Candida albicans, the most prevalent human fungal pathogen, was considered an obligate diploid that carried recessive lethal mutations throughout the genome. Here, we demonstrate that C. albicans has a viable haploid state that can be derived from diploid cells under in vitro and in vivo conditions and appears to arise via a concerted chromosome loss mechanism. Haploids undergo morphogenetic changes like those of diploids including the yeast-hyphal transition, chlamydospore formation, and a white-opaque switch that facilitates mating. Haploid opaque cells of opposite mating type mate efficiently to regenerate the diploid form, restoring heterozygosity and fitness. Homozygous diploids arise spontaneously by auto-diploidization and both haploids and auto-diploids display a similar reduction in fitness, in vitro and in vivo, relative to heterozygous diploids, suggesting that homozygous cell types are transient in mixed populations. Finally, we constructed stable haploid strains with multiple auxotrophies that will facilitate molecular and genetic analyses of this important pathogen.
Background
The opportunistic fungal pathogen C. albicans has been studied extensively since the 1800’s and has been considered a strictly diploid organism with no haploid state. The diploid nature of the organism has complicated genetic and genomic analyses of C. albicans biology and virulence. Studies in the 1960’s through the early 1980’s debated the ploidy of C. albicans, and proposed that a haploid state existed based on cell size heterogeneity1, parasexual genetics2 and estimates of DNA content by fluorimetry1,3. Haploid clinical isolates were reported2,3, but were later found to be diploid in genome content or were shown to be non-albicans Candida species (T. Suzuki, personal comm. and Fig. S1). Research supporting the diploid nature of the organism included DNA content measurements that suggested a genome size similar to that of diploid Saccharomyces cerevisiae4,5, molecular studies that demonstrated the need to delete two gene copies to produce a null mutant6,7, genetic studies that demonstrated heterozygosity of specific alleles8,9, and ultimately the complete genome sequence, which revealed heterozygosity throughout much of the genome10. This, together with the predominantly clonal nature of C. albicans within an individual host11,12, was interpreted as evidence that the organism spent most, if not all, of its life cycle in a diploid state.
The ‘obligate diploid’ nature of C. albicans was proposed to be due to recessive lethal mutations dispersed throughout the genome13,14. However, studies of C. albicans chromosome monosomy15,16, recombination17 and haplotype mapping18 demonstrated that homozygosis of certain chromosomes can occur, arguing against this balanced lethal mutation hypothesis. A diploid-tetraploid parasexual cycle was also discovered in C. albicans and shown to involve a switch to the ‘opaque’ physiological state that renders cells mating-competent19,20, conjugation between opaque diploids to form tetraploids21,22, and subsequent ploidy reduction resulting in diploid progeny that often carry multiple aneuploid chromosomes18,23. Importantly, specific aneuploid chromosomes can provide a selective advantage under stressful conditions such as exposure to antifungal drugs24. Parasexual ploidy reduction in tetraploids occurs via a non-meiotic process termed ‘concerted chromosome loss’. This process can facilitate the rapid generation of diversity through the production of homozygous and aneuploid progeny, under conditions where outcrossing in the host is unlikely25. If a haploid state for C. albicans were to exist, it could facilitate the elimination of lethal alleles from the population. Furthermore, mating between different haploids could promote adaptation to changing conditions within the mammalian host. With the advent of whole genome approaches that can distinguish ploidy states, it is now possible to ask if C. albicans exists, even transiently, in the tetraploid or the haploid state.
Detection of haploid C. albicans cells
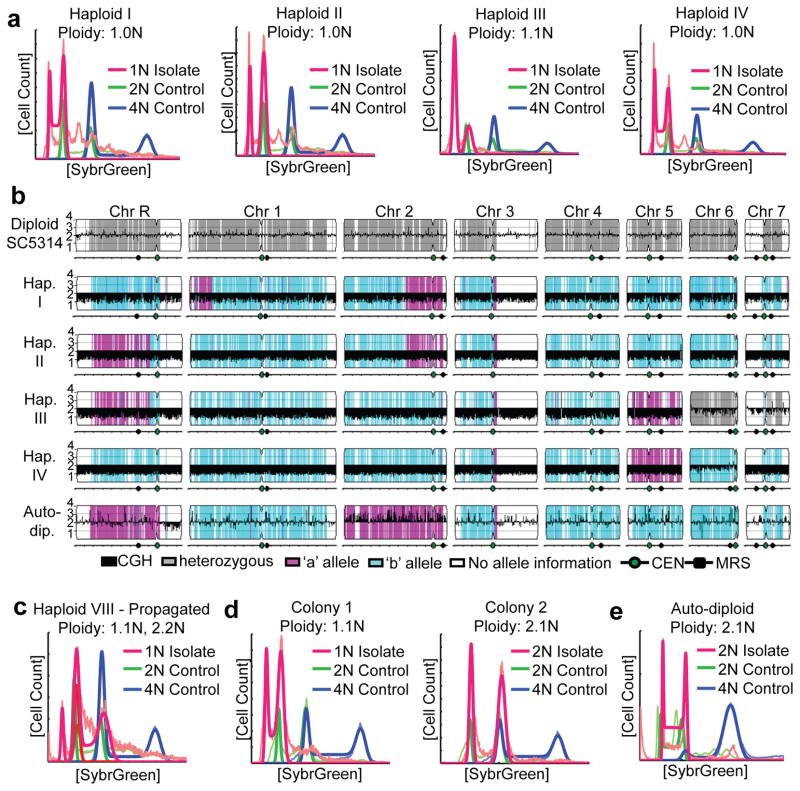
Haploid C. albicans was serendipitously discovered during experiments to follow loss of heterozygosity (LOH) at multiple independent loci. Using a multiply marked derivative of SC5314, the laboratory reference strain10, we selected for an initial LOH event at the GAL1 locus by growth on 2-deoxygalactose (2-DOG) and subsequently screened for additional LOH events at four other heterozygous loci. Importantly, growth in 2-DOG does not affect LOH rates (Fig. S2) and as such, the selection for cells with an LOH event does not artificially induce LOH. Amongst the ~2500 Gal− isolates, 42 exhibited additional LOH events and intriguingly one strain, Haploid I, was homozygous for all of the markers tested, as well as for multiple SNPs on every chromosome. Analysis of DNA content in Haploid I by flow cytometry indicated that the genome had half the amount of DNA of that in a diploid control (Fig. 1a).
