Abstract
Lewis Y (LeY) antigen is an oligosaccharide that is highly expressed at the cell surface in various human cancers. Increased LeY expression activates epidermal growth factor receptor (EGFR) and human epidermal growth factor receptor 2 (HER2) and promotes cell proliferation in EGFR-overexpressing cells. However, the effect of downregulation of LeY expression on cell proliferation in HER2-overexpressing cells remains unknown. FUT1 encodes α1,2-fucosyltransferase, a key enzyme for LeY synthesis. We knocked down FUT1 by short interfering RNA (siRNA) in four HER2-overexpressing human cancer cell lines, including NCI-N87, MKN7, SKBr3 and BT474. We investigated whether downregulation of LeY and alteration in the glycosylation status of these cells affect cell proliferation and HER2 activation. Knocking down FUT1 expression markedly inhibited proliferation of NCI-N87, which highly expressed EGFR and was sensitive to EGFR deprivation. Furthermore, FUT1 siRNA downregulated the total amount of HER2 protein, phosphorylation of HER2 and EGFR, and phosphorylation of extracellular signal-regulated kinase (ERK) in this cell line. Moreover, the marked downregulation of phosphorylation of HER2 and ERK was observed following short-time EGF-stimulation. These effects were not observed in the other three cell lines. Our results suggest that knockdown of FUT1 downregulates HER2 signaling via EGFR downregulation. FUT1 may serve as a new molecular target for HER2-overexpressing human cancers with activated EGFR signaling.
Keywords: α1,2-fucosyltransferase; Lewis Y antigen; HER2-overexpression; epidermal growth factor receptor; proliferation; short interfering RNA
Introduction
The cell membrane of mammalian cells comprises glycolipids, glycoproteins and proteoglycans; these carbohydrate structures undergo conformational changes during cellular differentiation and transformation (1). In particular, protein glycosylation accounts for the vast majority of post-translational processes that affect protein folding, stability, solubility and function (2). Glycosylation is catalyzed by glycosyltransferases. Among them, fucosyltransferases transfer an L-fucose sugar from a GDP-fucose donor substrate to an acceptor substrate (3). The fucosyltransferase gene family encodes enzymes that transfer fucose from α(1,2), α(1,3/4) and α(1,6) linkages to various glycans. FUT1 and FUT2 encode α(1,2)-fucosyltransferases, which transfer a terminal fucose residue from an α(1,2)-linkage to an existing galactose Type 1 or 2 precursor substance and form the H1 or H2 antigen as precursors of soluble ABH antigens, respectively (4). FUT1 is ubiquitously expressed in the human body and preferentially expressed in erythroid tissues and vascular endothelial cells. FUT2 is mainly expressed in the epithelial cells of the digestive and respiratory tracts (4). FUT3-FUT7 and FUT9 encode α(1,3)-fucosyltransferases, and their gene products transfer a fucose residue from an α(1,3)-linkage to galactose in H1 and H2 antigens to produce various Lewis antigens (5,6).
Antigens of the ABH and Lewis histo-blood group family can be found on the cell surface of various normal cells, mainly epithelial cells. However, the expression of various carbohydrate epitopes of this family is altered in carcinomas (7). For example, Lewis Y antigen (LeY), a Lewis antigen, is expressed in various cancer cells, including breast, ovarian and colorectal cancer; its expression is often associated with poor prognosis (8–12). In addition, forced FUT1 and FUT2 expression in human ovarian carcinoma-derived RMG-I cells increases activity of α(1,2)-fucosyltransferase and LeY antigen and promotes cell proliferation and resistance against anticancer drugs, such as 5-FU and carboplatin (13,14).
The molecular mechanisms through which overexpression of LeY antigen induces a malignant phenotype remain to be elucidated. However, increased LeY expression induced by FUT1 and FUT2 overexpression activates the epidermal growth factor receptor (EGFR) signaling and induces increase in mRNA expression and protein levels of human epidermal growth factor receptor 2 (HER2), a member of EGFR family (15). Furthermore, an FUT1- and FUT2-overexpressing cell line proliferates more aggressively than the parent cell line (15), suggesting that activation of EGFR and HER2 induces a malignant phenotype in human cancer cells.
HER2 is a transmembrane glycoprotein that is fucosylated by fucosyltransferase. It is involved in transmitting signals that stimulate cell division (16). HER2-overexpression is caused by amplification of the HER2 gene and it is observed in various cancers (17,18), including breast and gastric cancer. Clinically, it is a molecular target of trastuzumab, a monoclonal antibody against HER2 (18,19). However, whether alteration of glycosylation affects cell proliferation in HER2-overexpressing cancer cells remains to be investigated. In this study, we examined the effect of FUT1 suppression on the HER2 pathway and cell proliferation.
Materials and methods
Cell lines and cell culture
In this study, we used four HER2-overexpressing human cancer cell lines, NCI-N87 and MKN7 (derived from gastric cancer) and SKBr3 and BT474 (derived from breast cancer), and two cell lines, A431 and VMRC-LCD (derived from lung cancers), that do not overexpress HER2. NCI-N87, SKBr3, BT474 and VMRC-LCD were obtained from the American Type Culture Collection (Manassas, VA, USA). MKN7 and A431 were provided by the Cell Resource Center for Biomedical Research (Institute of Development, Aging and Cancer, Tohoku University, Sendai, Japan). All cell lines were maintained in RPMI-1640 medium (Sigma, St. Louis, MO, USA) supplemented with 10% heat-inactivated FBS (Gibco, Grand Island, NY, USA) and incubated at 37°C in a 5% CO2 humidified atmosphere.
Preparation of siRNA and transfection
The following three pairs of siRNA oligomers were designed according to the sequence of human FUT1 (GenBank accession number: NM_000148): FUT1-1 siRNA, 5′-AAAGGAUCUCUCAAGUC CGCGTT-3′ and 5′-CGCGGACUUGAGAGAUCCUUUTT-3′; FUT1-2 siRNA, 5′-GCUACACCGUGGAAAGACUTT-3′ and 5′-AGUCUUUCCACGGUGUAGCTT-3′; FUT1-3 siRNA, 5′-UCGAUGUUUUCUUUACACCAC-3′ and 5′-GGUGUAA AGAAAACAUCGACA-3′.
