Abstract
Carbohydrates exhibit many physiologically and pharmacologically important activities, yet their complicated structure and sequence pose major analytical challenges. Although their structural complexity makes analysis of carbohydrate difficult, mass spectrometry (MS) with high sensitivity, resolution and accuracy has become a vital tool in many applications related to analysis of carbohydrates or oligosaccharides. This application is essentially based on soft ionization technique which facilitates the ionization and vaporization of large, polar and thermally labile biomolecules. Electrospray-ionization (ESI), one of the soft ionization technique, tandem MS has been used in the sequencing of peptides, proteins, lipids, nucleic acids and more recently carbohydrates. The development of the ESI and tandem MS has begun to make carbohydrate analysis more routine. This review will focus on the application of the ESI tandem MS for the sequence analysis of native oligosaccharides, including neutral saccharides with multiple linkages, and the uronic acid polymers, alginate and glycosaminoglycans structures containing epimers.
Keywords: ESI tandem MS, Neutral oligosaccharide, Uronic acid oligosaccharides, Glycosaminoglycan oligosaccharides, Sequence
1. INTRODUCTION
Glycosylation is a common post-translational modification to cell surface and extracellular matrix proteins as well as lipids. As a result, cells carry a dense coat of carbohydrate on their surface that mediates a wide variety of cell-cell and cell-matrix interactions that are crucial to development and functions. Carbohydrates are biomolecules having very complicated structures. Their diverse monosaccharide residues, different modes of linkage (α-and β-anomers at multiple positions), their linear or branched structures with different length and presence of various substituent groups (sulfo, phospho, acetyl and methyl etc.) contribute to the chemical heterogeneity, and structural complexity of carbohydrate. So it is essential to determine these biomolecular structures in order to understand their biological functions. Recent development in biological mass spectrometry makes structural determination much easier. This is primarily the result of the invention of the “soft ionization” techniques, [1, 2] which permit the ionization and vaporization of the large biomolecules such as peptides, proteins, lipids, nucleic acids and carbohydrates. Soft ionization include both matrix assisted laser-desorption-ionization (MALDI) [3] and electrospray ionization (ESI) [4, 5] widely used to detect biological samples and to analyze unknowns in the combination with tandem MS (MS/MS) techniques [6]. In addition, improvements in mass spectrometer performance, including sensitivity, resolution, and mass accuracy have opened the door for many novel analytical applications related to large biomolecules. ESI can be coupled with the “nanospray”, [7] which vastly improved the sensitivity, and with HPLC or CE separation methods [8] to enhance the sensitive analysis of polar molecules.
Since the initial development of the ESI and tandem MS these methods have been greatly enhanced resulting in their acceptance as a powerful techniques for the analysis of the structure and sequence of carbohydrates or oligosaccharides in industrial and academic settings. Fragment-ions are critical for the sequence of carbohydrates [9-11]. The nomenclature for oligosaccharide fragmentation was established by Domon and Costello [12]. Based on these rules the fragment ions that contain a non-reducing terminus are labeled with uppercase letter from beginning of the alphabet (A, B, C), and those that contain a non-reducing terminus of the oligosaccharide or the aglycon are labeled with the letters from the end of the alphabet (X, Y, Z). Subscripts are used to indicate the cleaved ions. The A and X ions are produced by the cleavage across the glycosidic ring, and are labeled by assigning each ring bond a number and counting clockwise from 0 (which is between the oxygen and the anomeric carbon) to 5, superscripts indicate the ring bond number. Ions produced from the cleavage of successive residues are labeled: Am, Bm, and Cm with m = 1 for the non-reducing end and Xn, Yn, and Xn with n = 1 for the reducing-end residues. Y0 and Z0 refer to the fragmentation of the bond to the aglycon. The fragments produced by double glycosidic cleavage are labeled with D. The subscripts indicate the two cleavage glycosidic bonds counting from non-reducing end like B and C-type [13].
The mass spectrometric analysis of proteins, peptides [14-17], lipids [18-21] and DNA [22-25] is much more highly developed relative to those for carbohydrates, in which tandem mass spectrometric product-ion patterns are more complex, and the results depend on the types of derivative and precursor-ion used. Thus, the sequence analysis of the carbohydrate is considerably difficult than for proteins, lipids and DNA. In most cases, one analyzes the protonated forms of underivatized proteins and peptides and tandem MS results in definition of a complete sequence or of a partial sequence that is useful in the identification of peptides coupled with data base reaching. In sequencing oligodeoxynucleic acid by ESI tandem MS, mass ladders are identified by sequentially adding each of the four possible nucleotide masses, and by searching the spectrum for the best match with expected ions. In carbohydrate mass spectral analysis, native structure, fully derivatized structure, or reducing terminal derivatives, or of analyzing native structures are often examined. An analyst must choose behave protonated, metal-cationized ions. In addition of glycosidic cleavage fragment ions, ions produced by cleaving glycosidic bond between two neighbored monosaccharide residues, crossring cleavage fragment ions present as a series of crucial information of sequence of carbohydrates. A great number of epimer or isomer occurs in oligosaccharides. Though all of these challenges make carbohydrate analysis more complicated than other biomacromolecules in ESI tandem MS, many analysts work on it and delineate a hopeful future. As a result in recent years there has been a divergence of published structural analysis methods in carbohydrate mass spectrometry field. This review focuses on using ESI tan-dem MS in negative mode to analyze the native carbohydrate with diverse structural properties, including neutral oligosaccharides, oligouronic acids and sulfated oligosaccharides.
2. SEQUENCE ANALYSIS OF NEUTRAL OLIGOSACCHARIDES
ESI tandem MS has been widely used in the analysis of the neutral oligosaccharides following derivatization, e.g., permethylation [26-29] or reducing terminal derivation [30-34]. Most recently, ESI tandem MS had been successfully used to analyze underivatized neutral oligosaccharides, in which the sequence, both linkage information and difference of the isomeric structures can be confirmed through a series of A-, B-, C- and D-type cleavage ion peaks in the resulting spectrum.
In one example, a series of neutral oligosaccharides, extracted and purified from human urine [13], were analyzed by ESI-CID-MS/MS in negative-mode. In these neutral glycans, there are fourteen linear and mono-, di-, and trifucosylated oligosaccharides. The CID spectra of these oligosaccharides demonstrated that sequence and partial linkage information can be derived and isomeric structures can be differentiated.
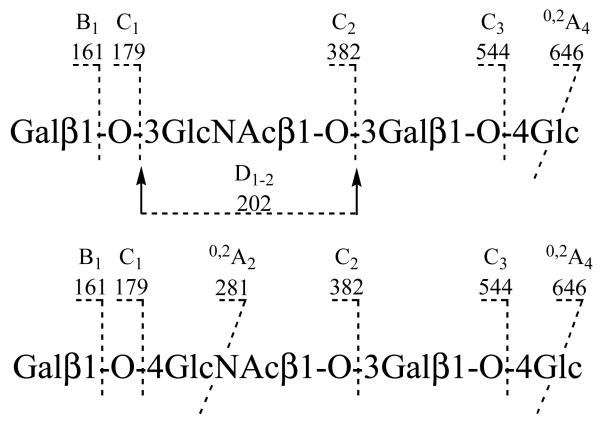
Two linear tetramers of LNT (Galβ1-3GlcNAcβ1-3Galβ1-4Glc) and LNnT (Galβ1-4GlcNAcβ1-3Galβ1-4Glc) showed some differences in their spectra. The fragmentations were assigned in Scheme 1. Besides the glycosidic cleavage fragment ions (C1 at m/z 179, C2 at m/z 382, C3 at m/z 544 and B1 at m/z 161), an A-type cross-ring fragmentation occurred at 4-linked HexNAc or 4-linked Hex residues. Both LNT and LNnT give 0,2A4 ions at m/z 646 from cleavage of the reducing terminal -4 Hex. More usefully, a 0,2A2 ion (m/z 281, together with its dehydrated ion m/z 263) is produced from the -4 HexNAc in LNnT but not from the -3 HexNAc in the spectrum of LNT. However, this 3-linked HexNAc residue in LNT gives a unique ion at m/z 202. This is assigned as a C2-Z2 double cleavage, designated as D1-2, due to favorable fragmentation at the reducing end of the glycosidic oxygen. Thus, the -3 HexNAc linkage in LNT and the -4 HexNAc linkage in LNnT can be readily differentiated by the 0,2A2 ion (m/z 281) and D1-2 ion (m/z 202), respectively.
Scheme 1.
Fragmentation patterns of two linear neutral oligosaccharides under ESI-CID-MS/MS.
