Abstract
We are in the midst of a worldwide epidemic of type 2 diabetes (T2D) and obesity. Understanding the mechanisms underlying these diseases is critical if we are to halt their progression and ultimately prevent their development. The advent and widespread implementation of microarray technology has allowed analysis of small samples of human skeletal muscle, adipose, liver, pancreas and blood. While patterns differ in each tissue, several dominant themes have emerged from these studies, including altered expression of genes indicating increased inflammation and altered lipid and mitochondrial oxidative metabolism and insulin signaling in patients with T2D, and in some cases, in those at risk for disease. Unraveling which changes in gene expression are primary, and which are secondary to an insulin resistant or diabetes metabolic milieu remains a scientific challenge but we are one step closer.
Keywords: Insulin resitance, type 2 diabetes, genomic analyses in humans
Introduction
Type 2 diabetes (T2D) in the United States and around the world has reached epidemic proportions. At present, 18.8 million people in the United States have been diagnosed with diabetes, with an additional 7 million undiagnosed (1). An additional 79 million Americans suffer from prediabetes. Unfortunately, the incidence of T2D is also increasing at an alarming rate, with more than tripling between 1980 and 2009 (1) and a projected increase of 165% from 2000 to 2050 (2). Intimately linked with the rise in diabetes prevalence is the burgeoning epidemic of obesity, particularly in developed societies (3). These alarming patterns will likely translate into increases in cardiovascular and other health risks associated with insulin resistance and diabetes.
An important public health goal is to better understand the complex pathophysiology of T2D and to determine which individuals are at highest risk, in order to target prevention and early intervention strategies. Fortunately, the incidence of T2D can be modified by alterations in lifestyle, including exercise, modest weight loss, and specific medication (4–7). Despite the many advances in behavioral and pharmaceutical interventions, the vast majority of individuals with diabetes continue to have suboptimal metabolic control (8), emphasizing the need for improved understanding of disease pathophysiology and the identification of new and more effective targets for treatment and prevention.
Pathophysiology of Type 2 Diabetes
Multiple physiologic defects have been identified in T2D, including insulin resistance in muscle, adipose tissue, and liver, impaired insulin secretion, impaired incretin secretion and action, and altered balance of CNS pathways controlling food intake and energy expenditure. While the relative contribution of each of these defects continues to be debated, longitudinal studies in high-risk individuals suggest that insulin resistance is a very early phenomenon, occurring years before any evidence of glucose intolerance or β-cell failure and predicting the development of type 2 diabetes (9). In turn, both genetic and environmental factors, including family history, early-life development, obesity, inactivity, and aging, all contribute to insulin resistance. Strategies to identify genetic determinants have included genome-wide association studies; these have identified novel candidate loci linked to diabetes risk, but the majority of those identified thus far appear related to β-cell function and not insulin resistance, and together, identified loci explain only about 10% of diabetes heredity (10). At a cellular level, mitochondrial oxidative dysfunction, endoplasmic reticulum stress, oxidative stress, and alterations in insulin signaling have all been linked to insulin resistance. However, which of these are critical for diabetes risk in humans remains unknown (11).
Microarray techniques have been widely utilized over the past decade for the study of obesity and type 2 diabetes. In this review, we will focus on results of gene expression analysis in accessible human tissues which have yielded new insights into transcriptional patterns associated with type 2 diabetes. For more details on individual studies, the reader is encouraged to refer to Supplementary Table 1.
Skeletal Muscle
Skeletal muscle is the largest insulin-sensitive organ in humans, accounting for more than 80% of insulin-stimulated glucose disposal (12). Muscle also contributes to systemic energy expenditure, both directly and via secretion of myokines regulating oxidation in adipose tissue (13). Multiple metabolic defects have been observed in muscle from insulin-resistant but normoglycemic subjects at high risk for diabetes development, including reduced insulin-stimulated glycogen synthesis (14;15), altered insulin signaling (16;17), and increased muscle lipid accumulation (18). Although it remains unclear whether any of these defects play a causal role in insulin resistance, intramyocellular lipid excess strongly correlates with the severity of insulin resistance (18;19). Moreover, muscle lipid excess has been linked experimentally to induction of insulin resistance (20–22).
Effects of insulin on gene expression patterns in muscle
Insulin can regulate gene expression directly, via both PI 3-kinase and ERK-dependent signaling pathways modulating transcription, or effects on transcript stability/turnover (23). Additional in vivo insulin effects may be indirect, reflecting the impact of insulin on systemic metabolism (e.g. suppression of lipolysis and proteolysis) which could secondarily influence transcription. To isolate primary effects, Hansen and colleagues performed a detailed time course of insulin effects in cultured human myotubes (24). Insulin robustly stimulated gene expression in as little as 30 minutes (e.g. FOS, EGR1, JUNB, SRF, BHLHB2). Additional genes emerged during later time points up to 24 hours, including proinflammatory, chemokine, angiogenic, metabolic, and cell cycle regulatory genes.
In vivo transcriptional effects of insulin in healthy individuals have been analyzed by comparing gene expression in muscle biopsies obtained in the fasting state and after 3–4 hours of insulin infusion (25–27). Despite different experimental protocols, these studies identified similar robust effects of insulin on genes regulating transcription and translation, protein metabolism, and muscle development. Similar to effects in myotubes, insulin stimulation in vivo also upregulated genes related to inflammation and early transcriptional responses (e.g. FOS/JUN, BHLHB2, HES1, NR4A1). Insulin infusion also altered miRNA (28), potentially via SREBP1c and MEF2c-dependent mechanisms. Interestingly, expression of the insulin receptor substrate IRS1 was decreased, and that of p85 regulatory subunit of PI 3-kinase increased – findings which would be predicted to induce insulin resistance. Thus, insulin may induce transcriptional effects which limit insulin action in the face of sustained hyperinsulinemia.
Chronic resistance to insulin could have a major impact on differential gene expression patterns in patients with T2D. For example, insulin resistance might attenuate the impact of surges of insulin (e.g. in postprandial state); in addition, the fasting hyperinsulinemia characteristic of insulin resistance might also influence transcriptional responses to insulin, particularly if insulin resistance is pathway-specific. Not surprisingly, differences in insulin-responsive gene expression in both myotubes (24) and muscle biopsy samples (29) from patients with T2D vs. controls are quite robust. One study suggested that BHLHB2, a repressive basic helix-loop-helix transcription factor, could regulate about 10% of genes regulated by insulin and could contribute to differential gene expression in insulin resistant individuals and altered oxidative metabolism (29).
To identify effects of chronic insulin therapy in vivo, Sreekumar and colleagues treated T2D patients with uncontrolled hyperglycemia with insulin for 10 days and assessed gene expression (30). In parallel with improved metabolism, insulin normalized differences in gene expression between patients and controls. These included genes linked to structural and contractile function, growth, tissue development, stress response, and energy metabolism. Eleven transcripts differentially expressed in patients with T2D were unresponsive to insulin (remaining differentially expressed), including multiple muscle fiber proteins, complex I NADH dehydrogenase, cadherin, and HSP70.
Together, these studies demonstrate that both acute and chronic insulin can potently regulate gene expression in muscle. It remains unclear whether in vivo differences in insulin-responsive gene expression between controls and patients with T2D reflect basal differences in transcriptional patterns (which could influence the ability of insulin to subsequently modulate them), differences in global or pathway-specific insulin action, or the time course or mechanisms by which insulin signals are terminated.
Differences in muscle gene expression in T2D and insulin resistance
Multiple studies have analyzed global gene expression patterns in skeletal muscle from patients with T2D vs. controls. Although the specific genes dysregulated vary from study to study, ontology and pathway analysis led to a consistent finding in both Caucasians and Mexican-Americans with T2D - a consistent 20-30% reduction in expression of multiple nuclear-encoded genes of the OXPHOS pathway and the PGC1-ERR family of coactivators (31–33). These patterns were observed both in the fasting state (31) and following insulin stimulation (32). Similar reductions in OXPHOS expression have also been observed in some populations of insulin resistant, but completely normoglycemic, individuals (31;34) and even at the protein level in aged individuals (35). Furthermore, these patterns were recapitulated experimentally by inactivity (36), lipid infusion (37), or high-fat feeding (38). Conversely, these patterns could be reversed by exercise training (35;39;40) or treatment with the insulin sensitizer rosiglitazone (41;42). Given that OXPHOS gene expression typically paralleled insulin sensitivity changes in these studies, and was linked to both genetic and epigenetic control (43–46), these data provided support for the hypothesis that reductions in OXPHOS gene expression might be causally linked to the impaired substrate oxidation and metabolic flexibility associated with insulin resistance.
Despite the initial enthusiasm for this hypothesis, not all studies demonstrated similar patterns, potentially due to differences in methodology (47), cohort size (48), differences in degree of insulin resistance and/or deficiency, and the impact of age, obesity, physical fitness, and ethnicity (43;44;49;50). For example, Asian Indian individuals with insulin resistance have increased expression of OXPHOS genes (51). OXPHOS gene expression or function is similarly increased in some high fat diet-fed rodent strains (52–54). Our own studies in Caucasian individuals revealed downregulation of OXPHOS gene expression in established T2D, but no change in isolated insulin resistance (55). Thus, insulin resistance is not always accompanied by downregulation of OXPHOS genes. Moreover, experimental disruption of OXPHOS in several rodent models does not induce insulin resistance, but rather enhances insulin sensitivity, perhaps via adaptation in AMPK-dependent pathways (56).
Taken together, these data suggest that transcriptional dysregulation of PGC1-directed OXPHOS gene expression is unlikely to play a dominant causal role in the pathogenesis of insulin resistance. Rather, these patterns may be secondary to other aspects of the diabetes metabolic environment, including insulin deficiency or insulin resistance. Several lines of evidence provide support for this concept. Firstly, reduced OXPHOS gene expression in either mice or patients with established diabetes can be partially normalized by insulin treatment (30;57;58). Secondly, during acute high-fat feeding in humans, hepatic insulin resistance precedes dysregulation of muscle energetics and gene expression (54). Finally, defects in OXPHOS expression are not observed during prolonged ex vivo culture of primary myotubes, despite persistence of insulin resistance (24;59). Together, these data indicate that dysregulation of OXPHOS gene expression are not likely to be a primary defect. Rather, downregulation of OXPHOS may be a “secondary” response to limit oxidative stress in settings of sustained overnutrition and/or inactivity. This perspective does not exclude a role for other genes and pathways regulating oxidative energy metabolism (e.g. TCA cycle, lipid oxidation, anaplerotic pathways, responses to oxidative stress) nor for dysregulation mediated at post-transcriptional levels.