Figure 1. C. albicans haploid and auto-diploid genotypes.
a) Flow cytometry analysis of DNA content of haploid strains (red) compared to diploid (SC5314, green) and tetraploid (RBY18, blue) control strains. Thin lines, raw data; bold lines, ‘best fit’ data as described in the Methods Summary. b) SNP/CGH array analysis of indicated strains (left) showing copy number (log2 ratio, black) and SNP allele information (Grey, heterozygous; magenta, allele ‘a’; cyan, allele ‘b’; white, no SNP data.) c) Flow cytometry after prolonged propagation of a haploid revealed a mixture of ploidies within a single population (c), while single colonies from this haploid (d) exhibit distinct haploid (left) and diploid (right) ploidy. e) Flow cytometry of a homozygous auto-diploid.
Once we became aware that haploid C. albicans cells could be detected, we used flow cytometry to screen isolates from many sources, including in vitro stresses,26 from which one haploid isolate (Haploid IV) was identified amongst small colonies growing in the presence of fluconazole (in the halo of an E-test strip), a commonly used antifungal drug. We also screened C. albicans cells isolated from in vivo mouse models of candidemia27 and candidiasis28 and discovered several additional isolates with 1N DNA content. The overall population of post-in vivo isolates screened for ploidy included ~300 isolates from YPD plates (Gal+), and ~740 isolates from the 2-DOG selection plates (Gal−) and only the DOG-resistant colonies contained isolates that were haploid. We found that 3.2% of DOG-resistant colonies from the OPC model were haploid (10 out of 312), while 1.2% of DOG-resistant colonies from the systemic model were haploid (5 out of 431). Since LOH frequencies in vivo are ~10−3, 29, this indicates that detectable haploids appeared at a frequency of 1–3 per 100,000 cells. Loss of GAL1 is often, but not always associated with the haploid state (e.g. Haploid IV), especially in diploid progenitors that are heterozygous for this locus. In total, we characterized eleven strains with haploid (or close to haploid) flow cytometry profiles (Fig. 1a, S3 and Table S1) that were derived from multiple independent and varied sources.
Since previous reports of haploid C. albicans3 included misidentified species (Fig. S1), we determined if these newly identified isolates were bona fide C. albicans by PCR amplifying and sequencing the mating-type-like (MTL) locus. All isolates contained sequences identical to either the MTLa or MTLα alleles of SC5314 (Fig. S4 and data not shown). Next, we analyzed the haploid genomes by hybridization to SNP/CGH arrays30 which measure allelic ratios and also infer relative chromosome copy numbers. Six isolates were completely euploid (Fig. 1b and S5); in the remaining five haploids, one or two chromosomes (Chr6 and Chr7) were disomic. These disomic chromosomes were heterozygous except in one isolate (Haploid IV) where the disomic chromosome (Chr6) was homozygous.
Based upon the C. albicans haplotype map30, each haploid chromosome primarily contained alleles from only one parental homolog with few, if any, obvious crossovers detectable (Fig. 1b and S5). This implies that haploids did not arise through conventional meiosis, which usually requires at least one crossover per chromosome31. The few crossovers detected likely arose by mitotic recombination prior to selection for LOH events. For example, Haploid I was isolated following selection for loss of GAL1, which is within the crossover region on Chr1. Furthermore, in C. albicans haploids (Fig. 1b and S5), the majority of disomic chromosomes were heterozygous, which would appear following segregation of homologs in meiosis I. Such meiosis I segregation is not seen in Drosophila male meioses, which lack recombination32; it is seen in Candida lusitaniae meiotic progeny, which undergo high levels of recombination33. The low level of genetic recombination together with the presence of heterozygous disomic chromosomes remains most consistent with random homolog segregation in C. albicans. Accordingly, we propose that haploids arise via a concerted chromosome loss mechanism akin to that described for the diploid-tetraploid C. albicans parasexual cycle23, although a non-conventional meiotic program cannot be completely discounted.