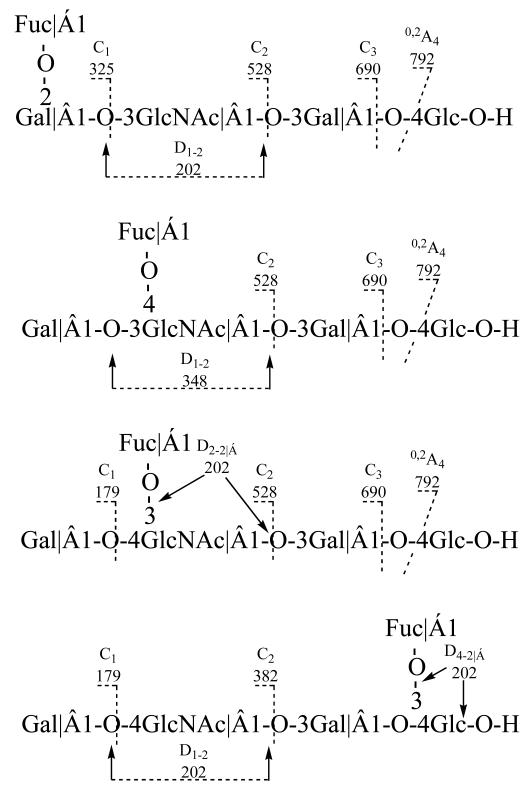
Isomeric monofucosylated pentasaccharides LNFP I (Fucα1-2Galβ1-3GlcNAc 1-3Galβ1-4Glc), II (Galβ1-3 (Fucα1-4) GlcNAc 1-3Galβ1-4Glc), III (Galβ1-4 (Fucα1-3) GlcNAcβ1-3Galβ1-4Glc), and IV (Galβ1-3GlcNAcβ1-3Galβ1-4 (Fucα1-3)Glc) each gave unique CID fragment ion spectra. The fragmentations were shown in Scheme 2. All four pentasaccharides contain a 3-linked HexNAc that readily undergoes double cleavage to produce D-type fragments. LNFP I has an unbranched -3 HexNAc, and hence, m/z 202 is observed as in the spectra of LNT. In the spectrum of LNFP II, the major fragment at m/z 348 results from D1-2 double cleavage of the 3-linked HexNAc, indicating a deO-Hex residue linked at the 4-position of the -3 HexNAc (202 + 146). The D2-2 at m/z 364 in LNFP III indicates a Gal at the 4-position (202 + 162). Similarly, the double cleavage D1-2 ion at m/z 202 in LNFP V indicates a nonsubstituted and 3-linked HexNAc. In LNFP V the deOHex at the reducing terminal Hex can be inferred by the mass difference of 308 (146 + 162, deOHex + Hex) between the ion C3 and [M - H]. Interestingly, the deOHex linkage at the 3-position of the terminal -4 Hex is also labile and can undergo fragmentation consistent with a double cleavage of D-type similar to a 3-linked HexNAc. The resulting D4-4α fragment ion at m/z 688 serves to define the deOHex 3-linked to the Hex. As HexNAc and Hex have the same stereoconfiguration, the favorable D-type fragmentation occurs in both 3-linked HexNAc(GlcNAc) and Hex (Glc) but not in a 3-linked Hex (Gal). Thus, LNFP I, II, III, and V are readily differentiated by the distinctive ions at m/z 202, 348, 364, and 688, respectively, allowing the sequence of these oligosaccharides to be deduced. Others di-, and tri-fucosylated oligosaccharides showed ESI-CID-MS/MS spectra, that could be similarly interpreted [13].
Scheme 2.
Fragmentation patterns of fucose substituted neutral oligosaccharides under ESI-CID-MS/MS.
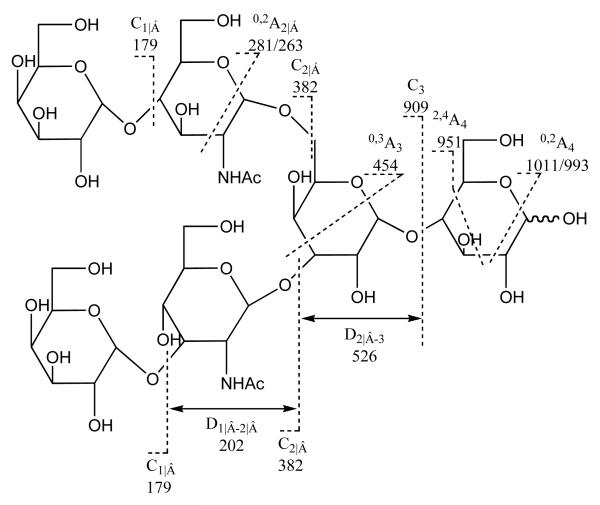
In another example, the spectra of nine branched oligosaccharides prepared from human milk [35], LNH, Galβ1-4GlcNAcβ1-6 (Galβ1-3GlcNAcβ1-3) Galβ1-4Glc, showed some sequence information, which include the product-ion spectra of singly charged and doubly charged ion. The fragmentations of the precursor display different patterns as shown in the Scheme 3. The product-ion spectrum of [M-H]− of LNH was dominated by fragment ions from the 6-linked branch, a feature of all the spectra of oligosaccharides in this series. Information on the 3-linked branch is missing. In contrast, the product-ion spectrum of the doubly charged molecular ion [M-2H]2− (m/z 535) in showed that fragments are produced from both branches, not only the same ions as in the [M-H]− spectrum but also D1-2 (m/z 202) from the -3 HexNAc- in the 3-linked branch. In addition, a doubly charged 2,4A4 ion (m/z 475) is intense when compared with its corresponding singly charged ion m/z 951 in the production spectrum of [M-H ]−. Hence, the product-ion spectra of [M-H]− and [M-2H]2− of LNH give complementary information, as details as the 6-linked branch and the disaccharide core can be obtained from the [M-H]− spectrum, and the sequence of the 3-linked branch can be derived from the additional fragmentation in the [M-2H]2− spectrum.
Scheme 3.
Fragmentation patterns of branched neutral oligosaccharide under ESI-CID-MS/MS.
These two studies show ESI-CID-MS/MS in negative mode for the sequence analysis of neutral oligosaccharides with diverse linkages, linear and branched structures, and with different saccharide residues. The author summarize as follows: (1) Sequence can be unambiguously deduced from a complete set of C-type fragments. (2) Partial linkage information comes from the D- and A-type fragments. If the 3-linked GlcNAc was unsubstituted, a D-ion was produced at m/z 202. If the 3-linked GlcNAc was substituted, a D-ion having an m/z 202 + m/z of substituent was observed. It is similar that branching pattern can be derived from the characteristic D2-3 ion comprising the 6-linked chain and the core branching Gal residue in the product-ion spectrum of [M-H]−. (3) Structural information of the 3-linked branch was deduced from additional ions in the product-ion spectrum of [M-2H]2−. A-type and D-type ions are diagnostic for linkage pattern identification.
ESI multistage MS was also used for sequencing these oligosaccharides by Karas and his collaborators [36, 37]. For example, in analysis of LNT (Galβ1-3GlcNAcβ1-3Galβ1-4Glc) and LNnT (Galβ1-4GlcNAcβ1-3Galβ1-4Glc), the MS3 were performed on the C2 m/z 382 produced in MS2. The product ions at m/z 202, 179 and m/z 281, 263, were observed in the MS3 spectra of both LNT and LNnT. The presence of A-type cross-ring cleavage ions at m/z 281, 263 in MS3 spectrum of LNnT results 1-4 linkage between Gal and GlcNAc residues, while the sequence of LNT was reflected by the cleavage ions at m/z 202, 179. This results confirmed the sequence analysis only using MS/MS [13]. For other more complicated structures, MS2, MS3 and even MS4 were applied to obtain the sequence information.
Linear and branched neutral oligosaccharides could be sequenced in linkage and substituent pattern by ESI tandem MS. However, the identification of epimer in neutral oligosaccharides is still a major challenge for this method.
3. SEQUENCE ANALYSIS OF URONIC ACID OLIGOSACCHARIDES FROM POLYSACCHARIDES
ESI-SID-MS/MS has been applied to sequence oligogalacturonic acid [38] and alginate [39]. Polygalacturonic acid is a kind of homo-polymer that can be broken down to homo-oligosaccharides. Multi-stage ESI-MS and isotopic labeling (O18) at the position 1 of the reducing end [39-41] were used to confirm the fragmentation pattern in the MS spectrum of the oligouronic acid. Alginate, [42, 43] is an acid linear polysaccharide, consisting of α-L-guluronic and β-D-mannuronic acid exclusively containing 1-4 linkages. Homo-oligomeric regions include a mannuronic acid (M) block and guluronic acid (G) block. Alginate also contains a hetero-oligomeric region, the (MG) block. The alginate sequencing requires that the mannuronate and guluronate epimers be distinguished, one of the most challenging tasks in MS analysis. Using ESI-CID-MS/MS, it was possible to distinguish epimers of mannuronate and guluronate along the chains, affording the sequences of a series of homo- and hetero-oligosaccharides of alginate.