Given these findings, our group and others have aimed to identify additional genes and pathways transcriptionally dysregulated in both insulin resistance and T2D. For example, we recently reported that the top-ranking gene set in skeletal muscle, enriched in both insulin resistant subjects with family history of T2D (FH+) and those with established T2D, was a group of genes regulated by the transcription factor SRF, its coactivator MKL1, and the upstream protein STARS (60). This pathway may play a role in insulin resistance, as expression of STARS correlates with insulin resistance, and genetic and pharmacological inhibition of this pathway improves insulin action, oxidative metabolism, and glucose tolerance in obese mice (60).
Additional genes and pathways, many related to metabolism, have also been identified in insulin resistant individuals. For example, offspring of parents with T2D have decreased expression of lipoprotein lipase (LPL) in muscle, paralleling decreased mitochondrial density (61). Experimental decreases in LPL expression in myotubes also decrease mitochondrial content and reduce fatty acid-mediated activation of PPARδ (61). Conversely, alterations in lipid partitioning may also promote insulin resistance, via accumulation of potentially deleterious lipids (62). Expression of stearoyl CoA desaturase (SCD1), promoting lipogenesis, is markedly increased in obese subjects compared to lean (63). This pattern correlates with triglyceride content and inversely with fatty acid oxidation, and is maintained in cultured myotubes derived from obese donors, indicating potential genetic or epigenetic mechanisms. Similarly, we recently reported that downregulation of SFRS10 in muscle in obese patients with T2D may contribute to altered patterns of splicing, enhanced lipogenesis, and triglyceride accumulation (64). Despite increases in lipogenic expression, it remains unclear whether these responses remain inadequate relative to lipid supply, thus favoring the accumulation of deleterious species (e.g. DAG, ceramides) and promoting further insulin resistance (65).
Additional genes identified in microarray analysis have subsequently been assigned a functional role in insulin resistance phenotypes. For example, thioredoxin interacting protein (TXNIP) is repressed by insulin in healthy humans and upregulated in T2D (66). Overexpression of TXNIP reduces glucose uptake (66). Additional examples of genes dysregulated in T2D and linked to insulin resistance phenotypes include heat shock protein 72 (decreased) (67), cathepsin L (decreased) (68), and Rad (increased) (69;70).
As noted earlier, many changes in muscle metabolism and gene expression in insulin resistance and T2D are closely related to physical inactivity and markers of poor physical fitness (36;71). Exercise training increases expression of PGC1 and OXPHOS genes (39;40;72), malonyl CoA decarboxylase (73), changes expected to promote improved oxidative capacity. Conversely, inactivity reduces expression of genes regulating OXPHOS, TCA cycle, and lipid and BCAA oxidation, and increases Rad expression (36). Paradoxically, inactivity increases the effect of insulin to upregulate inflammation and ER stress genes (36), again highlighting the possibility that insulin resistance can induce pathway-specific and potentially deleterious transcriptional responses to insulin.
Adipose Tissue
White, brown, and recently-identified “beige” adipose tissues are recognized as important subtypes of adipose tissue which may play distinct roles in regulation of systemic metabolism (74). White adipose accounts for the greatest mass, and is widely distributed throughout subcutaneous and intraabdominal regions. White adipose is a highly heterogeneous tissue dominated by adipocytes with unilocular lipid droplets. Adipocytes play a dominant role in storing triglycerides during positive energy balance while promoting lipolysis during negative energy balance. Additional cell types, including preadipocytes, fibroblasts, vascular cells, and immune cells, also contribute to adipose regulation of systemic metabolism via the collective secretion of adipocytokines (e.g. leptin, adiponectin, TNFα, IL-6, and others). By contrast, brown adipose tissue functions in energy and heat dissipation and is characterized not only by high numbers of mitochondria, but also by expression of UCP1 in the inner mitochondrial membrane. While recent studies have clearly demonstrated that brown adipose tissue can be functional in adults (75;76), and potentially less active in obesity (75), brown or beige fat is difficult to access in humans, and studies to date have largely focused on developmental origins (76;77). Thus, we will focus our discussion in this section on studies of white adipose tissue.
Given the heterogeneity of white adipose tissue, gene expression studies in this tissue must take into consideration whether there are differences in proportions of cell types, or of phenotypes within in each cell type. For example, obesity itself is typically associated with increased proportions of large cells, which have higher expression of immune-related genes; similar patterns are also observed in large cells from lean individuals (78;79). Thus, it is important to consider experimental methods, as sample processing could introduce a bias if a smaller or larger population of cells is selected (e.g. lysis of larger cells or loss of smaller cells during adipocyte isolation). Moreover, adipose needle biopsy samples differ from surgical biopsies, with decreases in stromal-vascular fractions, increased blood cell contamination, and differential expression of genes within inflammatory, extracellular matrix and metabolic pathways (80). Thus, comparison of samples obtained using different methods must be avoided.
Adipose depot-related differences in gene expression (e.g. subcutaneous vs. visceral) are also prominent. Accumulation of visceral adipose correlates with insulin resistance, glucose intolerance, and other components of the metabolic syndrome (81), while subcutaneous adipose is generally viewed as protective for metabolic disease. Many genes are more highly expressed in subcutaneous adipose tissue (SAT), with enrichment of genes linked to lipid and carbohydrate metabolism, cell structure, and homeobox transcription factors. By contrast, visceral adipose tissue (VAT) is enriched for genes involved in immune response, cell proliferation/differentiation, cell cycle, cell death and angiogenesis (82–91). These pathways are also generally altered in obesity (92–95). In obese humans, inflammatory genes in VAT correlate with plasma glucose (94). Additionally, depot-specific differences in splicing isoforms have been observed for TCF7L2, a gene linked to genetic risk for T2D (96). Whether depot-specific differences in gene expression simply reflect differences in developmental origin (97) or functional differences in metabolism, or both, remains uncertain.
Analysis of gene expression patterns in lean vs. obese humans have nevertheless demonstrated several key features, including increased expression of immune-related genes and widespread dysregulation of metabolic genes in both SAT and visceral depots (92). Adipose from obese individuals is also characterized by increased expression of genes related to long-chain fatty acid uptake, MAP kinases, and markers of early adipocyte differentiation (e.g. CDKN1A, GADD45B, LMNA, ANGPTL4) with parallel reductions in expression of genes related to lipid metabolism (e.g. AKAP1, APOE), growth factors and final-stage adipocyte differentiation (e.g. CDKN2C, LPL, PPARγ and PCK1) (90;98–100). Similar patterns suggesting impaired adipogenesis are seen in obese adolescents (e.g. downregulation of PIK3R3, PIK3CD, PIK3R2, SREBP1, upregulation of PIK3CB, FBP1, PRKAA1) (101). Obese individuals also have reduced expression of genes associated with lipid oxidation (e.g. ACADM, HADHSC, ETFDH), fatty acid biosynthesis (e.g. FASN, SCD1) and glucose metabolism (e.g. PCK1) in SAT (102). Similarly, mitochondrial OXPHOS genes (e.g. NDUFB5, SDHB, UQCRFS1) are also downregulated in VAT from T2D patients (56). Interestingly, studies of monozygotic twin pairs demonstrate that the more obese co-twin has reduced expression of genes linked to oxidative metabolism, suggesting a potential role for epigenetic or postnatal diet in mediating these differences (103).
Upregulation of inflammatory (e.g MMP3, MCP-1) genes has also been observed in isolated adipocytes from obese Pima Indians (104–106). Such changes appear to be even more prominent in individuals consuming a diet rich in saturated fatty acids, as compared with the more anti-inflammatory profiles linked to monounsaturated fatty acids (107).
Expression of microRNA species is also altered in obese VAT. For example, miR-17-5p and miR-132 are reduced in both VAT and in the circulation of obese subjects (108). In an independent study, miRNA which are downregulated during adipocyte differentiation and maturation (miR-221, miR-125b and miR-100) were found to be upregulated in obese subjects, while miR-185, which is upregulated in mature adipocytes, was downregulated in obese men (109). Overexpression of miR-519d has also been associated with obesity and influence on PPARγ expression (110). This inverse mRNA profile suggests that miRNA could also be involved in dysregulation of adipogenesis in the setting of obesity.
Once obesity is established, it is difficult to discern which changes are primary, and which are secondary to the metabolic effects of obesity. Analysis of time course responses to overfeeding, the effects of insulin resistance, and cell type-specific responses may all be useful to dissect these components. One study examined subcutaneous adipose gene expression patterns in humans during the course of high-fat feeding over a two month period (17). Genes related to lipid metabolism were altered by day 14; by day 56, an enrichment of genes related to lipid metabolism, extracellular matrix, angiogenesis and renin-angiotensin signaling was observed, in parallel with downregulation of Wnt/β-catenin signaling (17). These data suggest that changes in lipid metabolism, vascularization, and tissue remodeling are among the earliest changes in adipose tissue under conditions of nutrient excess.
A second approach to dissecting the potential contribution of insulin resistance itself to expression changes in obesity is to examine adipocytes isolated from non-obese insulin resistant (IR) subjects with family history of diabetes. Interestingly genes linked to cell cycle regulation were upregulated, in parallel with genes related to lipid and protein metabolism (111). Moreover, genes regulating adipogenic signaling (WNT1, FZD1, DVL1, TCF1) and early stages of adipogenesis (C/EBPα/β/δ, PPARγ, SREBP1, aP2) were decreased in adipose tissue from non-obese IR subjects. Whether these patterns simply reflected the larger adipocyte size in these individuals or the underlying pathophysiology of adipocyte insulin resistance remains unclear. Nevertheless, these findings suggest that early insulin resistance is accompanied by adipose dysfunction (impared adipogenesis or cell differentiation) even before the onset of obesity or T2D (111). Insulin resistance in the setting of obesity is accompanied by additional changes, including altered expression of insulin signaling genes (increased IRS-1, PI3Kγ, Akt2, GLUT4, GLUT1; decreased PI3Kα, PDK2, mTor, Rheb, 4EBP, eIF members, Ras and MEK1/2 kinase) when compared to either lean or obese, insulin sensitive subjects. These changes are even more prominent in VAT (100). Thus, insulin resistance, rather than isolated obesity, is associated with altered expression of insulin signaling genes.
Thirdly, given the heterogeneity of adipose tissue, studies examining the effect of obesity and insulin resistance on expression patterns in isolated preadipocytes may also provide clues. Preadipocytes from obese T2D patients (112) demonstrate downregulation of genes related to adipogenesis, extracellular matrix remodeling, insulin signaling and lipid metabolism (e.g. CPT1, IL-8, COL4a, IGF1, ALDOC, PFKP, GAPDH, ACSL3), with parallel increases in genes related to inflammation and apoptosis (e.g. ASS1, TWIST2, CPE, C3, PPL). Similar changes have been observed in obese individuals of Pima heritage (113). More specific sorting of macrophages (CD34-/CD14+) from both SAT and VAT of individuals with a spectrum of adiposity demonstrated that macrophage marker expression correlated positively with BMI, WHR, and plasma insulin (114).
