Abstract
Proper hepatocyte function is vital for survival; thus, unrepaired destruction of the parenchymal tissue leading to liver decompensation is devastating. Therefore, understanding the homeostatic process regulating liver regeneration is clinically important, and evidence that the aryl hydrocarbon receptor (AhR) can promote cell survival after intrinsic apoptotic stimuli is integral to the regenerative process. The current study uses primary hepatocytes to identify survival mechanisms consistent with normal AhR biology. Taking advantage of the Cre-lox system to manipulate AhR status, we designed a comprehensive microarray analysis to identify immediate and direct changes in the transcriptome concomitant with the loss of the AhR. As a result, we identified a unique data set with minimal overlap, compared with previous array studies, culminating in the identification of Stanniocalcin 2 (Stc2) as a novel receptor target gene previously reported to have a cytoprotective role in endoplasmic reticulum stress. The Stc2 promoter contains multiple putative xenobiotic response elements clustered in a 250-bp region that was shown to recruit the AhR by chromatin immunoprecipitation. Of interest, Stc2 gene expression is refractory to classic exogenous AhR agonists, but responds to cellular stress in an AhR-dependent mechanism consistent with a process promoting cell survival.
Introduction
The liver is an organ of immense complexity essential for survival, because other tissues and organs are unable to compensate for its myriad functions. Liver damage is common in clinical practice because of the prevalent use of alcohol, exposure to pharmacological agents, hepatotropic pathogens, and various other disease states. Because most forms of liver injury target the hepatocytes, either directly or indirectly, uncovering the hepatocyte cellular response mechanisms to injury is essential for understanding the molecular events involved in liver regeneration and for the development of therapeutic strategies designed to ameliorate liver function resulting from injury or disease. The aryl hydrocarbon receptor (AhR) is highly expressed in hepatocytes and implicated in physiologic liver homeostasis (Mitchell et al., 2006), including cell cycle control during liver regeneration.
The AhR is a cytosolic, ligand-activated transcription factor that functions in concert with the AhR nuclear translocator (Arnt) to regulate the expression of several genes in response to halogenated aromatic hydrocarbon ligands, such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (Okey et al., 1994; Schmidt and Bradfield, 1996). Ligand binding triggers AhR nuclear translocation, Arnt protein dimerization, and binding of the AhR/Arnt dimer to xenobiotic response elements (XRE) to modulate target gene expression (Lees and Whitelaw, 1999), although evidence for additional mechanisms has emerged (Ohtake et al., 2003; Marlowe et al., 2004; Vogel et al., 2007; Huang and Elferink, 2012). Numerous studies, designed to examine gene expression after AhR activation by an exogenous ligand, have repeatedly reported AhR-dependent transcriptional regulation of CYP1 genes associated with AhR’s traditional role in the adaptive metabolism of xenobiotics (Nebert et al., 2004). More recently, microarray studies have proved to be paramount in identifying expression changes in TCDD-dependent gene batteries associated with the toxicities observed after AhR activation (Vezina et al., 2004; Boverhof et al., 2005; Hayes et al., 2007; Lo et al., 2011). AhR activation by exogenous ligands is associated with several toxicities affecting multiple tissues and biologic systems, including hepatotoxicity, cardiotoxicity, teratogenesis, endocrine disruption, and diabetes (Denison et al., 2011). However, the exact mechanism for the toxicities associated with AhR activation still eludes investigators. Microarray analysis using HepG2 cells treated with TCDD alone or in combination with cycloheximide highlighted the complexity associated with AhR signaling after TCDD treatment (Puga et al., 2000). The investigators reported a total of 310 significant changes in the transcript profile. However, 65% of the transcriptional changes were blocked by cycloheximide treatment, suggesting that changes in gene expression were attributable to downstream or secondary effects of TCDD treatment rather than to direct AhR regulation. In addition, AhR activation and function has proven to be species and tissue dependent (Chen et al., 1998; Boutros et al., 2009; Flaveny et al., 2010), further complicating transcriptional networks and physiologic AhR pathways.
Collectively, the aforementioned observations suggest that the AhR regulates a wide variety of genes after exogenous activation. However, AhR gene regulation in the absence of an exogenous ligand is far less well described, although a handful of reports are now beginning to show that the AhR is capable of regulating gene transcription in response to endogenous cues (Tijet et al., 2006; Wang et al., 2007; Boutros et al., 2009). In these studies, transcript levels of AhR-null and C57BL/6J wild-type mice are compared using gene array technologies. According to Tijet el al., AhR status alone altered the expression of 392 genes in the liver. Comparison of kidney and liver transcriptomes between wild-type and AhR-null mice concluded that basal gene expression in the absence of TCDD were similar with 379 and 471 AhR-dependent, TCDD-independent changes in transcript levels, respectively (Boutros et al., 2009). The similarities in gene expression between the kidney and liver of the AhR-null mice suggest that AhR has an important role in regulating gene expression to maintain homeostasis. In addition, AhR’s role in development is evident through generation of the AhR-null mouse, in which these mice exhibit multiple physiologic abnormalities independent of xenobiotic exposure, including decreased liver size and impaired vasculature formation (Schmidt et al., 1996).
Although consensus of AhR transcriptional control in the absence of an exogenous ligand is coalescing, precise targets and mechanisms of normal AhR biology remain to be resolved. To this end, we embarked on studies designed to identify immediate and direct changes in the transcriptome concomitant with the loss of the AhR. Using the Cre-lox system in primary hepatocytes, we were able to perform a microarray analysis in a physiologically relevant model focusing on direct changes that respond to endogenous cues. Analysis of the microarray data identified changes in the transcriptome dependent on AhR expression, culminating in the identification of Stanniocalcin 2 (Stc2) as a novel AhR target gene uniquely responsive to endogenous AhR signaling processes. Because mammalian Stc2 is a secreted glycoprotein thought to impart a protective function in the unfolded protein response (UPR) and apoptosis (Ito et al., 2004), it is conceivable that AhR-regulated Stc2 expression contributes to hepatocyte survival.
Materials and Methods
Materials.
Antibodies were obtained from various commercial sources: AhR (Western; Enzo Life Sciences, Farmingdale, NY), AhR and histone H3 chromatin immunoprecipitation (ChIP) grade (AbCam, Cambridge, MA), IgG for ChIP (Cell Signaling Technology, Danvers, MA), P4501A1 and CHOP (Santa Cruz Biotechnology, Santa Cruz, CA), Actin (Millipore, Billerica, MA), all horseradish peroxidase (HRP)-conjugated secondary antibodies (Invitrogen Life Sciences, Carlsbad, CA), and all fluorescent secondary antibodies (GE Healthcare, Piscataway, NJ); 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) was purchased from Cerilliant (Round Rock, TX), and 3-methylcholanthrene (3-MC) and β-napthoflavone (BNF) were purchased from Sigma-Aldrich (St. Louis, MO). The fluorogenic caspase-3 substrate Ac-DEVD-AFC was purchased from BD Biosciences Pharmingen (San Diego, CA).
Adenovirus Construction.
Generation of AdGFP (control virus) and AdrAhRFL was described previously (Elferink et al., 2001; Park et al., 2005). To generate AdCreGFP, the Cre-recombinase–expressing Ad5 vector, AdCreM2, was purchased from Microbix (Mississauga, Ontario, Canada) and amplified using human embryonic kidney 293 packaging cells. The Ad5 vector was collected and then purified by phenol/chloroform extraction. Cre-recombinase was then polymerase chain reaction (PCR) amplified from the AdCreM2 vector with use of oligonucleotide primers (5′-GCGGCCGCATGCCCAAGAAGAAGAGG-3′ and 5′-GCGGCCGCCTAATCGCCATCTTCCAG-3′) and subsequently ligated into the PCR4-TOPO vector using the TOPO TA Cloning Kit, as described by the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Successful cloning was verified by sequencing, and the correct clone was designated Cre-TA. Next, the Cre sequence from Cre-TA was cloned into the NotI site of the shuttle vector pAdTrack-CMV, and the resulting synthesis and analysis of the recombinant adenovirus was performed as described elsewhere (He et al., 1998). Viral stocks were prepared as previously described (Elferink et al., 2001).
Animals.
For experiments, 8–10-week-old AhRfx/fx (AhR floxed) and AhRfx/fx/CreAlb (AhR conditional knockout [CKO]) female mice were used in accordance with the Institutional Animal Care and Use Committee at the University of Texas Medical Branch. The mice were maintained on a 12-hour light/dark cycle and allowed free access to water and chow. AhR-floxed mice were purchased from the Jackson Laboratory (Bar Harbor, ME), and AhR CKO mice were previously described and verified (Mitchell et al., 2010).