FUT1-1 and FUT1-2 siRNAs were designed based on a previous report (20) and the resource of Open Biosystems (http://www.openbiosystems.com), respectively. FUT1-3 siRNA was designed using a web-based online software system (siDirect version 2.0, http://sidirect2.rnai.jp). These siRNAs were chemically synthesized by Hokkaido System Science, Co., Ltd. (Hokkaido, Japan).
Signal Silence EGF Receptor siRNA 1 and 2 (Santa Cruz Biotechnology, Inc., CA, USA) were used as EGFR-siRNA1 and 2, respectively. Negative control siRNA (Silencer Negative Control no. 1 siRNA) was obtained from Ambion, Inc. (Austin, TX, USA). Cells were seeded in a 6- or 96-well plate. After 24 h, the cells were transfected with siRNA (100 nM final concentration) using Dharmafect 2 reagent (Dharmacon, Lafayette, CO, USA).
Measurement of mRNA expression by real-time PCR
Real-time polymerase chain reaction (PCR) was used to measure the mRNA expression of FUT1 in the cells. Cells were plated at 1.5×104 in a 96-well plate and transfected with siRNA (100 nM final concentration) as described above. Twenty-four hours post-transfection, the total-RNA from the cells was extracted using the RealTime ready cell lysis kit (Roche Diagnostics GmbH, Mannheim, Germany) and reverse transcribed using the Transcriptor First Strand cDNA synthesis kit (Roche Diagnostics GmbH) with oligo(dt) primer. Real-time PCR was performed using SsoFast™ EvaGreen Supermix (Bio-Rad, Richmond, CA, USA) and gene-specific primers in a thermal cycler CFX96 real-time PCR detection system (Bio-Rad). The primers used for amplification were FUT1 F, 5′-AACGCCTCCTCTTCCTGTC-3′ and R, 5′-TGGGG TAGACAGTCCAGGTG-3′; glyceraldehyde 3-phosphate dehydrogenase (GAPDH) F, 5′-GAAGGTGAAGGTCG GAGTC-3′ and R, 5′-GAAGATGGTGATGGGATTTC-3′ (GenBank accession no. NM_002046). The designs of both primers have been previously described (21). Quantified data were normalized to GAPDH. The PCR program included 45 cycles of 95°C for 1 sec and 60°C for 5 sec. After the PCR reaction was completed, a melting curve analysis was performed. Each primer pair produced a single and sharp peak, thereby indicating that the primers amplified only one specific PCR product. No primer dimers were observed. All samples were amplified in triplicate.
Western blotting
Cells were plated at 2.0×105 in a 6-well plate and transfected with siRNA (100 nM final concentration) as described above. After 72 h, the cells were washed with cold PBS and harvested with lysis buffer [50 mM Tris-HCl (pH 8.0), 150 nM NaCl, 5 nM EDTA, 1% NP-40, protease inhibitor cocktail (Roche Diagnostics GmbH) and phosphatase inhibitor (Roche Diagnostics GmbH)]. In the short-time EGF stimulation experiment, 72 h post-transfection, the cells were starved in serum-free medium for 12 h and stimulated with EGF (10 ng/ml) for 10 min. The cells were washed immediately and harvested as described above.
Cell lysates were separated by 12.5% SDS-PAGE and blotted onto a PVDF membrane. The membranes were blocked with the Odyssey blocking buffer (Li-Cor Biosciences, Lincoln, NE, USA) and then probed with polyclonal anti-HER2 antibody (Dako, Carpinteria, CA, USA), monoclonal anti-phosphorylated HER2 antibody (Tyr1248; Santa Cruz Biotechnology, Inc.), monoclonal anti-EGFR antibody (Santa Cruz Biotechnology, Inc.), monoclonal anti-phosphorylated EGFR antibody (Tyr1068), monoclonal anti-GAPDH antibody (Santa Cruz Biotechnology, Inc.), anti-ERK1/2 antibody (Cell Signaling Technology, Inc.), anti-phosphorylated ERK1/2 antibody (Thr202/Tyr204; Cell Signaling Technology, Inc.) and anti-LeY antibody (Abcam, Cambridge, UK), followed by incubation with a goat anti-rabbit or a goat anti-mouse Alexa Fluor 680 IgG secondary antibody (Invitrogen, Carlsbad, CA, USA). Protein bands were detected and quantified using the Odyssey system (Li-Cor Biosciences).
Cell proliferation assay
Cells were plated at 3.0×103 in a 96-well plate and transfected with siRNA (100 nM final concentration) as described above. At 0, 72 and 120 h the cells were harvested and cell viability was determined using the Cell Counting kit-8 (Dojin Laboratories, Kumamoto, Japan), which measures mitochondrial succinate dehydrogenase activity. Briefly, 10 μl of 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium monosodium salt (WST-8) solution was added to each well. After a 2-h incubation at 37°C, the resulting water-soluble formazan dye was assayed by a microplate autoreader SpectraMax (Molecular Devices, Sunnyvale, CA, USA) at a wavelength of 450 nm with a reference of 630 nm. All experiments were performed in triplicate.
Cell cycle analysis using fluorescence-activated cell sorter (FACS)
Cells were plated at 2.0×105 in a 6-well plate and transfected with siRNA (100 nM final concentration) as described above. After a 72-h incubation, cells were treated with trypsin and fixed with 70% ethanol in PBS overnight. The cells were then washed once with PBS, incubated in the presence of RNase A (0.25 mg/ml) for 30 min at 37°C, collected by centrifugation at 200 × g for 5 min and stained with propidium iodide (50 μl/ml). The cells were filtered through a 50-μm pore size nylon mesh and analyzed for cell cycle using a FACS system (Beckman Coulter, Miami, FL, USA).