Many of the changes observed in adipose tissue or its fractions are quite similar to those induced by LPS injection in vivoor incubation ex vivo (115;115). Given the increased recognition that hypoxia may be a limiting factor in adipose expansion and remodeling during nutrient excess (116), the convergence of inflammatory and hypoxic stimuli may both contribute to obesity and T2D-linked expression patterns.
Many intervention protocols designed to minimize obesity and insulin resistance also profoundly alter adipose gene expression. Even modest weight reduction (e.g. 5%) is associated with beneficial changes, including downregulation of genes related to proinflammatory, extracellular matrix, and cell death-related genes (25;90). Caloric restriction also alters expression of genes involved in adipogenesis and lipogenesis, protein synthesis, and development (80;117–119). Similarly, a very low caloric diet intervention for 28 days yielded adipose expression patterns more similar to that of lean controls (120). During the weight loss phase, the majority of genes involved in lipid, carbohydrate, and mitochondrial energy pathways were downregulated; during weight stabilization, lipid and carbohydrate metabolism were upregulated but expression of genes involved in immunity, defense, and tissue remodeling remained decreased. Interestingly, genes controlling both cellular proliferation and cell death were overrepresented in SAT from those subjects who regained weight (e.g. ACLY, CFL1, ACSL5, LCP1, MAPK3, MEST, ADIPOR1, EIF4A, LASS2, CES1, CDK2AP1, RNF5, E2F4), while sustained improvements in mitochondrial oxidative genes was associated with continued weight loss (e.g. PLAT, NDUFA9, BTBD7, IGF1, VGLL3, ATP10A) (121). Even greater degrees of weight loss, as with bariatric surgery, can also normalize expression patterns (122), including downregulation of inflammatory (e.g. MCP1, PLAUR, CSF3) and HIF-related pathways in SVF (123). Furtherrmore, treatment with the PPARγ ligand rosiglitazone decreased expression of the cell cycle regulator E2F4 and in parallel, increased expression of the preadipocyte marker necdin, suggesting enhanced differentiation (124).
In summary, gene expression patterns support the concept that obesity and insulin resistance are associated with impaired adipocyte metabolic function, perhaps in part due to altered differentiation. With continued nutrient excess, the capacity for lipid storage is overwhelmed, unrestrained lipolysis ensues, and proinflammatory cascades are engaged (125). This cascade is accompanied by dysfunctional adipocytokine secretion which may then promote deleterious changes in systemic metabolism facilitating accumulation of excess lipids in non-adipose (ectopic) locations, such as muscle and liver.
Liver
The liver plays a central, unique role in carbohydrate, protein, and fat metabolism. It is critical for maintaining glucose homeostasis (1) during fuel availability, via storage of glucose as glycogen or conversion to lipid for export and storage in adipose tissue, and (2) in the fasting state, via catabolism of glycogen, synthesis of glucose from noncarbohydrate sources such as amino acids (gluconeogenesis), and ketogenesis. In turn, these responses are regulated by the key hormones insulin and glucagon, which modulate signaling pathways and gene expression, leading to inhibition or stimulation of glucose production, respectively.
Recent human data have highlighted the importance of disordered hepatic metabolism, including inappropriately increased hepatic glucose production, hyperlipidemia, and lipid accumulation, in both obesity and type 2 DM. Furthermore, fatty liver and non-alcoholic steatohepatitis (NASH) are intimately linked to the metabolic syndrome and risk for type 2 DM (126). Similar to muscle and adipose, hepatic lipid excess in NASH is paralleled by insulin resistance, increased lipid oxidation and peroxidation, increased lipogenesis (127) and abnormalities in mitochondrial structure (128) and ATP metabolism (129). While these data in humans cannot demonstrate causality, rodent data also support an important role for the liver in diabetes pathogenesis. For example, liver-specific insulin receptor knockout (LIRKO) mice develop glucose intolerance, impaired insulin suppression of hepatic glucose production, dyslipidemia, and altered hepatic gene expression (130;131).
Unfortunately, despite the critical role of the liver in mediating whole-body glucose and lipid metabolism, we know little about transcriptional responses in the liver in early stages of diabetes pathogenesis, given that the liver remains relatively inaccessible to tissue sampling in otherwise healthy humans. The majority of the available (though limited) data has derived from individuals with obesity undergoing abdominal surgery, with or without coexisting T2D. Selection and recruitment of an appropriate healthy control group matched for key demographics is a difficult task in these circumstances. Moreover, interpretation is complicated by several factors, including individual variability in the extent of inflammation and fibrosis, differences in the proportion of specific cell types, subject diet (not typically controlled), relatively small sample sizes, and variation in experimental approaches and microarray methodology.
One of the earliest microarray studies of human liver was performed by Sreekumar and colleagues, who analyzed liver samples from patients with NASH (BMI 30) and age-matched healthy controls (BMI 27). Interestingly, among the 12 downregulated genes were genes related to lipid metabolism and β-oxidation (e.g. long-chain CoA synthase, HADHA), ketone metabolism (HMGCS2), gluconeogenesis (glucose-6-phosphatase), and ROS defenses (e.g. SOD1, catalase) (132). Conversely, upregulation of complement C3 and α-1 antitrypsin hints at increased inflammation in NASH.
Subsequent studies of transcriptional patterns associated with obesity-linked liver disease have attempted to segregate effects of inflammation and lipid accumulation by studying subjects with NASH in comparison with isolated steatosis or healthy controls. Patterns linked to NASH are relatively consistent across studies, including dysregulation of genes regulating lipid metabolism and antioxidant responses, inflammation, and fibrosis. Individuals with NASH have increased expression of genes related to lipid metabolism (e.g. ACSL4, HMGCS2), matrix remodeling (e.g. MMP2, CPSG2, SDC4), and antioxidant responses (e.g. CAT, GSTA4), but downregulation of other genes linked to β-oxidation (ACADSB) or mitogenic signaling (e..g JUN, ETS2) (133;134). Even higher levels of expression of ACSL4 and glutathione transferases were observed in an African-American cohort with NASH (135). Similar patterns of increased expression of genes regulating lipogenesis (136), inflammation (e.g. MHC class II, chemokines) and fibrosis (e.g. collagen, TGFβ) have also been observed (137).
Since hepatic steatosis is also associated with insulin resistance, analysis of isolated steatosis may give clues to early transcriptional patterns linked to insulin resistance and diabetes risk, in the absence of inflammation-related signals which cloud interpretation. Even in isolated steatosis, there is substantial variability between studies, perhaps related to the heterogeneity of subject metabolic phenotypes and tissue inflammation not appreciated on histology. For example, Chiappini et al demonstrated upregulation of several genes related to mitochondrial and peroxisomal lipid metabolism, OXPHOS, IGF binding proteins, cell cycle regulation, inflammation, and cell adhesion, and increased mitochondrial DNA in isolated steatosis (mean BMI 27) (138), while downregulation of IGFBP1 and upregulation of ACSL4 and multiple antioxidant response genes (e.g. GSTM) was the dominant pattern in patients with NASH (134). Similar patterns have been observed even in patients with well-controlled diabetes, with (BMI 27) and without (BMI 22) modest obesity (139). Even higher levels of obesity are associated with increased steatosis, as expected, together with upregulation of genes regulating OXPHOS, β-oxidation, gluconeogenesis, and the antioxidant response. Interestingly, this pattern was also correlated with measures of insulin resistance and linked to expression of PGC1β, PPARγ, and the thyroid receptors TRα and TRβ. Taken together, these studies hint that disordered lipid metabolism and oxidative stress may be common features of hepatic steatosis.
Transcriptional patterns may differ in severe obesity and steatosis. Pihlajamaki and colleagues (140) analyzed gene expression in severely obese individuals with or without coexisting T2D (mean BMI 52) as compared with lean controls (BMI 27). Single gene analysis again highlighted increased expression of genes regulating lipogenesis (e.g. FASN, DGAT1/2) in severe obesity. In contrast to studies evaluating patients with lower degrees of obesity, gene set analysis in this cohort demonstrated that thyroid hormone-responsive genes regulating mitochondrial oxidative metabolism were downregulated in obesity. In turn, this pattern was potentially linked to reduced expression of the coactivator PGC1α. A similar pattern was produced experimentally by high-fat feeding in rodents. Together, these data raise the possibility that severe obesity and insulin resistance may alter nuclear receptor transcriptional activity and potentially induce hepatic thyroid hormone resistance. While this could potentially limit excessive lipid oxidation and ROS production, it might also contribute to a vicious cycle of reduced oxidative capacity.
Additional patterns of liver gene expression are observed in patients with coexisting severe obesity and T2D (140). Expression of genes regulating glycolysis (ALDOA, PFKL), inflammation (HLA class II), and fibrosis (TGFβ1) was increased in individuals with coexisting obesity/T2D. These patterns correlated with fasting glucose levels.
Additional insights may be drawn from analysis of patients undergoing therapies associated with improvement in hepatic metabolism. For example, Elam and colleagues (141;142) compared gene expression patterns in severely obese women undergoing gastric bypass surgery with individuals who had already experienced surgical weight loss. Although this was not a paired analysis, there were robust differences in gene expression between severely obese and post-weight loss subjects, particularly within genes related to lipid metabolism (e.g. reduced fatty acid elongation and increased lipogenic gene expression in severely obese), inflammation (e.g. SOCS3, JAK/STAT), oxidative, and signaling (e.g. reduced leptin receptor and SOCS2). Similarly, expression of genes related to protein and lipid metabolism and inflammation is reduced in individuals losing weight with hypocaloric diet, as compared with weight-stable individuals (143).
In summary, microarray technology has allowed us to probe transcriptional patterns associated with insulin resistance and hepatic steatosis. Despite differences in study design and study participants, several patterns are consistent in the majority of studies, including increased expression of genes linked to lipogenesis, lipid oxidation, response to oxidative stress, and inflammation. These patterns may be further disrupted in severe obesity, with downregulation of oxidative metabolism, potentially contributing further to disordered lipid catabolism. Further transcriptional dysregulation may promote inflammation and fibrosis associated with progressive liver disease.
Such cross-sectional studies in human populations clearly cannot identify mechanisms responsible for these patterns with certainty. However, potential mediators include cellular insulin resistance itself, transcriptional changes associated with insulin resistance, obesity-associated proinflammatory adipocytokines, pathogenic lipid accumulation, or by-products of lipid metabolism (e.g. ROS). Several experimental approaches have been taken to unravel the relative contributions of each of these potential regulatory mechanisms. For example, short-term exposure of human liver slices (144) or hepatic cell lines (145) to LPS or the saturated fatty acid palmitate, respectively, can recapitulate some features of the steatosis-NASH pattern. These include including upregulation of proinflammatory, lipid oxidation, and oxidative stress genes and dysregulation of cell cycle control.