The existence of haploids refutes the argument that recessive lethal alleles are present on each C. albicans homolog13,14. Nonetheless, the absence of some chromosomal homologs from the haploid progeny suggests that recessive lethal alleles may exist. Indeed, only the ‘b’ homologs for Chr3, Chr4, Chr6, Chr7 and most of Chr1 were detected in the haploids analyzed (Fig. 1b and S5). Interestingly, a similar homolog bias was seen in diploid parasexual progeny derived from SC531418. In contrast, homologs of Chr5, which carries the MTL, appeared in equal numbers (six MTLa and five MTLα) and were entirely of one or the other parental haplotype. Similarly, both homologs of ChrR and Chr2 were observed. As such, homologs from more than half the chromosomes (Chr3, Chr4, Chr6, Chr7 and most of Chr1) potentially carry at least one recessive lethal mutation and may limit the frequency with which viable haploids appear. Accordingly, we propose that haploidization may provide an effective mechanism for eliminating recessive lethal mutations from the predominantly diploid populations of C. albicans.
Auto-diploidization of haploids
In the course of these studies, prolonged propagation of haploid isolates yielded cultures with a mixed population of haploid and diploid cells (Fig. 1c). Subsequent colony purification yielded distinct haploid and/or diploid populations (Fig. 1d). Surprisingly, some haploid isolates also diploidized during the process of strain shipping, which involved storage, transit, and revival from partial dehydration on a solid surface. For example, a potential haploid isolate identified in Taiwan was sent to Minnesota on sterile filter paper. Once revived and grown in liquid culture, the genome was diploid (Fig. 1e) and homozygous for all SNPs (Fig. 1b, ‘Auto-dip.’), suggesting auto-diploidization from a haploid phase. While homozygous diploids could arise through mitotic defects or by self-mating34, mating between cells of the same mating-type was not detectable, as discussed below. Thus, we suggest that auto-diploids arise through mitotic defects that may be analogous to the auto-diploidization events in vertebrate haploid stem cell cultures35–37.
Haploid Morphogenesis and Mating
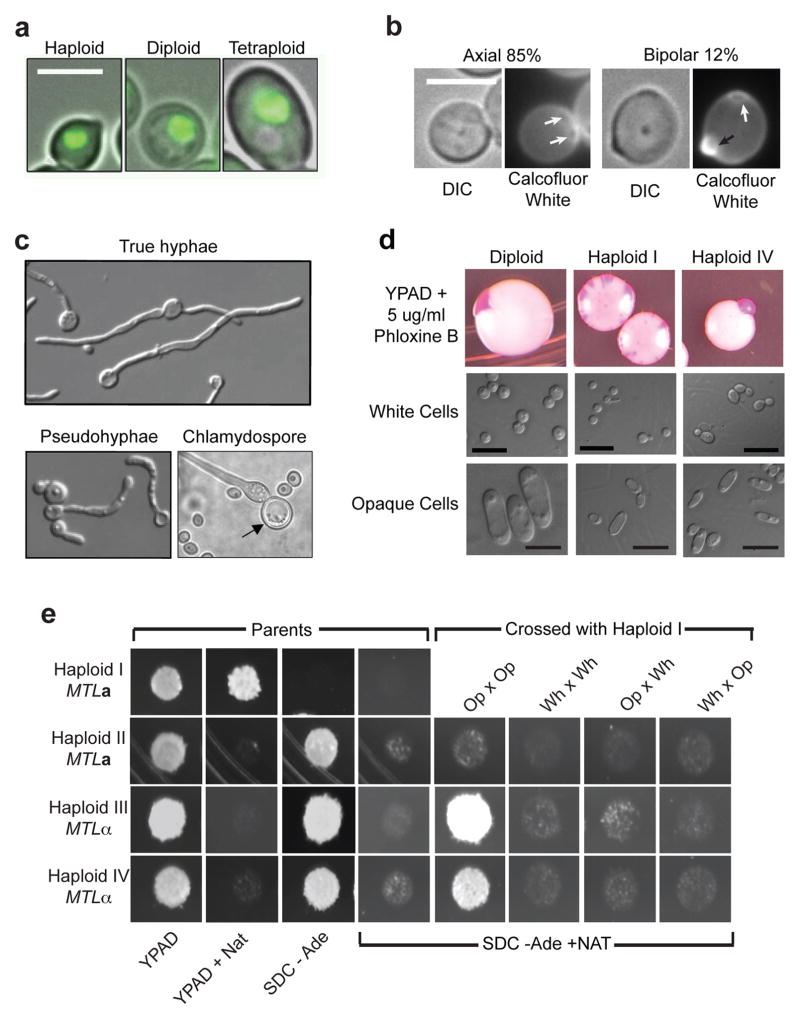
Consistent with reduced ploidy content in other yeast species38, haploid C. albicans cells were smaller, on average, for both cell and nuclear size compared to diploid cells (Fig. 2a and S6). The bud site selection patterns of haploids, like diploid C. albicans were predominantly axial, with a small fraction of cells displaying a bipolar budding pattern (Fig. 2b)39. Significantly, under conditions that induce morphogenesis of diploid C. albicans to form true hyphae, pseudohyphae, and chlamydospores40,41, similar morphogenetic events occurred in haploids (Fig. 2c).