Statistical analysis
All experiments were performed independently and in triplicate. Data are expressed as means ± standard error. Statistical data were analyzed using Student’s t-test. Significance was set at P<0.05.
Results
FUT1 knockdown by siRNA decreases LeY antigen expression
To detect the effects of FUT1 siRNAs, we performed real-time PCR analysis (Fig. 1A) and western blotting of LeY antigen (Fig. 1B). FUT1 mRNA and LeY antigen expression were reduced in all cells transfected with FUT1 siRNAs, but not in those transfected with control siRNA. Compared with the cells transfected with control siRNA, the level of LeY expression in the cells transfected with FUT1-1, FUT1-2 or FUT1-3 siRNAs was FUT1-1 67.9, FUT1-2 44.5 and FUT1-3 46.9% in NCI-N87 cells; FUT1-1 50.5, FUT1-2 58.6 and FUT1-3 51.7% in MKN7 cells; FUT1-1 49.2, FUT1-2 33.3 and FUT1-3 62.3% in SKBr3 cells; and FUT1-1 54.1, FUT1-2 41.1 and FUT1-3 58.8% in BT474 cells. These results indicated that FUT1 siRNAs efficiently reduced the levels of FUT1 mRNA and LeY antigen expression.
Figure 1.
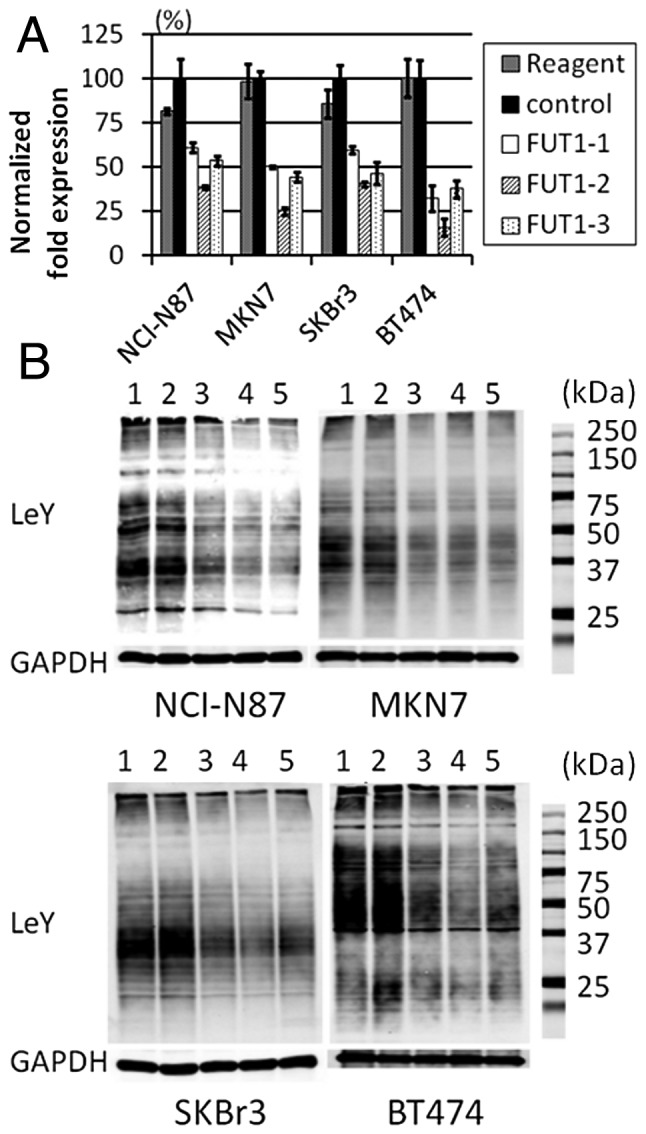
(A) Suppression of FUT1 mRNA expression after transfection of siRNAs was analyzed by real-time PCR. The level of FUT1 mRNA expression was determined and plotted as fold change relative to the control. Data were normalized to the amount of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA. (B) Alteration of LeY expression after transfection of siRNAs in each whole cell lysate. GAPDH is shown as a loading control. Lane 1, reagent; lane 2, control; lane 3, FUT1-1; lane 4, FUT1-2; and lane 5, FUT1-3. A molecular weight standard is shown at the right side.
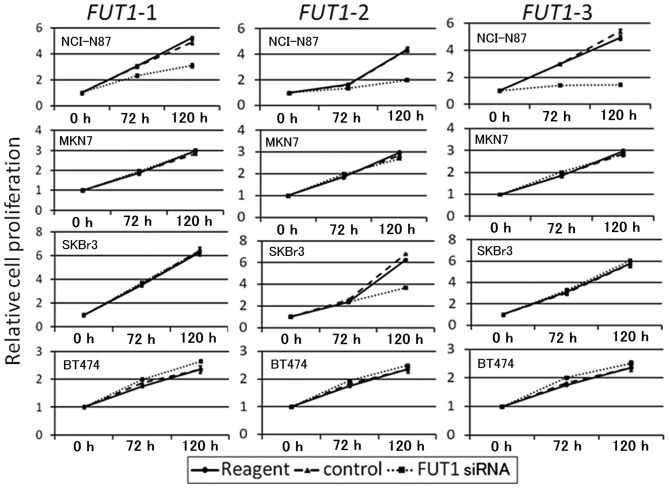
FUT1 knockdown inhibits cell proliferation in NCI-N87 cells, but not in other cell lines
To examine the effect of siRNA-mediated FUT1 knockdown on cell growth, cell proliferation assays were performed for the four HER2-overexpressing cell lines. Data are shown in Fig. 2. FUT1 siRNAs inhibited NCI-N87 cell proliferation 120 h post-transfection, whereas they did not inhibit proliferation in MKN7 or BT474 cells. Although FUT1-2 suppressed proliferation in SKBr3 cells, FUT1-1 and FUT1-3 did not. Therefore this was considered to be an off-target siRNA effect.