Isolation, Culture, and Treatment of Primary Hepatocytes or AML-12 Cells.
AhR floxed and AhR CKO primary hepatocytes were isolated using a collagenase perfusion method as previously described (Park et al., 2005). Hepatocytes were plated at a density of 8.5 × 104–1.0 × 105 cells/cm2 in Williams E medium containing penicillin (100 U/ml), streptomycin (100 μg/ml), and 5% fetal bovine serum and replaced every 24 hours with fresh medium. Hepatocytes were infected with the appropriate adenovirus at a multiplicity of infection of 20–60 at the time of plating and maintained in culture for the indicated times; 100% infection was ensured via expression of green fluorescent protein with use of a Zeiss Axiovert 200 inverted microscope (Zeiss Microscopy, Thornwood, NY). Hepatocytes treated with TCDD after infection were allowed to culture for 6 hours with 6nM TCDD after the indicated times. Noninfected hepatocytes were allowed to attach for 3 hours and then were treated with dimethylsulfoxide, TCDD, 3-MC, or BNF at 0.2%, 6 nM, 5 μM, and 10 μM respectively. The hepatocytes were collected 24 hours after treatment. The murine immortalized AML-12 cell line was cultured as described previously (Wu et al., 1994) and infected with appropriate virus at a multiplicity of infection of 100 because of poor infection rates as determined empirically.
RNA Isolation.
Total RNA was isolated from AhR floxed and AhR CKO primary hepatocytes with use of the Chomezynski and Sacchi method (Chomczynski and Sacchi, 1987) or Trizol (Invitrogen), according to the manufacturers’ directions. Total RNA yield was assessed using a Nanodrop ND-1000 (Thermo Scientific, Wilmington, DE), and RNA integrity was confirmed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
Reverse-Transcription PCR Analysis.
First-strand cDNA was generated from 1μg of total RNA with use of an oligo(dT) primer (New England BioLabs, Ipswich, MA) and Superscript II reverse transcriptase (Invitrogen). PCR using Taq polymerase (Fisher Scientific, Pittsburgh, PA) was performed using oligonucleotide primers for mouse AhR designed to produce different size PCR products depending on the presence of Exon 2 in the AhR transcript (5′-CGCAAGCCGGTGCAGAAAAC-3′ and 5′-ATGGAGGGTGGCTGAAGTGGAGTA-3′). All PCR products were analyzed by fractionation on a 0.8% (w/v) agarose gel and visualized by ethidium bromide staining.
Western Blot Analysis.
Whole-cell lysates were prepared from cultured primary hepatocytes or AML-12 cells after infection or treatment by washing with phosphate-buffered saline (PBS), then scraping directly in 2× SDS-PAGE loading buffer, and denatured for 10 minutes at 100°C. Protein was fractionated by SDS-PAGE, transferred to Amersham Hybond-P membranes (Amersham Biosciences, Piscataway, NJ), and blocked with Tris-buffered saline containing 0.1% (v/v) Tween 20 and 5% (w/v) nonfat dry milk. Membranes were incubated with primary antibodies for 3 hours at room temperature or overnight at 4°C, followed by incubation with fluorescent or HRP–conjugated secondary antibodies for 1 hour at room temperature. HRP-conjugated antibodies were then visualized using enhanced chemiluminescence (Amersham Biosciences), and both HRP- and fluorescent secondaries were imaged using the Typhoon Trio Variable Mode Imager (GE Healthcare). Quantification of Western blot analysis was performed using ImageQuant TL software (version 7.0; GE Healthcare).
Microarray Analysis.
Total RNA was isolated from AhR floxed primary hepatocyte cultures after infection with appropriate adenovirus as described. Total RNA prepared from three independent experiments was analyzed using the UTMB Molecular Genomics Core with use of GeneChip Mouse Genome 430 2.0 Arrays (Affymetrix, Santa Clara, CA). In brief, total RNA (500 ng) was converted to cRNA with use of the Ambion MessageAmp Premier RNA Amplification Kit (Life Technologies Corporation, Grand Island, NY) according to the manufacturer’s instructions. Total fragmented cRNA (10 µg) was hybridized to the Affymetrix GeneChip array according to the manufacturer’s conditions. The chips were washed and stained in a GeneChip Fluidics Station 450, and fluorescence was detected with an Affymetrix-7G Gene Array scanner using the Affymetrix GeneChip Command Console software (AGCC1.1). Gene expression changes were identified using Partek Genomics Suite (Partek, St. Louis, MO), according to the default gene expression workflow. The resulting values were then filtered for P values ≤0.05 with use of one-way analysis of variance (ANOVA). Fold change was determined by comparing AdCreGFP normalized expression with AdGFP normalized expression.
Ingenuity Pathways Analysis.
The microarray data set was analyzed using Ingenuity Pathways Analysis (IPA; Ingenuity Systems, www.ingenuity.com). The data set containing gene identifiers from all 24 hour statistically significant microarray genes and their corresponding expression values was uploaded into in the application. Each identifier was mapped to its corresponding object in the Ingenuity Knowledge Base. All Network Eligible molecules were overlaid onto a global molecular network developed from information contained in the Ingenuity Knowledge Base. Networks of Network Eligible Molecules were then algorithmically generated on the basis of their connectivity.
SYBR Array and Single Assay Analysis.
Total RNA was isolated from AhR floxed primary hepatocyte cultures and analyzed by the UTMB Molecular Genomics Core. In brief, quantitative reverse-transcription PCR (qRT-PCR) assays were designed from the coding sequence (CDS) of the gene of interest (NCBI), and exon-exon junctions were mapped via BLAT (Kent, 2002). Whenever possible, at least one of the two PCR primers was designed to transcend an exon-intron junction to reduce the impact of potential genomic DNA contamination in the surveyed RNA samples. Primers were synthesized (Integrated DNA Technologies, Coralville, IA) and reconstituted to a final concentration of 100 mM (master stock) before dilution to a working stock of 2 mM. Reverse transcription was performed on 1μg of total RNA with random primers, using TaqMan reverse transcription reagents (Applied Biosystems), as recommended by the manufacturer. The reverse transcription reaction was used as a template for the subsequent PCR, consisting of SYBR Green PCR Master Mix, template cDNA, and assay primers in a total reaction volume of 25μl. Thermal cycling was performed using an ABI prism 7000 sequence detection system (Life Technologies) under factory defaults (50°C, 2 minutes; 95°C, 10 minutes; and 40 cycles at 95°C, 15 seconds; 60°C, 1 minute). Threshold cycle numbers (Ct) were defined as fluorescence values, generated by SYBR green binding to double-stranded PCR products, exceeding baseline. Relative transcript levels were quantified as a comparison of measured Ct values for each reaction to a designated control via the ∆∆Ct method (Livak and Schmittgen, 2001). To normalize for template input, glyceraldehyde 3-phosphate dehydrogenase (endogenous control) transcript levels were measured for each sample and used in these calculations. Student’s t test was applied to glyceraldehyde 3-phosphate dehydrogenase Ct values to rule out any change in expression of the endogenous control resulting from treatment.
Taqman qRT-PCR Analysis.
Taqman qRT-PCR was performed by the UTMB Molecular Genomics Core Facility using an Stc2 primer and probe set purchased from Applied Biosystems. All single qRT-PCR assays were normalized against the internal control ribosomal RNA 18S.
ChIP.
Liver tissue from AhR floxed and AhR CKO female mice were extracted 2 hours after vehicle or TCDD treatment. Livers were rinsed with ice cold PBS, finely minced, and cross-linked with 1% formaldehyde in PBS at room temperature for 10 minutes. Cross-linking was stopped using 0.5 M glycine solution. Samples were homogenized in Dounce homogenizer and centrifuged at 3200g for 5 minutes at 4°C. Pellet was resuspended in 4 ml of cell lysis buffer (150 mM NaCl, 25 mM Tris, 5 mM EDTA, 1% Triton X, 0.1% SDS, 0.5% Na deoxycholate, protease inhibitors) and homogenized with Dounce homogenizer. Samples were incubated on ice for 15 minutes and centrifuged at 3200g for 5 minutes at 4°C, and pellet was further processed using ChIP-IT Express Enzymatic Kit (Active Motif) according to the manufacturer’s instructions. The sheared chromatin (input) was incubated overnight with appropriate antibodies AhR, histone H3 as positive control, or IgG as negative control. Input and immunoprecipitated DNA were PCR amplified using CYP1A1 (forward 5′-CTATCTCTTAAACCCCACCCCAA-3′, reverse 5′-CTAAGTATGGTGGAGGAAAGGGTG-3′) primers and primers specific to the XRE’s in the STC2 promoter (forward 5′-CTCAGTCCATTCGGCCATTGC-3′, reverse 5′-ACTTCTACGGGAGGAAGCGGAG-3′). PCR product was run on 5% polyacrylamide gel, stained with SYBR Green I (Invitrogen), and imaged on Typhoon Trio.