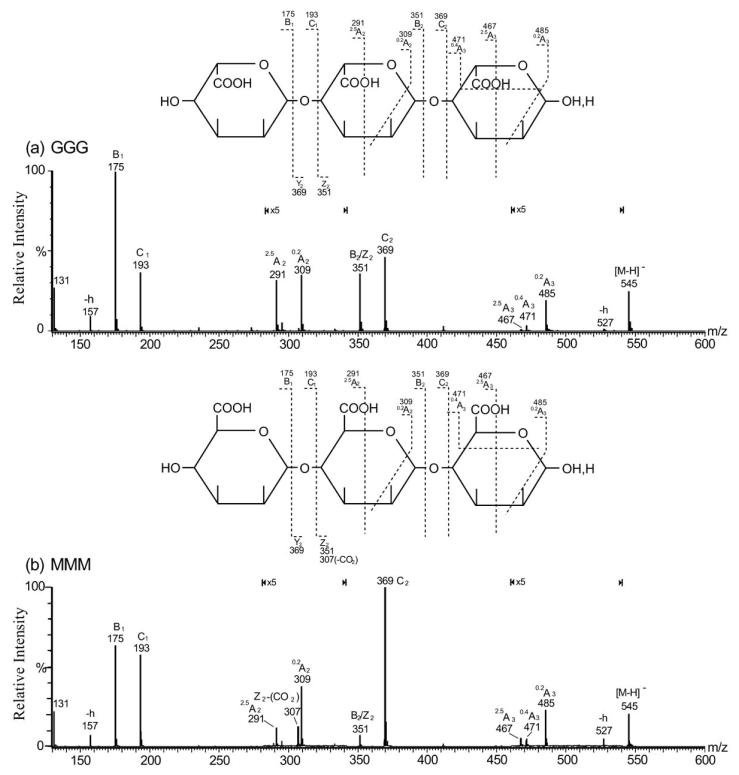
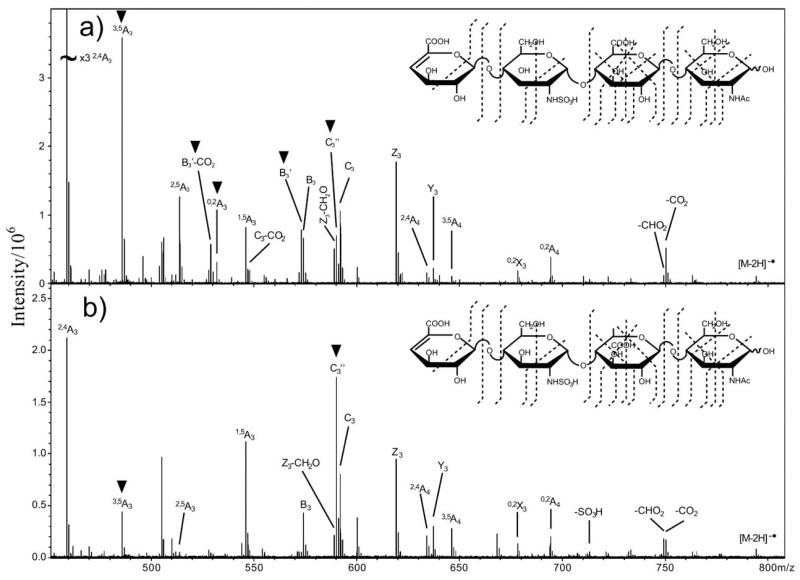
The product-ion spectra of homo-trisaccharides (G3 and M3) (Fig. 1) showed the major ions at m/z 369, 193 and 175. These ions are produced from single glycosidic bond cleavage and can be assigned as C2/Y2, C1/Y1 and B1/Z1, respectively. These fragments are derived from either the reducing or non-reducing termini, indicating linear sequences HexA-HexA-HexA. Weak A-type ions, e.g. 0,2A (m/z 309 and 485; note the 5-fold magnification factor applied to specific regions), 2,5A (m/z 291 and 467) and 0,4A3 ions (m/z 471) are also observed in both spectra.
Fig. (1).
Negative-ion ES-CID-MS/MS product-ion spectra of G3 and M3. (a) G3, (b) M3. Structures are shown to indicate the proposed fragmentation (-h: dehydrated ion). Figure taken with permission from ref. [42].
Non-reducing terminal M and G give identical ions C1 (m/z 193), B1 (m/z 175) together ions at m/z 157 and m/z 131 associated with the dehydration and decarboxylation (Figs. 1a and 1b). Thus it is impossible to differentiate the two residues at a non-reducing terminus. However, identification of the two residues at an internal or reducing terminal position can be made by careful comparison of some specific, albeit weak, fragment ions. An internal M produces a unique ion at m/z 307, whereas an internal G does not afford this ion. The ion m/z 307 can be explained by a decarboxylated Z2(Z2 – CO2). Decarboxylation cannot be used as a criterion to differentiate M/G at either the reducing or non-reducing termini, as decarboxylation is not observed for M at a reducing terminus while a decarboxylated B1 (m/z 131) is observed for both M and G at the non-reducing termini. A reducing terminal M can be assigned by careful comparison of the intensity ratio of two weak reducing terminal ions, 2,5A3 (m/z 467) and 0,4A3 (m/z 471). The higher [2,5A3]/[0,4A3] ratio suggests a reducing terminal M in Fig. (1b), while a lower ratio in Fig. (1a) suggests the presence of reducing terminal G. Hence, it is not unexpected that 0,4Ared fragmentation is more favorable for a reducing terminal G and 2,5Ared fragmentation is more favorable for a reducing terminal M.
In the analysis of large hetero-oligomeric oligosaccharides, the same rules can also be applied. However, for longer oligosaccharide chains, the M residue next to the reducing terminus produces the strongest Zint – CO2 ion, while an internal M next to the non-reducing terminus produces the weakest. Using this method, the sequence of various alginate oligosaccharides, even very large oligomers can be established using ESI-CID-MS/MS.
Different uronic acids (glucuronic and iduronic acid) also occur in glycosaminoglycans (GAGs). Amster and his coworkers [44] used ESI-electron detachment dissociation (EDD)-MS/MS to distinguish the glucuronic (GlcA) and iduronic acid (IdoA) in two pairs of tetrasaccharide epimers of heparan sulfate (ΔUA-GlcNS-GlcA-GlcNAc, ΔUA-GlcNS-IdoA-GlcNAc and ΔUA-GlcNH2-GlcA-GlcNAc, ΔUA-GlcNH2-IdoA-GlcNAc) The diagnostic ions for GlcA are the 0,2A3, [B-H]-. and [B-H-CO2]− product ions. IdoA can be distinguished by the absence of these ions in the EDD mass spectrum. The spectra of ΔUA-GlcNS-GlcA-GlcNAc and ΔUA-GlcNS-IdoA-GlcNAc are shown in Fig. (2).
Fig. (2).
Expansion of the m/z 450-800 region of EDD tandem MS spectra of the [M-2H]2− precursor-ions of HS epimers: (a) ΔUA-GlcNS-GlcA-GlcNAc, (b) ΔUA-GlcNS-IdoA-GlcNAc. Figure taken with permission from ref. [41].
4. SEQUENCE ANALYSIS OF GLYCOSAMINO-GLYCANS-DERIVED SULFATED OLIGOSACCHARIDES
Sulfated polysaccharides and oligosaccharides show a great variety of bioactivities important in physiological and pathophysiological processes [45]. Glycosaminoglycans (GAGs) are a family of structurally complex, polydisperse linear polysaccharides, composed of the disaccharide repeating units. The GAG, heparin, is used as a pharmaceutical agent to control coagulation, and is representative of the bioactivity associated with such complex, and large biomolecules [45-47]. The structural character of GAGs are often determined by disaccharide compositional analysis of the relying on HPLC [48] and CE [49]. Recently MS has been recognized as a powerful technique for both the disaccharide analysis and sequencing of GAGs [50]. The sequence analysis of GAG oligosaccharides by ESI tandem MS is a relatively new approach. The acidity and lability of the sulfo groups, as well as the different linkage type and presence of uronic acid make analysis by mass spectrometry extremely challenge. Recently, much effort has been applied to GAG analysis using ESI tandem MS. ESI is generally recognized as mild ionization method useful for polar compounds and is known to result in less sulfo group bond cleavage than MALDI [51]. Successful ionization and detection of GAG-derived oligosaccharides by ESI tandem MS, including hyaluronan (HA), [52-55] keratan sulfate (KS), [56-58] chondroitin sulfate (CS), [59-65] dermatan sulfate (DS), [66] heparin/heparin sulfate (Hep/HS), [66-73] oligosaccharides have been widely reported.
Hyaluronan (HA)
The chemical structure of hyaluronan was determined in the 1950s in the laboratory of Karl Meyer [74, 75]. HA is a homocopolymer of D-glucuronic acid and D-acetylglucosamine. This disaccharide repeating unit is linked through alternating β-1,4 and β-1,3 glycosidic bonds. HA is structurally simplest member of the GAG family and is unique among the GAGs in the absence of sulfo groups. ESI tandem MS has been performed hyaluronan oligosaccharides containing both an even- and odd- number of saccharide residences [52, 53], and glycosidic fragment ions are observed with single or multiple charges. Since there is no sequence heterogeneity in HA, the structure is confirmed by chain length. Fragment ions can be useful for determining the terminal sugars in the HA oligosaccharides. Fragment ions show difference of m/z=174.8 (GlcA-H2O) or m/z=201.8 (GlcNAc-H2O).
Keratan Sulfate (KS)
Keratan sulfate is a GAG that does not contain an uronic acid. Its disaccharide repeating units is composed of alternating residues of D-Gal and D-GlcNAc with alterative β1-4 and β1-3 linkages. KS has variable chain length and variable degree of substitution with sulfo groups at 6-position of both Gal and GlcNAc [76]. The ESI tandem MS has recently been employed to analyze KS oligosaccharides. Oguma, [57] Zhang [58] and Hirabayashi [56] analyzed KS oligosaccharides using ESI tandem MS, and obtained similar results.
A series of keratan sulfate oligosaccharides were sequenced by Hirabayashi and coworkers [56] using multistage ESI tandem MS. A tetrasaccharide, D-Gal β (1-4) GlcNAc β (6S) β (1-3) Gal (6S) β (1-4) GlcNAc (6S), was applied here as an example. Their fragmentation is listed in Table 1. MS1 shows a triply-charged molecular ion [M-3H]3−. MS2 performed on [M-3H]3− ion obtained in MS1 showed few fragment-ions corresponding to glycosidic cleavages. This is in contrast to the MS2 analysis of CS or HS oligosaccharides (see below). MS2 shows peaks for [0,2A4-3H]3− at 294.6 m/z, [M-H2O-3H]3− at 322.2 m/z, [B3-2H]2− at 342.4 m/z, [C3-2H]2− at 351.4 and [0,2 A4-2H]2− at 372.4 m/z which provide little information on the sequence of the KS oligosaccharide. MS3 was first performed on the [M-H2O-3H] 3− and the spectrum, afforded fragments associated with glycosidic cleavages including [Y1-H2O-OSO3H]− at 183.9 m/z, [Z1-H2O]− at 264.0 m/z, [Y3-H2O-3H]3− at 268.2 m/z, [Y1-H2O]− at 281.7 m/z, [B3-2H]2− at 342.5 m/z, [C3-2H] 2− at 351.4 m/z, and C2 at 461.9 m/z, in which [Y1-H2O-OSO3H]− at 183.9 m/z was the diagnostic ion for the GlcNAc6S at the reducing end. Next MS3 was performed on the 0,2A4 ion, which is the precursor ion produced in MS2. Two interesting features were observed. First, an intense peak corresponding to 2,4A4 ion was observed. This type of reducing end fragment-ion is characteristic of MS3 targeting 0,2A in reducing end. Second, a [OCHCH2OSO3]− ion at 139 m/z, although not very intense, was observed, the appearance of which is indicative of a sulfo group at the reducing end C-6. MS4 was performed on the 2,4A4 ion. Complete distinctive fragmentation was observed in MS4 spectrum consistent with extensive cleavage of glycosidic linkages. This resulted in abundant information on the oligosaccharide sequence. The [Y3/B3-2H]2− at 261.4 m/z, [Y3-2H]2− at 291.4 m/z, Y2 at 300.8 m/z, [0,2X3-2H]2− at 312.3 m/z, [B3-2H]2− at 342.3 m/z, B2 at 443.8 m/z, and [2,4A4-OSO3H]− at 647.9 m/z were present in the spectrum. The Y2 ion was consistent with Hex6S-HexNAc6S sequence at reducing end, B2 ion was consistent with a monosulfated disaccharide sequence at the non-reducing end.
