y19F:kanMX spt10Δ::LEU2 ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2961 | MATa hsl7Δ::HIS3 cdc28-T18A Y19F:kanMX lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2962 | MATa cln3Δ::HIS3 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2963 | MATa cln3Δ::HIS3 spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2964 | MATa pat1Δ::kanMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2965 | MATa pat1Δ::kanMX lsm1Δ::natMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2966 | MATa pat1Δ::kanMX4 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2967 | MATa mec1Δ::LEU2 sml1Δ::HIS3 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2816 | MATa spt21Δ::HIS3 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2817 | MATα spt21Δ::HIS3 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2968 | MATα nap1Δ::kanMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2969 | MATα bck2Δ::hphMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2970 | MATα lsm1Δ::natMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 + pFW217 (SPT10-URA3-CEN) |
| FY2971 | MATα hsl7Δ::HIS3 lsm1Δ::natMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2972 | MATα hsl7Δ::HIS3 bck2Δ::hphMX lsm1Δ::natMX spt10Δ::LEU2 lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY1924 | MATα hsl7Δ::HIS3 ura3Δ0 his3Δ200 leu2Δ0 trp1Δ63 |
| FY2973 | MATα nap1Δ::kanMX lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2974 | MATα bck2Δ::hphMX lys2-128δ ura3Δ0 his3Δ200 leu2Δ0 |
| FY2975 | MATα lsm1Δ::natMX lys2-128δ ura3Δ0 his3D200 leu2Δ0 |
| FY2978 | MATa spt10Δ::KanMX leu2Δ1 ura3-52 lys2-128δ his3Δ200 + pFW217 (SPT10-URA3-CEN) |
| FY2979 | MATα asf1Δ::HIS3 leu2Δ0 ura3Δ0 lys2-128δ his3Δ200 |
| FY2980 | MATa hir1Δ::LEU2 his4-912δ HIS3 ura3Δ0 or ura3-52 lys2-128d leu2Δ0 or leu2Δ1 |
| FY2981 | MATa spt21Δ::HIS3 ura3Δ0 leu2Δ0 lys2-128δ his3Δ200 |
| FY2982 | MATα asf1Δ::HIS3 ura3Δ0 leu2Δ0 lys2-128δ his3Δ200 |
| FY2903 | MATa cac1Δ::KanMX leu2Δ0 ura3Δ0 lys2-128δ his3Δ200 |
| FY2933 | MATα spt21Δ::HIS3 ura3Δ0 leu2Δ0 lys2-128δ his3Δ200 |
| FY1235 | MATα hir1Δ::LEU2 leu2Δ1 ura3-52 lys2-128δ his4-912δ trp1Δ63 |
Transposon mutagenesis screen
The transposon mutagenesis screen was performed as described (Burns et al. 1994). In summary, the LEU2-marked library DNA was digested with NotI, then used to transform strain FY2191. Transformant colonies were selected on SC-Leu-Ura medium then replica plated to 5-FOA medium to select for cells that had lost pFW217 (SPT10-URA3), leaving colonies containing the library insertion in an spt10Δ genetic background. Colonies that failed to grow were designated synthetic lethal candidates, and colonies growing more quickly than FY2191 were designated growth suppressor candidates. All candidates were purified to single colonies, which were then individually patched on SC-Leu medium followed by replica plating to verify the growth phenotype. All candidates remaining after this rescreening were purified and tested a third time. Each candidate was then crossed to an spt10Δ leu2 strain to test whether the mutant phenotype cosegregated with the LEU2 marker on the transposon. For the confirmed mutants, genomic DNA was isolated, and vectorette PCR was used to identify the location of each transposon insertion (Arnold and Hodgson 1991). As one growth suppressor candidate was tightly linked to the SPT10 locus, instead of vectorette PCR, we used a candidate gene approach and by a combination of PCR and sequencing, demonstrated the insertion to be within LSM1.
Synthetic genetic array (SGA) screen
A collection of yeast strains containing deletions of every nonessential gene was screened for phenotypes in an spt10Δ background using an SGA screen (Tong et al. 2001). The collection was spotted onto YPD plates with deletion set strains hoΔ::KanMX, lys2Δ::KanMX, and lys12Δ::KanMX spotted separately at the top and bottom of each plate as controls that do not affect spt10Δ growth. The array was mated by replica plating to a lawn with an spt10Δ strain (FY2923) containing a can1::STE2pr-HIS3 allele and carrying the pFW217 (SPT10-URA3) plasmid. Diploids were selected on SC-Leu-Ura and sporulated on solid 1% potassium acetate medium supplemented with histidine, uracil, leucine, and lysine. MATa haploids that contain the deletion set mutation, spt10Δ, and the SPT10 plasmid were selected by replica plating onto SC-Arg-His-Leu-Ura+canavanine+G418 medium. The cells were then replica plated to SC + 5-FOA medium to leave the mutant spt10Δ as the only SPT10 allele present. Strains with better or worse growth compared with the control strains were identified and retested, and then tetrads were dissected to assay for 2:2 segregation and cosegregation of the suppression phenotype with the kanamycin resistance marker.
Dilution spot tests
For dilution spot tests, unless noted otherwise, strains harboring the pFW217 (SPT10-URA3-CEN) plasmid were single colony purified on 5-FOA medium to select for plasmid loss, and single colonies were then patched to YPD media. After 2 d, the cells were resuspended in water to a density of 4 × 106 cells/mL (Figure 2) or 1 × 107 cells/mL (Figures 1, 3−6). Fivefold serial dilutions were spotted onto the media indicated. Plates were scanned after 2−3 d at 30°, unless otherwise indicated.
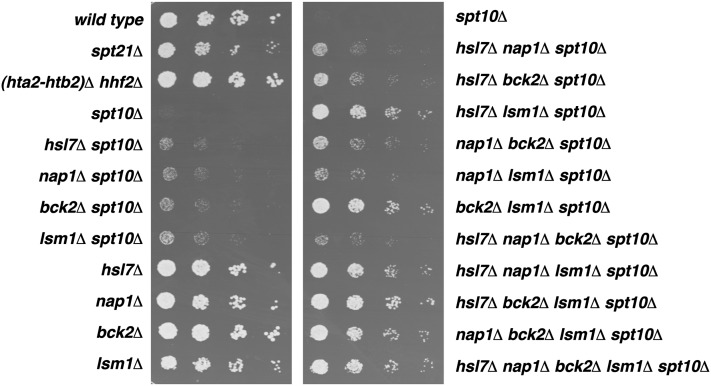
Figure 2 .