Figure 2. Morphology and mating competency of haploid C. albicans.
a) Representative DIC images of haploid, diploid, and tetraploid cells overlaid with fluorescence images of their nuclei. b) Calcofluor white staining revealed primarily cells with the axial budding pattern, 15% with a bipolar budding pattern (n = 72) and 3% that were difficult to resolve. White arrows, previous bud scars; black arrow, newest bud. Scale bars represent 5μm. c) Haploids form true hyphae, pseudohyphae, and chlamydospores in serum, RPMI, and corn meal agar media, respectively. d) White-opaque switching detected as pink colony sectoring (top) and by microscopy of cells from white and pink/opaque sectors. Diploid, MTLa/MTLα1Δα2Δ (YJB12234), Haploid I (MTLa), and Haploid IV (MTLα). e) Mating between haploid cells. ‘Parents’: Haploid I (MTLa NAT1 ade2Δ), Haploid II (MTLa ADE2), Haploids III and IV (MTLα ADE2) showing growth on media indicated. ‘Crossed with Haploid I’: opaque (Op) or white cells (Wh) cells from Haploid I were mixed with opaque and/or white cells from Haploids II, III or IV and plated to medium selective for mating products.
C. albicans diploid cells that are homozygous at the MTL can undergo a transcriptionally induced switch to the opaque state that renders them competent for mating19,42. Similarly, haploids readily switched from the white (Wh) to the opaque (Op) state, as detected by colony sectoring (Fig. 2d). Haploid opaque cells had elongated opaque-like cell morphology43, yet were considerably smaller than diploid opaque cells (Fig. 2d). Importantly, haploid opaque cells of opposite mating type were mating competent. Mating between cells with complementary genetic markers resulted in the formation of heterozygous diploids (Fig. 2e, Op x Op). In contrast, neither opaque cells of the same mating type, nor white cells of opposite mating type, mated efficiently (Fig. 2e). This is consistent with opposite mating types and the opaque state being prerequisites for conventional mating19,42. Thus, we propose that haploids participate in a non-meiotic haploid-diploid parasexual cycle.
Reduced growth and virulence of haploids
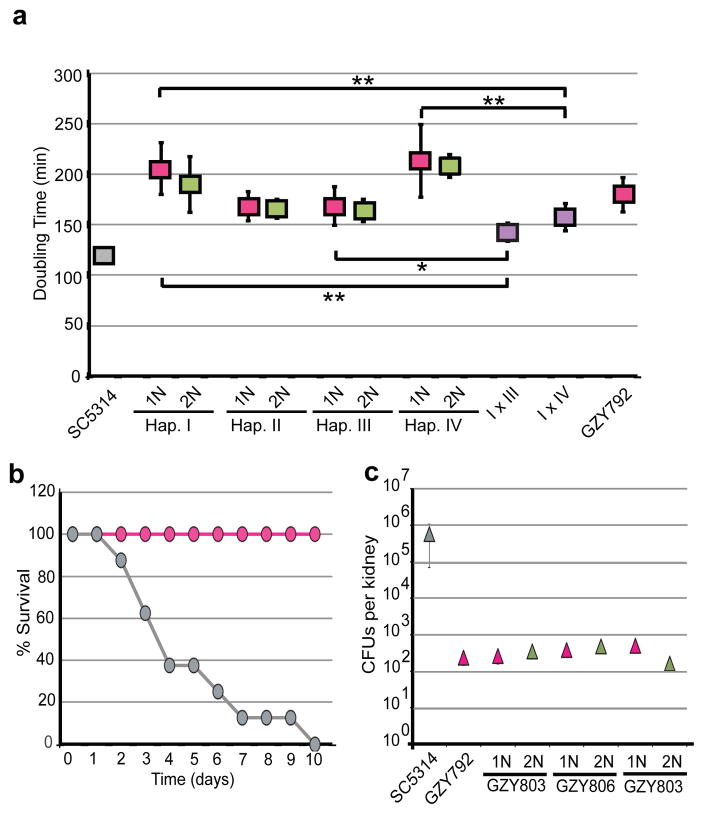
The products from haploid a and α mating were heterozygous for MTL (Fig. S7), near-diploid in genome content (Fig. S7), and likely inherited the extra aneuploid copies of Chr6 and/or Chr7 from their MTLα parent. All the haploid isolates grew significantly slower than SC5314 (Fig. 3a and S8, p < 0.001 for all haploids). However, the mating products grew significantly faster than either of their haploid parents (Fig. 3a), as expected if heterozygosity restores fitness through the complementation of recessive alleles44. Consistent with this, auto-diploids exhibited growth rates indistinguishable from their haploid progenitors (Fig. 3a and S8), with the single exception of Haploid VIII (p = 0.02). We propose that the low fitness of haploids, as well as their corresponding auto-diploids, is a consequence of unmasking recessive alleles that reduce growth potential. Furthermore, the variability in fitness between haploids is likely due to the specific combination of alleles inherited by each haploid isolate.
Figure 3. Haploid growth in vitro and in vivo.
a) Growth (doubling times in YPAD) of control diploid, haploids (pink), their auto-diploid derivatives (green), and heterozygous, mating products I × III and I × IV (purple). Error bars reflect one standard deviation from the mean. * p < 0.01, ** p < 0.001, Student’s t-test. b) Survival of mice (tail vein systemic candidiasis model) following inoculation with Haploid II (pink) or its diploid progenitor, YJB12419 (grey). c) Recovery of colony forming units (CFUs) from mouse kidneys (three mice per yeast strain) 48 hours post-infection.