Caspase-3 Activity Assays.
AhR floxed primary hepatocytes were infected with the appropriate adenovirus and allowed to culture for 24 or 48 hours. Cultures were washed once with cold PBS, and hepatocytes were harvested by scraping in cold PBS on ice. The hepatocytes were then pelleted by centrifugation at 1000g for 5 minutes at 4°C, washed with cold PBS, and centrifuged again to pellet the hepatocytes. The pellet was then resuspended in lysis buffer (10 mM Tris [pH, 7.5], 10 mM NaH2PO4/Na2HPO4 [pH, 7.5], 130 mM NaCl, 1% [v/v] Triton X-100, 10 mM NaPPi), and cellular membranes were disrupted using a dounce homogenizer. Cell lysates were then clarified by centrifugation at 10,000g for 10 minutes at 4°C, and supernatants were transferred to fresh ice-cold microcentrifuge tubes. A total of 50 μl of each sample was added to 50 μl assay buffer/dithiothreitol Mix (20 mM PIPES, 100 mM NaCl, 10 mM dithiothreitol, 1 mM EDTA, 0.1% [w/v] CHAPS, 10% [w/v] sucrose [pH, 7.2]); 5 μl of 1 mM fluorogenic caspase-3 substrate, Ac-DEVD-AFC, was added to each sample and allowed to incubate at 37°C for 1–3 hours. Fluorescence resulting from the cleavage of 7-amino-4-trifluoromethylcoumarin was quantified fluorometrically using a Fluoroskan Ascent fluorometer (Thermo Fisher Scientific, Waltman, MA) with a 400-nm excitation filter and 505-nm emission filter.
Statistical Analysis.
Where appropriate, the data are represented as mean ± S.E.M. GraphPad Prism 4 (GraphPad Software, San Diego, CA) was used to calculate statistical significance (P ≤ 0.05) and is denoted using brackets to show comparison. Comparison of basal caspase-3 activity in AdGFP- and AdCreGFP-infected hepatocytes and comparison of CHOP protein expression used a two-way ANOVA, followed by a Bonferroni’s post hoc test. Statistical significance in the caspase-3 activity assay 48 hours after infection used a one-way ANOVA, followed by a Bonferroni’s post hoc test. All qRT-PCR assays compared the fold change values calculated as a log2 ratio of the ∆∆Ct averages of the biologic replicates with the control sample, with use of Student’s t test to evaluate statistical significance.
Results
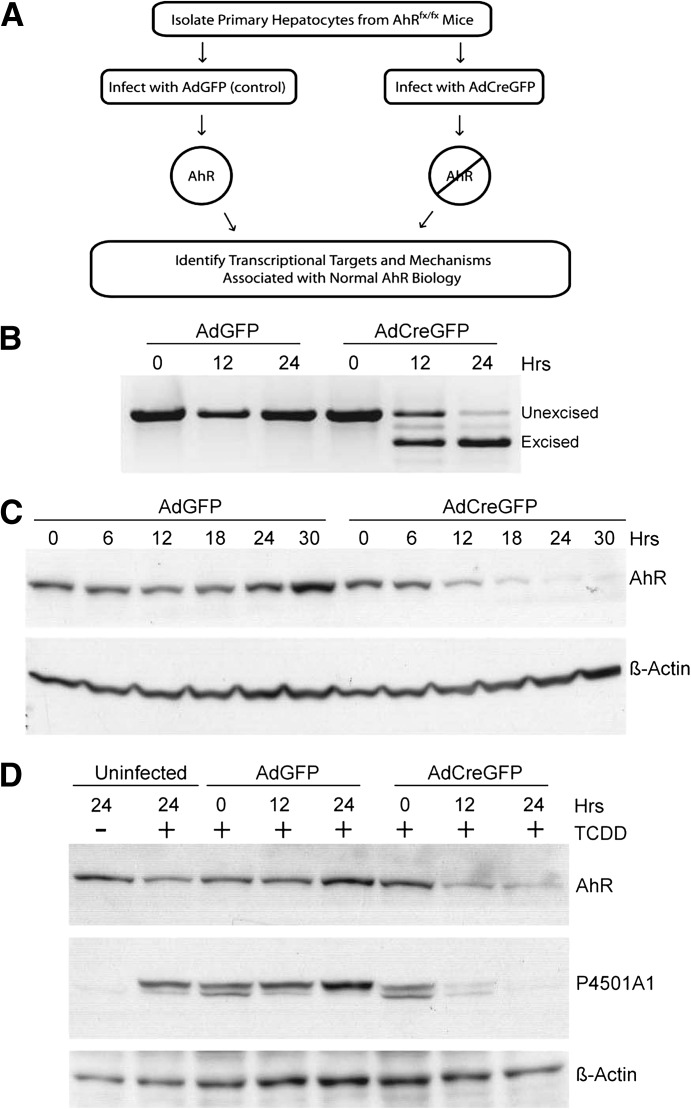
To identify AhR transcriptional targets associated with normal AhR biology, we used the Cre-lox system to manipulate receptor expression in primary hepatocytes isolated from the AhRfx/fx mouse (Huang and Elferink, 2012). The general strategy used for many of the experiments described in this study is depicted in Fig. 1A. The ability to monitor changes in signaling concurrent with AhR loss represents an unprecedented opportunity to evaluate direct AhR processes in the absence of an exogenous agonist. Analysis of the AhR transcript demonstrates that loss of the mature mRNA in hepatocytes infected with the Cre recombinase-expressing virus (AdCreGFP) was essentially complete within 24 hours (Fig. 1B). The advent of the nonfunctional excised transcript identified by RT-PCR reflects removal of the intervening genetic element encoding exon 2 that is flanked by the loxP sites (Walisser et al., 2005). Accordingly, AhR protein expression disappears over the same period (Fig. 1C), indicative of a very short protein half-life in the primary hepatocytes. To verify that the loss of AhR is functionally relevant, uninfected hepatocytes and those infected with the control virus (AdGFP) or virus expressing the Cre recombinase (AdCreGFP) were treated with vehicle (dimethylsulfoxide) or 6 nM TCDD for 6 hours, and P4501A1 expression was determined immunologically (Fig. 1D). The result shows that loss of the AhR was sufficiently robust to completely abolish induction of the Cyp1a1 target gene.
Fig. 1.
Monitoring loss of the AhR in primary hepatocytes. (A) Schematic depicting the general strategy used to generate AhR-positive and AhR-negative primary hepatocytes. (B) RT-PCR was performed to monitor excision of the AhR gene loxP-flanked exon 2 with use of total RNA isolated from AhRfx/fx primary hepatocytes infected with AdGFP (control) or AdCreGFP (Cre recombinase) for the indicated times. PCR primers were designed to amplify different size PCR products distinguishing the unexcised from excised transcript. (C) Whole-cell lysate from AhRfx/fx primary hepatocytes infected with AdGFP or AdCreGFP were analyzed using Western blotting for AhR protein. Actin was used as a loading control. (D) AhRfx/fx primary hepatocytes were either uninfected or infected with AdGFP or AdCreGFP for the indicated times, followed by treatment with DMSO (lane 1) or 6 nM TCDD (lanes 2–8) for 6 hours. AhR and P4501A1 protein expression was monitored using Western blot analysis. Actin was used as a loading control.