It is likely that more chronic transcriptional mechanisms contribute to the consistent upregulation of lipogenic gene expression (e.g. FASN, SREBP1, lipin) in association with lipid accumulation. Elam and colleagues (142) demonstrated that the SREBP1 promoter was positively regulated by insulin and negatively by STAT3; increases in SOCS3, which inhibit the STAT pathway, could promote increased expression of SREBP-1c (142). Recent data implicate alterations in expression of miRNA (136) in mediating increased lipogenic gene expression. For example, miR-122 expression is decreased in NASH; experimental silencing of miR-122 recapitulated increases in lipogenic genes (136).
Our own studies recently elucidated a novel mechanism contributing to increased lipogenesis in hepatic steatosis (64). Among the top-ranking genes downregulated in steatosis was the family of SR proteins regulating RNA splicing. To evaluate the potential role of SR proteins in dysregulation of lipid metabolism, we assessed the impact of experimental modulation of SFRS10, a representative member of this family. Interestingly, SFRS10-specific siRNA induces expression of lipogenic genes, and increases lipogenesis and lipid accumulation in both hepatocytes and myotubes. Similarly, Sfrs10 heterozygous mice have increased hepatic lipogenic gene expression and VLDL secretion. Effects of SFRS10 appear to be mediated by alternative splicing of LPIN1, favoring the lipogenic β isoform of LPIN1 (64).
Clearly, we have much to learn about the role of the liver in contributing to systemic insulin resistance and T2D risk, and the cellular mechanisms mediating patterns of gene expression linked to hepatic lipid accumulation. While it is likely that such patterns are maintained by distinct transcriptional regulation in response to genetic, epigenetic, nutritional and metabolic signals, it is encouraging that these transcriptional patterns appear to be potentially modifiable in response to caloric deficit and weight loss. Further understanding of these mechanisms will be critical if we are to improve strategies to not only reduce risk for obesity-associated insulin resistance and T2D, but also reduce the hepatic consequences of these diseases.
Pancreas
Not surprisingly, it has been difficult to assess gene expression in human islets, given the relative inaccessibility of pancreas for sampling and differences in cell populations between individuals (variable numbers of β-cells, non-β cells and contaminating exocrine cells). Gunton and colleagues (146) used microarrays to study islets isolated from 5 donors with type 2 diabetes (mean duration 6 years) and 7 controls. The authors found decreased expression of HNF4α, insulin receptor, IRS2, Akt2, and other genes regulating glucose metabolism. Genes related to RNA processing were also altered, an interesting finding given that this pathway is also downregulated in liver and muscle (64). The top-ranking individual gene, downregulated by over 80% in the patients with T2D, was ARNT/HIF1β. Reduction in ARNT expression in both cells and mice reduced glucose-stimulated insulin secretion and yielded expression patterns similar to the T2D patients.
Results from genome-wide association studies have pointed to the possibility that loci associated with genetic risk for diabetes may influence insulin secretion. This was supported by the finding that expression of TCF7L2 was increased 5-fold in T2D islets, particularly in carriers of the diabetes risk genotype, and that overexpression of TCF7L2 reduced glucose-stimulated insulin secretion (147). To more broadly test this hypothesis, Taneera and colleagues (148) isolated human islets to examine expression of genes in the vicinity of single-nucleotide polymorphisms (SNPs) linked to genetic risk for T2D. Of 48 SNP-associated candidate genes, expression of 5 was reduced in islets from donors with T2D, paralleling defects in insulin secretion and hyperglycemia. Some of these genes modulated insulin secretion in cultured β- cells; whether additional candidates contribute to β-cell mass or development remains an unanswered but important question.
Laser capture microdissection can permit examination of relatively purified β-cell populations. Using this technique, Marselli et al (149) examined gene expression in samples from individuals with T2D vs. controls. Differential expression was noted for genes linked to glucotoxicity, regeneration-associated genes (e.g. upregulation of REG1/3), and some GWAS-associated genes (e.g. upregulation of IGF2BP2, TSPAN8, HNF1β, downregulation of JAZF1 and SLC30A8).
Together, these data indicate that T2D is associated with changes in islet gene expression, but specific genes and pathways vary from study to study, potentially due to confounding factors associated with cadaveric sampling and islet heterogeneity. What remains unknown is whether these changes are primary, and potentially linked to genetic risk, or result from attempts to compensate for reductions in β-cell function or mass with long-standing T2D, or potentially from environmental or developmentally-induced epigenetic changes (150).
Blood
Blood samples can be easily accessed in clinical practice. Thus, the identification of disease markers in blood would be very helpful to identify high-risk individuals for targeted prevention strategies or to tailor treatment approaches to metabolic stage. Grayson and colleagues evaluated gene expression in peripheral blood from subjects with metabolic syndrome or T2D, vs. healthy controls (151). Metabolic syndrome patients had an upregulation of proinflammatory genes, and genes regulating T-cell activation and signaling were overexpressed in T2D subjects (151). Finally, a study comparing white blood cells from patients with T2D to controls demonstrated upregulation of genes linked to immune response, lipid metabolism, and organismal injury (152).
While analysis of whole blood samples may be confounded by alterations in populations of leukocytes at the time of sampling, it is interesting that the pathways identified in the limited number of analyses to date demonstrated altered expression within proinflammatory and lipid metabolism – the same pathways also dysregulated in liver, adipose, and muscle in insulin resistance and diabetes. Thus, blood-derived gene expression patterns may not only serve as markers of systemic disease but also underscore the importance of both inflammation and dysregulation of lipid metabolism in many tissues.
Meta-analyses
Given the abundance of microarray data linked to T2D, computational analyses to discern potential transcription factors responsible for gene expression patterns have been initiated. For example, Zelezniak (153) integrated gene expression data from skeletal muscle with known metabolic pathways to identify key regulatory enzymes and associated metabolites; this method highlighted not only TCA cycle, OXPHOS, lipid metabolism, and NAD+ and ATP-dependent pathways, but also enrichment of binding sites for CREB, NRF1, and PPAR in regulated genes. Similarly, Ptitsyn (154) used principal component analysis to demonstrate dysregulation of fiber type control and glycolytic metabolism in insulin resistant muscle Using seven independent gene prioritization methods to integrate both genetic and genomic data, Tiffin and colleagues (155) identified 9 genes potentially linked to T2D, including BCKDHA, OAT (amino acid metabolism), ACAA2, ECHS1, LPL (lipid metabolism), PRKCSH, PGM1 (glycogen metabolism), CSF1 (cytokine signaling), and TGFBR2 (transforming growth factor-β receptor). Of these, BCKDHA and LPL were also linked to obesity risk.
Summary and Perspectives
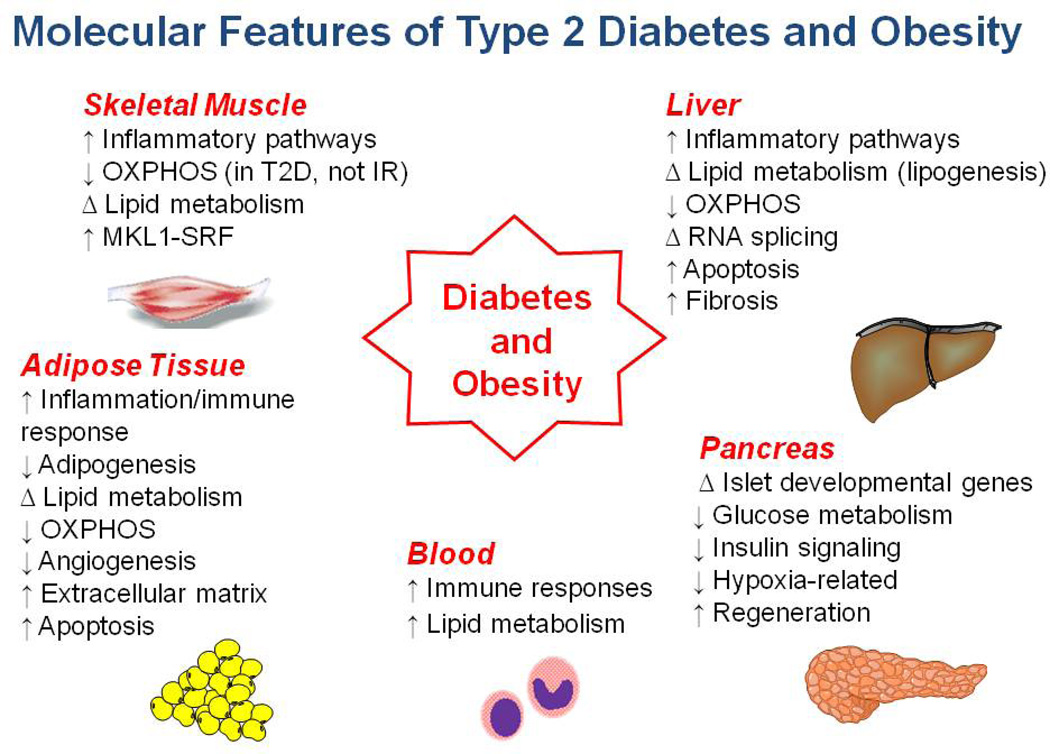
In summary, gene expression analysis in many human populations has identified novel genes and pathways linked to insulin resistance and diabetes risk (Figure 1 and Supplementary Table 1). Several of these findings are concordant across multiple tissues (e.g. relationship between inflammation and insulin resistance, altered expression of genes regulating oxidative metabolism), providing potential support for their role in pathophysiology. Given the need for robust animal models of disease, it is encouraging that many of these patterns are also observed in overnourished rodent models. For example inflammatory gene expression patterns emerge in adipose tissue early in the development of obesity and insulin resistance in obesity-prone mice (156). Whether similar temporal patterns are present in humans is an unanswered question. Still other patterns in humans are more tissue-specific (e.g. STARS-SRF pathway activation in muscle of T2D)(157), potentially reflecting the unique contributions of individual tissues to whole-body metabolism.
Figure 1.
Summary of tissue-specific findings in insulin resistance and T2D.