Table 1. Fragmentation of Tetrasaccharide, D-Gal β (1-4) GlcNAc (6S) β (1-3) Gal (6S) β (1-4) GlcNAc (6S) in Multi-Stage Tandem MS. (m/z Values, Charge States and Assignments).
| MW | MS1 | MS2
(on [M-nH]n−) |
MS3 (on [M-H2O-nH]n− |
MS3 (on0,2A4) |
MS4
(on2,4A4) |
|---|---|---|---|---|---|
| 988.2 | 328.1 [M-3H]3− |
294.6 0,2A43− |
183.9 [Yi-H2O-OSO3H]− |
138.8 [OCHCH2OSO3]− |
261.4 Y3/B32− |
| 322.2 [M-H2O-3H]3− |
264.0 [Z1-H2O]− |
372.4 2,4A42− |
291.4 Y32− |
||
| 342.4 B32− |
268.2 [Y3-H2O]3− |
300.8 Y2− |
|||
| 351.4 C32− |
281.7 [Y1-H2O]− |
312.3 0,2X32− |
|||
| 372.4 2,4A42− |
342.5 B32− |
342.3 B32− |
|||
| 351.4 C32− |
443.8 B2− |
||||
| 461.9 C2− |
647.9 [2,4A4-OSO3H]− |
Nine additional KS oligosaccharides were analyzed using this multistage MS process. The fragmentation rules and diagnostic ion m/z values for KS are summarized in the Table 2. Using these fragmentation rules the sequence of KS oligosaccharides can be determined.
Table 2. Summary of the Fragmentation Rules and Diagnostic m/z Values for Keratan Sulfates.
| MSn Stage | Fragmentation Rule | Diagnostic m/z |
|---|---|---|
| MS1 | Molecular ion ([M-nH]n−) is given without considerable loss of sulfate. | |
| The number of sulfate and/or carboxylate (n), as well as molecular weight (M) is determinable. | ||
| MS2 (on [M-nH]n−) | Dehydration ([M-H2O-nH]n−) and 0,2Ar cleavage occur in most oligosaccharides. | |
| The dehydration is ascertained to take place between C-2 and C-3 of reducing GlcNAc. | ||
| Desialylation occurs preferentially in sialylated oligosaccharides. | m/z 290 | |
| MS3 (on [M-H2O- nH]n−) |
[Y1 - H2O - OSO3H]− ion is detected for GlcNAc6S at reducing end. | m/z 184 |
| B1 ion is detected for Gal6S at nonreducing end. | m/z 241 | |
| MS3 (on 0,2Ar) | 2,4Ar ion appears intensely (except for L2 and SL1). | |
| [OCHCH2OSO3]− ion is detected for 6S at reducing end. | m/z 139 | |
| [0,2Ar-H2O]n− ion is detected for Gal-GlcNAc6S (L2) sequence at reducing end. | ||
| MS4 (on 2,4Ar) | Extensive fragment ions resulting from glycosidic cleavages are obtained, which give valuable information for sequence determination. |
|
| B1 ion is detected for GlcNAc6S or Ga16S at nonreducing end. |
m/z 282 (GlcNAc6S) m/z 241 (Gal6S) |
|
| B2 ion is detected for monosulfated disaccharide sequence at nonreducing end. | m/z 444 | |
| Doubly charged B2 ion is detected for disulfated disaccharide sequence at nonreducing end. | m/z 261.5 | |
| Y2 ion is detected for Gal6S-GlcNAc6S (L4) sequence at reducing end. | m/z 301 | |
| [OSO3H]- and/or [2,4Ar-OSO3H]n− ions are detected for Gal6S at any position in the sequence. | m/z 97 |
Chondroitin Sulfate (CS)
Chondroitin sulfate GAGs are unbranched polysaccharides of variable length containing D-GlcA (1-3) D-GalNAc a repeating disaccharide unit. Some GlcA residues are epimerized into L-IdoA in dermatan sulfate also known as CS-B. Each monosaccharide residue in CS may be unsulfated, mono-sulfated, or di-sulfated. Most commonly the hydroxyls of the 4 and 6 positions of the GlcNAc residues contain O-sulfo groups [77-79]. CS-A contains GlcA and most (often 90%) of the GalNAc residue contain minor amount of unsulfated or 6-O-sulfo substituted GalNAc residues. CS-B and CS-C contain IdoA or GlcA, respectively and most (often 90%) of the GalNAc residues contain 4-O-sulfo groups with minor amounts of unsulfated and 6-O-sulfo group containing GlcNAc residues. CS-D, -E, and -K are oversulfated, containing 2-, 6-di-O-sulfo, 4-, 6-di-O-sulfo and 3-, 4-di-O-saulfo group substitution, respectively [80].
The ESI tandem MS was used to confirm the sequence of the CS oligosaccharides with respect to the position of sulfo groups in GalNAc sufation [64, 81]. For example, various collision energies were used to dissociate the precursor molecules in CID cell, and tandem MS was performed on precursor molecules having diverse charge states by Zaia and collaborators [64]. Based on the data obtained from various experiments, the sequence and cleavage mechanism of the CS oligosaccharides analyzed by the ESI tandem MS was established. Unsaturated reduced, unsaturated, and saturated di-, tetra-, hexa-, and octasaccharides of CS-A and CS-C were each investigated.
The fragment ions from a saturated tetrasaccharide (GlcA1-3GalNAc4S1-4GlcA1-3GalNAc4S) were produced at collision energy −10, −20, and −30 V, in which the tandem MS performed on the multiply charged molecular ion m/z 467, [M-2H]2−(Table 3). The data shows increasing complexity of fragmentation as collision energy is decreased. The precursor-ion minimally fragments at −10V collisional energy. At −20V, ions corresponding to Y11−, Y32−, B31−, and precursor are observed in moderate relative abundance. The Y11− ion is the most abundant ion in the spectrum at −30V collision energy, and the precursor-ion is not detected. The unsaturated reduced oligosaccharide was analyzed under identical conditions. The fragment-ions produced by glycosidic cleavage (Y11−, Y32−, B31−) were also observed at the −20V collision energy, although the m/z values shifted by 18 and 2 amu as the result of the double bond at nonreducing termini and alditol at the reducing termini. The best collision energy for the dissociation of the precursor to produce the glycosidic cleavage ions, important for oligosaccharide sequencing is −20V. This collision energy also is useful in locating the sulfo group with in the GlcNAc residue. Ions at m/z 282 and 458 were observed in spectra of both unsaturated reduced and saturated oligosaccharide. Since the former differs from the latter in structure at both reducing and nonreducing termini, they contain neither terminus and result from multiple bond cleavage. These ions are at low abundances between −10 and −20 V collision energy, and their percent of total ion abundance further diminishes between collision energies of −30 and −40V. The ions observed at m/z 282 and 458 in the CID product-ion spectra of CS-A tetrasaccharide correspond to internal product ions. This type of internal fragment ions is useful for confirmation of the linkage configuration in sequencing [13, 35, 54]. Internal cleavages are observed with the appearance of the residues of -3GlcNAc in the tandem MS spectra of the multiply charged molecular ions. Thus, ions at m/z 458 first observed at collision energy of −20V correspond to fragment ions of D1-3, which undergoes further fragmentation. Higher collision energies increase in the percent total ion abundance observed for the ions corresponding to 2821− (D1-2), Y1, and B1. The same fragmentation was observed in the analysis of tetrasaccharides of CS-C, hexa- and octasaccharides of CS-A and CS-C. Glycosidic cleavage fragment ions Bn, Cn, Yn, and Zn (where the n=odd), were produced of CID on multiply charged molecular ions.
Table 3. Fragmentations of Saturated Tetrasaccharides of CS-A.
| m/z | MS/MS (on doubly charged precursor) |
MS/MS (on singly charged precursor) |
|---|---|---|
| Y11− | 300 | |
| Y21− | 476 | |
| Y32− | 379 | |
| B21− | 458 | |
| B31− | 654 | |
| D1-2 | 282 | |
| D1-3 | 458 | |
| [B2+SO3]1− | 538 | |
| [Y2+SO3]1− | 556 | |
| [M-SO3-H]1− | 855 | |
| [M-SO3-H2O-H]1− | 837 |
ESI tandem MS was also performed on the singly charged ions of this oligosaccharides of CS (Table 3). The product-ion mass spectrum of the saturated tetramer of CS-A shows abundant ions corresponding to [M-H-SO3]−, Y2, and B2. Other ions include sulfate-adducted Y2 and B2, but these ions are observed at reduced abundance. The identity of ions corresponding to glycosidic cleavage was also confirmed from the product-ions of corresponding alditols and unsaturated oligosacchardes. Differences of product-ion patterns for singly charged ions and doubly charged ions were observed. CID performed on singly charged precursor and produced a much greater abundance of ions produced from loss of SO3 or H2SO4 loss from the precursor ion, and formation of B2 and Y2 ions in place of odd numbered Bn and Yn ions, and the generation of product ions containing an additionally adducted sulfo group. Differences are also observed in the product-ion spectra of hexa- and octasaccharides. These differences imply the singly- and multiply-charged ions have significantly different conformations, producing different fragmentation patterns. The multiply-charged ion is expected to adopt an extended conformation keeping its charges separated. A more compact conformation is possible for singly-charged ions. The MS data are consistent with transfer of a protonated sulfo group to an oxygen atom elsewhere in the molecule prior to (or concurrent with) glycosidic bond cleavage [57].