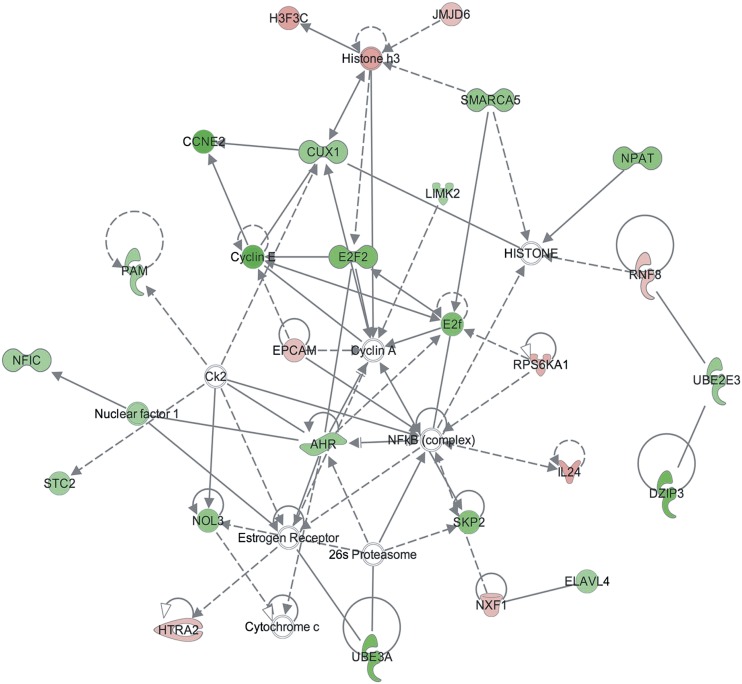
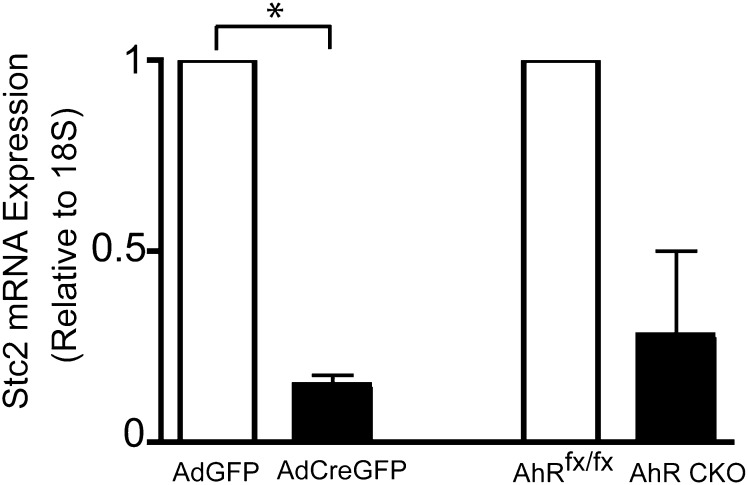
After validation of the Cre-lox system in the primary hepatocyte model, we initiated a comprehensive DNA microarray study to identify changes in the transcriptome with the loss of the AhR. Microarray analysis was performed in triplicate with use of independent RNA samples isolated from primary hepatocytes at 0, 12, and 24 hours after infection with the AdGFP or AdCreGFP virus. The experiment identified 97 and 246 statistically significant (P ≤ 0.05) changes in gene expression at 12 and 24 hours, respectively (Supplemental Table 1). Of the 97 changes detected at 12 hours, 88 genes increased expression and 9 decreased expression with loss of AhR expression. By 24 hours, 119 genes increased and 127 genes decreased expression. Of interest, there was little overlap between the sets of genes identified in this study and those identified in previous microarray studies that monitored changes in expression after exposure to an exogenous AhR agonist or changes in the steady-state transcriptome associated with prolonged AhR loss in the AhR knockout mouse (unpublished data). This most likely reflects the strategy used here that was designed to identify immediate changes in expression concomitant with loss of AhR in the absence of an exogenous agonist. Custom qRT-PCR SYBR arrays were performed to validate expression of selected genes identified in the microarrays. The rationale used in choosing the validation set included focusing on well-annotated genes and those linked to relevant signaling pathways identified by IPA. Figure 2 depicts a network identified by IPA using the statistically significant data obtained by the 24 hour sample microarrays, indicating both decreases and increases in the AdCreGFP-infected cells. A total of 31 and 145 genes were chosen for validation at 12 hours and 24 hours after viral infection, respectively. None of the expression changes detected in the microarray at 12 hours could be verified by qRT-PCR. However, qRT-PCR identified 36 genes that reproducibly changed by 24 hours: 3 increased and 33 decreased expression after loss of the receptor (Supplemental Table 1). Consistent with the experimental design, expression of the full-length AhR transcript decreased. Of interest, the expression of Stc2, one of the genes identified by the IPA and validated by qRT-PCR as being consistently decreased concomitant with loss of the AhR in primary hepatocytes, also decreased (Fig. 3). Of note, the decrease in Stc2 was also observed in vivo in conditional knockout mice lacking AhR expression specifically in the hepatocytes (Fig. 3). These data suggest that the Stc2 gene is a direct AhR target and that steady state constitutive expression is predominantly AhR dependent.
Fig. 2.
Network generated in IPA using the 24-hour microarray data comparing AdGFP- and AdCreGFP-infected hepatocytes. Green and red symbols denote a decrease or increase in expression, respectively, accompanying the loss of AhR. Solid lines represent a direct relationship as reported by the Ingenuity Knowledge database, whereas dashed lines represent an indirect relationship.
Fig. 3.
Validation of Stc2 expression by qRT-PCR. qRT-PCR was performed on total RNA isolated from AhRfx/fx primary hepatocytes infected with AdGFP or AdCreGFP for 24 hours or primary hepatocytes isolated from AhRfx/fx and AhR CKO mice. The data presented for Stc2 represents the mean ± S.E.M. of three (AdGFP vs AdCreGFP) or two (AhRfx/fx vs AhR CKO) independent experiments. Asterisk indicates statistical significance (P < 0.01) when compared to control infected hepatocytes. Quantification of mRNA expression is relative to ribosomal RNA 18S.
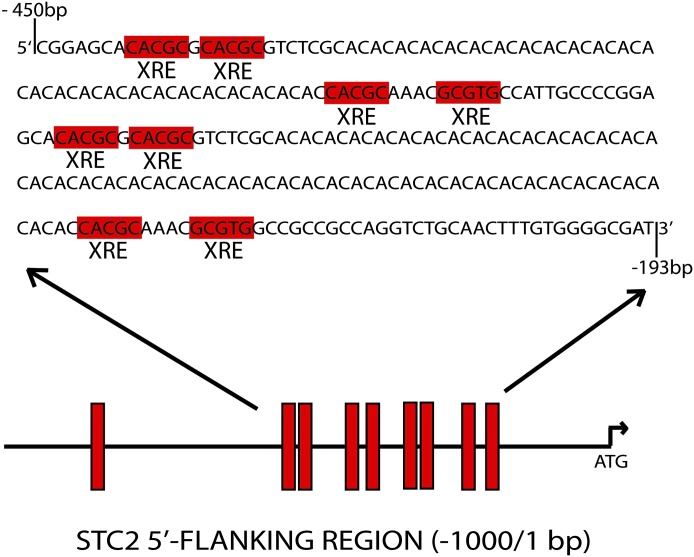
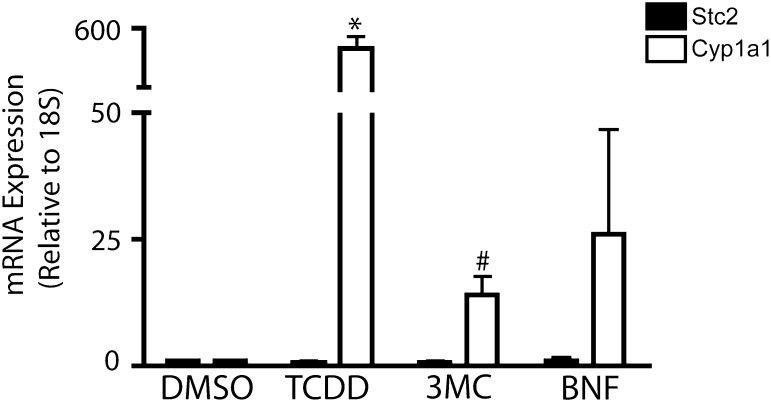
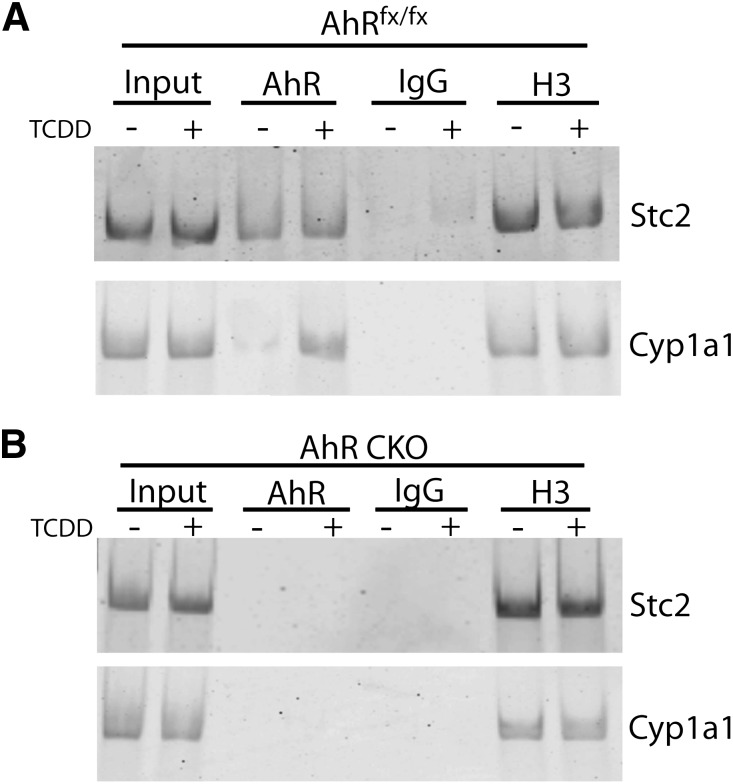