Figure 2.
The growth curves after transfection with FUT1 siRNA. Cells were assayed for growth 0, 72 and 120 h after transfection. The absorbance at 450 nm at 0 h was normalized to one. Absorbance at subsequent time points was plotted relative to the initial value.
FUT1 knockdown leads to apoptosis in NCI-N87 cells
To examine whether FUT1 knockdown changes the proportion of cells in each cell cycle phase, FACS analysis was performed for NCI-N87 cells (Fig. 3). All siRNAs significantly increased the subG1 fraction (P<0.01). The G2/M fraction also increased, but not significantly (P=0.09). These results indicate that the downregulation of FUT1 mRNA and LeY antigen expression leads to apoptosis in NCI-N87 cells.
Figure 3.
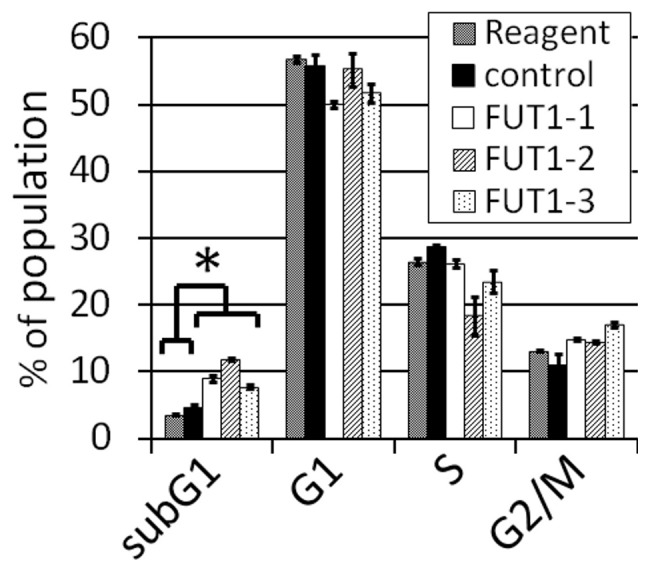
Analysis of the proportion of cells in each cell cycle phase after transfection with each FUT1 siRNA in NCI-N87 cells. Values shown are average ± SE (n=3). Values of the sub-G1, G1, S and G2/M fractions are portions of the total population. *P<0.01.
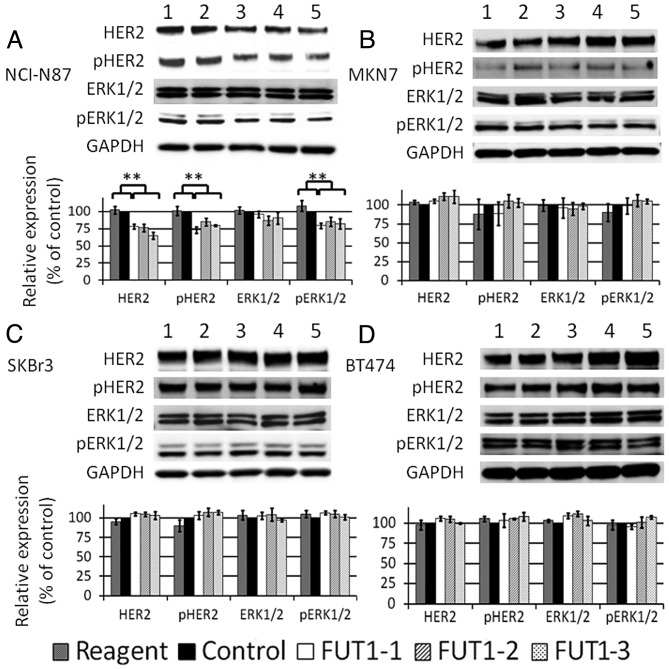
FUT1 knockdown downregulates the expression of HER2 and the phosphorylated HER2 (pHER2) and phosphorylated ERK1/2 (pERK) in NCI-N87 cells
To elucidate the mechanism of cell growth inhibition induced by FUT1 knockdown, HER2, pHER2, ERK1/2 and pERK were assessed by western blotting 72 h after transfection (we called it ‘normal cultural conditon’).
Representative western blotting data and bar charts by triplicate experiments are shown in Fig. 4. FUT1 knockdown significantly downregulated the total amount of HER2 and pHER2 in NCI-N87 cells (Fig. 4A). The amount of pERK also decreased, but the total amount of ERK remained unchanged. In contrast to NCI-N87, no significant changes were observed in MKN7, SKBr3 or BT474 cells (Fig. 4B-D).
Figure 4.
The effect of downregulating FUT1 on protein expression and tyrosine phosphorylation of HER2 and ERK1/2 in normal culture condition. The bar chart shows the relative expression ratio of each protein by western blotting. Each value is calculated as the ratio of signal intensity compared to that of control. Data were normalized to signal intensity of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Lane 1, reagent; lane 2, control; lane 3, FUT1-1; lane 4, FUT1-2; and lane 5, FUT1-3. (A) NCI-N87, (B) MKN7, (C) SKBr3 and (D) BT474 cells. **P<0.05.