When CID was performed on the multiply-charged ions of the CS oligosaccharide, internal cleavage ions are observed. These confirm the presence of a 1-3 linkage. Odd numbered, glycosidic cleavage ions are observed where the charge equals number of sulfo group. When CID was performed on the singly-charged ions of the CS oligosaccharides, the even numbered glycosidic cleavage ions are observed in addition to those with odd numbered ions. Thus, the sequence of the CS oligosaccharides could be confirmed with the ESI tandem MS.
Heparin and Heparan Sulfate (Hep/HS)
Heparin and heparan sulfate are two closely related GAGs and are the most structurally complex members of this family of molecules. The main disaccharide unit that occurs in heparin is composed of a 2-O-sulfo IdoA and 6-O-sulfo, N-sulfo GlcN linked in alternatively α 1→4 linkage. Minor sequences contain GlcA, a reduced number of sulfo groups as well as N-acetyl groups. The main disaccharide unit in heparan sulfate is composed of GlcA 1→4 linked to GlcNAc, this disaccharide typically making up approximately 50% of the total disaccharide units in heparan sulfate [82, 83].
Tandem mass spectra are applied to afford patterns of glycosidic bond and cross-ring cleavages from which saccharide sequence can be deduced. For example, multi-stage ESI tandem mass spectrometric analysis was performed on a series of heparin-like glycosaminoglycans [84], in which abundant glycosidic bond cleavages result, despite the high degree of sulfation. CID production mass spectra generated from the multiply charged molecular ion peaks, those combined with sodium cations and those combined with calcium cations in a tetrasulfated trisaccharide, a pentasulfated tetrasaccharide, and an octasulfated pentasaccharide result in abundant ions corresponding to glycosidic bond even cross-ring cleavage. These spectral data demonstrate that abundant glycosidic bond cleavages can be obtained, provided that most of the sulfo groups are charged. Repulsions from such charges are likely to destabilize glycosidic bond, and to make their rupture energetically favorable when compared loss of SO3 from the precursor ion. The nomenclature used assumes the protons are displaced with metal cations, and only the loss of protons confers charge on the ion. More sequence information is produced from the CID mass spectra of metal combined precursor ions than from ions without metal cations. This sequence information takes the form of glycosidic-bond and cross-ring cleavages. The cross-ring fragment ions 0,2A1 and 0,4X4 (2Ca) are observed in the tandem mass spectrum of [M (3Ca)—4H] 4−, as well as the glycosidic fragment ions were observed in both of [M (3Ca)—4H]4− and [M–6H]6− of the pentasaccharide. Furthermore, it was observed that lability of sulfo groups follow the order SO3− > SO3Na > SO3 (1/2) Ca+. Pairing the sulfated oligosaccharide anions with metal cations serve to increase sulfo group stability so that abundant backbone cleavage fragments are observed, allowing more complete sequence related data to be obtained in a single MS stage. This result has also confirmed in the MS analysis of sulfated peptides [85].
In addition to the direct analysis of oligosaccharides, the composition analysis of disaccharides units was an effective strategy for the characterization of the structure of GAGs. Recently the 8 disaccharide units of Hep/HS were distinguished quantitatively by Leary and coworkers [71, 72, 86] using ESI tandem MS. The 8 disaccharide standards were listed in the Table 4, include nonsulfated, monosufated (O- or N-), disulfated (O- and O- or N-), and trisulfated (O-, O- and N-), saccharide were distinguished by ESI-MS based on the differences in their molecular mass. The O-monosulfo isomers (ΔUA-GlcNAc6S and ΔUA2S-GlcNAc) and O, N-disulfo isomers (ΔUA2S-GlcNS and ΔUA2S-GlcNS6S) were differentiated by tandem MS using production spectra generated by MS2. Because the sulfo group can be located on the either GlcA/IduA or GlcN moieties, the glycosidic fragment ion m/z 237.0 was observed in the MS/MS spectrum of the ΔUA2S-GlcNAc as a diagnostic ion, as well as 0,2 A2 at m/z 357.1 was used as the diagnostic ion for ΔUA-GlcNAc6S. The 0,2 A2 ion is observed in the MS/MS spectra of both ΔUA-GlcNAc6S and ΔUA2S-GlcNAc isomers, the relative abundance of the ion differs substantially and reproductively for each isomers (Table 5). ΔUA2S-GlcNS and ΔUA2S-GlcNS6S were differentiated by the selected diagnostic ions of 0,4A2 and 0,2A2-H2O at m/z 297.1 and 339.1 in the MS3 spectra of 0,2 A2 at m/z 357.1, respectively (Table 5). The roles and mechanisms of dissociation for isomeric heparin disaccharides was confirmed using isotope labeling and ion-trap tandem mass spectrometry [51]. Quantitative analysis were performed by calculation of their contribution in the relative intensity of the diagnostic fragment ions in the total ion current (TIC) of the disaccharide mixtures [50]. The quantitative analysis of nonisomeric disaccharides was done with an internal standard ΔUA2S-GlcNCOEt6S, which is a semi-synthetic analogue of the heparin disaccharides with different molecular weight, not found in heparin or heparan sulfate. The linearity of an equal molar mixture of the 8 disaccharides ranged from 1 to 100 pmol. The response factor ( R ) for each disaccharide was calculated at each concentration of disaccharide and found to be constant over the range from 1 to 100 pmol/L. The disaccharide mixture with different concentrations of each disaccharide could be quantitatively analyzed by MS with the internal standard ΔUA2S-GlcNCOEt6S and tandem MS was used to distinguish the isomers.
Table 4. Chemical Structures of the Eight Most Prevalent, Biologically Relevant Heparin/Heparan Sulfate Disaccharides Produced by the Action of Heparin Lyases I, II, and III (A-H) and Disaccharide I-P, the Internal Standard Used for Quantification.
| Disaccharides | m/z | |
|---|---|---|
| A | ΔUA→GlcNAc | 378.1 |
| B | ΔUA2S→GlcNAc | 458.1 |
| C | ΔUA→GlcNAc6S | 458.1 |
| D | ΔUA→GlcNS | 416.1 207.7 |
| E | ΔUA2S→GlcNAc6S | 268.7 |
| F | ΔUA2S→GlcNS | 247.7 |
| G | ΔUA→GlcNS6S | 247.7 |
| H | ΔUA2S→GlcNS6S | 191.5 287.6 |
| I | ΔUA2S→GlcNCOEt6S | 275.7 |
Table 5. Reproducibility of Contributions for Disaccharide Standards from MS2 and MS3 Spectra.
| % Ion Contribution | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CID | Dimer | m/z | 1 | 2 | 3 | 4 | 5 | 6 | Av | SD |
| MS2 | ΔUA2S-GlcNAc | 237.0 | 22.84 | 21.03 | 22.29 | 21.03 | 21.61 | 21.84 | 21.77 | 0.71 |
| 357.1 | 1.92 | 2.13 | 2.87 | 2.03 | 2.10 | 2.11 | 2.19 | 0.34 | ||
| ΔUA-GlcNAc6S | 237.0 | 0.02 | 0.02 | 0.01 | 0.02 | 0.02 | 0.02 | 0.02 | 0.00 | |
| 357.1 | 23.07 | 23.60 | 23.31 | 23.57 | 23.32 | 24.06 | 23.49 | 0.34 | ||
| MS3 | ΔUA2S-GlcNS | 297.1 | 29.60 | 30.21 | 31.08 | 30.40 | 29.12 | 31.04 | 30.24 | 0.78 |
| 339.1 | 9.38 | 10.11 | 9.30 | 11.03 | 8.88 | 9.49 | 9.70 | 0.76 | ||
| ΔUA-GlcNS6S | 297.1 | 0.00 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | |
| 339.1 | 62.29 | 63.41 | 63.24 | 63.10 | 62.65 | 62.47 | 62.86 | 0.45 | ||
Quantitative analysis of disaccharide composition of Hep/HS is an effective method in partially sequencing. In conjunction with ESI tandem MS analysis of Hep/HS oligosaccharides an effective strategy can be designed for sequencing. Software, known as HOST (heparin oligosaccharide sequencing tool), was developed to analyzed sequence [86]. Each Hep/HS oligosaccharide can be subjected to two experiments. One is digestion and analysis of disaccharide composition; another is ESI-MS and MSn analysis directly without digestion. The disaccharide compositional analysis of the heparin oligosaccharide provides the identities of the building block constituents of the saccharide, as well as their relative ratios. From the tandem MS experiments on the heparin oligosaccharide, a wealth of information might be obtained. The main types of dissociations observed in the product-ion spectra were neutral loss of a small molecule (SO3, H2O, CO2), glycosidic cleavage (B, C, Y, Z-type), and cross-ring cleavage (0,2A, X-type). The productions generated by these cleavages can provide information on sequence and identity of positions of the sulfo groups. Based on the two experiments, the program (HOST) was applied to integrate the information obtained for the sequence analysis of heparin tetra-, and hexa-saccharide. HOST lists all possible structures generating a scoring system that puts the more likely structures at the top of the list and relates subsequent structural sequences to the best match of the series [71]. The sequences of Hep/HS were determined by enzymatic digestion, followed by ESI-MS and MSn analysis of the in tact oligosaccharide species. The information obtained was integrated using the HOST.