Representative suppressors of the spt10Δ slow growth phenotype. Shown are fivefold dilution spot tests. spt10Δ strains were cured of the pFW217 SPT10-URA3-CEN plasmid and grown as described in Materials and Methods, then resuspended to 4 × 106 cells/mL. They were subjected to fivefold dilutions, spotted onto YPD medium, and photographed after 2 d. Strains were wild type (FY2200), spt21Δ (FY2482), (hta2-htb2)Δ hhf2Δ (FY2929), spt10Δ (FY2924), hsl7Δ spt10Δ (FY2930), nap1Δ spt10Δ (FY2931), bck2Δ spt10Δ (FY2932), lsm1Δ spt10Δ (FY2933), hsl7Δ (FY2934), nap1Δ (FY2935), bck2Δ (FY2936), lsm1Δ (FY2937), spt10Δ (FY2938), hsl7Δ nap1Δ spt10Δ (FY2939), hsl7Δ bck2Δ spt10Δ (FY2940), hsl7Δ lsm1Δ spt10Δ (FY2941), nap1Δ bck2Δ spt10Δ (FY2942), nap1Δ lsm1Δ spt10Δ (FY2943), bck2Δ lsm1Δ spt10Δ (FY2944), hsl7Δ nap1Δ bck2Δ spt10Δ (FY2945), hsl7Δ nap1Δ lsm1Δ spt10Δ (FY2946), hsl7Δ bck2Δ lsm1Δ spt10Δ (FY2947), nap1Δ bck2Δ lsm1Δ spt10Δ (FY2948), and hsl7Δ nap1Δ bck2Δ lsm1Δ spt10Δ (FY2949).
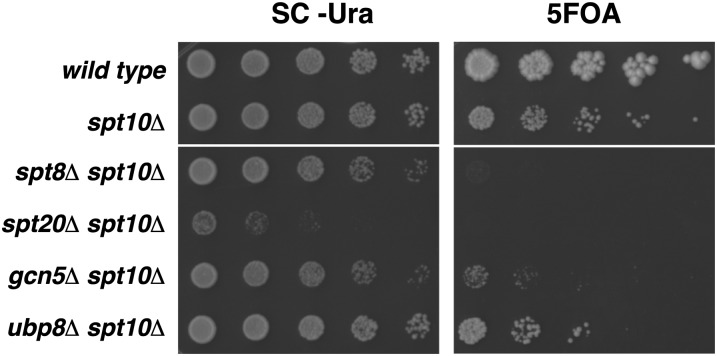
Figure 1 .
Mutations in genes encoding SAGA subunits lead to lethality or poor growth in an spt10Δ background. Shown are fivefold dilution spot tests. All strains were grown to saturation in SC-Ura medium in the presence of the pFW217 SPT10-URA3-CEN plasmid. They were serially diluted fivefold and spotted onto SC-Ura and 5-FOA plates to select for cells that have maintained or lost the SPT10 plasmid, respectively. The SC-Ura plate is shown after 2 d of incubation at 30° and the 5-FOA plate after 5 d. Upper and lower panels are from the same plate. The strains were wild type (FY2200), spt10Δ (FY2924), spt8Δ spt10Δ (FY2925) spt20Δ spt10Δ (FY2926), gcn5Δ spt10Δ (FY2927), and ubp8Δ spt10Δ (FY2928).
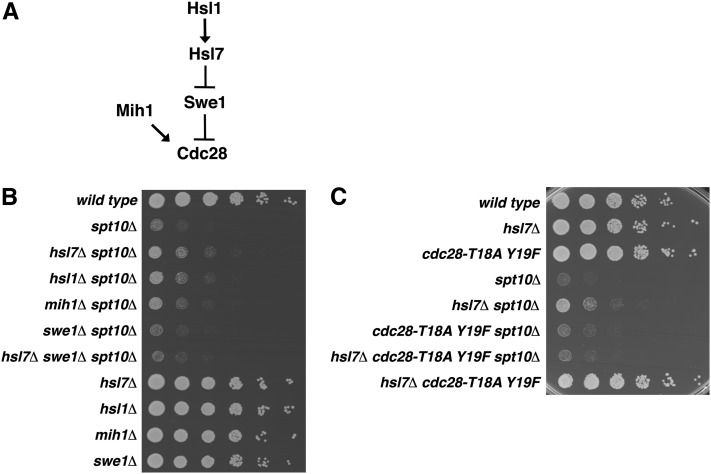
Figure 3 .
Perturbed progression through the bud morphogenesis checkpoint can suppress the spt10Δ growth defect. (A) Diagram of the Hsl−Swe1−Cdc28 pathway. (B, C) Fivefold dilution spot tests. Each strain was grown to saturation and diluted to 1.0 × 107 cells/mL for the densest spot. Strains in (B) were wild type (FY2200), spt10Δ (FY2924), hsl7Δ spt10Δ (FY2930), hsl1Δ spt10Δ (FY2951), mih1Δ spt10Δ (FY2952), swe1Δ spt10Δ (FY2953), hsl7Δ swe1Δ spt10Δ (FY2954), hsl7Δ (FY2934), hsl1Δ (FY2955), mih1Δ (FY2956), and swe1Δ (FY2957). Strains in (C) were wild type (FY2200), hsl7Δ (FY2934), cdc28-T18A Y19F (FY2958), spt10Δ (FY2924), hsl7Δ spt10Δ (FY2930), cdc28-T18A Y19F spt10Δ (FY2959), hsl7Δ cdc28-T18A Y19F spt10Δ (FY2960), and hsl7Δ cdc28-T18A Y19F (FY2961). Pictures were taken after 2 d.
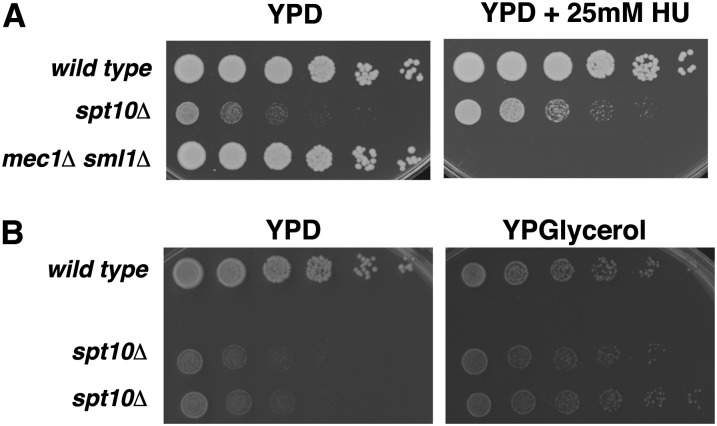
Figure 6 .
Nongenetic means of suppressing the spt10Δ slow growth phenotype. (A) Fivefold dilutions were made as in Figure 3, then spotted onto YPD medium or YPD + 25 mM HU. Pictures were taken after 2 d. Strains were WT (FY2200), spt10Δ (FY2924), and mec1Δ sml1Δ (FY2967). mec1Δ sml1Δ mutants are hypersensitive to HU. (B) Wild-type (FY2200) and spt10∆ (FY2924) strains were subjected to fivefold serial dilutions as in Figure 3 and grown on YPD medium for two days or on YP + 3% glycerol medium for 5 d.
cDNA synthesis and real-time PCR
RNA was extracted from 10 mL of yeast cultures in exponential growth as described (Ausubel et al. 1988; Swanson et al. 1991). Then, 10 μg of RNA was treated with 2 units of DNase (TURBO DNA free kit, Ambion) and reverse transcribed with Superscript III reverse transcriptase (Invitrogen) using an oligo-dT primer. Real-time PCR was performed with a Stratagene MX3000P machine using 50 ng of cDNA and 1 μg of each primer per 50 μL of reaction, with each reaction performed in triplicate. Primer sequences (Table 2) were provided by Neil McLaughlin and David Clark (personal communication). The specificity of each primer pair was confirmed using the corresponding deletion mutant. Thermocycling parameters were: 10:00 at 94°, then 35−40 cycles of (0:30 at 94°, 0:30 at 52°, 1:00 at 72°), followed by a melting curve to assay product specificity. Linearity and efficiency was confirmed for each primer pair on each plate.