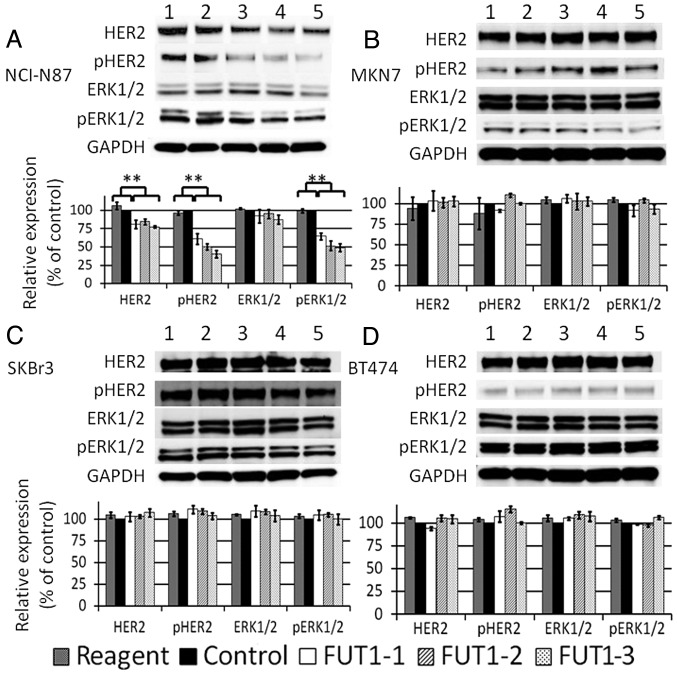
FUT1 knockdown strongly downregulates pHER2 and pERK following short-time EGF stimulation in NCI-N87 cells
To examine whether short-time EGF-stimulation alters downregulation of HER2 and ERK1/2 by FUT1 knockdown, we administered EGF for 10 min after starvation of the cells and assessed the amount of HER2, pHER2, ERK1/2 and pERK. The amount of pHER2 and pERK was markedly reduced in NCI-N87 cells (Fig. 5A). This reduction was more apparent than that of normal culture condition (Fig. 4A). Alterations in HER2 and ERK1/2 levels were similar to those observed in normal culture condition. In contrast, no significant changes were observed following EGF stimulation in MKN7, SKBr3 or BT474 cells (Fig. 5B-D).
Figure 5.
The effect of downregulating FUT1 on protein expression and tyrosine phosphorylation of HER2 and ERK1/2 under EGF-stimulated condition. The bar chart shows the relative expression ratio of each protein found by western blotting. Each value is calculated as the ratio of signal intensity compared to that of control. Data were normalized to signal intensity of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Lane 1, reagent; lane 2, control; lane 3, FUT1-1; lane 4, FUT1-2; and lane 5, FUT1-3. (A) NCI-N87, (B) MKN7, (C) SKBr3 and (D) BT474 cells. **P<0.05.
FUT1 knockdown downregulates EGFR signaling in NCI-N87
To examine whether FUT1 suppression affects EGFR signaling, first, western blotting for EGFR expression was performed (Fig. 6A). EGFR expression in each cell line was lower than that in the EGFR-overexpressing A431 cell line, whereas EGFR expression in NCI-N87 was higher than that in other HER2-overexpressing cell lines. Then, we investigated EGFR signaling of NCI-N87 after FUT1 suppression. The result showed that phosphorylation of EGFR was downregulated in both normal cultural and EGF-stimulating conditions. The total amount of EGFR did not change in either of the conditions (Fig. 6B and C).
Figure 6.
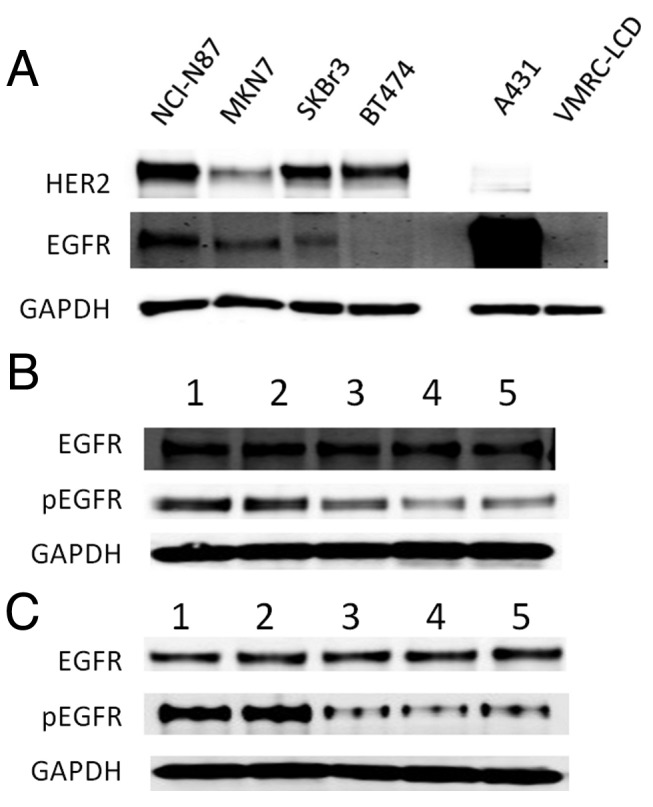
(A) HER2 and EGFR expression in each cell line was analyzed by western blotting. A431 is shown as a positive control for EGFR-overexpression and VMRC-LCD is shown as a negative control. (B) The effect of downregulating FUT1 on protein expression and tyrosine phosphorylation of EGFR under normal cultural condition in NCI-N87 cells. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is shown as the loading control. (C) The effect of downregulating FUT1 on protein expression and tyrosine phosphorylation of EGFR under EGF-stimulated condition in NCI-N87 cells. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is shown as the loading control. Lane 1, reagent; lane 2, control; lane 3, FUT1-1; lane 4, FUT1-2; and lane 5, FUT1-3.
Suppression of EGFR signaling downregulates HER2 signaling and proliferation of NCI-N87 cells
To confirm the proliferation dependence of EGFR signaling the cell lines were transfected with EGFR siRNA. EGFR siRNA-1 and EGFR siRNA-2 suppressed the proliferation of NCI-N87 cells by 23.6 and 44.7%, respectively. However the proliferation of other cell lines were not changed by EGFR knockdown (Fig. 7). Next, to investigate whether EGFR suppression affects HER2 signaling in NCI-N87 cells, western blotting was performed. EGFR suppression downregulated HER2, pHER2 and pERK as well as FUT1 suppression in NCI-N87 cells (Fig. 8). The results indicated that the proliferation of NCI-N87 cells was also dependent on EGFR signaling and EGFR suppression resulted in downregulation of HER2 signaling in this cell line.