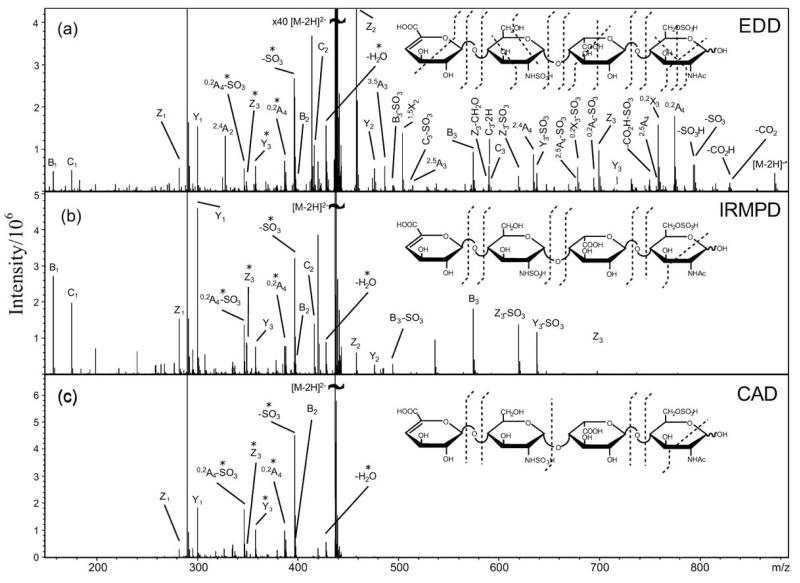
Most recently, EDD has been applied to analyze the sequence of HS oligosaccharides, such as distinguishing GlcA and IdoA in the oligosaccharides of HS [44]. In addition, Amster and coworkers [87] used EDD to sequence four tetrasaccharide (ΔUA-GlcNH2-GlcA-GlcNAc, ΔUA-GlcNH-Ac-GlcA-GlcNAc, ΔUA-GlcNS-GlcA-GlcNAc, and ΔUA-GlcNS-IdoA-GlcNAc6S) of HS determining both substituent position, and epimerized residues. Compared to collisionally activated dissociation (CAD) and infrared multiphoton dissociation (IRMPD) MS, EDD provides improved cross-ring fragmentation important for determining the pattern of sulfation, acetylation, and hexuronic acid stereochemistry on a GAG oligosaccharide. The MS/MS spectrum on [M-2H]− precursor ion of UA-GlcNS-IdoA-GlcNAc6S is showed in Fig. (3). In this spectrum, predominantly singly charged even-electron product ions are observed. Some doubly charged product-ions are observed such as [M-2H-SO3]2− and 0,2A42−, as well as the Y32− and Z32− glycosidic cleavages. Some product-ions are observed both as product-ions with two sulfo groups as well as with the loss of one molecule of SO3. For example, the Z3 and Y3 product ions are also observed as Z3-SO3 and Y3-SO3. This EDD spectrum exhibits peaks that result from cleavage of all glycosidic bonds as well as from abundant cross-ring cleavages. The cross-ring fragmentation of the reducing end sugar places the sulfate on C6 and identifies this sugar as GlcNAc6S. Glycosidic fragmentation identifies the sugar next to the nonreducing end as a sulfated GlcN residue. The cross-ring fragmentation places the sulfate on either the C2 amino group or C6. The IdoA residue was identified by absence of 0,2A3, [B3-H]− and [B3-H-CO2]− [44]. Based on the extensive fragmentation observed in EDD, these complicated sulfated oligosaccharides were sequenced unambiguously.
Fig. (3).
Tandem mass spectra of the [M-2H]2− precursor ion of tetrasaccharide (ΔUA-GlcNS-IdoA-GlcNAc6S), obtained by using (a) EDD, (b) IRMPD, and (c) CAD. Figure taken with permission from ref. [84].
CONCLUSIONS
Although the analysis of complex oligosaccharides, even polysaccharides remains a challenge, recent researches to neutral, uronic acid and sulfated carbohydrate by ESI tandem MS in negative-mode suggests one possible approach. Multiply charged ions observed in the spectra of structures with highly charged species. ESI MS can complicate interpretation. But these ions also can afford many glycosidic and cross-ring cleavage fragment-ions, which contribute to the sequence analysis. Furthermore, multiply-charged ions can be utilized as precursor-ions for MS/MS analysis to successfully produce new diagnostic ions. Although the carbohydrates have different linkages and substitutes, their sequence and structure can often be confirmed based on glycosidic and cross-ring fragment-ions produced in ESI tandem MS. In these applications, ESI tandem MS in negative-mode represents a powerful, effective and sensitive technique to analyze and sequence neutral and acid, branched and linear chains, sulfated and unsulfated oligosaccharides. Using ESI tandem MS directly without carbohydrate derivatization of makes sequencing approach rapid and require only small amounts of sample. Recently, more advanced fragmentation methods such as EDD have been shown to provide complete sequence information including information on saccharide chirality. The flexibility of the methods described should facilitate the development of a fully automated sequencing methodology. Once it is possible to quickly establish the sequence of a carbohydrate, dramatic progress will be made in correlating carbohydrate structure to the biological functions.
REFERENCES
- [1].Baldwin MA. Mass spectrometers for the analysis of biomolecules. Methods Enzymol. 2005;402:3–48. doi: 10.1016/S0076-6879(05)02001-X. [DOI] [PubMed] [Google Scholar]
- [2].Griffiths WJ, Jonsson AP, Liu S, Rai DK, Wang Y. Electrospray and tandem mass spectrometry in biochemistry. Biochem. J. 2001;355(Pt 3):545–561. doi: 10.1042/bj3550545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 1988;60(20):2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- [4].Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246(4926):64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- [5].Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization-principles and practice. Mass Spectrom. Rev. 1990;9:37–70. [Google Scholar]
- [6].Mclafferty FWE. Tandem mass spectrometry. Wiley; 1983. [Google Scholar]
- [7].Wilm M, Mann M. Analytical properties of the nanoelectrospray ion source. Anal. Chem. 1996;68(1):1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- [8].Barcelo-Barrachina E, Moyano E, Galceran MT. State-of-the-art of the hyphenation of capillary electrochromatography with mass spectrometry. Electrophoresis. 2004;25(13):1927–1948. doi: 10.1002/elps.200305908. [DOI] [PubMed] [Google Scholar]
- [9].Harvey DJ. Fragmentation of negative ions from carbohydrates: part 3. Fragmentation of hybrid and complex N-linked glycans. J. Am. Soc. Mass Spectrom. 2005;16(5):647–659. doi: 10.1016/j.jasms.2005.01.006. [DOI] [PubMed] [Google Scholar]
- [10].Harvey DJ. Fragmentation of negative ions from carbohydrates: part 2. Fragmentation of high-mannose N-linked glycans. J. Am. Soc. Mass Spectrom. 2005;16(5):631–646. doi: 10.1016/j.jasms.2005.01.005. [DOI] [PubMed] [Google Scholar]
- [11].Harvey DJ. Fragmentation of negative ions from carbohydrates: part 1. Use of nitrate and other anionic adducts for the production of negative ion electrospray spectra from N-linked carbohydrates. J. Am. Soc. Mass Spectrom. 2005;16(5):622–630. doi: 10.1016/j.jasms.2005.01.004. [DOI] [PubMed] [Google Scholar]
- [12].Domon B, Costello CE. Oligosaccharide fragmentation nomenclature. Glycoconjug. J. 1988;5:397–409. [Google Scholar]
- [13].Chai W, Piskarev V, Lawson AM. Negative-ion electrospray mass spectrometry of neutral underivatized oligosaccharides. Anal. Chem. 2001;73(3):651–657. doi: 10.1021/ac0010126. [DOI] [PubMed] [Google Scholar]
- [14].Johnson RS, Martin SA, Biemann K, Stults JT, Watson JT. Novel fragmentation process of peptides by collision-induced decomposition in a tandem mass spectrometer: differentiation of leucine and isoleucine. Anal. Chem. 1987;59(21):2621–2625. doi: 10.1021/ac00148a019. [DOI] [PubMed] [Google Scholar]
- [15].Mann M, Wilm M. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem. 1994;66(24):4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- [16].Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- [17].Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379(6564):466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- [18].Griffiths WJ, Yang Y, Lindgren JÅ, Sjövall J. Electrospray-collision-induced dissociation mass spectrometry of mono-, di- and tri-hydroxylated lipoxygenase products, including leukotrienes of the B-series and lipoxins. Rapid Commun. Mass Spectrom. 1996;10(2):183–196. doi: 10.1002/(SICI)1097-0231(19960131)10:2<183::AID-RCM456>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- [19].Kerwin JL, Wiens AM, Ericsson LH. Identification of fatty acids by electrospray mass spectrometry and tandem mass spectrometry. J. Mass Spectrom. 1996;31(2):184–192. doi: 10.1002/(SICI)1096-9888(199602)31:2<184::AID-JMS283>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- [20].Jensen NJ, Tomer KB, Gross ML. FAB MS/MS for phosphatidylinositol, -glycerol, -ethanolamine and other complex phospholipids. Lipids. 1987;22(7):480–489. doi: 10.1007/BF02540363. [DOI] [PubMed] [Google Scholar]
- [21].Wysocki SJ, French NP, Grauaug A. Organic aciduria associated with isovaleric acidemia. Clin. Chem. 1983;29(5):1002–1003. [PubMed] [Google Scholar]
- [22].Beck JL, Colgrave ML, Ralph SF, Sheil MM. Electrospray ionization mass spectrometry of oligonucleotide complexes with drugs, metals, and proteins. Mass Spectrom. Rev. 2001;20(2):61–87. doi: 10.1002/mas.1003. [DOI] [PubMed] [Google Scholar]
- [23].Huber CG, Oberacher H. Analysis of nucleic acids by on-line liquid chromatography-mass spectrometry. Mass Spectrom. Rev. 2001;20(5):310–343. doi: 10.1002/mas.10011. [DOI] [PubMed] [Google Scholar]