While these findings are exciting and have initiated the search for novel targets for prevention and therapy targeting T2D, many important questions remain unanswered: (1) Are differential expression patterns mediated by DNA sequence (genetic) or by epigenetic mechanisms resulting from developmental or environmental factors which together influence chromatin structure and transcriptional regulation? (2) Do changes in gene expression mirror what is happening at a protein level or in key metabolic pathways? Thus far, few studies have examined the relationship between genomic and proteomic data (158), and metabolomics studies in human tissues are just now emerging. (3) How can we assess transcriptional response patterns in tissues not accessible for study in humans (e.g. brain, intestine), but clearly important for obesity and diabetes risk? (4) Finally, which patterns are initiated early in the course of isolated insulin resistance, and thus more likely to be primary to the pathogenesis of T2D? Conversely, which are secondary to the diabetes metabolic environment? To address these difficult, but key challenges, longitudinal translational studies which integrate detailed molecular phenotyping with in vivo metabolic measures in humans at risk for T2D are sorely needed.
Supplementary Material
Summary of studies of gene expression changes referenced in the review.
Acknowledgments
We gratefully acknowledge research support from CAPES (government of Brazil, to VS), NIH R56 DK096158 (to MEP), the American Diabetes Association (to MEP), and the Graetz Fund (to MEP).
Reference List
- 1.Centers for Disease Control and Prevention USDoHaHS. National Diabetes Fact Sheet. 2011 www.cdc.gov/diabetes
- 2.Boyle JP, Honeycutt AA, Narayan KM, et al. Projection of diabetes burden through 2050: impact of changing demography and disease prevalence in the U.S. Diabetes Care. 2001;24(11):1936–1940. doi: 10.2337/diacare.24.11.1936. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. WHO. 2008
- 4.Tuomilehto J, Lindstrom J, Eriksson JG, et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med. 2001;344(18):1343–1350. doi: 10.1056/NEJM200105033441801. [DOI] [PubMed] [Google Scholar]
- 5.Hu FB, Manson JE, Stampfer MJ, et al. Diet, lifestyle, and the risk of type 2 diabetes mellitus in women. N Engl J Med. 2001;345(11):790–797. doi: 10.1056/NEJMoa010492. [DOI] [PubMed] [Google Scholar]
- 6.Knowler WC, Barrett-Connor E, Fowler SE, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346(6):393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wing RR, Lang W, Wadden TA, et al. Benefits of Modest Weight Loss in Improving Cardiovascular Risk Factors in Overweight and Obese Individuals With Type 2 Diabetes. Diabetes Care. 2011;34(7):1481–1486. doi: 10.2337/dc10-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saydah SH, Fradkin J, Cowie CC. Poor control of risk factors for vascular disease among adults with previously diagnosed diabetes. JAMA. 2004;291(3):335–342. doi: 10.1001/jama.291.3.335. [DOI] [PubMed] [Google Scholar]
- 9.Tabak AG, Jokela M, Akbaraly TN, Brunner EJ, Kivimaki M, Witte DR. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet. 2009;373(9682):2215–2221. doi: 10.1016/S0140-6736(09)60619-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Billings LK, Florez JC. The genetics of type 2 diabetes: what have we learned from GWAS? Ann N Y Acad Sci. 2010;1212:59–77. doi: 10.1111/j.1749-6632.2010.05838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doria A, Patti ME, Kahn CR. The emerging genetic architecture of type 2 diabetes. Cell Metab. 2008;8(3):186–200. doi: 10.1016/j.cmet.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeFronzo RA, Gunnarsson R, Bjorkman O, Olsson M, Wahren J. Effects of insulin on peripheral and splanchnic glucose metabolism in noninsulin-dependent (type II) diabetes mellitus. J Clin Invest. 1985;76(1):149–155. doi: 10.1172/JCI111938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bostrom P, Wu J, Jedrychowski MP, et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012 doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eriksson J, Franssila-Kallunki A, Ekstrand A, et al. Early metabolic defects in persons at increased risk for non-insulin-dependent diabetes mellitus. N Engl J Med. 1989;321(6):337–343. doi: 10.1056/NEJM198908103210601. [DOI] [PubMed] [Google Scholar]
- 15.Vaag A, Henriksen JE, Beck-Nielsen H. Decreased insulin activation of glycogen synthase in skeletal muscles in young nonobese caucasion first-degree relatives of patients with non-insulin-dependent diabetes mellitus. J Clin Invest. 1992;89:782–788. doi: 10.1172/JCI115656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pratipanawatr W, Pratipanawatr T, Cusi K, et al. Skeletal muscle insulin resistance in normoglycemic subjects with a strong family history of type 2 diabetes is associated with decreased insulin-stimulated insulin receptor substrate-1 tyrosine phosphorylation. Diabetes. 2001;50(11):2572–2578. doi: 10.2337/diabetes.50.11.2572. [DOI] [PubMed] [Google Scholar]
- 17.Alligier M, Meugnier E, Debard C, et al. Subcutaneous adipose tissue remodeling during the initial phase of weight gain induced by overfeeding in humans. J Clin Endocrinol Metab. 2012;97(2):E183–E192. doi: 10.1210/jc.2011-2314. [DOI] [PubMed] [Google Scholar]
- 18.Jacob S, Machann J, Rett K, et al. Association of increased intramyocellular lipid content with insulin resistance in lean nondiabetic offspring of type 2 diabetic subjects. Diabetes. 1999;48(5):1113–1119. doi: 10.2337/diabetes.48.5.1113. [DOI] [PubMed] [Google Scholar]
- 19.Malenfant P, Joanisse DR, Theriault R, Goodpaster BH, Kelley DE, Simoneau JA. Fat content in individual muscle fibers of lean and obese subjects. Int J Obes Relat Metab Disord. 2001;25(9):1316–1321. doi: 10.1038/sj.ijo.0801733. [DOI] [PubMed] [Google Scholar]
- 20.Dresner A, Laurent D, Marcucci M, et al. Effects of free fatty acids on glucose transport and IRS-1-associated phosphatidylinositol 3-kinase activity. J Clin Invest. 1999;103:253–259. doi: 10.1172/JCI5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koves TR, Ussher JR, Noland RC, et al. Mitochondrial overload and incomplete Fatty Acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7(1):45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 22.Holland WL, Brozinick JT, Wang LP, et al. Inhibition of ceramide synthesis ameliorates glucocorticoid-, saturated-fat-, and obesity-induced insulin resistance. Cell Metab. 2007;5(3):167–179. doi: 10.1016/j.cmet.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 23.O'Brien RM, Granner DK. Regulation of gene expression by insulin. Physiol Rev. 1996;76(4):1109–1161. doi: 10.1152/physrev.1996.76.4.1109. [DOI] [PubMed] [Google Scholar]
- 24.Hansen L, Gaster M, Oakeley EJ, et al. Expression profiling of insulin action in human myotubes: induction of inflammatory and pro-angiogenic pathways in relationship with glycogen synthesis and type 2 diabetes. Biochem Biophys Res Commun. 2004;323(2):685–695. doi: 10.1016/j.bbrc.2004.08.146. [DOI] [PubMed] [Google Scholar]
- 25.Clement K, Viguerie N, Poitou C, et al. Weight loss regulates inflammation-related genes in white adipose tissue of obese subjects. FASEB J. 2004;18(14):1657–1669. doi: 10.1096/fj.04-2204com. [DOI] [PubMed] [Google Scholar]
- 26.Wu X, Wang J, Cui X, et al. The effect of insulin on expression of genes and biochemical pathways in human skeletal muscle. Endocrine. 2007;31(1):5–17. doi: 10.1007/s12020-007-0007-x. [DOI] [PubMed] [Google Scholar]
- 27.Coletta DK, Balas B, Chavez AO, et al. Effect of acute physiological hyperinsulinemia on gene expression in human skeletal muscle in vivo. Am J Physiol Endocrinol Metab. 2008;294(5):E910–E917. doi: 10.1152/ajpendo.00607.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granjon A, Gustin MP, Rieusset J, et al. The microRNA signature in response to insulin reveals its implication in the transcriptional action of insulin in human skeletal muscle and the role of a sterol regulatory element-binding protein-1c/myocyte enhancer factor 2C pathway. Diabetes. 2009;58(11):2555–2564. doi: 10.2337/db09-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rome S, Meugnier E, Lecomte V, et al. Microarray analysis of genes with impaired insulin regulation in the skeletal muscle of type 2 diabetic patients indicates the involvement of basic helix-loop-helix domain-containing, class B, 2 protein (BHLHB2) Diabetologia. 2009;52(9):1899–1912. doi: 10.1007/s00125-009-1442-4. [DOI] [PubMed] [Google Scholar]
- 30.Sreekumar R, Halvatsiotis P, Schimke JC, Nair KS. Gene expression profile in skeletal muscle of type 2 diabetes and the effect of insulin treatment. Diabetes. 2002;51(6):1913–1920. doi: 10.2337/diabetes.51.6.1913. [DOI] [PubMed] [Google Scholar]
- 31.Patti ME, Butte AJ, Crunkhorn S, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100(14):8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 33.Mootha VK, Handschin C, Arlow D, et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A. 2004;101(17):6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruce CR, Anderson MJ, Carey AL, et al. Muscle oxidative capacity is a better predictor of insulin sensitivity than lipid status. J Clin Endocrinol Metab. 2003;88(11):5444–5451. doi: 10.1210/jc.2003-030791. [DOI] [PubMed] [Google Scholar]
- 35.Ghosh S, Lertwattanarak R, Lefort N, et al. Reduction in reactive oxygen species production by mitochondria from elderly subjects with normal and impaired glucose tolerance. Diabetes. 2011;60(8):2051–2060. doi: 10.2337/db11-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alibegovic AC, Sonne MP, Hojbjerre L, et al. Insulin resistance induced by physical inactivity is associated with multiple transcriptional changes in skeletal muscle in young men. Am J Physiol Endocrinol Metab. 2010;299(5):E752–E763. doi: 10.1152/ajpendo.00590.2009. [DOI] [PubMed] [Google Scholar]
- 37.Richardson DK, Kashyap S, Bajaj M, et al. Lipid infusion decreases the expression of nuclear encoded mitochondrial genes and increases expression of extracellular matrix genes in human skeletal muscle. J Biol Chem. 2004;280(11):10290–10297. doi: 10.1074/jbc.M408985200. [DOI] [PubMed] [Google Scholar]