To examine the role of AhR in Stc2 expression further, we performed an in silico analysis of the murine Stc2 promoter region, in which we identified nine putative XRE consensus sequences (5′-GCGTC-3′) in 1 kb of the Stc2 transcriptional start site; eight of these were found to be clustered in a 250 bp cassette (Fig. 4). The clustering of eight XRE sequences in the Stc2 promoter region greatly exceeds the estimated random distribution of one XRE per ∼10 Kb, and connotes a functional role akin to the XRE repeats in the Cyp1a1 promoter. Therefore, we exposed primary hepatocytes to well-known AhR exogenous agonists to determine whether Stc2 expression parallels that of the Cyp1a1 locus (Fig. 5). The evidence shows that Stc2 gene induction is refractory to the classic receptor agonists 2,3,7,8-tetrachlorodibenzo-p-dioxin, 3-methylcolanthrene, or β-napthoflavone, despite efficient induction of the Cyp1a1 gene. However, ChIP detected direct binding of the AhR to the region encompassing the XRE cassette in the Stc2 promoter (Fig. 6). In contrast to the TCDD-dependent AhR binding observed at the Cyp1a1 promoter, the AhR interaction with the Stc2 promoter occurs independently of TCDD treatment (Fig. 6A). The lack of a Stc2 ChIP product in the CKO mouse liver nuclei further shows the capacity of AhR to bind to the Stc2 promoter region harboring the XRE cluster (Fig. 6B). The IgG and histone H3 ChIP results confirm the specificity of the protein-DNA binding events. The inability to induce Stc2 expression with use of the exogenous agonists may be reconciled with the constitutive AhR interaction detected at the Stc2 promoter. Whether this reflects the action of an endogenous receptor agonist is unknown.
Fig. 4.
Schematic representing 9 putative XRE binding sites (5′-GCGTG-3′) in 1 kb of the Stc2 transcriptional start site. The XRE cassette is located 226 bp upstream of the start site and spans a 218-bp region. XRE core consensus sequences are highlighted in red.
Fig. 5.
Stc2 gene induction is refractory to classic AhR agonists. qRT-PCR was performed using total RNA prepared from AhRfx/fx hepatocytes treated for 24 hours with the vehicle dimethylsulfoxide (DMSO), 6 nM TCDD, 5 µM 3-methylchloranthrene (3-MC), or 10 µM β-napthoflavone (BNF). The data presented represent the mean ± S.E.M. of three independent experiments. Asterisk or number sign indicates statistical significance of P < 0.01 or P < 0.05, respectively, when compared with vehicle-treated samples. Quantification of mRNA expression is relative to ribosomal RNA 18S and normalized to vehicle-treated samples.
Fig. 6.
The AhR binds the putative XRE cassette located upstream of the Stc2 transcriptional start site. ChIP analysis of AhR protein binding to the Stc2 promoter region in liver tissue from AhRfx/fx (A) or AhR CKO (B) mice treated with vehicle (-) or 20 μg/kg TCDD (+) for 2 hours. IgG and histone H3 antibodies were used as negative and positive controls, respectively. The primers used for ChIP analysis correspond to base pairs -473 to -453 (For) and -182 to -161 (Rev).
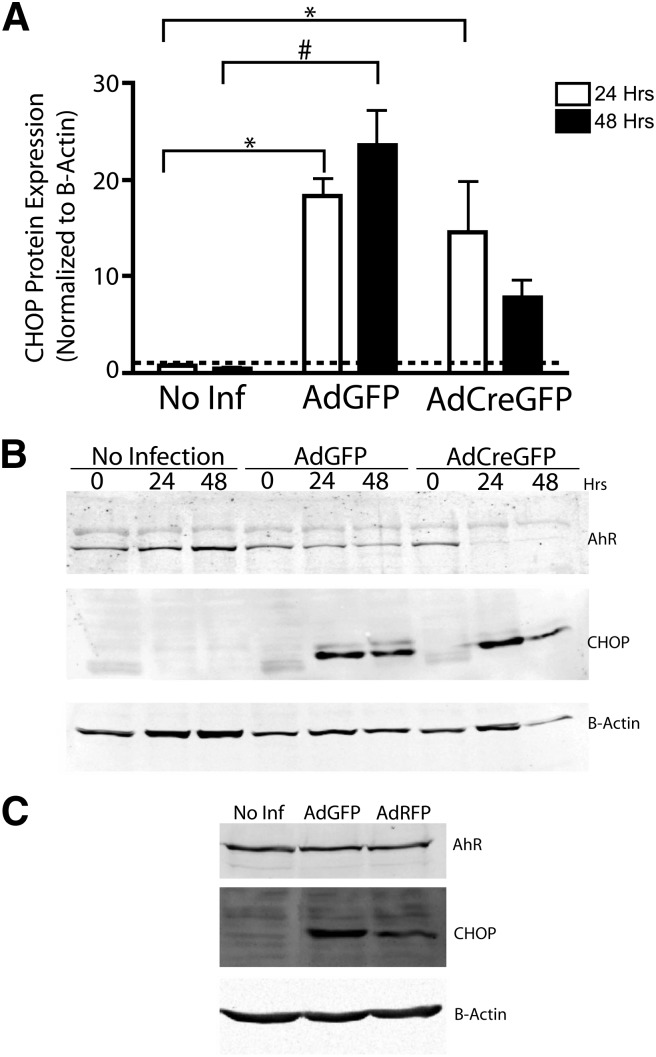
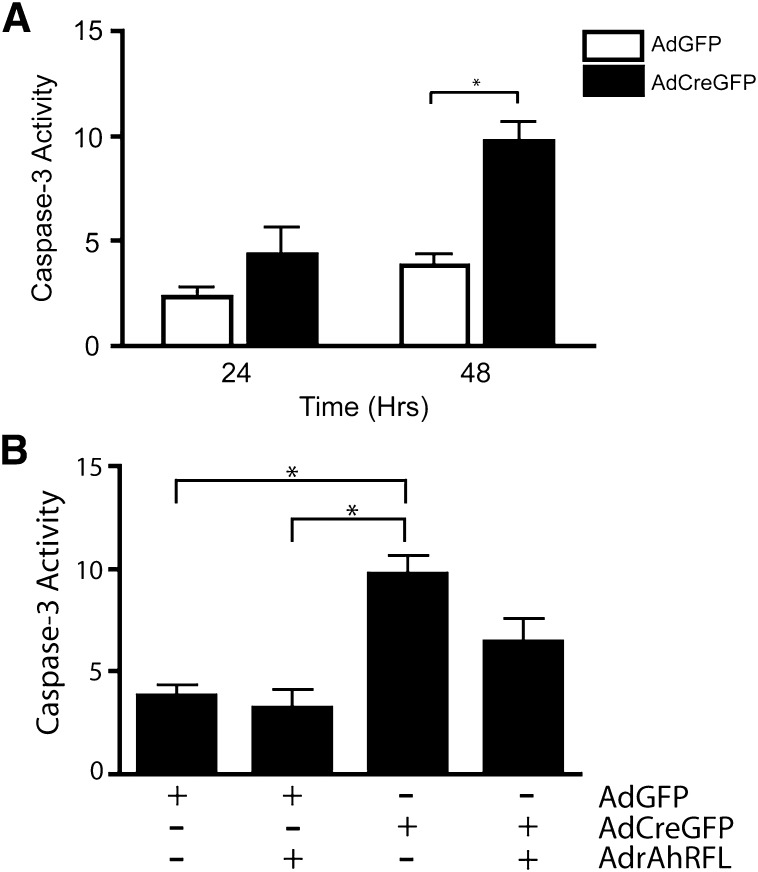
Mammalian Stc2 is a secreted glycoprotein believed to confer a protective function in the UPR and apoptosis (Ito et al., 2004). Because numerous cellular insults result in activation of the UPR, including viral infection (He, 2006), we investigated whether adenoviral infection in our primary hepatocyte model was capable of activating the UPR. Immunologic monitoring of the UPR marker CHOP suggests that viral infection of both AdGFP and AdCreGFP results in activation of the UPR, as indicated by a substantial increase in CHOP expression 24 hours after adenoviral infection (Fig. 7). Prolonged CHOP induction serves as a signal for the UPR and subsequent endoplasmic reticulum (ER) stress, to initiate apoptosis through the intrinsic pathway, ultimately resulting in caspase-3 cleavage and, eventually, cell death (McCullough et al., 2001; Ma et al., 2002). As revealed in Fig. 7, infection with the adenoviruses results in persistent CHOP induction in the hepatocytes. A similar CHOP induction was also observed in the AhR-positive immortalized murine hepatocyte AML-12 cell line (Fig. 7C), indicating that the response to viral infection is not limited to the primary liver cells. In keeping with persistent CHOP expression, direct measurement of caspase-3 activity detected progressive increases in hepatocytes infected with either virus, albeit substantially more so after infection with AdCreGFP (Fig. 8A). This suggested that loss of the AhR enhanced the apoptotic response to endoplasmic reticulum stress (ER stress). Accordingly, concomitant ectopic expression of the receptor