Table 2. Primers used to measure histone mRNA levels.
| Primer | Gene | Orientation | Sequence |
|---|---|---|---|
| FO6006 | HTA1 | Forward | TTCAAAACAAACAAATTTCA |
| FO6007 | HTA1 | Reverse | AAATACCAGAACCGATCTTA |
| FO6008 | HTA2 | Forward | GGAAAGTACAGAACAAGAGC |
| FO6009 | HTA2 | Reverse | CTTTGTTTCTTTTCAACTCAG |
| FO6010 | HTB1 | Forward | CAAACCACAAATAAACCATAC |
| FO6011 | HTB1 | Reverse | AGGAAGTGATTTCATTATGC |
| FO6012 | HTB2 | Forward | ACCAACAACAACTTACTCTACA |
| FO6013 | HTB2 | Reverse | AATCACAATACCTAGTGAGTGA |
| FO6014 | HHT1 | Forward | TATATAAACGCAAACAATGG |
| FO6015 | HHT1 | Reverse | AACTGATGACAATCAACAAA |
| FO6016 | HHT2 | Forward | TACTAAAGGATCCAAGCAAA |
| FO6017 | HHT2 | Reverse | AAAAATTCCCGCTTTATATT |
| FO6018 | HHF1 | Forward | AACAAACAAAAACAAGCAAC |
| FO6019 | HHF1 | Reverse | TTGTTGTTACCGTTTTCTTA |
| FO6020 | HHF2 | Forward | GTAGCAAAAACAACAATCAA |
| FO6021 | HHF2 | Reverse | ATAATTTCAAACACCGATTG |
| FO6145 | ACT1 | Forward | TTTTGTCCTTGTACTCTTCC |
| FO6146 | ACT1 | Reverse | CTGAATCTTTCGTTACCAAT |
Results
Identification of mutations that enhance or suppress the spt10Δ slow-growth phenotype
To study the basis of the spt10Δ slow growth phenotype, we screened for mutations that enhance or suppress the growth defect by using both transposon insertion mutagenesis (Burns et al. 1994) and the S. cerevisiae deletion set (Giaever et al. 2002), both as described in Materials and Methods. As spontaneous suppressors of the spt10Δ slow growth phenotype arise at a high frequency, we maintained a low-copy SPT10 plasmid (pFW217) in the spt10Δ strains until the final screening step for each method.
We began with a transposon insertion mutagenesis screen (Burns et al. 1994; Kumar and Snyder 2002) in which we tested 9000 independent transformants for improved or impaired growth compared with the spt10Δ parent (Materials and Methods). By this approach, we identified eight mutations in a total of six genes (Table 3). Three mutations that confer suppression of spt10Δ poor growth were in two genes and five mutations that cause lethality when combined with spt10Δ were identified in four genes. For all six genes, we tested a complete deletion of the identified gene and found the same suppression phenotype, suggesting that all of the insertion mutations cause null phenotypes. For all subsequent experiments, the deletion mutations were used.
Table 3. Genes identified by a transposon screen.
| Gene | Effect When Combined With spt10Δ | Insertion Location Relative to ATG | Description |
|---|---|---|---|
| HSL7 | Improved growth | +1232 | Arginine N-methyltransferase involved in regulation of Swe1 degradation |
| HSL7 | Improved growth | +1654 | Arginine N-methyltransferase involved in regulation of Swe1 degradation |
| LSM1 | Improved growth | −191 | Part of a complex involved in degradation of cytoplasmic mRNAs |
| ASF1 | Lethality | +102 | Histone chaperone |
| ASF1 | Lethality | +283 | Histone chaperone |
| YDR333C | Lethality | +530 | Unknown function |
| DBF2 | Lethality | +1475 | Ser/Thr kinase; exit from mitosis |
| LEA1 | Lethality | +361 | Component of U2 snRNP |
From this initial screen, a concern of bias arose, as we had obtained two different transposon insertions within ASF1 without obtaining any insertions in other genes whose deletions were previously shown to be lethal in combination with spt10Δ. These genes include HTA1, HTB1, HHF1, HIR1, ASF1, RKR1, and MBP1 (Braun et al. 2007; Fassler and Winston 1988; Hess 2004; Hess and Winston 2005; Sutton et al. 2001). Therefore, rather than saturate the transposon mutagenesis screen, which would require testing 30,000 transformants (Burns et al. 1994), we switched to the more systematic approach of screening the deletion set.
We screened the deletion set for mutations that either suppress or enhance the spt10Δ slow growth defect (Materials and Methods). Our screen yielded 44 mutations that cause lethality in combination with spt10Δ (Table 4) and 13 mutations that improve spt10Δ growth (Table 5). Interestingly, there was no overlap with the mutations identified from the transposon mutagenesis screen, although some functionally related genes were identified (LSM genes). The lack of overlap indicates that the deletion set screen had many false-negative results. There was also a class of 12 mutants that appeared to cause lethality during the original screen but showed little or no growth defect upon tetrad dissection (discussed in the section Genes involved in silencing show mutant phenotypes in combination with spt10Δ).
Table 4. Genes found by SGA analysis whose deletion causes double-mutant lethality or extreme sickness with spt10Δ.
| Gene | Description |
|---|---|
| BCK1 | MAP KKK in the protein kinase C signaling pathway |
| BUD20 | Protein involved in bud site selection |
| CAC2 | Component of chromatin assembly complex CAF-I |
| CTF19 | Component of the COMA complex |
| CYS3 | Cysteine biosynthesis |
| DOA1 | Ubiquitin-mediated protein degradation |
| ELP2 | Component of the Elongator complex |
| ELP4 | Component of the Elongator complex |
| ELP6 | Component of the Elongator complex |
| HHF1 | Histone H4 |
| HHT1 | Histone H3 |
| HIR2 | Component of the HIR complex |
| HIR3 | Component of the HIR complex |
| HIT1 | Function unknown |
| HPC2 | Component of the HIR complex |
| IES2 | Associates with the INO80 chromatin remodeling complex |
| IXR1 | Binds DNA containing intrastrand cross-links formed by cisplatin |
| MCM21 | Component of the COMA complex |
| MDM20 | Component of the NatB N-terminal acetyltransferase |
| MRPL38 | Mitochondrial ribosomal protein of the large component |
| MSD1 | Mitochondrial aspartyl-tRNA synthetase |
| NHX1 | Endosomal Na+/H+ exchanger |
| PEP7 | Facilitates vesicle-mediated vacuolar protein sorting |
| PGD1 | Component of the mediator complex |
| REG1 | Negative regulation of glucose-repressible genes |
| RMD8 | Cytosolic protein required for sporulation |
| SAM37 | Component of the mitochondrial SAM complex |
| SGF11 | Component of the SAGA complex |
| SGF29 | Component of the SAGA complex |
| SIN3 | Component of the Rpd3-Sin3 complex |
| SLX8 | Component of the Slx5-Slx8 SUMO-targeted ubiquitin ligase complex |
| SOD1 | Cytosolic copper-zinc superoxide dismutase |
| SPT3 | Component of the SAGA complex |
| SPT8 | Component of the SAGA complex |
| SWC3 | Component of the SWR1 complex |
| TAF14 | Component of TFIID, TFIIF, INO80, SWI/SNF, and NuA3 complexes |
| THR1 | Threonine synthesis |
| THR4 | Threonine synthase |
| UMP1 | Chaperone required for maturation of the 20S proteasome |
| VMA8 | Component of the peripheral membrane domain of the vacuolar H+-ATPase |
| VMS1 | Protein degradation and quality control |
| VPS54 | Component of the GARP complex |
| YAF9 | Component of both the NuA4 histone H4 and SWR1 complexes |
| YGL149W | Dubious open reading frame, overlaps INO80 |
Table 5. Genes found by SGA analysis whose deletion suppresses the spt10Δ poor growth phenotype.