- 38.Sparks LM, Xie H, Koza RA, et al. A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes. 2005;54(7):1926–1933. doi: 10.2337/diabetes.54.7.1926. [DOI] [PubMed] [Google Scholar]
- 39.Wisloff U, Najjar SM, Ellingsen O, et al. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science. 2005;307(5708):418–420. doi: 10.1126/science.1108177. [DOI] [PubMed] [Google Scholar]
- 40.Wright DC, Han DH, Garcia-Roves PM, Geiger PC, Jones TE, Holloszy JO. Exercise-induced mitochondrial biogenesis begins before the increase in muscle PGC-1alpha expression. J Biol Chem. 2006 doi: 10.1074/jbc.M606116200. [DOI] [PubMed] [Google Scholar]
- 41.Skov V, Glintborg D, Knudsen S, et al. Reduced expression of nuclear-encoded genes involved in mitochondrial oxidative metabolism in skeletal muscle of insulin-resistant women with polycystic ovary syndrome. Diabetes. 2007;56(9):2349–2355. doi: 10.2337/db07-0275. [DOI] [PubMed] [Google Scholar]
- 42.Skov V, Glintborg D, Knudsen S, et al. Pioglitazone enhances mitochondrial biogenesis and ribosomal protein biosynthesis in skeletal muscle in polycystic ovary syndrome. PLoS ONE. 2008;3(6):e2466. doi: 10.1371/journal.pone.0002466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ronn T, Poulsen P, Hansson O, et al. Age influences DNA methylation and gene expression of COX7A1 in human skeletal muscle. Diabetologia. 2008;51(7):1159–1168. doi: 10.1007/s00125-008-1018-8. [DOI] [PubMed] [Google Scholar]
- 44.Ronn T, Poulsen P, Tuomi T, et al. Genetic variation in ATP5O is associated with skeletal muscle ATP50 mRNA expression and glucose uptake in young twins. PLoS ONE. 2009;4(3):e4793. doi: 10.1371/journal.pone.0004793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ling C, Poulsen P, Carlsson E, et al. Multiple environmental and genetic factors influence skeletal muscle PGC-1alpha and PGC-1beta gene expression in twins. J Clin Invest. 2004;114(10):1518–1526. doi: 10.1172/JCI21889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barres R, Osler ME, Yan J, et al. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10(3):189–198. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 47.Yang X, Pratley RE, Tokraks S, Bogardus C, Permana PA. Microarray profiling of skeletal muscle tissues from equally obese, non-diabetic insulin-sensitive and insulin-resistant Pima Indians. Diabetologia. 2002;45(11):1584–1593. doi: 10.1007/s00125-002-0905-7. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen LL, Kriketos AD, Hancock DP, Caterson ID, Denyer GS. Insulin resistance does not influence gene expression in skeletal muscle. J Biochem Mol Biol. 2006;39(4):457–463. doi: 10.5483/bmbrep.2006.39.4.457. [DOI] [PubMed] [Google Scholar]
- 49.Ling C, Poulsen P, Carlsson E, et al. Multiple environmental and genetic factors influence skeletal muscle PGC-1alpha and PGC-1beta gene expression in twins. J Clin Invest. 2004;114(10):1518–1526. doi: 10.1172/JCI21889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petersen KF, Befroy D, Dufour S, et al. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300(5622):1140–1142. doi: 10.1126/science.1082889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nair KS, Bigelow ML, Asmann YW, et al. Asian Indians have enhanced skeletal muscle mitochondrial capacity to produce ATP in association with severe insulin resistance. Diabetes. 2008;57(5):1166–1175. doi: 10.2337/db07-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hancock CR, Han DH, Chen M, et al. High-fat diets cause insulin resistance despite an increase in muscle mitochondria. Proc Natl Acad Sci U S A. 2008;105(22):7815–7820. doi: 10.1073/pnas.0802057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crunkhorn S, Dearie F, Mantzoros C, et al. Peroxisome proliferator activator receptor gamma coactivator-1 expression is reduced in obesity: potential pathogenic role of saturated fatty acids and p38 mitogen-activated protein kinase activation. J Biol Chem. 2007;282(21):15439–15450. doi: 10.1074/jbc.M611214200. [DOI] [PubMed] [Google Scholar]
- 54.Brons C, Jensen CB, Storgaard H, et al. Impact of short-term high-fat feeding on glucose and insulin metabolism in young healthy men. J Physiol. 2009;587(Pt 10):2387–2397. doi: 10.1113/jphysiol.2009.169078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin W, Goldfine AB, Boes T, et al. Increased SRF transcriptional activity in human and mouse skeletal muscle is a signature of insulin resistance. J Clin Invest. 2011;121(3):918–929. doi: 10.1172/JCI41940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patti ME, Corvera S. The Role of Mitochondria in the Pathogenesis of Type 2 Diabetes. Endocr Rev. 2010 doi: 10.1210/er.2009-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karakelides H, Asmann YW, Bigelow ML, et al. Effect of insulin deprivation on muscle mitochondrial ATP production and gene transcript levels in type 1 diabetic subjects. Diabetes. 2007;56(11):2683–2689. doi: 10.2337/db07-0378. [DOI] [PubMed] [Google Scholar]
- 58.Yechoor VK, Patti ME, Saccone R, Kahn CR. Coordinated patterns of gene expression for substrate and energy metabolism in skeletal muscle of diabetic mice. Proc Natl Acad Sci U S A. 2002;99(16):10587–10592. doi: 10.1073/pnas.142301999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frederiksen CM, Hojlund K, Hansen L, et al. Transcriptional profiling of myotubes from patients with type 2 diabetes: no evidence for a primary defect in oxidative phosphorylation genes. Diabetologia. 2008;51(11):2068–2077. doi: 10.1007/s00125-008-1122-9. [DOI] [PubMed] [Google Scholar]
- 60.Jin W, Goldfine AB, Boes T, et al. Increased SRF transcriptional activity in human and mouse skeletal muscle is a signature of insulin resistance. J Clin Invest. 2011;121(3):918–929. doi: 10.1172/JCI41940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morino K, Petersen KF, Sono S, et al. Regulation of mitochondrial biogenesis by lipoprotein lipase in muscle of insulin-resistant offspring of parents with type 2 diabetes. Diabetes. 2012;61(4):877–887. doi: 10.2337/db11-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thrush AB, Brindley DN, Chabowski A, Heigenhauser GJ, Dyck DJ. Skeletal muscle lipogenic protein expression is not different between lean and obese individuals: a potential factor in ceramide accumulation. J Clin Endocrinol Metab. 2009;94(12):5053–5061. doi: 10.1210/jc.2008-2565. [DOI] [PubMed] [Google Scholar]
- 63.Hulver MW, Berggren JR, Carper MJ, et al. Elevated stearoyl-CoA desaturase-1 expression in skeletal muscle contributes to abnormal fatty acid partitioning in obese humans. Cell Metab. 2005;2(4):251–261. doi: 10.1016/j.cmet.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pihlajamaki J, Lerin C, Itkonen P, et al. Expression of the splicing factor gene SFRS10 is reduced in human obesity and contributes to enhanced lipogenesis. Cell Metab. 2011;14(2):208–218. doi: 10.1016/j.cmet.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chibalin AV, Leng Y, Vieira E, et al. Downregulation of diacylglycerol kinase delta contributes to hyperglycemia-induced insulin resistance. Cell. 2008;132(3):375–386. doi: 10.1016/j.cell.2007.12.035. [DOI] [PubMed] [Google Scholar]
- 66.Parikh H, Carlsson E, Chutkow WA, et al. TXNIP Regulates Peripheral Glucose Metabolism in Humans. PLoS Med. 2007;4(5):e158. doi: 10.1371/journal.pmed.0040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurucz I, Morva A, Vaag A, et al. Decreased expression of heat shock protein 72 in skeletal muscle of patients with type 2 diabetes correlates with insulin resistance. Diabetes. 2002;51(4):1102–1109. doi: 10.2337/diabetes.51.4.1102. [DOI] [PubMed] [Google Scholar]
- 68.Huang X, Vaag A, Carlsson E, Hansson M, Ahren B, Groop L. Impaired cathepsin L gene expression in skeletal muscle is associated with type 2 diabetes. Diabetes. 2003;52(9):2411–2418. doi: 10.2337/diabetes.52.9.2411. [DOI] [PubMed] [Google Scholar]
- 69.Reynet C, Kahn CR. Rad: A member of the ras family overexpressed in muscle of type II diabetic humans. Science. 1993;262:1441–1444. doi: 10.1126/science.8248782. [DOI] [PubMed] [Google Scholar]
- 70.Ilany J, Bilan PJ, Kapur S, et al. Overexpression of Rad in muscle worsens diet-induced insulin resistance and glucose intolerance and lowers plasma triglyceride level. Proc Natl Acad Sci U S A. 2006;103(12):4481–4486. doi: 10.1073/pnas.0511246103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu X, Patki A, Lara-Castro C, et al. Genes and biochemical pathways in human skeletal muscle affecting resting energy expenditure and fuel partitioning. J Appl Physiol. 2011;110(3):746–755. doi: 10.1152/japplphysiol.00293.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Short KR, Vittone JL, Bigelow ML, et al. Impact of aerobic exercise training on age-related changes in insulin sensitivity and muscle oxidative capacity. Diabetes. 2003;52(8):1888–1896. doi: 10.2337/diabetes.52.8.1888. [DOI] [PubMed] [Google Scholar]
- 73.Kuhl JE, Ruderman NB, Musi N, et al. Exercise training decreases the concentration of malonyl-CoA and increases the expression and activity of malonyl-CoA decarboxylase in human muscle. Am J Physiol Endocrinol Metab. 2006;290(6):E1296–E1303. doi: 10.1152/ajpendo.00341.2005. [DOI] [PubMed] [Google Scholar]
- 74.Cannon B, Nedergaard J. Cell biology: Neither brown nor white. Nature. 2012;488(7411):286–287. doi: 10.1038/488286a. [DOI] [PubMed] [Google Scholar]
- 75.Cypess AM, Lehman S, Williams G, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360(15):1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Virtanen KA, Lidell ME, Orava J, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360(15):1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 77.Wu J, Bostrom P, Sparks LM, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150(2):366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jernas M, Palming J, Sjoholm K, et al. Separation of human adipocytes by size: hypertrophic fat cells display distinct gene expression. FASEB J. 2006;20(9):1540–1542. doi: 10.1096/fj.05-5678fje. [DOI] [PubMed] [Google Scholar]
- 79.Olsson M, Olsson B, Jacobson P, et al. Expression of the selenoprotein S (SELS) gene in subcutaneous adipose tissue and SELS genotype are associated with metabolic risk factors. Metabolism. 2011;60(1):114–120. doi: 10.1016/j.metabol.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Franck N, Gummesson A, Jernas M, et al. Identification of adipocyte genes regulated by caloric intake. J Clin Endocrinol Metab. 2011;96(2):E413–E418. doi: 10.1210/jc.2009-2534. [DOI] [PubMed] [Google Scholar]