Significantly, the diploid mating products from haploid crosses grew more slowly than the highly heterozygous diploid SC5314. The generation of robust and diverse diploid progeny from haploid mating would require outcrossing, which is predicted to occur infrequently11,12, as many humans are colonized with a single strain of C. albicans. As it appears that some lethal mutations have accumulated on one of the two homologs for more than half the chromosomes (Fig. 1 and S5), mating between related haploids would not reestablish complete genome heterozygosity. Thus, even though the formation of haploids potentially eliminates lethal mutations, continued inbreeding between related haploids would perpetuate homozygosity of most chromosomes and consequently would reduce overall fitness.
Consistent with reduced growth rates in vitro, Haploid II was avirulent in a mouse model of systemic candidiasis (Fig. 3b) and was cleared from the mouse after ten days. However, after 48 hours of infection with a haploid strain, cells could be recovered (Fig. S9a) and the majority of these isolates remained haploid. Subsequent in vivo experiments compared the colony forming units (CFUs) of haploids, auto-diploids and SC5314 isolated from mouse kidneys 48 hours post-inoculation in individual and direct competition experiments (Fig. 3c and S9b). The number of CFUs recovered from mice inoculated with haploids and auto-diploids was several orders of magnitude lower than the number of CFUs recovered from mice inoculated with SC5314 (Fig. 3c). Furthermore, competition experiments indicated that haploids and auto-diploids exhibited similar fitness in vivo, whereas both of these forms were outcompeted by heterozygous diploids (Fig. S9b). Similar to growth rates in vitro, the low fitness of haploids and their corresponding auto-diploids in vivo, indicates that it is not the diploid state, per se, that is beneficial to growth but rather extensive allelic heterozygosity, as evidenced with SC5314. From these data, we hypothesize that haploid formation may not be rare, but that haploids likely represent a very small fraction of the overall population due to their low competitive fitness relative to heterozygous diploids.
Haploid strains as genetic tools
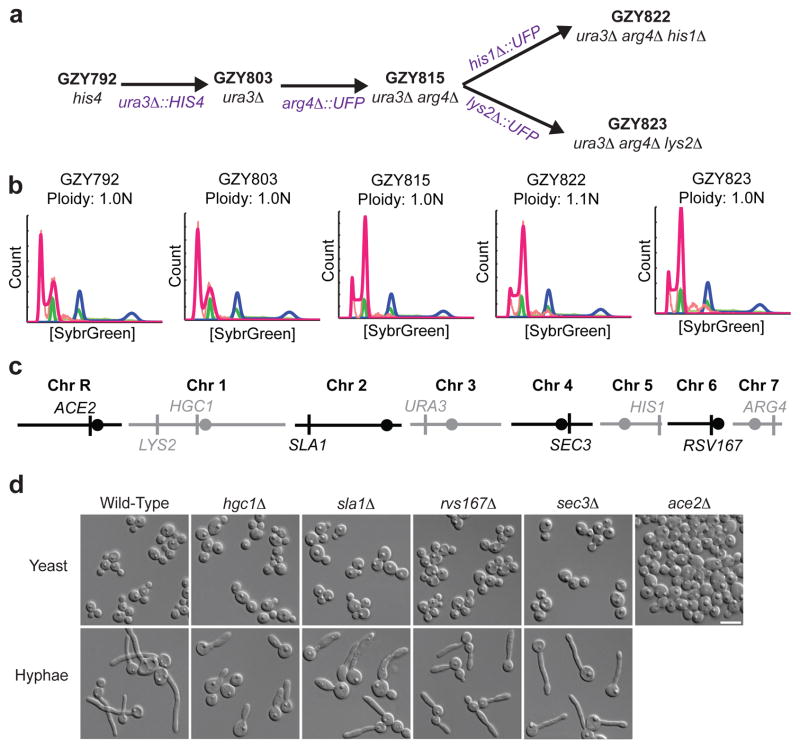
The haploid state greatly facilitates experimental approaches such as classical genetic screens for recessive alleles and a single-round of gene-knockout phenotyping. To illustrate this potential, a set of auxotrophic C. albicans strains was derived from a relatively stable haploid, GZY792 (Fig. 4a and S10) and genes important for morphogenesis in diploids (HGC1, RVS167, SLA1, SEC3 and ACE2) were deleted in a single step. The resulting mutants exhibited morphogenesis defects closely resembling those of the corresponding diploid null mutants (Fig. 4c)45–48. Furthermore, because the deleted genes map to all eight chromosomes (Fig. 4b), the ability to delete each gene in a single step confirms that all chromosomes were monosomic in the parental haploid.
Figure 4. Auxotrophic haploid strains enable one-step gene deletions.
a) Series of strains constructed from a stable haploid isolate, GZY792 (MTLα, his4) was isolated after propagation for 30 passages, screening for ploidy by flow cytometry and selection of isolates that were consistently haploid. GZY803 (ura3Δ) was constructed by disruption of URA3 with HIS4. Other auxotrophies were generated by the URA-flipper approach50. b) Flow cytometry of these auxotrophic strains. c) Genes disrupted in one-step map to all eight chromosomes. Circles, centromere position. d) Cell morphology phenotypes of haploid mutants grown in minimal media (yeast) or media supplemented with 20% FBS at 37° (hyphae) are similar to phenotypes seen for the corresponding diploid null mutants.