| Gene | Description |
|---|---|
| BCK2 | Protein kinase C signaling pathway and the G1/S transition |
| CLB2 | B-type cyclin involved in G2 to M progression |
| HAL5 | Putative protein kinase |
| HDA2 | Component of a class II histone deacetylase complex |
| IES3 | Component of the INO80 complex |
| ITR1 | Myo-inositol transporter |
| LAS21 | Synthesis of the glycosylphosphatidylinositol (GPI) core structure |
| LSM6 | Part of complexes involved in RNA processing, splicing, and decay |
| LSM7 | Part of complexes involved in RNA processing, splicing, and decay |
| NAP1 | Bud morphogenesis, microtubule dynamics, and transport of histones H2A and H2B |
| SIF2 | Component of the Set3C complex |
| SLM4 | Component of the EGO complex |
| SYH1 | Protein of unknown function, influences nuclear pore distribution |
The loss of specific classes of SAGA genes is lethal in combination with spt10Δ
Our screens identified four genes encoding components of the SAGA coactivator complex whose deletion is lethal when combined with spt10Δ: SPT3, SPT8, SGF11, and SGF29. These four factors are believed to be involved in distinct activities of the multifunctional SAGA complex, as Spt3 and Spt8 modulate the recruitment of the TATA-binding protein (TBP) to promoters (Bhaumik and Green 2001, 2002; Dudley et al. 1999; Larschan and Winston 2001), Sgf11 is part of the DUB module of SAGA (Kohler et al. 2010; Samara et al. 2010), and Sgf29 has recently been shown to bind to H3K4me2/3, to be required for Gcn5-dependent histone acetylation in vivo, and to help recruit TBP to promoters (Bian et al. 2011; Shukla et al. 2012). To test whether the double-mutant lethality with spt10Δ is general for all SAGA deletion mutants or specific for certain classes, we tested deletions of SPT20, encoding a core component of SAGA, UBP8, encoding a histone deubiquitylase, and GCN5, encoding the histone acetyltransferase. Our results (Figure 1) show that the spt20Δ spt10Δ double mutant is inviable, whereas both the ubp8Δ spt10Δ and gcn5Δ spt10Δ double mutants are viable but grow poorly, even worse than the spt10Δ single mutant. Our genetic analysis, then, demonstrates that Spt10 shares essential or important roles with distinct functions of the SAGA coactivator complex. In light of the spt10Δ-gcn5Δ genetic interaction, we note that we did not see a genetic interaction between spt10Δ and rtt109Δ (RTT109 encodes a histone acetyltransferase that has been implicated in histone gene transcription) (Fillingham et al. 2009).
Double-mutant lethality of spt10Δ with asf1Δ and hir/hpc2Δ mutations suggests functional overlaps
Among the genes identified as causing double-mutant lethality with spt10Δ were asf1Δ, hir2Δ, hir3Δ, and hpc2Δ. Previous studies also showed that spt10Δ asf1Δ double mutants are inviable (Sutton et al. 2001). Asf1 has been shown to be a histone chaperone (Munakata et al. 2000), the Hir complex (comprised of Hir1-3 and Hpc2) has been implicated in chaperone and nucleosome assembly activities (Green et al. 2005; Prochasson et al. 2005), and both Asf1 and the Hir complex have been shown to regulate histone gene transcription (Osley and Lycan 1987; Sutton et al. 2001; Xu et al. 1992). Furthermore, these factors are believed to function both physically and genetically with each other and with the Caf-1 complex (Green et al. 2005; Kaufman et al. 1998; Liu et al. 2012; Sutton et al. 2001).
The isolation of asf1Δ and hir/hpc2Δ mutations as causing lethality when combined with spt10Δ suggests that Spt10 participates in this set of functions. To test this further, we crossed spt10Δ by hir1Δ and by cac1/rlf2Δ (CAC1 encodes a component of the Caf-1 complex) to test for double mutant lethality. Our results (Table 6) show that spt10Δ causes inviability with asf1Δ and hir/hpc mutations, but not with cac1Δ. This pattern is reminiscent of earlier studies that showed that both asf1Δ and hir/hpc mutations cause double-mutant sickness with cac mutations, but not with each other (Kaufman et al. 1998; Sutton et al. 2001). We note that our screens did not identify mutations in RTT106, which encodes a histone chaperone that has been shown to regulate histone gene transcription by interactions with Asf1/Hir/Caf-1 (Fillingham et al. 2009; Huang et al. 2007; Kurat et al. 2011; Silva et al. 2012; Zunder and Rine 2012). Similarly, a screen for mutations that cause double-mutant lethality with rtt106Δ did not identify spt10Δ (Imbeault et al. 2008). In contrast to spt10Δ, an spt21Δ mutation allowed viability when combined with hir1Δ or asf1Δ (Table 6). Taken together, our results suggest that Spt10, but not Spt21, contributes to an essential function in collaboration with Asf1 and the Hir complex, likely either in histone gene activation or an aspect of chromatin assembly.
Table 6. spt10Δ is inviable with hir1Δ and asf1Δ.
| Double Mutant | Phenotypea |
|---|---|
| spt10Δ hir1Δ | Inviableb |
| spt10Δ asf1Δ | Inviablec |
| spt10Δ cac1Δ | Viabled |
| spt21Δ hir1Δ | Viablee |
| spt21Δ asf1Δ | Viablef |
| spt21Δ cac1Δ | Viableg |
The phenotype was determined by testing the ability of the double mutant to survive loss of plasmid pFW217 (SPT10-URA3-CEN) by assaying growth on 5FOA plates as described in Materials and Methods. The cross done for each combination is listed below.
FY2978 × FY1235.
FY2924 × FY2979.
FY2903 × FY2938.
FY2980 × FY2933.
FY2981 × FY2982.
FY2903 x FY2933.
Genes involved in silencing show mutant phenotypes in combination with spt10Δ
One notable class of mutants appeared to show lethality in combination with spt10Δ during our systematic screen. However, upon retesting by tetrad dissection, viable double mutant spores were obtained at the expected frequency, without substantial growth defects. This class of mutants included sir1Δ, ard1Δ, and pol32Δ, all of which have roles in silencing (Pillus and Rine 1989; van Welsem et al. 2008; Whiteway et al. 1987). Others have reported a similar pattern of apparent lethality for sir1Δ dot1Δ and pol32Δ dot1Δ in another deletion set screen (van Welsem et al. 2008). They discovered that the pattern actually resulted from mating type silencing defects, which prevent growth when the SGA screening method is used. Our studies of Spt10 have demonstrated it to be required for silencing (Chang and Winston 2011).