- 81.Wajchenberg BL. Subcutaneous and visceral adipose tissue: their relation to the metabolic syndrome. Endocr Rev. 2000;21(6):697–738. doi: 10.1210/edrv.21.6.0415. [DOI] [PubMed] [Google Scholar]
- 82.Svensson PA, Gabrielsson BG, Jernas M, Gummesson A, Sjoholm K. Regulation of human aldoketoreductase 1C3 (AKR1C3) gene expression in the adipose tissue. Cell Mol Biol Lett. 2008;13(4):599–613. doi: 10.2478/s11658-008-0025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Linder K, Arner P, Flores-Morales A, Tollet-Egnell P, Norstedt G. Differentially expressed genes in visceral or subcutaneous adipose tissue of obese men and women. J Lipid Res. 2004;45(1):148–154. doi: 10.1194/jlr.M300256-JLR200. [DOI] [PubMed] [Google Scholar]
- 84.Sjoholm K, Palming J, Olofsson LE, et al. A microarray search for genes predominantly expressed in human omental adipocytes: adipose tissue as a major production site of serum amyloid A. J Clin Endocrinol Metab. 2005;90(4):2233–2239. doi: 10.1210/jc.2004-1830. [DOI] [PubMed] [Google Scholar]
- 85.Quinkler M, Bujalska IJ, Tomlinson JW, Smith DM, Stewart PM. Depotspecific prostaglandin synthesis in human adipose tissue: a novel possible mechanism of adipogenesis. Gene. 2006;380(2):137–143. doi: 10.1016/j.gene.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 86.Poitou C, Divoux A, Faty A, et al. Role of serum amyloid a in adipocytemacrophage cross talk and adipocyte cholesterol efflux. J Clin Endocrinol Metab. 2009;94(5):1810–1817. doi: 10.1210/jc.2008-2040. [DOI] [PubMed] [Google Scholar]
- 87.Bujalska IJ, Quinkler M, Tomlinson JW, Montague CT, Smith DM, Stewart PM. Expression profiling of 11beta-hydroxysteroid dehydrogenase type-1 and glucocorticoid-target genes in subcutaneous and omental human preadipocytes. J Mol Endocrinol. 2006;37(2):327–340. doi: 10.1677/jme.1.02048. [DOI] [PubMed] [Google Scholar]
- 88.Lee YH, Tharp WG, Maple RL, Nair S, Permana PA, Pratley RE. Amyloid precursor protein expression is upregulated in adipocytes in obesity. Obesity (Silver Spring) 2008;16(7):1493–1500. doi: 10.1038/oby.2008.267. [DOI] [PubMed] [Google Scholar]
- 89.Taleb S, Lacasa D, Bastard JP, et al. Cathepsin S, a novel biomarker of adiposity: relevance to atherogenesis. FASEB J. 2005;19(11):1540–1542. doi: 10.1096/fj.05-3673fje. [DOI] [PubMed] [Google Scholar]
- 90.Kolehmainen M, Salopuro T, Schwab US, et al. Weight reduction modulates expression of genes involved in extracellular matrix and cell death: the GENOBIN study. Int J Obes (Lond) 2008;32(2):292–303. doi: 10.1038/sj.ijo.0803718. [DOI] [PubMed] [Google Scholar]
- 91.Marrades MP, Milagro FI, Martinez JA, Moreno-Aliaga MJ. Differential expression of aquaporin 7 in adipose tissue of lean and obese high fat consumers. Biochem Biophys Res Commun. 2006;339(3):785–789. doi: 10.1016/j.bbrc.2005.11.080. [DOI] [PubMed] [Google Scholar]
- 92.Klimcakova E, Roussel B, Marquez-Quinones A, et al. Worsening of obesity and metabolic status yields similar molecular adaptations in human subcutaneous and visceral adipose tissue: decreased metabolism and increased immune response. J Clin Endocrinol Metab. 2011;96(1):E73–E82. doi: 10.1210/jc.2010-1575. [DOI] [PubMed] [Google Scholar]
- 93.Zhang Y, Bosse Y, Marceau P, et al. Gene expression variability in subcutaneous and omental adipose tissue of obese men. Gene Expr. 2007;14(1):35–46. doi: 10.3727/000000007783991772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wolfs MG, Rensen SS, Bruin-Van Dijk EJ, et al. Co-expressed immune and metabolic genes in visceral and subcutaneous adipose tissue from severely obese individuals are associated with plasma HDL and glucose levels: a microarray study. BMC Med Genomics. 2010;3:34. doi: 10.1186/1755-8794-3-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baranova A, Collantes R, Gowder SJ, et al. Obesity-related differential gene expression in the visceral adipose tissue. Obes Surg. 2005;15(6):758–765. doi: 10.1381/0960892054222876. [DOI] [PubMed] [Google Scholar]
- 96.Prokunina-Olsson L, Kaplan LM, Schadt EE, Collins FS. Alternative splicing of TCF7L2 gene in omental and subcutaneous adipose tissue and risk of type 2 diabetes. PLoS ONE. 2009;4(9):e7231. doi: 10.1371/journal.pone.0007231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131(2):242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 98.Walewski JL, Ge F, Gagner M, et al. Adipocyte accumulation of longchain fatty acids in obesity is multifactorial, resulting from increased fatty acid uptake and decreased activity of genes involved in fat utilization. Obes Surg. 2010;20(1):93–107. doi: 10.1007/s11695-009-0002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marrades MP, Gonzalez-Muniesa P, Martinez JA, Moreno-Aliaga MJ. A dysregulation in CES1, APOE and other lipid metabolism-related genes is associated to cardiovascular risk factors linked to obesity. Obes Facts. 2010;3(5):312–318. doi: 10.1159/000321451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Maclaren R, Cui W, Simard S, Cianflone K. Influence of obesity and insulin sensitivity on insulin signaling genes in human omental and subcutaneous adipose tissue. J Lipid Res. 2008;49(2):308–323. doi: 10.1194/jlr.M700199-JLR200. [DOI] [PubMed] [Google Scholar]
- 101.Kursawe R, Eszlinger M, Narayan D, et al. Cellularity and adipogenic profile of the abdominal subcutaneous adipose tissue from obese adolescents: association with insulin resistance and hepatic steatosis. Diabetes. 2010;59(9):2288–2296. doi: 10.2337/db10-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Marrades MP, Gonzalez-Muniesa P, Arteta D, Martinez JA, Moreno- Aliaga MJ. Orchestrated downregulation of genes involved in oxidative metabolic pathways in obese vs. lean high-fat young male consumers. J Physiol Biochem. 2011;67(1):15–26. doi: 10.1007/s13105-010-0044-4. [DOI] [PubMed] [Google Scholar]
- 103.Mustelin L, Pietilainen KH, Rissanen A, et al. Acquired obesity and poor physical fitness impair expression of genes of mitochondrial oxidative phosphorylation in monozygotic twins discordant for obesity. Am J Physiol Endocrinol Metab. 2008;295(1):E148–E154. doi: 10.1152/ajpendo.00580.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee YH, Nair S, Rousseau E, et al. Microarray profiling of isolated abdominal subcutaneous adipocytes from obese vs non-obese Pima Indians: increased expression of inflammation-related genes. Diabetologia. 2005;48(9):1776–1783. doi: 10.1007/s00125-005-1867-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Traurig MT, Permana PA, Nair S, Kobes S, Bogardus C, Baier LJ. Differential expression of matrix metalloproteinase 3 (MMP3) in preadipocytes/stromal vascular cells from nonobese nondiabetic versus obese nondiabetic Pima Indians. Diabetes. 2006;55(11):3160–3165. doi: 10.2337/db06-0373. [DOI] [PubMed] [Google Scholar]
- 106.Dahlman I, Kaaman M, Olsson T, et al. A unique role of monocyte chemoattractant protein 1 among chemokines in adipose tissue of obese subjects. J Clin Endocrinol Metab. 2005;90(10):5834–5840. doi: 10.1210/jc.2005-0369. [DOI] [PubMed] [Google Scholar]
- 107.van Dijk SJ, Feskens EJ, Bos MB, et al. A saturated fatty acid-rich diet induces an obesity-linked proinflammatory gene expression profile in adipose tissue of subjects at risk of metabolic syndrome. Am J Clin Nutr. 2009;90(6):1656–1664. doi: 10.3945/ajcn.2009.27792. [DOI] [PubMed] [Google Scholar]
- 108.Heneghan HM, Miller N, McAnena OJ, O'Brien T, Kerin MJ. Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. J Clin Endocrinol Metab. 2011;96(5):E846–E850. doi: 10.1210/jc.2010-2701. [DOI] [PubMed] [Google Scholar]
- 109.Ortega FJ, Moreno-Navarrete JM, Pardo G, et al. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS ONE. 2010;5(2):e9022. doi: 10.1371/journal.pone.0009022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Martinelli R, Nardelli C, Pilone V, et al. miR-519d overexpression is associated with human obesity. Obesity (Silver Spring) 2010;18(11):2170–2176. doi: 10.1038/oby.2009.474. [DOI] [PubMed] [Google Scholar]
- 111.Yang X, Jansson PA, Nagaev I, et al. Evidence of impaired adipogenesis in insulin resistance. Biochem Biophys Res Commun. 2004;317(4):1045–1051. doi: 10.1016/j.bbrc.2004.03.152. [DOI] [PubMed] [Google Scholar]
- 112.Urs S, Smith C, Campbell B, et al. Gene expression profiling in human preadipocytes and adipocytes by microarray analysis. J Nutr. 2004;134(4):762–770. doi: 10.1093/jn/134.4.762. [DOI] [PubMed] [Google Scholar]
- 113.Nair S, Lee YH, Rousseau E, et al. Increased expression of inflammationrelated genes in cultured preadipocytes/stromal vascular cells from obese compared with non-obese Pima Indians. Diabetologia. 2005;48(9):1784–1788. doi: 10.1007/s00125-005-1868-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Klimcakova E, Roussel B, Kovacova Z, et al. Macrophage gene expression is related to obesity and the metabolic syndrome in human subcutaneous fat as well as in visceral fat. Diabetologia. 2011;54(4):876–887. doi: 10.1007/s00125-010-2014-3. [DOI] [PubMed] [Google Scholar]
- 115.Shah R, Lu Y, Hinkle CC, et al. Gene profiling of human adipose tissue during evoked inflammation in vivo. Diabetes. 2009;58(10):2211–2219. doi: 10.2337/db09-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun K, Kusminski CM, Scherer PE. Adipose tissue remodeling and obesity. J Clin Invest. 2011;121(6):2094–2101. doi: 10.1172/JCI45887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dankel SN, Fadnes DJ, Stavrum AK, et al. Switch from stress response to homeobox transcription factors in adipose tissue after profound fat loss. PLoS ONE. 2010;5(6):e11033. doi: 10.1371/journal.pone.0011033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee MJ, Gong DW, Burkey BF, Fried SK. Pathways regulated by glucocorticoids in omental and subcutaneous human adipose tissues: a microarray study. Am J Physiol Endocrinol Metab. 2011;300(3):E571–E580. doi: 10.1152/ajpendo.00231.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gummesson A, Jernas M, Svensson PA, et al. Relations of adipose tissue CIDEA gene expression to basal metabolic rate, energy restriction, and obesity: population-based and dietary intervention studies. J Clin Endocrinol Metab. 2007;92(12):4759–4765. doi: 10.1210/jc.2007-1136. [DOI] [PubMed] [Google Scholar]