Concluding remarks
In summary, C. albicans can no longer be considered to be an obligate diploid. Rather, it has the ability to form haploids that subsequently mate to form diploids or undergo auto-diploidization. The lower fitness of haploids and auto-diploids suggests they will not persist in the population and thus should be detected only rarely. Nonetheless, efficient mating between viable haploids can produce heterozygous diploids that have increased fitness, presumably due to the complementation of detrimental recessive alleles. Furthermore, the reduction to a haploid state can serve as a vehicle to eliminate recessive lethal alleles from a heterozygous diploid population. We propose that C. albicans rapidly generates genetic diversity by producing a broad range of different ploidy states, including haploid, diploid, tetraploid, and aneuploid25. Each of these ploidy states are mating competent and genetic outcrossing can introduce, albeit infrequently, further genetic diversity into the population. While the ploidy reduction mechanism(s) used by C. albicans remain an enigma, the discovery of a haploid form and the potential for a haploid-diploid parasexual cycle significantly expands our ability to manipulate, and thereby better understand, this opportunistic pathogen. Furthermore, this study reveals how whole-genome analyses can lead to a re-evaluation of common assumptions about genomic structure in microbial organisms.
Supplemental Methods
Haploid screening from the mouse models of infection
Bloodstream infections were performed by injecting 106 cells of parent strain AF7 (gal1Δ::URA3/GAL1), into the tail vein of 13 outbred ICR male mice (22–25 g, Harlan, Indianapolis)27. When moribund (at 5–7 days), mice were anesthetized using isofluorane, euthanized, and both kidneys were removed. Kidneys were combined, homogenized with 1 mL of water. 1:1000 dilutions of kidney homogenates were plated for total cell count onto YPD, and 1:10 dilutions of the same homogenate were plated onto 2-DOG medium to obtain Gal− colony counts at 3 days.
For the oropharyngeal model of infection, Balb/C mice were immunosuppressed with cortisone on days −1, 1 and 3 of infection. Calcium alginate swabs were saturated with a suspension of 106 cells/ml of YJB9318 (gal1Δ::URA3/GAL1) under their tongues for 75 min. Mice were sacrificed at 1, 2, 3, and 5 days post-infection. 1:1000 dilutions of tongue tissue homogenates were plated for total cell count onto YPD, and 1:10 dilutions of the same homogenate were plated onto 2-DOG medium to obtain Gal− colony counts at 3 days. Importantly, to confirm that 2-DOG resistant cells only arose during in vivo passage and were not selected for by plating on 2-DOG medium, a gal1/gal1 strain and a gal1/GAL1 heterozygote strain were plated onto 2-DOG medium and observed for growth. 2-DOG resistant colonies grew up from the gal1/gal1 mutant but not from the GAL1/gal1 heterozygote by day 3 after plating27. Therefore, we used a cut-off of day 3 for picking 2-DOG resistant colonies after plating.
Approximately 300 colonies were transferred from YPD plates and ~740 colonies from 2-DOG plates (312 from OPC and 431 BSI isolates) into 96-well plates containing 50% glycerol and stored at −80°. All C. albicans cells isolated following in vivo passaging were analyzed by flow cytometry to determine cell ploidy.
Flow cytometry preparation and analysis
Mid-log phase cells were harvested, washed and resuspended in 50:50 TE (50mM Tris pH8 : 50mM EDTA) and fixed with 95% ethanol. Cells were washed with 50:50 TE and treated with 1 mg/ml RNAse A and then 5 mg/ml Proteinase K. Cells were washed with 50:50 TE and resuspended in SybrGreenI (1:85 dilution in 50:50 TE) incubated overnight at 4°. Stained cells were collected and resuspended in 50:50 TE and analyzed using a FACScaliber. Whole genome ploidy was estimated by fitting DNA content data with a multi-Gaussian cell cycle model that assumes the G2 peak has twice the fluorescence of the G1 peak and that minimizes S-phase cell contribution to the error function. Ploidy values were calculated by comparing the ratio of peak locations in experimental samples to those of diploid and tetraploid controls.
Mating Assays
Opaque or white cells were mixed together in equal cell numbers and incubated on Spider media34 for 18 hours prior to replica-plating onto SDC −Ade + Nat (to select for mating products), as well as YPAD, YPAD + Nat, SDC −Ade and SDC −Ade + Nat to detect parental auxotrophies and then photographed 24 hours later.
In vitro growth assays
Strains were grown in YPD media supplemented with adenine, uridine and histidine in a 96-well microtiter plate and optical density was measured every 15 minutes with a plate reader (Tecan Sunrise) for 24 hours. Doubling times were calculated as previously described30.
In vivo assays
For virulence assays, eight mice per C. albicans strain were inoculated with ~6.0 × 105 CFUs by tail vein injection49. For survival assays, three mice per strain were inoculated by tail vein injection, both kidneys were harvested at 48h and CFUs were determined by plating on YPAD. Ploidy of randomly selected isolates was determined by flow cytometry.