Figure 7.
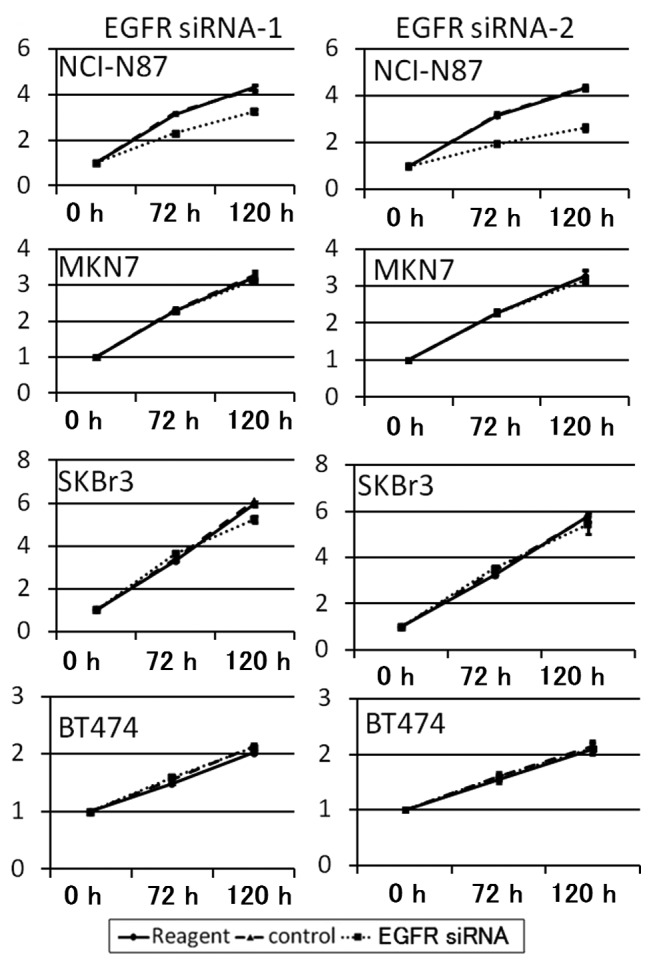
The growth curves after transfection with EGFR siRNA-1 and EGFR siRNA-2. Cells were assayed for growth 0, 72 and 120 h after transfection. The absorbance at 450 nm at 0 h was normalized to one. Absorbance at subsequent time points was plotted relative to the initial value.
Figure 8.
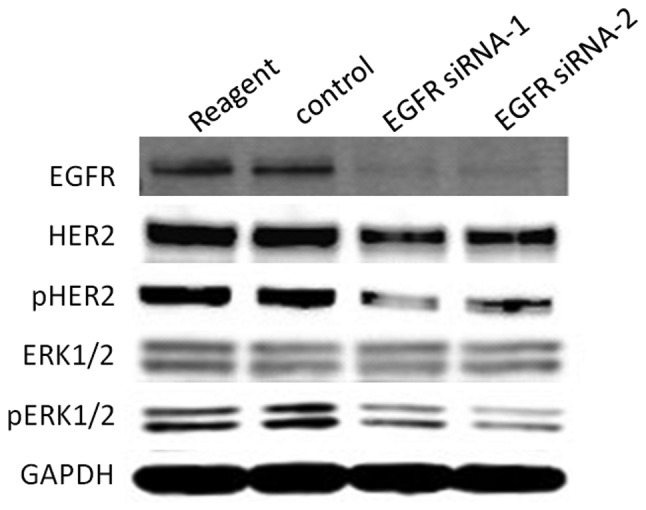
The effect of EGFR knockdown on protein expression and tyrosine phosphorylation of HER2 and ERK1/2 in NCI-N87 cells. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is shown as the loading control.
Discussion
LeY antigen belongs to the histo-blood group antigens and α1,2-fucosyltransferase is the key enzyme, which also FUT1 and FUT2 encode. Previous studies suggested that forced expression of α1,2-fucosyltransferase in RMG-I human ovarian cancer cell line caused overexpression of LeY antigen and promoted cell proliferation via activation of EGFR and HER2 (15). Furthermore, Palumberi et al(20) indicated that suppression of α1,2-fucosyltransferase inhibited the cell proliferation of the EGFR-overexpressing cell line A431. In the present study, we attempted to suppress FUT1 gene by its specific siRNA and observe whether FUT1 knockdown affected the cell proliferation of HER2-overexpressing cell lines.
Our results indicated that FUT1 siRNA downregulated FUT1 mRNA and altered fucosylation; it was shown by inhibition of LeY antigen expression, in four HER2-overexpressing cell lines. However, the effects on cell proliferation varied. In NCI-N87 cells, FUT1 suppression decreased the total amount of HER2, pHER2 and pERK, and inhibited cell proliferation. However, FUT1 suppression in MKN7, SKBr3 and BT474 did not alter HER2, pHER2 or pERK levels and did not affect cell proliferation.
In a previous study, HER2 inhibition led to suppression of cell proliferation in HER2-overexpressing cell lines (22–24). This observation is similar to our results and indicates that HER2 plays an important role in cell proliferation in HER2-overexpressing cells.
In addition, our study suggested that EGFR signaling was involved in FUT1-mediated inhibition of HER2 signaling in NCI-N87 cells. The experiment of EGFR siRNA transfection indicated that the proliferation of NCI-N87 cells was depentdent not only on HER2 signaling but also on EGFR signaling and EGFR suppression led to HER2 signaling inhibition. Previous studies indicated that cetuximab, a monoclonal antibody against EGFR, inhibits cell proliferation in NCI-N87 cells (25), but not in SKBr3 or BT474 cells (24). These results suggest that EGFR potently contributes to the proliferation of NCI-N87 cells.
We speculate that FUT1 suppression leads to HER2 inhibition and cell proliferation via EGFR signaling inhibition through one or both mechanisms described below.