The slow growth of spt10Δ mutants can be suppressed through multiple genetic pathways
The mutations that we identified that suppress the spt10Δ growth defect fall into several functional categories. For the remainder of our analysis, we focused on the four mutations that individually caused the strongest suppression of the spt10Δ growth defect: hsl7Δ, nap1Δ, bck2Δ, and lsm1Δ (Figure 2). Hsl7 is an arginine methyltransferase with a role in the bud morphogenesis checkpoint (Lew 2000). Nap1 is a histone chaperone involved in the nuclear import of histones, and it regulates cell-cycle progression in G2/M (Zlatanova et al. 2007). Bck2 regulates the transition from G1 to S phase of the cell cycle (Epstein and Cross 1994; Lee et al. 1993), and Lsm1 is part of a heteroheptameric complex involved in RNA decapping and processing (Tharun 2009). Lsm1 has recently been shown to control histone mRNA stability (Herrero and Moreno 2011). All of the deletion mutations are partial suppressors individually, but when lsm1Δ is combined with hsl7Δ or bck2Δ, strong additive effects are seen (Figure 2). Little or no additivity is seen with other combinations. This finding suggests that hsl7Δ and bck2Δ suppress the spt10Δ growth defect through a different genetic pathway than does lsm1Δ. To study these effects, we conducted a more detailed genetic analysis of each suppressor.
Perturbations of the G2/M transition allow spt10Δ mutants to grow faster
HSL7, along with HSL1, initially was isolated in a histone synthetic lethal screen, which identified genes that become essential when the tail of either histone H3 or histone H4 is deleted (Ma et al. 1996). Although the basis of this synthetic lethality remains unknown, Hsl1, a protein kinase, and Hsl7 have been shown to regulate the bud morphogenesis checkpoint through the Hsl−Swe1−Cdc28 pathway, which monitors whether cytoskeletal events have been properly completed prior to mitosis (Figure 3A) (Lew 2000). The cyclin-dependent kinase Cdc28 controls cell-cycle progression through the G2/M transition; its activity is inhibited by the kinase Swe1 and activated by the phosphatase Mih1. When an S. cerevisiae cell buds, Hsl1 recruits Hsl7 to the bud neck and phosphorylates both proteins. This recruits Swe1, leading to Swe1 degradation, causing decreased phosphorylation of Cdc28 and thereby promoting progression through G2/M. Thus, an hsl7Δ single mutant has increased Swe1 activity, resulting in decreased Cdc28 activity. We tested the effects of other mutations in the Hsl−Swe1−Cdc28 pathway on spt10Δ growth. Consistent with our findings for hsl7Δ, both hsl1Δ and mih1Δ, which also impair progression through the bud morphogenesis checkpoint, suppress the spt10Δ growth defect, whereas a mutation (swe1Δ) that promotes progression does not (Figure 3B). As additional evidence that impairment of G2/M progression suppresses the spt10Δ growth defect, we identified clb2Δ as a suppressor in our screen (Table 5).
To test whether suppression of the spt10Δ growth defect by hsl7Δ occurs within the Hsl−Swe1−Cdc28 pathway, we tested combinations of mutations in this pathway. First, we found that swe1Δ is epistatic to hsl7Δ with respect to suppression of the spt10Δ growth defect (Figure 3B), suggesting that suppression by hsl7Δ is mediated through Swe1 activity. Second, we tested whether the inhibitory phosphorylation of Cdc28 by Swe1 plays a role in hsl7Δ suppression of the spt10Δ growth defect. To do this, we used the cdc28-T18A Y19F allele (Amon et al. 1992; Sorger and Murray 1992), which makes cells insensitive to mutations upstream in the Hsl-Swe1-Cdc28 pathway, thus mimicking loss of Swe1. We found that hsl7Δ no longer suppresses the spt10Δ growth defect in the presence of the cdc28-T18A Y19F allele (Figure 3C), further supporting that hsl1Δ- and hsl7Δ-mediated suppression occurs through the Hsl−Swe1−Cdc28 pathway. Taken together, our genetic analysis suggests that mutations that activate the bud morphogenesis checkpoint can confer improved growth of spt10Δ cells.
Perturbations at the G1/S transition also suppress the spt10Δ growth defect
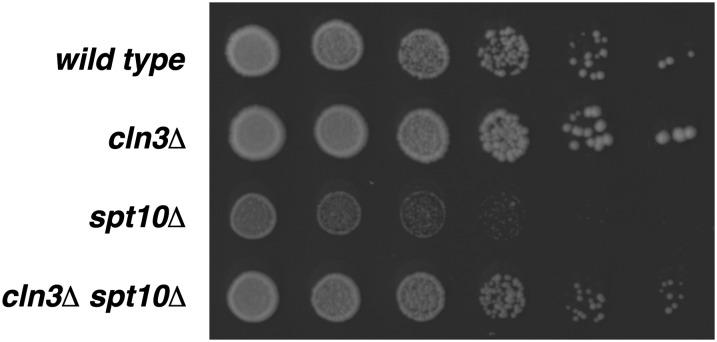
Bck2 was originally isolated as a factor important in protein kinase C signaling, and it has been found to be important in controlling the G1/S transition of the cell cycle (Epstein and Cross 1994; Lee et al. 1993). A related protein involved in regulating the G1/S transition is Cln3, a cyclin that binds to Cdc28 to regulate the transition through START (Richardson et al. 1989). We asked whether a cln3Δ mutation can also suppress the spt10Δ growth defect. Spot tests demonstrate that cln3Δ spt10Δ mutants grow better than spt10Δ single mutants (Figure 4), suggesting that different perturbations in the G1/S transition can suppress the spt10Δ growth defect. Taken together with the hsl7Δ suppression data, our genetic analysis demonstrates that the spt10Δ slow growth can be suppressed by mutations that delay cell cycle progression at either the G1/S transition or the bud morphogenesis G2/M checkpoint.
Figure 4 .
A mutation perturbing the G1/S transition can partially suppress the spt10Δ growth defect. Fivefold dilution spot assays were performed as in Figure 3. Strains were wild type (FY2200), cln3Δ (FY2962), spt10Δ (FY2924), and cln3Δ spt10Δ (FY2963). Pictures were taken after 2 d.