Supplementary Material
Acknowledgments
We would like to thank M. McClellan, K. Matter, E. Voigt and F.Y. Chan for technical assistance, S. Filler and J. Becker for work involving mouse models of infection, F.M. Chang and T.Y. Ou for contributing to the isolation of the progenitor of GZY792, and L. Burrack, J. Heitman, M. Kupiec, K. Nielsen and N. Pavelka, for comments on the manuscript. M.A.H. is supported by a NRSA post-doctoral fellowship (F32GM096536-02). A.F. is supported by the National Institute of Allergy and Infectious Diseases (NIAID) (R15-AI090633-01A1 and R01 AI0624273). M.P.H. is supported by a training grant for Graduate Assistance in Areas of National Need (P200A100100). B.D.H. is supported by the National Institute of Dental & Craniofacial Research (T32DE007288). R.J.B. is supported by NIAID (AI081560 and AI081704) and a PATH Award from the Burroughs Wellcome Fund. Y.W. is supported by Agency for Science, Technology, & Research, Singapore. J.B. is supported by the NIAID (AI0624273).
Footnotes
Author Contributions
M.A.H. performed flow cytometry analysis, SNP/CGH hybridizations, species identification, white-opaque switching and mating assays, and in vitro growth assays. Y.W. and G.Z. designed and analyzed auxotrophs, morphogenesis mutants and in vivo growth experiments; GZ constructed the mutants; Y.M.W. collected isolates post-in vivo. A.F. and H.C.S. initially isolated haploid/homozygous isolates. D.A. developed the flow cytometry analysis and SNP/CGH pipelines. M.P.H. performed virulence and in vivo competition assays. B.D.H. collected and analyzed cell and nuclear size data and budding patterns. M.A.H. and J.B. assembled the data and wrote the manuscript with editorial input from A.F., R.J.B. and Y.W.
Literature Cited
- 1.van der Walt JP. Sexually active strains of Candida albicans and Cryptococcus albidus. Antonie Van Leeuwenhoek. 1967;33:246–256. doi: 10.1007/BF02045570. [DOI] [PubMed] [Google Scholar]
- 2.Sarachek A, Rhoads DD, Schwarzhoff RH. Hybridization of Candida albicans through fusion of protoplasts. Arch Microbiol. 1981;129:1–8. doi: 10.1007/BF00417169. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki T, Nishibayashi S, Kuroiwa T, Kanbe T, Tanaka K. Variance of ploidy in Candida albicans. J Bacteriol. 1982;152:893–896. doi: 10.1128/jb.152.2.893-896.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olaiya AF, Sogin SJ. Ploidy determination of Candida albicans. J Bacteriol. 1979;140:1043–1049. doi: 10.1128/jb.140.3.1043-1049.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riggsby WS, Torres-Bauza LJ, Wills JW, Townes TM. DNA content, kinetic complexity, and the ploidy question in Candida albicans. Mol Cell Biol. 1982;2:853–862. doi: 10.1128/mcb.2.7.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly R, Miller SM, Kurtz MB, Kirsch DR. Directed mutagenesis in Candida albicans: one-step gene disruption to isolate ura3 mutants. Mol Cell Biol. 1987;7:199–208. doi: 10.1128/mcb.7.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurtz MB, Marrinan J. Isolation of hem3 mutants from Candida albicans by sequential gene disruption. Mol Gen Genet. 1989;217:47–52. doi: 10.1007/BF00330941. [DOI] [PubMed] [Google Scholar]
- 8.Magee PT, Kakar S, Kwon-Chung K. Genetic analysis of Candida albicans by complementation. In: Schessinger DM, editor. Microbiology. American Society of Microbiology; Washington DC: 1983. pp. 230–233. [Google Scholar]
- 9.Whelan WL, Partridge RM, Magee PT. Heterozygosity and segregation in Candida albicans. Mol Gen Genet. 1980;180:107–113. doi: 10.1007/BF00267358. [DOI] [PubMed] [Google Scholar]
- 10.Jones T, et al. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci USA. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gräser Y, et al. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc Natl Acad Sci USA. 1996;93:12473–12477. doi: 10.1073/pnas.93.22.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bougnoux ME, et al. Mating is rare within as well as between clades of the human pathogen Candida albicans. Fungal Genet Biol. 2008;45:221–231. doi: 10.1016/j.fgb.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whelan WL, Soll DR. Mitotic recombination in Candida albicans: recessive lethal alleles linked to a gene required for methionine biosynthesis. Mol Gen Genet. 1982;187:477–485. doi: 10.1007/BF00332632. [DOI] [PubMed] [Google Scholar]
- 14.Sarachek A, Weber DA. Segregant-defective heterokaryons of Candida albicans. Curr Genet. 1986;10:685–693. doi: 10.1007/BF00410917. [DOI] [PubMed] [Google Scholar]
- 15.Barton RC, Gull K. Isolation, characterization, and genetic analysis of monosomic, aneuploid mutants of Candida albicans. Mol Microbiol. 1992;6:171–177. doi: 10.1111/j.1365-2958.1992.tb01998.x. [DOI] [PubMed] [Google Scholar]
- 16.Janbon G, Sherman F, Rustchenko E. Monosomy of a specific chromosome determines L-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc Natl Acad Sci USA. 1998;95:5150–5155. doi: 10.1073/pnas.95.9.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andaluz E, et al. Rad52 function prevents chromosome loss and truncation in Candida albicans. Mol Microbiol. 2011;79:1462–1482. doi: 10.1111/j.1365-2958.2011.07532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forche A, et al. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 2008;6:e110. doi: 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 20.Soll DR, Lockhart SR, Zhao R. Relationship between switching and mating in Candida albicans. Eukaryotic Cell. 2003;2:390–397. doi: 10.1128/EC.2.3.390-397.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magee BB, Magee PT. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science. 2000;289:310–313. doi: 10.1126/science.289.5477.310. [DOI] [PubMed] [Google Scholar]