- 120.Capel F, Klimcakova E, Viguerie N, et al. Macrophages and adipocytes in human obesity: adipose tissue gene expression and insulin sensitivity during calorie restriction and weight stabilization. Diabetes. 2009;58(7):1558–1567. doi: 10.2337/db09-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Marquez-Quinones A, Mutch DM, Debard C, et al. Adipose tissue transcriptome reflects variations between subjects with continued weight loss and subjects regaining weight 6 mo after caloric restriction independent of energy intake. Am J Clin Nutr. 2010;92(4):975–984. doi: 10.3945/ajcn.2010.29808. [DOI] [PubMed] [Google Scholar]
- 122.Behre CJ, Gummesson A, Jernas M, et al. Dissociation between adipose tissue expression and serum levels of adiponectin during and after dietinduced weight loss in obese subjects with and without the metabolic syndrome. Metabolism. 2007;56(8):1022–1028. doi: 10.1016/j.metabol.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 123.Cancello R, Henegar C, Viguerie N, et al. Reduction of macrophage infiltration and chemoattractant gene expression changes in white adipose tissue of morbidly obese subjects after surgery-induced weight loss. Diabetes. 2005;54(8):2277–2286. doi: 10.2337/diabetes.54.8.2277. [DOI] [PubMed] [Google Scholar]
- 124.Goldfine AB, Crunkhorn S, Costello M, et al. Necdin and E2F4 Are Modulated by Rosiglitazone Therapy in Diabetic Human Adipose and Muscle Tissue. Diabetes. 2006;55(3):640–650. doi: 10.2337/diabetes.55.03.06.db05-1015. [DOI] [PubMed] [Google Scholar]
- 125.Kosteli A, Sugaru E, Haemmerle G, et al. Weight loss and lipolysis promote a dynamic immune response in murine adipose tissue. J Clin Invest. 2010;120(10):3466–3479. doi: 10.1172/JCI42845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kotronen A, Seppala-Lindroos A, Bergholm R, Yki-Jarvinen H. Tissue specificity of insulin resistance in humans: fat in the liver rather than muscle is associated with features of the metabolic syndrome. Diabetologia. 2008;51(1):130–138. doi: 10.1007/s00125-007-0867-x. [DOI] [PubMed] [Google Scholar]
- 127.Diraison F, Dusserre E, Vidal H, Sothier M, Beylot M. Increased hepatic lipogenesis but decreased expression of lipogenic gene in adipose tissue in human obesity. Am J Physiol Endocrinol Metab. 2002;282(1):E46–E51. doi: 10.1152/ajpendo.2002.282.1.E46. [DOI] [PubMed] [Google Scholar]
- 128.Sharma A, Sharma VK, Horn-Saban S, Lancet D, Ramachandran S, Brahmachari SK. Assessing natural variations in gene expression in humans by comparing with monozygotic twins using microarrays. Physiol Genomics. 2005 doi: 10.1152/physiolgenomics.00228.2003. [DOI] [PubMed] [Google Scholar]
- 129.Cortez-Pinto H, Chatham J, Chacko VP, Arnold C, Rashid A, Diehl AM. Alterations in liver ATP homeostasis in human nonalcoholic steatohepatitis: a pilot study. JAMA. 1999;282(17):1659–1664. doi: 10.1001/jama.282.17.1659. [DOI] [PubMed] [Google Scholar]
- 130.Michael MD, Kulkarni RN, Postic C, et al. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol Cell. 2000;6:87–97. [PubMed] [Google Scholar]
- 131.Biddinger SB, Hernandez-Ono A, Rask-Madsen C, et al. Hepatic insulin resistance is sufficient to produce dyslipidemia and susceptibility to atherosclerosis. Cell Metab. 2008;7(2):125–134. doi: 10.1016/j.cmet.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sreekumar R, Rosado B, Rasmussen D, Charlton M. Hepatic gene expression in histologically progressive nonalcoholic steatohepatitis. Hepatology. 2003;38(1):244–251. doi: 10.1053/jhep.2003.50290. [DOI] [PubMed] [Google Scholar]
- 133.Younossi ZM, Baranova A, Ziegler K, et al. A genomic and proteomic study of the spectrum of nonalcoholic fatty liver disease. Hepatology. 2005;42(3):665–74. doi: 10.1002/hep.20838. [DOI] [PubMed] [Google Scholar]
- 134.Younossi ZM, Gorreta F, Ong JP, et al. Hepatic gene expression in patients with obesity-related non-alcoholic steatohepatitis. Liver Int. 2005;25(4):760–771. doi: 10.1111/j.1478-3231.2005.01117.x. [DOI] [PubMed] [Google Scholar]
- 135.Stepanova M, Hossain N, Afendy A, et al. Hepatic gene expression of Caucasian and African-American patients with obesity-related nonalcoholic fatty liver disease. Obes Surg. 2010;20(5):640–650. doi: 10.1007/s11695-010-0078-2. [DOI] [PubMed] [Google Scholar]
- 136.Cheung O, Puri P, Eicken C, et al. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48(6):1810–1820. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Baker SS, Baker RD, Liu W, Nowak NJ, Zhu L. Role of alcohol metabolism in non-alcoholic steatohepatitis. PLoS ONE. 2010;5(3):e9570. doi: 10.1371/journal.pone.0009570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chiappini F, Barrier A, Saffroy R, et al. Exploration of global gene expression in human liver steatosis by high-density oligonucleotide microarray. Lab Invest. 2006;86(2):154–65. doi: 10.1038/labinvest.3700374. [DOI] [PubMed] [Google Scholar]
- 139.Takamura T, Misu H, Matsuzawa-Nagata N, et al. Obesity upregulates genes involved in oxidative phosphorylation in livers of diabetic patients. Obesity (Silver Spring) 2008;16(12):2601–2609. doi: 10.1038/oby.2008.419. [DOI] [PubMed] [Google Scholar]
- 140.Pihlajamaki J, Boes T, Kim EY, et al. Thyroid hormone-related regulation of gene expression in human fatty liver. J Clin Endocrinol Metab. 2009;94(9):3521–3529. doi: 10.1210/jc.2009-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Elam MB, Yellaturu C, Howell GE, et al. Dysregulation of sterol regulatory element binding protein-1c in livers of morbidly obese women is associated with altered suppressor of cytokine signaling-3 and signal transducer and activator of transcription-1 signaling. Metabolism. 2010;59(4):587–598. doi: 10.1016/j.metabol.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Elam MB, Yellaturu C, Howell GE, et al. Dysregulation of sterol regulatory element binding protein-1c in livers of morbidly obese women is associated with altered suppressor of cytokine signaling-3 and signal transducer and activator of transcription-1 signaling. Metabolism. 2010;59(4):587–598. doi: 10.1016/j.metabol.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hietaniemi M, Jokela M, Rantala M, et al. The effect of a short-term hypocaloric diet on liver gene expression and metabolic risk factors in obese women. Nutr Metab Cardiovasc Dis. 2009;19(3):177–183. doi: 10.1016/j.numecd.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 144.Szalowska E, Dijkstra M, Elferink MG, et al. Comparative analysis of the human hepatic and adipose tissue transcriptomes during LPS-induced inflammation leads to the identification of differential biological pathways and candidate biomarkers. BMC Med Genomics. 2011;4:71. doi: 10.1186/1755-8794-4-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Swagell CD, Henly DC, Morris CP. Expression analysis of a human hepatic cell line in response to palmitate. Biochem Biophys Res Commun. 2005;328(2):432–441. doi: 10.1016/j.bbrc.2004.12.188. [DOI] [PubMed] [Google Scholar]
- 146.Gunton JE, Kulkarni RN, Yim S, et al. Loss of ARNT/HIF1beta mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell. 2005;122(3):337–349. doi: 10.1016/j.cell.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 147.Lyssenko V, Lupi R, Marchetti P, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest. 2007;117(8):2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Taneera J, Lang S, Sharma A, et al. A systems genetics approach identifies genes and pathways for type 2 diabetes in human islets. Cell Metab. 2012;16(1):122–134. doi: 10.1016/j.cmet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 149.Marselli L, Thorne J, Dahiya S, et al. Gene expression profiles of Betacell enriched tissue obtained by laser capture microdissection from subjects with type 2 diabetes. PLoS ONE. 2010;5(7):e11499. doi: 10.1371/journal.pone.0011499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ling C, Del Guerra S, Lupi R, et al. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 2008 doi: 10.1007/s00125-007-0916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Grayson BL, Wang L, Aune TM. Peripheral blood gene expression profiles in metabolic syndrome, coronary artery disease and type 2 diabetes. Genes Immun. 2011;12(5):341–351. doi: 10.1038/gene.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mao J, Ai J, Zhou X, et al. Transcriptomic profiles of peripheral white blood cells in type II diabetes and racial differences in expression profiles. BMC Genomics. 2011;12(Suppl 5):S12. doi: 10.1186/1471-2164-12-S5-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zelezniak A, Pers TH, Soares S, Patti ME, Patil KR. Metabolic network topology reveals transcriptional regulatory signatures of type 2 diabetes. PLoS Comput Biol. 2010;6(4):e1000729. doi: 10.1371/journal.pcbi.1000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ptitsyn A, Hulver M, Cefalu W, York D, Smith SR. Unsupervised clustering of gene expression data points at hypoxia as possible trigger for metabolic syndrome. BMC Genomics. 2006;7:318. doi: 10.1186/1471-2164-7-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tiffin N, Adie E, Turner F, et al. Computational disease gene identification: a concert of methods prioritizes type 2 diabetes and obesity candidate genes. Nucleic Acids Res. 2006;34(10):3067–3081. doi: 10.1093/nar/gkl381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mori MA, Liu M, Bezy O, et al. A systems biology approach identifies inflammatory abnormalities between mouse strains prior to development of metabolic disease. Diabetes. 2010;59(11):2960–2971. doi: 10.2337/db10-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jin W, Goldfine A, Boes T, et al. Increased SRF transcriptional activity in human and mouse skeletal muscle is a signature of insulin resistance. J Biol Chem. 2011;121:918–929. doi: 10.1172/JCI41940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yi Z, Bowen BP, Hwang H, et al. Global relationship between the proteome and transcriptome of human skeletal muscle. J Proteome Res. 2008;7(8):3230–3241. doi: 10.1021/pr800064s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of studies of gene expression changes referenced in the review.