Impairment of the Lsm1-7−Pat1 complex suppresses the spt10Δ slow growth phenotype
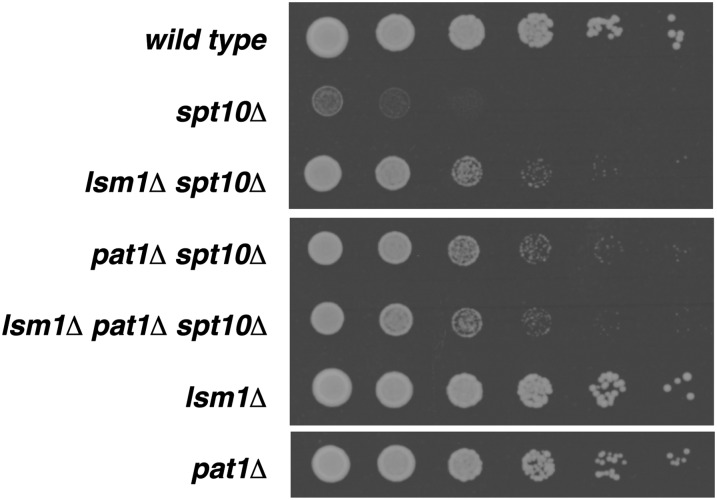
Next we conducted a more detailed genetic analysis of three closely related suppressors: lsm1Δ, lsm6Δ, and lsm7Δ. The eight S. cerevisiae LSM (like Sm) genes form two distinct, ring-shaped, heteroeptameric complexes (Tharun 2009). The first complex, containing Lsm2-8, localizes to the nucleus and regulates pre-mRNA splicing. The second complex, containing Lsm1-7, is localized to the cytoplasm and regulates the decapping of polyadenylated mRNAs, in conjunction with Pat1 (protein associated with Topoisomerase II). We note that in both larger eukaryotes (Tharun 2009) and in yeast (Herrero and Moreno 2011), the Lsm1-7−Pat1 complex has been implicated in promoting the degradation of histone mRNAs.
The result that lsm1Δ suppresses the spt10Δ slow growth phenotype suggests that it is the Lsm1-7−Pat1 complex, rather than the Lsm2−Lsm8 complex that is related to spt10Δ growth. We therefore also tested whether pat1Δ suppresses the spt10Δ growth phenotype. Our results (Figure 5) show that pat1Δ does suppress the spt10Δ growth defect and, furthermore, that suppression by lsm1Δ and pat1Δ is not additive, suggesting that lsm1Δ and pat1Δ suppress the spt10Δ growth defect through the same pathway. The other LSM genes in the complex are essential for viability and could not be tested.
Figure 5 .
Suppression of the spt10Δ growth defect by mutations in the Lsm1-7-Pat1 complex. Dilution spot assays were performed as in Figure 3 with the following strains: wild type (FY2200), spt10Δ (FY2924), lsm1Δ spt10Δ (FY2933), pat1Δ spt10Δ (FY2964), lsm1Δ pat1Δ spt10Δ (FY2965), lsm1Δ (FY2937), and pat1Δ (FY2966). Pictures were taken after 2 d.
Environmental conditions that slow cell division also suppress the spt10Δ slow growth phenotype
Considering that genetic means of slowing cell-cycle progression can suppress the spt10Δ slow growth phenotype, we asked whether altered growth conditions that slow cell cycle progression will also suppress this phenotype. First, we assayed the growth of spt10Δ strains on medium containing 25 mM hydroxyurea (HU), a ribonucleotide reductase inhibitor that impedes S-phase progression. We found that addition of 25 mM HU causes modest suppression of the spt10Δ growth defect relative to wild-type growth (Figure 6A).
Second, we slowed growth using medium that contains glycerol rather than glucose as a carbon source. Relative to wild-type, spt10Δ growth modestly improves on this medium (Figure 6B). These findings are consistent with the possibility that slowing cell cycle progression through multiple means improves spt10Δ growth.
Suppressors of the spt10Δ growth phenotype do not restore histone mRNA levels
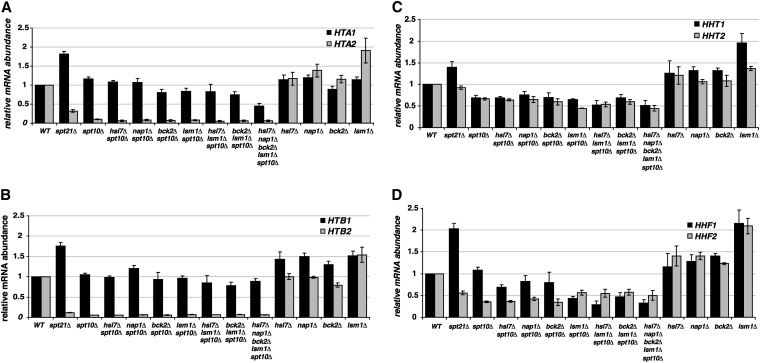
Because Spt10 binds to histone gene promoters and regulates histone gene transcription (Dollard et al. 1994; Eriksson et al. 2011; Hess et al. 2004; Sherwood and Osley 1991; Xu et al. 2005), we wanted to test whether the suppressors improve spt10Δ growth by increasing histone gene mRNA levels. We therefore measured mRNA levels for all eight histone genes in the suppressor strains, using reverse transcription and real-time PCR. We used primer pairs highly specific for their corresponding transcripts (Table 2; N. McLaughlin and D. Clark, personal communication) to distinguish the two nearly identical copies of each histone gene.
Our results (Figure 7) show that the suppressors do not restore histone mRNA levels in an spt10Δ background. First, in agreement with previous results (Dollard et al. 1994; Hess et al. 2004), we found that, in asynchronously growing cultures, HTA2 and HTB2 mRNA levels are decreased approximately 20-fold, with more modest decreases of HHT1, HHT2, and HHF2 mRNA levels. In an spt10Δ background, no single suppressor mutation or multiple suppressor combination restores mRNA levels for any histone gene. The only substantial change with any suppressor mutation is a decrease in HHF1 mRNA levels in spt10Δ mutants when LSM1 is deleted. This is in spite of the finding that some of the suppressor mutations cause modest changes in histone mRNA levels in a wild-type SPT10 background. The increased level of histone mRNAs observed for lsm1Δ agrees with previous results (Herrero and Moreno 2011). Overall, our results suggest that restoration of normal histone mRNA levels is not necessary for suppression of the spt10Δ slow growth phenotype.
Figure 7 .
mRNA abundance for the core histone genes in growth suppressor strains. RNA was isolated and reverse transcribed, and real-time PCR with gene-specific primers (Table 2) was used to quantitate histone mRNA levels for (A) HTA1 and HTA2; (B) HTB1 and HTB2; (C) HHT1 and HHT2; and (D) HHF1 and HHF2. All values were normalized to ACT1 mRNA levels and are shown relative to wild type, which was assigned a value of 1. Shown is the mean ± SEM for at least three independent experiments. Strains were wild type (FY2200 and FY1856), spt10Δ (FY2924 and FY2938), spt21Δ (FY2816 and FY2817), hsl7Δ spt10Δ (FY2930 and FY2950), nap1Δ spt10Δ (FY2931 and FY2968), bck2Δ spt10Δ (FY2932 and FY2969), lsm1Δ spt10Δ (FY2933 and FY2970), hsl7Δ lsm1Δ spt10Δ (FY2941 and FY2971), bck2Δ lsm1Δ spt10Δ (FY2944), hsl7Δ nap1Δ bck2Δ lsm1Δ spt10Δ (FY2949 and FY2972), hsl7Δ (FY2934 and FY1924), nap1Δ (FY2935, FY2973), bck2Δ (FY2936, FY2974), and lsm1Δ (FY2937, FY2975).
We note that, like spt10∆ mutants, spt21∆ mutants show decreased levels of HTA2, HTB2, and HHF2 mRNA, but unlike spt10∆ mutants or the suppressor strains, the spt21∆ mutants show modest increases in mRNA levels for HTA1, HTB1, HHF1, and to a lesser degree HHT1. These results suggest that Spt10 and Spt21 have some non-overlapping roles in histone gene regulation.