using a second virus encoding a functional AhR (AdrAhRFL) (Elferink et al., 2001) successfully diminished the increase in caspase-3 activity, consistent with the receptor’s protective role in apoptosis. These data highlight the receptor’s involvement in protecting primary hepatocytes from undergoing apoptosis in response to intrinsic signals. Previously, the ability of AhR to confer cell survival in response to intrinsic cues was largely attributed to activation of the PI3K-Akt/PKB pathway in hepatoma cell lines (Wu et al., 2007). However, we were unable to verify this finding in the primary hepatocytes (data not shown), nor did we detect reproducible changes in the expression of 84 genes with a well-documented role in apoptosis with use of an apoptosis gene array (Supplemental Fig. 1). A modest increase in p73 expression detected in the array could not be verified by subsequent qRT-PCR (Supplemental Fig. 1). Therefore, the link among the AhR, Stc2 expression, and its cytoprotective function provides a potentially novel mechanism by which the AhR confers cell survival.
Fig. 7.
Adenoviral infection induces the UPR marker CHOP in primary hepatocytes. AhRfx/fx primary hepatocytes were infected with AdGFP or AdCreGFP or were allowed to remain uninfected for the indicated period, after which whole cell lysates were prepared. Western blot analysis was performed on AhR and CHOP. Actin was used as a loading control. (A) Quantification of CHOP protein expression 24 and 48 hours after adenoviral infection. The data presented for CHOP represents the mean ± S.E.M. of three independent experiments. Asterisk or number sign indicates statistical significance of P < 0.01 or P < 0.001, respectively, when compared with uninfected samples. (B) Representative Western blot showing loss of AhR and CHOP induction after infection with the adenovirus. (C) Induction of CHOP in murine immortalized hepatocyte AML-12 cells after infection with AdGFP and AdRFP.
Fig. 8.
Loss of the AhR significantly increases caspase-3 activity, and ectopic expression of AhR decreases caspase-3 activity. (A) AhRfx/fx primary hepatocytes were infected with AdGFP or AdCreGFP for the indicated times and caspase-3 activity assayed. Asterisks indicate statistical significance (P < 0.01), when compared with AdGFP-infected hepatocytes. (B) AhRfx/fx primary hepatocytes were infected with either AdGFP or AdCreGFP alone or in combination with AdrAhRFL for 48 hours and assayed for caspase-3 activity. Asterisks indicate statistical significance (P < 0.01).
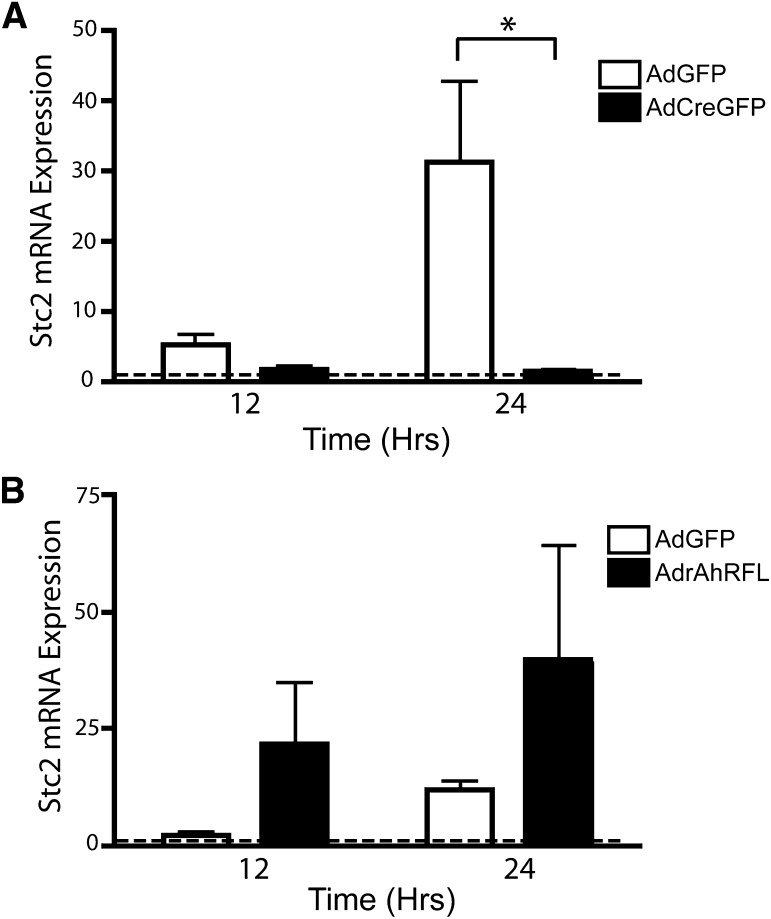
The initial microarray analysis and subsequent SYBR validation showed that Stc2 expression decreased in AdCreGFP-infected hepatocytes concomitant with loss of the AhR, implying that the AhR is necessary for basal Stc2 expression. Subsequent studies, however, revealed that the Stc2 transcript is nearly undetectable in uninfected cells and is increased by ~30-fold after AdGFP infection but not after infection with the AdCreGFP virus (Fig. 9A). Correspondingly, the increase in Stc2 expression is attenuated in primary hepatocytes isolated from the CKO mice lacking the AhR after AdGFP viral infection, whereas pronounced Stc2 expression is observed following infection with the AdrAhRFL to restore receptor expression (Fig. 9B). Collectively, these data show that Stc2 is a novel AhR target gene that has been implicated in promoting cell survival. Consequently, the evidence is consistent with the hypothesis that the AhR mediates cell survival by inducing Stc2 after an intrinsic apoptotic signal.
Fig. 9.
Adenoviral infection promotes Stc2 expression in an AhR-dependent manner. (A) qRT-PCR was performed on AhRfx/fx primary hepatocytes infected with AdGFP or AdCreGFP for the indicated times. The data presented for Stc2 represent the mean ± S.E.M. of five independent experiments. Asterisk indicates statistical significance (P < 0.01). (B) qRT-PCR was performed on AhR CKO primary hepatocytes infected with AdGFP or AdrAhRFL for the indicated times. The data presented for Stc2 represent the mean ± S.E.M. of three independent experiments. Quantification of mRNA expression is relative to ribosomal RNA 18S and normalized to uninfected hepatocytes at each time point and set to a value of one (dashed line).