First, downregulation of EGFR by FUT1 suppression may attenuate HER2 transcription. Liu et al(15) reported that FUT1-overexpression upregulated EGFR signaling and increased mRNA expression and protein levels of HER2. In our study, FUT1 knockdown decreased the total amount of HER2 in NCI-N87 cells. Therefore, FUT1 knockdown may have decreased HER2 levels by downregulating EGFR signaling. Since the level of attenuation of the total amount of HER2 was similar to that of pHER2, a reduction of the total amount of HER2 may cause downregulation of pHER2 and pERK in normal culture condition.
Second, FUT1 suppression may attenuate EGFR and HER2 heterodimer formation. HER2 forms homodimers or heterodimers with other EGFR family proteins, undergoes autophosphorylation at specific tyrosine residues of its intracellular domain and mediates signal transduction (17). In addition, EGFR forms homodimers or heterodimers with other EGFR family proteins following ligand stimulation (25).
Following starvation and short-time EGF stimulation, phosphorylation of HER2 and ERK1/2 was markedly reduced in FUT1-suppressed NCI-N87 cells. Zhang et al(26) reported that the suppression of FUT1 and FUT4 reduced LeY antigen, decreased binding of EGF to EGFR and resulted in inhibition of cell proliferation. In addition, some reports have shown that fucosylation on EGFR alters the binding affinity of EGF to EGFR and affects EGFR dimerization (2,27,28).
Hence, we propose that FUT1 suppression caused an alteration of fucosylation and attenuated EGF-mediated EGFR and HER2 heterodimerization.
Besides, our results indicated that apoptosis occurred in FUT1-mediated growth inhibition in NCI-N87 cells. G2/M fraction also tended to increase but not significantly. Previous studies revealed that HER2 inhibition by trastuzumab caused apoptosis in some HER2-overexpressing cell lines, e.g. SKBr3 or Calu-3 (29). However, it did not cause apoptosis in SKOV-3 which had HER2-overexpression (30). Hence, it is possible that HER2 suppression causes various effects on cell proliferation among each cell line.
Lapatinib is a dual tyrosine kinase inhibitor for EGFR and HER2 and is used to treat trastuzumab-resistant HER2 positive cancers. Redundant signaling from other EGFR family members is one of the molecular mechanisms of drug resistance to trastuzumab (31). Inhibition of EGFR and HER2 signaling is one strategy for treating trastuzumab-resistant HER2 positive cancers.
The role of fucosylation in cell proliferation is not completely understood. However, our results demonstrate that FUT1 knockdown results in the inhibition of cell proliferation and reduction of HER2, pHER2 and pERK in NCI-N87 cells. The reduction of pHER2 and pERK seems to depend on the reduction of EGFR signaling caused by inhibition of fucosylation. Further studies are necessary to identify a biomarker to predict which HER2-positive cancer cells are sensitive to FUT1 inhibition. The development of a fucosyltransferase inhibitor may constitute a novel drug for trastuzumab-resistant HER2 positive cancers.
Acknowledgements
The authors thank Satoko Aoki for her technical assistance.
References
- 1.Roseman S. Reflections on glycobiology. J Biol Chem. 2001;276:41527–41542. doi: 10.1074/jbc.R100053200. [DOI] [PubMed] [Google Scholar]
- 2.Liu YC, Yen HY, Chen CY, et al. Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proc Natl Acad Sci USA. 2011;108:11332–11337. doi: 10.1073/pnas.1107385108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Javaud C, Dupuy F, Maftah A, et al. The fucosyltransferase gene family: an amazing summary of the underlying mechanisms of gene evolution. Genetica. 2003;118:157–170. [PubMed] [Google Scholar]
- 4.Matzhold EM, Helmberg W, Wagner T, et al. Identification of 14 new alleles at the fucosyltransferase 1, 2, and 3 loci in Styrian blood donors, Austria. Transfusion. 2009;49:2097–2108. doi: 10.1111/j.1537-2995.2009.02293.x. [DOI] [PubMed] [Google Scholar]
- 5.Dettke M, Pálfi G, Loibner H. Activation-dependent expression of the blood group-related Lewis Y antigen on peripheral blood granulocytes. J Leukoc Biol. 2000;68:511–514. [PubMed] [Google Scholar]
- 6.Hokke CH, Neeleman AP, Koeleman CA, van den Eijinden DH. Identification of an α3-fucosyltransferase and a novel α2-fucosyltransferase activity in cercariae of the schistosome Trichobilharzia ocellata: biosynthesis of the Fucα1-->2Fucα1-->3[Gal(NAc)β1-->4] GlcNAc sequence. Glycobiology. 1998;8:393–406. doi: 10.1093/glycob/8.4.393. [DOI] [PubMed] [Google Scholar]
- 7.Nakagoe T, Fukushima K, Itoyanagi N, et al. Expression of ABH/Lewis-related antigens as prognostic factors in patients with breast cancer. J Cancer Res Clin Oncol. 2002;128:257–264. doi: 10.1007/s00432-002-0334-5. [DOI] [PubMed] [Google Scholar]
- 8.Tsuboi K, Asao T, Ide M, et al. α1,2-fucosylation is a superior predictor of postoperative prognosis for colorectal cancer compared with blood group A, B, or sialyl Lewis X antigen generated within colorectal tumor tissue. Ann Surg Oncol. 2007;14:1880–1889. doi: 10.1245/s10434-007-9363-2. [DOI] [PubMed] [Google Scholar]