Discussion
In this work, we have identified a broad spectrum of mutations that either cause lethality when combined with spt10Δ or that suppress the slow growth phenotype caused by spt10Δ. The first set of genes suggests that the function of Spt10 partially overlaps with the SAGA coactivator complex as well as with two factors involved in chromatin assembly and histone gene transcription, Asf1 and the Hir complex. Given the pleiotropic nature of mutants lacking these functions, as well as the documented role of Asf1 and the Hir complex in histone gene regulation (Osley and Lycan 1987; Sutton et al. 2001; Xu et al. 1992), these double mutant lethalities are not surprising. Several additional genes were identified in the screen for double-mutant lethality (Tables 3 and 4), and the results suggest that functional overlaps also exist between Spt10 and both the Elongator complex and the Ino80 complex. As there are no known roles for SAGA, Elongator, or Ino80 in histone gene expression, further studies of these interactions will be required to understand whether the essential process in which Spt10 and these other factors participate involves histone gene expression or a previously uncharacterized role for Spt10.
The suppressors of the spt10Δ growth defect led us to conclude that perturbations at multiple points of the cell cycle can suppress the slow growth of spt10Δ mutants. Although it seems paradoxical that an impairment of cell-cycle progression would enhance growth, there is precedent for a defect in one process suppressing a defect in a related process. For example, a cold-sensitive spt5 mutation is suppressed with 6-azauracil, which decreases the rate of transcription elongation (Hartzog et al. 1998). Furthermore, perturbations in multiple different cell cycle phases can suppress a silencing defect at the S. cerevisiae silent mating type loci and telomeres (Laman et al. 1995).
One model to explain our findings is that spt10Δ mutants grow slowly due to the shortage of a factor or factors necessary for normal growth, and that cell cycle perturbations compensate for this growth-limitation, either by allowing more time for the factor to be produced, or by adjusting the relative levels of factors with which it interacts. Considering the well-characterized role of Spt10 in activating histone gene transcription, obvious candidates for such factors are histone proteins. We note that histone levels are clearly a factor in spt10Δ growth, as a plasmid that encodes all four core histones (with the HTA1-HTB1 and HHT1-HHF1 loci) restores spt10Δ growth to nearly wild-type levels (Eriksson et al. 2005; Silva et al. 2012). However, we found that suppressors of the spt10Δ growth defect do not suppress the spt10Δ defect in histone mRNA levels, suggesting that the slow growth can be affected by other routes, possibly independent of histone gene transcription. Alternatively, the suppressors might partially alleviate the requirement for normal histone levels.
Left unresolved by these and other studies of Spt10 is the role of the Spt10 acetyltransferase domain. While it is required for Spt10 function (Hess et al. 2004), its target(s) remain unknown. The elucidation of these targets will go a long ways toward helping us understand the roles of Spt10 in growth.
Acknowledgments
We thank David Hess for providing the initial evidence for the genetic relationships between Spt10, Asf1, Hir1, and Cac1. We also thank Neil McLaughlin and David Clark for providing primer sequences, Angelika Amon for providing plasmid p433, and Dan Spatt for help with strain constructions. This work was supported by National Institutes of Health grant GM32967 to F.W.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Amin A. D., Vishnoi N., Prochasson P., 2012. A global requirement for the HIR complex in the assembly of chromatin. Biochim. Biophys. Acta 1819: 264–276 [DOI] [PubMed] [Google Scholar]
- Amon A., Surana U., Muroff I., Nasmyth K., 1992. Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae. Nature 355: 368–371 [DOI] [PubMed] [Google Scholar]
- Arnold C., Hodgson I. J., 1991. Vectorette PCR: a novel approach to genomic walking. PCR Methods Appl. 1: 39–42 [DOI] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., et al. , 1988. Current Protocolos in Molecular Biology, John Wiley & Sons, Inc., New York [Google Scholar]
- Bhaumik S. R., Green M. R., 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15: 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S. R., Green M. R., 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22: 7365–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian C., Xu C., Ruan J., Lee K. K., Burke T. L., et al. , 2011. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 30: 2829–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M. A., Costa P. J., Crisucci E. M., Arndt K. M., 2007. Identification of Rkr1, a nuclear RING domain protein with functional connections to chromatin modification in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 2800–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns N., Grimwade B., Ross-Macdonald P. B., Choi E. Y., Finberg K., et al. , 1994. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 8: 1087–1105 [DOI] [PubMed] [Google Scholar]
- Chang J. S., Winston F., 2011. Spt10 and Spt21 are required for transcriptional silencing in Saccharomyces cerevisiae. Eukaryot. Cell 10: 118–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho R. J., Campbell M. J., Winzeler E. A., Steinmetz L., Conway A., et al. , 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2: 65–73 [DOI] [PubMed] [Google Scholar]
- Denis C. L., Malvar T., 1990. The CCR4 gene from Saccharomyces cerevisiae is required for both nonfermentative and spt-mediated gene expression. Genetics 124: 283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollard C., Ricupero-Hovasse S. L., Natsoulis G., Boeke J. D., Winston F., 1994. SPT10 and SPT21 are required for transcription of particular histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 5223–5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley A. M., Rougeulle C., Winston F., 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13: 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein C. B., Cross F. R., 1994. Genes that can bypass the CLN requirement for Saccharomyces cerevisiae cell cycle START. Mol. Cell. Biol. 14: 2041–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson P. R., Mendiratta G., McLaughlin N. B., Wolfsberg T. G., Marino-Ramirez L., et al. , 2005. Global regulation by the yeast Spt10 protein is mediated through chromatin structure and the histone upstream activating sequence elements. Mol. Cell. Biol. 25: 9127–9137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson P. R., Ganguli D., Clark D. J., 2011. Spt10 and Swi4 control the timing of histone H2A/H2B gene activation in budding yeast. Mol. Cell. Biol. 31: 557–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson P. R., Ganguli D., Nagarajavel V., Clark D. J., 2012. Regulation of histone gene expression in budding yeast. Genetics 191: 7–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassler J. S., Winston F., 1988. Isolation and analysis of a novel class of suppressor of Ty insertion mutations in Saccharomyces cerevisiae. Genetics 118: 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J., Kainth P., Lambert J. P., van Bakel H., Tsui K., et al. , 2009. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol. Cell 35: 340–351 [DOI] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H., 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553 [DOI] [PubMed] [Google Scholar]
- Green E. M., Antczak A. J., Bailey A. O., Franco A. A., Wu K. J., et al. , 2005. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 15: 2044–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog G. A., Wada T., Handa H., Winston F., 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12: 357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A. B., Moreno S., 2011. Lsm1 promotes genomic stability by controlling histone mRNA decay. EMBO J. 30: 2008–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess D., 2004. Genetic and Molecular Analysis of Spt10 and Spt21 of Saccharomyces Cerevisiae: Roles in Histone Gene Transcription and Other Chromatin-Related Processes. Harvard University, Cambridge, MA [Google Scholar]