Discussion
The AhR has historically been a protein of interest when investigating liver injury because of its hepatotoxic mechanism(s) after activation by halogenated aromatic hydrocarbons, such as TCDD. However, it was only after the generation of the AhR knockout mouse that the field identified the importance of physiologic AhR activities in the absence of an exogenous ligand. In the present study, we aimed to extend the understanding of normal AhR biology through the use of a comprehensive DNA microarray screen designed to identify immediate and direct changes in the transcriptome concomitant with the loss of the AhR. Several independent groups have previously performed microarray analyses designed to link changes in the transcriptome with AhR status with use of knockout models or function with use of exogenous AhR agonists (Tijet et al., 2006; Wang et al., 2007; Franc et al., 2008; Boutros et al., 2009). Specifically, Tijet et al. identified several novel AhR target genes independent of TCDD activation with use of AhR knockout mice generated previously (Schmidt et al., 1996). However, the transcriptome in the AhR knockout mouse liver constitutes a steady state response to prolonged receptor loss affecting myriad indirect changes. This confounds identification of changes in gene expression attributable to direct AhR control. The observation that livers in the AhR knockout mice are abnormal is suggestive of robust secondary compensatory effects associated with long-term AhR loss during development. In contrast, our study differs from the previous ones, by monitoring immediate changes in the transcriptome concomitant with loss of AhR protein to preferentially identify transcriptional responses reflective of direct AhR target genes. Because the changes in expression occurred in the absence of any exogenous agonist, we conclude that the target genes identified are those involved in normal AhR-mediated physiologic processes. In this context, it is not surprising that the Cyp1a1 gene, nor several of the other AhR responsive phase I and phase II target genes typically associated with an exogenous agonist, went undetected in the microarrays. In fact, the finding that most of the altered transcripts are encoded by genes hitherto not associated with AhR regulation suggests that receptor biology responding to endogenous cues differs markedly from that after exposure to exogenous agonists and is reminiscent of AhR activation by selective AhR modulators (SAhRMs) (Safe et al., 2000).
The lingering AhR activity stemming from the residual expression after 12 hours of AdCreGFP infection prompted our focus on the 24-hour data set for analysis with IPA, coincident with complete loss of AhR expression and function by this time (Fig. 1). The purpose of using IPA was 2-fold: first, we wanted to determine what interactions existed between the targets identified in our data set and, second, what interactions were known between these targets and the AhR. The Ingenuity Knowledge Base created a network that included the AhR and several cell growth and survival proteins known to be functionally linked to the AhR, including the Estrogen Receptor, NFkB, E2F, and the Cyclins A and E ( Tian et al., 1999; Strobeck et al., 2000; Ohtake et al., 2003; Marlowe et al., 2004; Mitchell et al., 2006). Moreover, IPA identified Stc2 as a component of the network shown in Fig. 2, based on the connectivity of all Network Eligible Molecules included in our data set. The finding that Stc2 expression was decreased in both the AdCreGFP-infected primary hepatocytes and in the conditional knockout mouse liver (Fig. 3) is entirely consistent with the AhR playing a central role in Stc2 expression. Given the presence of the XRE cluster located in the Stc2 promoter, we sought to characterize its responsiveness to AhR agonists in greater detail (Fig. 5). The evidence indicates that Stc2 is completely refractory to the classic exogenous AhR agonists TCDD, 3-methylcholanthrene, and β-naphthoflavone. This nonresponsiveness may explain why previous expression studies failed to identify Stc2 as an AhR target gene. In addition, ChIP analysis identified a specific albeit constitutive AhR interaction with the Stc2 promoter encompassing the XRE cassette (Fig. 6). The implication is that AhR-mediated Stc2 expression may be dependent on additional regulatory events possibly involving coactivator recruitment reminiscent of the type II nuclear hormone receptors. Alternatively, higher-order chromatin rearrangements or other epigenetic modifications may contribute to Stc2 regulation. Our inability to recapitulate endogenous Stc2 expression using a transiently transfected luciferase reporter system harboring either the Stc2 XRE cassette or a 1 Kb region of the Stc2 promoter sequence, including the XRE cassette, suggests that appropriate chromatin architecture may indeed be crucial for Stc2 expression (data not shown).
Although few studies to date have examined Stc2 function (Ito et al., 2004; Chang et al., 2008), it is considered to be a secreted glycophosphoprotein that acts in an autocrine or paracrine manner (Wagner and Dimattia, 2006). Stc2 has been reported previously to have a cytoprotective role during tunicamycin- and thapsigargin-induced ER stress using the neuroblastoma cell line N2a (Ito et al., 2004). Likewise, Fazio et al. (Fazio et al., 2011) reported that Stc2 reduced cellular injury in cerulein-induced pancreatitis involving a process associated with altered PERK signaling. Of note, PERK is a pivotal ER sensor that reduces general protein translation during acute stress, but selectively induces transcription and translation of CHOP during prolonged or severe stress (Gregersen and Bross, 2010). We show here that adenovirus infection induced prolonged CHOP expression (a hallmark of ER stress) in both primary hepatocytes and the AML-12 cell line, regardless of AhR status (Fig. 7). However, loss of AhR resulted in enhanced caspase-3 activity (Fig. 8) and increased apoptosis after the intrinsic trigger, consistent with the previous finding that the AhR promotes cell survival by upregulating PI3K-Akt/PKB activity (Wu et al., 2007). The precise mechanism responsible for AhR-mediated survival in the primary hepatocytes remains unclear. This property could not be attributed to PI3K-Akt/PKB signaling, nor was it associated with a reproducible change in expression among any of 84 established apoptotic genes (Supplemental Fig. 1). Instead, resistance to apoptosis correlated with Stc2 expression, which was shown to be AhR dependent. Efforts to detect Stc2 protein immunologically with use of commercial reagents proved to be unsuccessful (unpublished data), a difficulty also encountered by others (Chang et al., 2008); therefore, we were unable to verify that the changes in Stc2 mRNA are reflected at the protein level.
From a pathophysiological perspective, chronic viral infection by hepatitis viruses B or C results in ER stress and can lead to many liver disease states, including hepatitis, cirrhosis, and hepatocellular carcinoma (Malhi and Kaufman, 2011). Here, we report that adenovirus infection leads to cellular stress, presumably in the ER, as evidenced by the induction of CHOP, and a concomitant induction of Stc2 in an AhR-dependent manner. However, the inability to induce Stc2 expression with canonical exogenous receptor agonists and the evidence for constitutive AhR binding to the Stc2 promoter suggests that receptor control of the gene differs significantly from the well-characterized mechanism regulating Cyp1a1 expression. Accordingly, our observed AhR-dependent virally mediated induction of Stc2 represents a novel insight into normal AhR biology. These findings merit further investigation into the AhR-Stc2 relationship, with focus on ER stress regulation during times of liver dysfunction and insult.
Supplementary Material
Acknowledgments
The authors thank Dr. Istvan Boldogh for the AML-12 cell line.
Abbreviations
- AhR
aryl hydrocarbon receptor
- ANOVA
analysis of variance
- ARNT
aryl hydrocarbon nuclear translocator
- BNF
β-napthoflavone
- CKO
conditional knockout
- ER stress
endoplasmic reticulum stress
- HRP
horseradish peroxidase
- IPA
Ingenuity Pathways Analysis
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- qRT-PCR
quantitative reverse-transcription polymerase chain reaction
- Stc2
stanniocalcin 2
- TCDD
tetrachlorodibenzo-p-dioxin
- 3-MC
3-methylcholanthrene
- UPR
unfolded protein response
- XRE
xenobiotic response element
Authorship Contributions
Participated in research design: Harper, Elferink.
Conducted experiments: Harper, Joshi.
Performed data analysis: Harper, Elfeirnk.
Wrote or contributed to the writing of the manuscript: Harper, Elferink, Joshi.
Footnotes
This work was supported by the National Institutes of Health National Institute of Environmental Health Sciences [Grants ES012018, ES06676, and ES07254].