- [24].Meng Z, Simmons-Willis TA, Limbach PA. The use of mass spectrometry in genomics. Biomol. Eng. 2004;21(1):1–13. doi: 10.1016/j.bioeng.2003.08.001. [DOI] [PubMed] [Google Scholar]
- [25].Ni J, Pomerantz C, Rozenski J, Zhang Y, McCloskey JA. Interpretation of oligonucleotide mass spectra for determination of sequence using electrospray ionization and tandem mass spectrometry. Anal. Chem. 1996;68(13):1989–1999. doi: 10.1021/ac960270t. [DOI] [PubMed] [Google Scholar]
- [26].Reinhold VN, Reinhold BB, Costello CE. Carbohydrate molecular weight profiling, sequence, linkage, and branching data: ES-MS and CID. Anal. Chem. 1995;67(11):1772–1784. doi: 10.1021/ac00107a005. [DOI] [PubMed] [Google Scholar]
- [27].Viseux N, de Hoffmann E, Domon B. Structural analysis of permethylated oligosaccharides by electrospray tandem mass spectrometry. Anal. Chem. 1997;69(16):3193–3198. doi: 10.1021/ac961285u. [DOI] [PubMed] [Google Scholar]
- [28].Weiskopf AS, Vouros P, Harvey DJ. Characterization of oligosaccharide composition and structure by quadrupole ion trap mass spectrometry. Rapid Commun. Mass Spectrom. 1997;11(14):1493–1504. doi: 10.1002/(SICI)1097-0231(199709)11:14<1493::AID-RCM40>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- [29].Weiskopf AS, Vouros P, Harvey DJ. Electrospray ionizationion trap mass spectrometry for structural analysis of complex N-linked glycoprotein oligosaccharides. Anal. Chem. 1998;70(20):4441–4447. doi: 10.1021/ac980289r. [DOI] [PubMed] [Google Scholar]
- [30].Ahn YH, Yoo JS. Malononitrile as a new derivatizing reagent for high-sensitivity analysis of oligosaccharides by electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 1998;12(24):2011–2015. doi: 10.1002/(SICI)1097-0231(19981230)12:24<2011::AID-RCM429>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- [31].Charlwood J, Langridge J, Tolson D, Birrell H, Camilleri P. Profiling of 2-aminoacridone derivatised glycans by electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 1999;13(2):107–112. doi: 10.1002/(SICI)1097-0231(19990130)13:2<107::AID-RCM463>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- [32].Li DT, Her GR. Structural analysis of chromophore-labeled disaccharides and oligosaccharides by electrospray ionization mass spectrometry and high-performance liquid chromatography/electrospray ionization mass spectrometry. J. Mass Spectrom. 1998;33(7):644–652. doi: 10.1002/(SICI)1096-9888(199807)33:7<644::AID-JMS667>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- [33].Shen X, Perreault H. Electrospray ionization mass spectrometry of 1-phenyl-3-methyl-5-pyrazolone derivatives of neutral and N-acetylated oligosaccharides. J. Mass Spectrom. 1999;34(5):502–510. doi: 10.1002/(SICI)1096-9888(199905)34:5<502::AID-JMS800>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- [34].Yoshino K, Takao T, Murata H, Shimonishi Y. Use of the derivatizing agent 4-aminobenzoic acid 2-(diethylamino)ethyl ester for high-sensitivity detection of oligosaccharides by electrospray ionization mass spectrometry. Anal. Chem. 1995;67(21):4028–4031. doi: 10.1021/ac00117a034. [DOI] [PubMed] [Google Scholar]
- [35].Chai W, Piskarev V, Lawson AM. Branching pattern and sequence analysis of underivatized oligosaccharides by combined MS/MS of singly and doubly charged molecular ions in negativeion electrospray mass spectrometry. J. Am. Soc. Mass Spectrom. 2002;13(6):670–679. doi: 10.1016/S1044-0305(02)00363-X. [DOI] [PubMed] [Google Scholar]
- [36].Pfenninger A, Karas M, Finke B, Stahl B. Structural analysis of underivatized neutral human milk oligosaccharides in the negative ion mode by nano-electrospray MS(n) (part 1: methodology) J. Am. Soc. Mass Spectrom. 2002;13(11):1331–1340. doi: 10.1016/S1044-0305(02)00645-1. [DOI] [PubMed] [Google Scholar]
- [37].Pfenninger A, Karas M, Finke B, Stahl B. Structural analysis of underivatized neutral human milk oligosaccharides in the negative ion mode by nano-electrospray MS(n) (part 2: application to isomeric mixtures) J. Am. Soc. Mass Spectrom. 2002;13(11):1341–1348. doi: 10.1016/S1044-0305(02)00646-3. [DOI] [PubMed] [Google Scholar]
- [38].Quemener B, Desire C, Debrauwer L, Rathahao E. Structural characterization by both positive and negative electrospray ion trap mass spectrometry of oligogalacturonates purified by high-performance anion-exchange chromatography. J. Chromatogr. A. 2003;984(2):185–194. doi: 10.1016/s0021-9673(02)01729-6. [DOI] [PubMed] [Google Scholar]
- [39].Zhang Z, Yu G, Zhao X, Liu H, Guan H, Lawson AM, Chai W. Sequence analysis of alginate-derived oligosaccharides by negative-ion electrospray tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2006;17(4):621–630. doi: 10.1016/j.jasms.2006.01.002. [DOI] [PubMed] [Google Scholar]
- [40].Asam MR, Ray KL, Glish GL. Collision-induced signal enhancement: a method to increase product ion intensities in MS/MS and MSn experiments. Anal. Chem. 1998;70(9):1831–1837. doi: 10.1021/ac971142n. [DOI] [PubMed] [Google Scholar]
- [41].Gaucher SP, Morrow J, Leary JA. STAT: a saccharide topology analysis tool used in combination with tandem mass spectrometry. Anal. Chem. 2000;72(11):2331–2336. doi: 10.1021/ac000096f. [DOI] [PubMed] [Google Scholar]
- [42].Haug A, Larsen B. Biosynthesis of alginate. Epimerisation of D-mannuronic to L-guluronic acid residues in the polymer chain. Biochim. Biophys. Acta. 1969;192(3):557–559. doi: 10.1016/0304-4165(69)90414-0. [DOI] [PubMed] [Google Scholar]
- [43].Haug A, Larsen B, Smidsrod O. A study of the constitution of alginic acid by partial acid hydrolysis. Acta Chem. Scand. 1966;20:183–190. [Google Scholar]
- [44].Wolff JJ, Chi L, Linhardt RJ, Amster IJ. Distinguishing glucuronic from iduronic acid in glycosaminoglycan tetrasaccharides by using electron detachment dissociation. Anal. Chem. 2007;79(5):2015–2022. doi: 10.1021/ac061636x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Capila I, Linhardt RJ. Heparin-protein interactions. Angew. Chem. Int. Ed. Engl. 2002;41(3):391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- [46].Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- [47].Wu ZL, Zhang L, Beeler DL, Kuberan B, Rosenberg RD. A new strategy for defining critical functional groups on heparan sulfate. FASEB J. 2002;16(6):539–545. doi: 10.1096/fj.01-0807com. [DOI] [PubMed] [Google Scholar]
- [48].Kim YS, Liu J, Han XJ, Pervin A, Linhardt RJ. Analysis of fluorescently labeled sugars by reversed-phase ion-pairing high-performance liquid chromatography. J. Chromatogr. Sci. 1995;33(4):162–167. doi: 10.1093/chromsci/33.4.162. [DOI] [PubMed] [Google Scholar]
- [49].Ampofo SA, Wang HM, Linhardt RJ. Disaccharide compositional analysis of heparin and heparan sulfate using capillary zone electrophoresis. Anal. Biochem. 1991;199(2):249–255. doi: 10.1016/0003-2697(91)90098-e. [DOI] [PubMed] [Google Scholar]
- [50].Chi L, Amster J, Linhardt RJ. Mass spectrometry for the analysis of highly charged sulfated carbohydrates. Current Anal. Chem. 2005;1:223–240. doi: 10.2174/157341105774573929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zaia J. Mass spectrometry of oligosaccharides. Mass Spectrom. Rev. 2004;23(3):161–227. doi: 10.1002/mas.10073. [DOI] [PubMed] [Google Scholar]
- [52].Kuhn AV, Raith K, Sauerland V, Neubert RH. Quantification of hyaluronic acid fragments in pharmaceutical formulations using LC-ESI-MS. J. Pharm. Biomed. Anal. 2003;30(5):1531–1537. doi: 10.1016/s0731-7085(02)00544-7. [DOI] [PubMed] [Google Scholar]
- [53].Kuhn AV, Ruttinger HH, Neubert RH, Raith K. Identification of hyaluronic acid oligosaccharides by direct coupling of capillary electrophoresis with electrospray ion trap mass spectrometry. Rapid Commun. Mass Spectrom. 2003;17(6):576–582. doi: 10.1002/rcm.950. [DOI] [PubMed] [Google Scholar]
- [54].Mahoney DJ, Aplin RT, Calabro A, Hascall VC, Day AJ. Novel methods for the preparation and characterization of hyaluronan oligosaccharides of defined length. Glycobiology. 2001;11(12):1025–1033. doi: 10.1093/glycob/11.12.1025. [DOI] [PubMed] [Google Scholar]