- Hess D., Winston F., 2005. Evidence that Spt10 and Spt21 of Saccharomyces cerevisiae play distinct roles in vivo and functionally interact with MCB-binding factor, SCB-binding factor and Snf1. Genetics 170: 87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess D., Liu B., Roan N. R., Sternglanz R., Winston F., 2004. Spt10-dependent transcriptional activation in Saccharomyces cerevisiae requires both the Spt10 acetyltransferase domain and Spt21. Mol. Cell. Biol. 24: 135–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Zhou H., Tarara J., Zhang Z., 2007. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. EMBO J. 26: 2274–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbeault D., Gamar L., Rufiange A., Paquet E., Nourani A., 2008. The Rtt106 histone chaperone is functionally linked to transcription elongation and is involved in the regulation of spurious transcription from cryptic promoters in yeast. J. Biol. Chem. 283: 27350–27354 [DOI] [PubMed] [Google Scholar]
- Kaufman P. D., Cohen J. L., Osley M. A., 1998. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 18: 4793–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A., Zimmerman E., Schneider M., Hurt E., Zheng N., 2010. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Snyder M., 2002. Protein complexes take the bait. Nature 415: 123–124 [DOI] [PubMed] [Google Scholar]
- Kurat C. F., Lambert J. P., van Dyk D., Tsui K., van Bakel H., et al. , 2011. Restriction of histone gene transcription to S phase by phosphorylation of a chromatin boundary protein. Genes Dev. 25: 2489–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laman H., Balderes D., Shore D., 1995. Disturbance of normal cell cycle progression enhances the establishment of transcriptional silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 3608–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E., Winston F., 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15: 1946–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. S., Hines L. K., Levin D. E., 1993. A pair of functionally redundant yeast genes (PPZ1 and PPZ2) encoding type 1-related protein phosphatases function within the PKC1-mediated pathway. Mol. Cell. Biol. 13: 5843–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D. J., 2000. Cell-cycle checkpoints that ensure coordination between nuclear and cytoplasmic events in Saccharomyces cerevisiae. Curr. Opin. Genet. Dev. 10: 47–53 [DOI] [PubMed] [Google Scholar]
- Liu W. H., Roemer S. C., Port A. M., Churchill M. E., 2012. CAF-1-induced oligomerization of histones H3/H4 and mutually exclusive interactions with Asf1 guide H3/H4 transitions among histone chaperones and DNA. Nucleic Acids Res. 40: 11229–11239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. J., Lu Q., Grunstein M., 1996. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 10: 1327–1340 [DOI] [PubMed] [Google Scholar]
- Mendiratta G., Eriksson P. R., Shen C. H., Clark D. J., 2006. The DNA-binding domain of the yeast Spt10p activator includes a zinc finger that is homologous to foamy virus integrase. J. Biol. Chem. 281: 7040–7048 [DOI] [PubMed] [Google Scholar]
- Mendiratta G., Eriksson P. R., Clark D. J., 2007. Cooperative binding of the yeast Spt10p activator to the histone upstream activating sequences is mediated through an N-terminal dimerization domain. Nucleic Acids Res. 35: 812–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munakata T., Adachi N., Yokoyama N., Kuzuhara T., Horikoshi M., 2000. A human homologue of yeast anti-silencing factor has histone chaperone activity. Genes Cells 5: 221–233 [DOI] [PubMed] [Google Scholar]
- Natsoulis G., Dollard C., Winston F., Boeke J. D., 1991. The products of the SPT10 and SPT21 genes of Saccharomyces cerevisiae increase the amplitude of transcriptional regulation at a large number of unlinked loci. New Biol. 3: 1249–1259 [PubMed] [Google Scholar]
- Natsoulis G., Winston F., Boeke J. D., 1994. The SPT10 and SPT21 genes of Saccharomyces cerevisiae. Genetics 136: 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald A. F., Landsman D., 1997. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22: 154–155 [DOI] [PubMed] [Google Scholar]
- Osley M. A., Lycan D., 1987. Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol. Cell. Biol. 7: 4204–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillus L., Rine J., 1989. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell 59: 637–647 [DOI] [PubMed] [Google Scholar]
- Prelich G., Winston F., 1993. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics 135: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochasson P., Florens L., Swanson S. K., Washburn M. P., Workman J. L., 2005. The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev. 19: 2534–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson H. E., Wittenberg C., Cross F., Reed S. I., 1989. An essential G1 function for cyclin-like proteins in yeast. Cell 59: 1127–1133 [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P., 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Samara N. L., Datta A. B., Berndsen C. E., Zhang X., Yao T., et al. , 2010. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328: 1025–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood P. W., Osley M. A., 1991. Histone regulatory (hir) mutations suppress delta insertion alleles in Saccharomyces cerevisiae. Genetics 128: 729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A., Lahudkar S., Durairaj G., Bhaumik S. R., 2012. Sgf29p facilitates the recruitment of TATA box binding protein but does not alter SAGA’s global structural integrity in vivo. Biochemistry 51: 706–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. C., Xu X., Kim H. S., Fillingham J., Kislinger T., et al. , 2012. The replication-independent histone H3–H4 chaperones HIR, ASF1, and RTT106 co-operate to maintain promoter fidelity. J. Biol. Chem. 287: 1709–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P. K., Murray A. W., 1992. S-phase feedback control in budding yeast independent of tyrosine phosphorylation of p34cdc28. Nature 355: 365–368 [DOI] [PubMed] [Google Scholar]
- Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K., et al. , 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9: 3273–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A., Bucaria J., Osley M. A., Sternglanz R., 2001. Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics 158: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M. S., Malone E. A., Winston F., 1991. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol. Cell. Biol. 11: 3009–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S., 2009. Lsm1–7-Pat1 complex: a link between 3′ and 5′-ends in mRNA decay? RNA Biol. 6: 228–232 [DOI] [PubMed] [Google Scholar]
- Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., et al. , 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- van Welsem T., Frederiks F., Verzijlbergen K. F., Faber A. W., Nelson Z. W., et al. , 2008. Synthetic lethal screens identify gene silencing processes in yeast and implicate the acetylated amino terminus of Sir3 in recognition of the nucleosome core. Mol. Cell. Biol. 28: 3861–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M., Freedman R., Van Arsdell S., Szostak J. W., Thorner J., 1987. The yeast ARD1 gene product is required for repression of cryptic mating-type information at the HML locus. Mol. Cell. Biol. 7: 3713–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F. D., Dollard C., Ricupero-Hovasse S. L., 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55 [DOI] [PubMed] [Google Scholar]
- Xu H., Kim U. J., Schuster T., Grunstein M., 1992. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 5249–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Zhang K., Grunstein M., 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121: 375–385 [DOI] [PubMed] [Google Scholar]
- Yamashita I., 1993. Isolation and characterization of the SUD1 gene, which encodes a global repressor of core promoter activity in Saccharomyces cerevisiae. Mol. Gen. Genet. 241: 616–626 [DOI] [PubMed] [Google Scholar]
- Zlatanova J., Seebart C., Tomschik M., 2007. Nap1: taking a closer look at a juggler protein of extraordinary skills. FASEB J. 21: 1294–1310 [DOI] [PubMed] [Google Scholar]
- Zunder R. M., Rine J., 2012. Direct interplay among histones, histone chaperones, and a chromatin boundary protein in the control of histone gene expression. Mol. Cell. Biol. 32: 4337–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]