- 9.Madjd Z, Parsons T, Watson NF, Spendlove I, Ellis I, Durrant LG. High expression of Lewis y/b antigens is associated with decreased survival in lymph node negative breast carcinomas. Breast Cancer Res. 2005;7:R780–787. doi: 10.1186/bcr1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arai Y, Nishida M. Differential diagnosis between normal endometrium and endometrial hyperplasia with immunostaining cytology using anti-LeY monoclonal antibody. Int J Gynecol Cancer. 2003;13:42–46. doi: 10.1046/j.1525-1438.2003.13009.x. [DOI] [PubMed] [Google Scholar]
- 11.Kim YS, Yuan M, Itzkowitz SH, et al. Expression of LeY and extended LeY blood group-related antigens in human malignant, premalignant, and non-malignant colonic tissues. Cancer Res. 1986;46:5985–5992. [PubMed] [Google Scholar]
- 12.Kitamura K, Stockert E, Garin-Chesa P, et al. Specificity analysis of blood group Lewis-y (Le(y)) antibodies generated against synthetic and natural Le(y) determinants. Proc Natl Acad Sci USA. 1994;91:12957–12961. doi: 10.1073/pnas.91.26.12957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwamori M, Tanaka K, Kubushiro K, et al. Alterations in the glycolipid composition and cellular properties of ovarian carcinoma-derived RMG-1 cells on transfection of the α1, 2-fucosyltransferase gene. Cancer Sci. 2005;96:26–30. doi: 10.1111/j.1349-7006.2005.00005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Lin B, Hao YY, et al. The effects of Lewis (y) antigen content on drug resistance to carboplatin in ovarian cancer line RMG-I. Prog Biochem Biophys. 2008;35:1175–1182. [Google Scholar]
- 15.Liu JJ, Lin B, Hao YY, et al. Lewis(y) antigen stimulates the growth of ovarian cancer cells via regulation of the epidermal growth factor receptor pathway. Oncol Rep. 2010;23:833–841. [PubMed] [Google Scholar]
- 16.Brennan PJ, Kumogai T, Berezov A, et al. HER2/Neu: mechanisms of dimerization/oligomerization. Oncogene. 2000;19:6093–6101. doi: 10.1038/sj.onc.1203967. [DOI] [PubMed] [Google Scholar]
- 17.Slamon DJ, Godolphin W, Jones LA. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 18.Gravalos C, Jimeno A. HER2 in Gastric Cancer: A New Prognostic Factor and a Novel Therapeutic Target. Ann Oncol. 2008;19:1523–1529. doi: 10.1093/annonc/mdn169. [DOI] [PubMed] [Google Scholar]
- 19.Owens MA, Horten BC, Da Silva MM. HER2 amplification ratios by fluorescence in situ hybridization and correlation with immunohistochemistry in a cohort of 6556 breast cancer tissues. Clin Breast Cancer. 2004;5:63–69. doi: 10.3816/cbc.2004.n.011. [DOI] [PubMed] [Google Scholar]
- 20.Palumberi D, Aldi S, Ermini L, et al. RNA-mediated gene silencing of FUT1 and FUT2 influences expression and activities of bovine and human fucosylated nucleolin and inhibits cell adhesion and proliferation. J Cell Biochem. 2010;111:229–238. doi: 10.1002/jcb.22692. [DOI] [PubMed] [Google Scholar]
- 21.Chang WW, Lee CH, Lee P, et al. Expression of Globo H and SSEA3 in breast cancer stem cells and the involvement of fucosyl transferases 1 and 2 in Globo H synthesis. Proc Natl Acad Sci USA. 2008;105:11667–11672. doi: 10.1073/pnas.0804979105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mittendorf EA, Liu Y, Tucker SL, et al. A novel interaction between HER2/neu and cyclin E in breast cancer. Oncogene. 2010;29:3896–3907. doi: 10.1038/onc.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanner M, Hollmén M, Junttila TT, et al. Amplification of HER-2 in gastric carcinoma: association with Topoisomerase IIa gene amplification, intestinal type, poor prognosis and sensitivity to trastuzumab. Ann Oncol. 2005;16:273–278. doi: 10.1093/annonc/mdi064. [DOI] [PubMed] [Google Scholar]
- 24.Brockhoff G, Heckel B, Schmidt-Bruecken E, et al. Differential impact of Cetuximab, Pertuzumab and Trastuzumab on BT474 and SK-BR-3 breast cancer cell proliferation. Cell Prolif. 2007;40:488–507. doi: 10.1111/j.1365-2184.2007.00449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel D, Bassi R, Hooper A, et al. Anti-epidermal growth factor receptor monoclonal antibody cetuximab inhibits EGFR/HER-2 heterodimerization and activation. Cancer Sci. 2008;99:1611–1617. [PubMed] [Google Scholar]
- 26.Zhang Z, Sun P, Liu J, et al. Suppression of FUT1/FUT4 expression by siRNA inhibits tumor growth. Biochim Biophys Acta. 2008;1783:287–296. doi: 10.1016/j.bbamcr.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Miyoshi E, Moriwaki K, Nakagawa T. Biological function of fucosylation in cancer biology. J Biochem. 2008;143:725–729. doi: 10.1093/jb/mvn011. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Gu J, Ihara H, et al. Core fucosylation regulates epidermal growth factor receptor-mediated intracellular signaling. J Biol Chem. 2006;281:2572–2577. doi: 10.1074/jbc.M510893200. [DOI] [PubMed] [Google Scholar]
- 29.Dogan I, Cumaoglu A, Aricioglu A, Ekmekci A. Inhibition of ErbB2 by herceptin reduces viability and survival, induces apoptosis and oxidative stress in Calu-3 cell line. Mol Cell Biochem. 2011;347:41–51. doi: 10.1007/s11010-010-0610-7. [DOI] [PubMed] [Google Scholar]
- 30.Bijman MN, van Berkel MP, Kok M, Janmaat ML, Boven E. Inhibition of functional HER family members increases the sensitivity to docetaxel in human ovarian cancer cell lines. Anticancer Drugs. 2009;20:450–460. doi: 10.1097/CAD.0b013e32832afc24. [DOI] [PubMed] [Google Scholar]
- 31.Kruser TJ, Wheeler DL. Mechanisms of resistance to HER family targeting antibodies. Exp Cell Res. 2010;316:1083–1100. doi: 10.1016/j.yexcr.2010.01.009. [DOI] [PubMed] [Google Scholar]