- [55].Tawada A, Masa T, Oonuki Y, Watanabe A, Matsuzaki Y, Asari A. Large-scale preparation, purification, and characterization of hyaluronan oligosaccharides from 4-mers to 52-mers. Glycobiology. 2002;12(7):421–426. doi: 10.1093/glycob/cwf048. [DOI] [PubMed] [Google Scholar]
- [56].Minamisawa T, Suzuki K, Hirabayashi J. Multistage mass spectrometric sequencing of keratan sulfate-related oligosaccharides. Anal. Chem. 2006;78(3):891–900. doi: 10.1021/ac051359e. [DOI] [PubMed] [Google Scholar]
- [57].Oguma T, Toyoda H, Toida T, Imanari T. Analytical method for keratan sulfates by high-performance liquid chromatography/turbo-ionspray tandem mass spectrometry. Anal. Biochem. 2001;290(1):68–73. doi: 10.1006/abio.2000.4940. [DOI] [PubMed] [Google Scholar]
- [58].Zhang Y, Kariya Y, Conrad AH, Tasheva ES, Conrad GW. Analysis of keratan sulfate oligosaccharides by electrospray ionization tandem mass spectrometry. Anal. Chem. 2005;77(3):902–910. doi: 10.1021/ac040074j. [DOI] [PubMed] [Google Scholar]
- [59].Desaire H, Leary JA. Detection and quantification of the sulfated disaccharides in chondroitin sulfate by electrospray tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2000;11(10):916–920. doi: 10.1016/S1044-0305(00)00168-9. [DOI] [PubMed] [Google Scholar]
- [60].Desaire H, Sirich TL, Leary JA. Evidence of block and randomly sequenced chondroitin polysaccharides: sequential enzymatic digestion and quantification using ion trap tandem mass spectrometry. Anal. Chem. 2001;73(15):3513–3520. doi: 10.1021/ac010385j. [DOI] [PubMed] [Google Scholar]
- [61].McClellan JE, Costello CE, O’Connor PB, Zaia J. Influence of charge state on product ion mass spectra and the determination of 4S/6S sulfation sequence of chondroitin sulfate oligosaccharides. Anal. Chem. 2002;74(15):3760–3771. doi: 10.1021/ac025506+. [DOI] [PubMed] [Google Scholar]
- [62].Zaia J, Costello CE. Compositional analysis of glycosaminoglycans by electrospray mass spectrometry. Anal. Chem. 2001;73(2):233–239. doi: 10.1021/ac000777a. [DOI] [PubMed] [Google Scholar]
- [63].Zaia J, Li XQ, Chan SY, Costello CE. Tandem mass spectrometric strategies for determination of sulfation positions and uronic acid epimerization in chondroitin sulfate oligosaccharides. J. Am. Soc. Mass Spectrom. 2003;14(11):1270–1281. doi: 10.1016/S1044-0305(03)00541-5. [DOI] [PubMed] [Google Scholar]
- [64].Zaia J, McClellan JE, Costello CE. Tandem mass spectrometric determination of the 4S/6S sulfation sequence in chondroitin sulfate oligosaccharides. Anal. Chem. 2001;73(24):6030–6039. doi: 10.1021/ac015577t. [DOI] [PubMed] [Google Scholar]
- [65].Zamfir A, Seidler DG, Schonherr E, Kresse H, Peter-Katalinic J. On-line sheathless capillary electrophoresis/nanoelectrospray ionization-tandem mass spectrometry for the analysis of glycosaminoglycan oligosaccharides. Electrophoresis. 2004;25(13):2010–2016. doi: 10.1002/elps.200405925. [DOI] [PubMed] [Google Scholar]
- [66].Yang HO, Gunay NS, Toida T, Kuberan B, Yu G, Kim YS, Linhardt RJ. Preparation and structural determination of dermatan sulfate-derived oligosaccharides. Glycobiology. 2000;10(10):1033–1039. doi: 10.1093/glycob/10.10.1033. [DOI] [PubMed] [Google Scholar]
- [67].Chai W, Luo J, Lim CK, Lawson AM. Characterization of heparin oligosaccharide mixtures as ammonium salts using electrospray mass spectrometry. Anal. Chem. 1998;70(10):2060–2066. doi: 10.1021/ac9712761. [DOI] [PubMed] [Google Scholar]
- [68].Henriksen J, Ringborg LH, Roepstorrf P. On-line size-exclusion chromatography/mass spectrometry of low molecular mass heparin. J. Mass Spectrom. 2004;39(11):1305–1312. doi: 10.1002/jms.723. [DOI] [PubMed] [Google Scholar]
- [69].Pope RM, Raska CS, Thorp SC, Liu J. Analysis of heparan sulfate oligosaccharides by nano-electrospray ionization mass spectrometry. Glycobiology. 2001;11(6):505–513. doi: 10.1093/glycob/11.6.505. [DOI] [PubMed] [Google Scholar]
- [70].Ruiz-Calero V, Moyano E, Puignou L, Galceran MT. Pressure-assisted capillary electrophoresis-electrospray ion trap mass spectrometry for the analysis of heparin depolymerised disaccharides. J. Chromatogr. A. 2001;914(1-2):277–291. doi: 10.1016/s0021-9673(00)01181-x. [DOI] [PubMed] [Google Scholar]
- [71].Saad OM, Leary JA. Compositional analysis and quantification of heparin and heparan sulfate by electrospray ionization ion trap mass spectrometry. Anal. Chem. 2003;75(13):2985–2995. doi: 10.1021/ac0340455. [DOI] [PubMed] [Google Scholar]
- [72].Saad OM, Leary JA. Delineating mechanisms of dissociation for isomeric heparin disaccharides using isotope labeling and ion trap tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2004;15(9):1274–1286. doi: 10.1016/j.jasms.2004.05.008. [DOI] [PubMed] [Google Scholar]
- [73].Thanawiroon C, Rice KG, Toida T, Linhardt RJ. Liquid chromatography/mass spectrometry sequencing approach for highly sulfated heparin-derived oligosaccharides. J. Biol. Chem. 2004;279(4):2608–2615. doi: 10.1074/jbc.M304772200. [DOI] [PubMed] [Google Scholar]
- [74].Rapport MM, Linker A, Meyer K. The hydrolysis of hyaluronic acid by pneumococcal hyaluronidase. J. Biol. Chem. 1951;192(1):283–291. [PubMed] [Google Scholar]
- [75].Weissmann B, Meyer K, Sampson P, Linker A. Isolation of oligosaccharides enzymatically produced from hyaluronic acid. J. Biol. Chem. 1954;208(1):417–429. [PubMed] [Google Scholar]
- [76].Hopwood JJ, Robinson HC. The structure and composition of cartilage keratan sulphate. Biochem. J. 1974;141(2):517–526. doi: 10.1042/bj1410517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Davidson EA, Meyer K. Chondroitin, a new mucopolysaccharide. J. Biol. Chem. 1954;211(2):605–611. [PubMed] [Google Scholar]
- [78].Hileman RE, Siegel MM, Tabei K, Balagurunathan K, Linhardt RJ. Isolation and characterization of beta-cyclodextrin sulfates by preparative gradient polyacrylamide gel electrophoresis, capillary electrophoresis and electrospray ionization - mass spectrometry. Electrophoresis. 1998;19(15):2677–2681. doi: 10.1002/elps.1150191517. [DOI] [PubMed] [Google Scholar]
- [79].Silbert JE. Organization of glycosaminoglycan sulfation in the biosynthesis of proteochondroitin sulfate and proteodermatan sulfate. Glycoconj. J. 1996;13(6):907–912. doi: 10.1007/BF01053185. [DOI] [PubMed] [Google Scholar]
- [80].Sugahara K, Mikami T, Uyama T, Mizuguchi S, Nomura K, Kitagawa H. Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr. Opin. Struct. Biol. 2003;13(5):612–620. doi: 10.1016/j.sbi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- [81].Mallis LM, Wang HM, Loganathan D, Linhardt RJ. Sequence analysis of highly sulfated, heparin-derived oligosaccharides using fast atom bombardment mass spectrometry. Anal. Chem. 1989;61(13):1453–1458. doi: 10.1021/ac00188a030. [DOI] [PubMed] [Google Scholar]
- [82].Gallagher JT, Walker A. Molecular distinctions between heparan sulphate and heparin. Analysis of sulphation patterns indicates that heparan sulphate and heparin are separate families of N-sulphated polysaccharides. Biochem. J. 1985;230(3):665–674. doi: 10.1042/bj2300665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Linhardt RJ. Heparin: an important drug enters its seventh decade. Chem. Ind. 1991;2:45–50. [Google Scholar]
- [84].Zaia J, Costello CE. Tandem mass spectrometry of sulfated heparin-like glycosaminoglycan oligosaccharides. Anal. Chem. 2003;75(10):2445–2455. doi: 10.1021/ac0263418. [DOI] [PubMed] [Google Scholar]
- [85].Yagami T, Kitagawa K, Aida C, Fujiwara H, Futaki S. Stabilization of a tyrosine O-sulfate residue by a cationic functional group: formation of a conjugate acid-base pair. J. Pept. Res. 2000;56(4):239–249. doi: 10.1034/j.1399-3011.2000.00746.x. [DOI] [PubMed] [Google Scholar]
- [86].Saad OM, Ebel H, Uchimura K, Rosen SD, Bertozzi CR, Leary JA. Compositional profiling of heparin/heparan sulfate using mass spectrometry: assay for specificity of a novel extracellular human endosulfatase. Glycobiology. 2005;15(8):818–826. doi: 10.1093/glycob/cwi064. [DOI] [PubMed] [Google Scholar]
- [87].Wolff JJ, Amster IJ, Chi L, Linhardt RJ. Electron detachment dissociation of glycosaminoglycan tetrasaccharides. J. Am. Soc. Mass Spectrom. 2007;18(2):234–244. doi: 10.1016/j.jasms.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]