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Boutros PC, Bielefeld KA, Pohjanvirta R, Harper PA. (2009) Dioxin-dependent and dioxin-independent gene batteries: comparison of liver and kidney in AHR-null mice. Toxicol Sci 112:245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boverhof DR, Burgoon LD, Tashiro C, Chittim B, Harkema JR, Jump DB, Zacharewski TR. (2005) Temporal and dose-dependent hepatic gene expression patterns in mice provide new insights into TCDD-Mediated hepatotoxicity. Toxicol Sci 85:1048–1063 [DOI] [PubMed] [Google Scholar]
- Chang AC, Hook J, Lemckert FA, McDonald MM, Nguyen MA, Hardeman EC, Little DG, Gunning PW, Reddel RR. (2008) The murine stanniocalcin 2 gene is a negative regulator of postnatal growth. Endocrinology 149:2403–2410 [DOI] [PubMed] [Google Scholar]
- Chen I, McDougal A, Wang F, Safe S. (1998) Aryl hydrocarbon receptor-mediated antiestrogenic and antitumorigenic activity of diindolylmethane. Carcinogenesis 19:1631–1639 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159 [DOI] [PubMed] [Google Scholar]
- Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. (2011) Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol Sci 124:1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferink CJ, Ge NL, Levine A. (2001) Maximal aryl hydrocarbon receptor activity depends on an interaction with the retinoblastoma protein. Mol Pharmacol 59:664–673 [DOI] [PubMed] [Google Scholar]
- Fazio EN, Dimattia GE, Chadi SA, Kernohan KD, Pin CL. (2011) Stanniocalcin 2 alters PERK signalling and reduces cellular injury during cerulein induced pancreatitis in mice. BMC Cell Biol 12:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaveny CA, Murray IA, Perdew GH. (2010) Differential gene regulation by the human and mouse aryl hydrocarbon receptor. Toxicol Sci 114:217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franc MA, Moffat ID, Boutros PC, Tuomisto JT, Tuomisto J, Pohjanvirta R, Okey AB. (2008) Patterns of dioxin-altered mRNA expression in livers of dioxin-sensitive versus dioxin-resistant rats. Arch Toxicol 82:809–830 [DOI] [PubMed] [Google Scholar]
- Gregersen N, Bross P. (2010) Protein misfolding and cellular stress: an overview. Methods Mol Biol 648:3–23 [DOI] [PubMed] [Google Scholar]
- Hayes KR, Zastrow GM, Nukaya M, Pande K, Glover E, Maufort JP, Liss AL, Liu Y, Moran SM, Vollrath AL, et al. (2007) Hepatic transcriptional networks induced by exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Chem Res Toxicol 20:1573–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B. (2006) Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ 13:393–403 [DOI] [PubMed] [Google Scholar]
- He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. (1998) A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA 95:2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Elferink CJ. (2012) A novel nonconsensus xenobiotic response element capable of mediating aryl hydrocarbon receptor-dependent gene expression. Mol Pharmacol 81:338–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D, Walker JR, Thompson CS, Moroz I, Lin W, Veselits ML, Hakim AM, Fienberg AA, Thinakaran G. (2004) Characterization of stanniocalcin 2, a novel target of the mammalian unfolded protein response with cytoprotective properties. Mol Cell Biol 24:9456–9469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees MJ, Whitelaw ML. (1999) Multiple roles of ligand in transforming the dioxin receptor to an active basic helix-loop-helix/PAS transcription factor complex with the nuclear protein Arnt. Mol Cell Biol 19:5811–5822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Lo R, Celius T, Forgacs AL, Dere E, MacPherson L, Harper P, Zacharewski T, Matthews J. (2011) Identification of aryl hydrocarbon receptor binding targets in mouse hepatic tissue treated with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Appl Pharmacol 257:38–47 [DOI] [PubMed] [Google Scholar]
- Ma Y, Brewer JW, Diehl JA, Hendershot LM. (2002) Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J Mol Biol 318:1351–1365 [DOI] [PubMed] [Google Scholar]
- Malhi H, Kaufman RJ. (2011) Endoplasmic reticulum stress in liver disease. J Hepatol 54:795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlowe JL, Knudsen ES, Schwemberger S, Puga A. (2004) The aryl hydrocarbon receptor displaces p300 from E2F-dependent promoters and represses S phase-specific gene expression. J Biol Chem 279:29013–29022 [DOI] [PubMed] [Google Scholar]
- McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. (2001) Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol 21:1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KA, Lockhart CA, Huang G, Elferink CJ. (2006) Sustained aryl hydrocarbon receptor activity attenuates liver regeneration. Mol Pharmacol 70:163–170 [DOI] [PubMed] [Google Scholar]
- Mitchell KA, Wilson SR, Elferink CJ. (2010) The activated aryl hydrocarbon receptor synergizes mitogen-induced murine liver hyperplasia. Toxicology 276:103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. (2004) Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem 279:23847–23850 [DOI] [PubMed] [Google Scholar]
- Ohtake F, Takeyama K, Matsumoto T, Kitagawa H, Yamamoto Y, Nohara K, Tohyama C, Krust A, Mimura J, Chambon P, et al. (2003) Modulation of oestrogen receptor signalling by association with the activated dioxin receptor. Nature 423:545–550 [DOI] [PubMed] [Google Scholar]
- Okey AB, Riddick DS, Harper PA. (1994) Molecular biology of the aromatic hydrocarbon (dioxin) receptor. Trends Pharmacol Sci 15:226–232 [DOI] [PubMed] [Google Scholar]
- Park KT, Mitchell KA, Huang G, Elferink CJ. (2005) The aryl hydrocarbon receptor predisposes hepatocytes to Fas-mediated apoptosis. Mol Pharmacol 67:612–622 [DOI] [PubMed] [Google Scholar]
- Puga A, Maier A, Medvedovic M. (2000) The transcriptional signature of dioxin in human hepatoma HepG2 cells. Biochem Pharmacol 60:1129–1142 [DOI] [PubMed] [Google Scholar]
- Safe S, Wormke M, Samudio I. (2000) Mechanisms of inhibitory aryl hydrocarbon receptor-estrogen receptor crosstalk in human breast cancer cells. J Mammary Gland Biol Neoplasia 5:295–306 [DOI] [PubMed] [Google Scholar]
- Schmidt JV, Bradfield CA. (1996) Ah receptor signaling pathways. Annu Rev Cell Dev Biol 12:55–89 [DOI] [PubMed] [Google Scholar]
- Schmidt JV, Su GH, Reddy JK, Simon MC, Bradfield CA. (1996) Characterization of a murine Ahr null allele: involvement of the Ah receptor in hepatic growth and development. Proc Natl Acad Sci USA 93:6731–6736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobeck MW, Fribourg AF, Puga A, Knudsen ES. (2000) Restoration of retinoblastoma mediated signaling to Cdk2 results in cell cycle arrest. Oncogene 19:1857–1867 [DOI] [PubMed] [Google Scholar]
- Tian Y, Ke S, Denison MS, Rabson AB, Gallo MA. (1999) Ah receptor and NF-kappaB interactions, a potential mechanism for dioxin toxicity. J Biol Chem 274:510–515 [DOI] [PubMed] [Google Scholar]
- Tijet N, Boutros PC, Moffat ID, Okey AB, Tuomisto J, Pohjanvirta R. (2006) Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol Pharmacol 69:140–153 [DOI] [PubMed] [Google Scholar]
- Vezina CM, Walker NJ, Olson JR. (2004) Subchronic exposure to TCDD, PeCDF, PCB126, and PCB153: effect on hepatic gene expression. Environ Health Perspect 112:1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel CF, Sciullo E, Li W, Wong P, Lazennec G, Matsumura F. (2007) RelB, a new partner of aryl hydrocarbon receptor-mediated transcription. Mol Endocrinol 21:2941–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GF, Dimattia GE. (2006) The stanniocalcin family of proteins. J Exp Zoolog A Comp Exp Biol 305:769–780 [DOI] [PubMed] [Google Scholar]
- Walisser JA, Glover E, Pande K, Liss AL, Bradfield CA. (2005) Aryl hydrocarbon receptor-dependent liver development and hepatotoxicity are mediated by different cell types. Proc Natl Acad Sci USA 102:17858–17863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhang R, Shi S, Hankinson O. (2007) The effect of aromatic hydrocarbon receptor on the phenotype of the Hepa 1c1c7 murine hepatoma cells in the absence of dioxin. Gene Regul Syst Bio 1:49–56 [PMC free article] [PubMed] [Google Scholar]
- Wu JC, Merlino G, Fausto N. (1994) Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc Natl Acad Sci USA 91:674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Zhang L, Hoagland MS, Swanson HI. (2007) Lack of the aryl hydrocarbon receptor leads to impaired activation of AKT/protein kinase B and enhanced sensitivity to apoptosis induced via the intrinsic pathway. J Pharmacol Exp Ther 320:448–457 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.