- 22.Hull CM, Raisner RM, Johnson AD. Evidence for mating of the ‘asexual’ yeast Candida albicans in a mammalian host. Science. 2000;289:307–310. doi: 10.1126/science.289.5477.307. [DOI] [PubMed] [Google Scholar]
- 23.Bennett RJ, Johnson AD. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003;22:2505–2515. doi: 10.1093/emboj/cdg235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berman J, Hadany L. Does stress induce (para)sex? Implications for Candida albicans evolution. Trends Genet. 2012;28:197–203. doi: 10.1016/j.tig.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forche A, et al. Stress alters rates and types of loss of heterozygosity in Candida albicans. MBio. 2011;2 doi: 10.1128/mBio.00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forche A, et al. A system for studying genetic changes in Candida albicans during infection. Fungal Genet Biol. 2003;39:38–50. doi: 10.1016/s1087-1845(02)00585-6. [DOI] [PubMed] [Google Scholar]
- 28.Solis NV, Filler SG. Mouse model of oropharyngeal candidiasis. Nat Protoc. 2012;7:637–642. doi: 10.1038/nprot.2012.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forche A, Magee PT, Selmecki A, Berman J, May G. Evolution in Candida albicans populations during a single passage through a mouse host. Genetics. 2009;182:799–811. doi: 10.1534/genetics.109.103325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abbey D, Hickman M, Gresham D, Berman J. High-resolution SNP/CGH microarrays reveal the accumulation of loss of heterozygosity in commonly used Candida albicans strains. G3 (Bethesda) 2011;1:523–530. doi: 10.1534/g3.111.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai JH, McKee BD. Homologous pairing and the role of pairing centers in meiosis. J Cell Sci. 2011;124:1955–1963. doi: 10.1242/jcs.006387. [DOI] [PubMed] [Google Scholar]
- 33.Reedy JL, Floyd AM, Heitman J. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr Biol. 2009;19:891–899. doi: 10.1016/j.cub.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alby K, Schaefer D, Bennett RJ. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature. 2009;460:890–893. doi: 10.1038/nature08252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi M, Hong N, Hong Y. Generation of medaka fish haploid embryonic stem cells. Science. 2009;326:430–433. doi: 10.1126/science.1175151. [DOI] [PubMed] [Google Scholar]
- 36.Freed JJ, Mezger-Freed L. Stable haploid cultured cell lines from frog embryos. Proc Natl Acad Sci USA. 1970;65:337–344. doi: 10.1073/pnas.65.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elling U, et al. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell. 2011;9:563–574. doi: 10.1016/j.stem.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mortimer RK. Radiobiological and genetic studies on a polyploid series (haploid to hexaploid) of Saccharomyces cerevisiae. Radiat Res. 1958;9:312–326. [PubMed] [Google Scholar]
- 39.Chaffin WL. Site selection for bud and germ tube emergence in Candida albicans. Microbiology. 1984;130:431–440. [Google Scholar]
- 40.Berman J, Sudbery PE. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat Rev Genet. 2002;3:918–930. doi: 10.1038/nrg948. [DOI] [PubMed] [Google Scholar]
- 41.Citiulo F, Moran GP, Coleman DC, Sullivan DJ. Purification and germination of Candida albicans and Candida dubliniensis chlamydospores cultured in liquid media. FEMS Yeast Res. 2009;9:1051–1060. doi: 10.1111/j.1567-1364.2009.00533.x. [DOI] [PubMed] [Google Scholar]
- 42.Lockhart SR, et al. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics. 2002;162:737–745. doi: 10.1093/genetics/162.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slutsky B, et al. White-opaque transition: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crow J, Kimura M. Evolution in sexual and asexual populations. American Naturalist. 1965;99:439–450. [Google Scholar]
- 45.Wang Y. CDKs and the yeast-hyphal decision. Curr Opin Microbiol. 2009;12:644–649. doi: 10.1016/j.mib.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Kelly MT, et al. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol Microbiol. 2004;53:969–983. doi: 10.1111/j.1365-2958.2004.04185.x. [DOI] [PubMed] [Google Scholar]
- 47.Zeng G, Wang Y-M, Wang Y. Cdc28-Cln3 phosphorylation of Sla1 regulates actin patch dynamics in different modes of fungal growth. Mol Biol Cell. 2012 doi: 10.1091/mbc.E12-03-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Douglas LM, Martin SW, Konopka JB. BAR domain proteins Rvs161 and Rvs167 contribute to Candida albicans endocytosis, morphogenesis, and virulence. Infect Immun. 2009;77:4150–4160. doi: 10.1128/IAI.00683-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morschhäuser J, Michel S, Staib P. Sequential gene disruption in Candida albicans by FLP-mediated site-specific recombination. Mol Microbiol. 1999;32:547–556. doi: 10.1046/j.1365-2958.1999.01393.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.